Abstract
Listeria monocytogenes, an important food-borne pathogen, causes listeriosis and is widely distributed in many different environments. In a previous study, we developed a novel enrichment broth containing D-allose that allows better isolation of L. monocytogenes from samples. However, the mechanism of D-allose utilization by L. monocytogenes remains unclear. In the present study, we determined the metabolism of D-allose in L. monocytogenes and found that lineage II strains of L. monocytogenes can utilize D-allose as the sole carbon source for growth, but lineage I and III strains cannot. Transcriptome analysis and sequence alignment identified six genes (lmo0734 to 0739) possibly related to D-allose metabolism that are only present in the genomes of lineage II strains. Recombinant strain ICDC-LM188 containing these genes showed utilization of D-allose by growth assays and Biolog phenotype microarrays. Moreover, lmo0734 to 0736 were verified to be essential for D-allose metabolism, lmo0737 and 0738 affected the growth rate of L. monocytogenes in D-allose medium, while lmo0739 was dispensable in the metabolism of D-allose in L. monocytogenes. This is the first study to identify the genes related to D-allose metabolism in L. monocytogenes, and their distribution in lineage II strains. Our study preliminarily determined the effects of these genes on the growth of L. monocytogenes, which will benefit the isolation and epidemiological research of L. monocytogenes.
Keywords: Listeria monocytogenes, lineage II, D-allose, metabolism, genes
Introduction
Listeria monocytogenes is a gram-positive foodborne pathogen of humans that causes listeriosis (Low and Donachie, 1997). Clinical symptoms include meningitis, septicemia, abortion, perinatal infections, and gastroenteritis. The elderly, newborns, pregnant women, and immunocompromised patients are more susceptible (Low and Donachie, 1997; Mead et al., 1999; Kathariou, 2002). Based on the serological reactions of somatic (O) and flagellar antigens (H), L. monocytogenes is divided into 13 serotypes, comprising 1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4a, 4ab, 4b, 4c, 4d, 4e, and 7. Previous study has been reported that a multiplex PCR assay has been developed to identify the serotype, which can separate four major serotype groups (Ward et al., 2008). Furthermore, 1/2b, 3b, 4b, 4d, 4e, and 7 belong to lineage I; serotypes 1/2a, 1/2c, 3a, and 3c belong to lineage II; while serotypes 4a and 4c belong to lineage III (Roberts et al., 2006). Some 4a, 4c, and atypical 4b strains were assigned to lineage IV based on sequence data analysis (Orsi et al., 2011). Lineage I strains are mostly isolated from human listeriosis cases, lineage II mostly exist in food and the environment, and lineage III and IV are rare and mainly found in animal hosts (Liu, 2008). Nevertheless, Zilelidou et al. (2015) have reported that multiply L. monocytogenes strains could isolate from a single food sample, suggesting that highly invasive L. monocytogenes strains might show stronger growth than low or modestly invasive strains. However, studies did not suggest a link between the competitive advantage and strain origin, serotype, or sequence type (Zilelidou et al., 2015, 2016).
L. monocytogenes is ubiquitous in the environment and is often isolated from contaminated environments and foods. Notably, L. monocytogenes has the ability to grow at refrigeration temperatures and survive at high salt concentrations, making it difficult to control (Zhang, 2007). L. monocytogenes also can adhere to the surfaces of food processing equipment and resist adverse conditions. It has been reported that L. monocytogenes was persistently isolated from raw pork in open markets in China (Luo et al., 2017). L. monocytogenes is aerobic, or facultatively anaerobic under some conditions. L. monocytogenes can metabolize different carbon sources, including D-glucose. Strains can utilize D-glucose to form lactate, acetate, and acetoin when they are grown aerobically, whereas, acetoin is not formed during anaerobic growth (Pine et al., 1989). In addition, cellobiose, fructose, mannose, galactose, lactose, salicin, maltose, dextrin, and glycerol can also be utilized to produce acid. The growth rate of L. monocytogenes increases if fermentable sugars are present (Siddiqi and Khan, 1982).
D-Allose, an aldohexose, is a rare monosaccharide in nature and its physiological functions are varied. D-allose inhibits tumor cell multiplication and active oxygen production (Murata et al., 2003). D-allose is used in a novel enrichment broth to improve the isolation of L. monocytogenes and reduce the growth of non-target organisms (Liu et al., 2017). However, the details of the utilization of D-allose in L. monocytogenes are unknown. D-allose has been reported to be a carbon source for Escherichia coli (Gibbins and Simpson, 1964). It could be converted to fructose-6-phosphate via D-allose-6-phosphate and D-allulose-6-phosphate in Aerobacter aerogenes and E. coli K12 (Gibbins and Simpson, 1964; Kim et al., 1997). Two operons are involved in D-allose metabolism in E. coli, one contains the alsI gene, which encodes allose 6-phosphate isomerase, and the other consists of six contiguous genes, alsR, B, A, C, E, and K. alsR is a negative regulator for the operon. alsB, A, and C comprise the transport system of D-allose, alsB encodes a D-allose-binding protein, alsA encodes an ATP-binding component, and alsC encodes a transmembrane protein. The allulose 6-phosphate epimerase is encoded by alsE and alsK is thought to encode allokinase. Furthermore, the two operons are adjacent in the genome of E. coli K12 (Poulsen et al., 1999). In L. monocytogenes, operons comprising the genes involved in D-allose are unknown. In this study, we investigated the utilization of D-allose in L. monocytogenes and analyzed the associated genes for D-allose metabolism.
Methods and materials
Strains, plasmids, media, and culture conditions
L. monocytogenes strains EGD-e and ICDC-LM188 were used as D-allose utilizing and D-allose non-utilizing control strains, respectively. A total of 278 L. monocytogenes strains, isolated from different areas and sample sources in China, were used for the D-allose utilization assay in this study (Supplementary Table 1). Lineage I and III strains (Table 1) were used for gene function analysis. These strains originated from American Type Culture Collection (ATCC, USA) or were isolated in China and stored in State Key Laboratory of Infectious Disease Prevention and Control, China CDC at −80°C (Table 1). Plasmid PIMK2 was used to construct the gene expression vectors (Table 1; Lauer et al., 2002). Bacterial cells were cultured in Brain Heart Infusion (BHI) Broth (BD, USA) with shaking at 220 rpm. Modified Welshimer's broth (MWB) medium was used in the growth assay (Premaratne et al., 1991), and Luria-Bertani (LB) medium (Oxoid, UK) was used in the transcriptome analysis.
Table 1.
Plasmids and strains used in this study.
| Strains or plasmids | Genotypes |
|---|---|
| PLASMIDS | |
| pIMK2 | pIMK rrnB T1 terminators Phelp |
| pAL1 | pIMK2 lmo0734 0735 0736 0737 0738 0739 |
| pAL2 | pIMK2 lmo0734 0735 0736 0737 0738 |
| pAL3 | pIMK2 lmo0734 0735 0736 0737 0739 |
| pAL4 | pIMK2 lmo0734 0735 0736 0738 0739 |
| pAL5 | pIMK2 lmo0734 0735 0737 0738 0739 |
| pAL6 | pIMK2 lmo0734 0736 0737 0738 0739 |
| pAL7 | pIMK2 lmo0735 0736 0737 0738 0739 |
| STRAINS (SEROTYPE) | |
| EGD-e (1/2a) | lmo0734, 0735, 0736, 0737, 0738, 0739 |
| ICDC-LM188 (1/2b) | None |
| RS801 (1/2b) | ICDC-LM188::pAL1 |
| RS802 (1/2b) | ICDC-LM188::pAL2 |
| RS803 (1/2b) | ICDC-LM188::pAL3 |
| RS804 (1/2b) | ICDC-LM188::pAL4 |
| RS805 (1/2b) | ICDC-LM188::pAL5 |
| RS806 (1/2b) | ICDC-LM188::pAL6 |
| RS807 (1/2b) | ICDC-LM188::pAL7 |
| ATCC 19114 (4a) | None |
| RS19114 (4a) | ATCC19114::pAL1 |
| ATCC 19115 (4b) | None |
| RS19115 (4b) | ATCC19115::pAL1 |
| ACTC 19116 (4c) | None |
| RS19116 (4c) | TCC19116::pAL1 |
| ATCC 19117 (4d) | None |
| RS19117 (4d) | ATCC19117::pAL1 |
| ATCC 19118 (4e) | None |
| RS19118 (4e) | ATCC19118::pAL1 |
D-allose utilization and growth curve analysis
Single colonies of L. monocytogenes strains EGD-e and ICDC-LM188 were incubated in BHI medium with constant shaking at 220 rpm overnight at 37°C, and then a 1% volume of the BHI cultures were transferred separately into BHI medium, and shaken at 220 rpm at 37°C until the OD600 values reached 0.6. The strains were then diluted 1:100 into 0.2% D-allose or D-glucose MWB medium (MWB medium containing 2 g/L D-allose or D-glucose as the sole carbohydrate). The strains were grown in a Bioscreen C microbiology reader (Growth Curves Ltd, Helsinki, Finland) at 37°C with shaking. The OD600 was monitored at 30 min intervals for 24 h. To obtain the maximum growth rate for each strain (Pontinen et al., 2017), the OD600 data were fitted to growth curves using GraphPad Prism software (GraphPad Software Inc., San Diego, CA, USA).
To test the universality of D-allose utilization in L. monocytogenes, a single colony of each of 278 L. monocytogenes strains was cultured in BHI medium at 37°C with shaking at 220 rpm overnight, and 1% volumes of the cultures were transferred into BHI with shaking 220 rpm at 37°C until the OD600 value reached 0.6. Subsequently, the strains were diluted 1:100 into 0.2% D-Allose MWB medium, and the cultures were shaken at 220 rpm for 24 h at 37°C. Finally, the growth status of the bacterial cells were detected.
Transcriptome analysis and D-allose metabolism related gene detection
L. monocytogenes EGD-e was selected as the reference for the transcriptome analysis. One colony of EGD-e was incubated in BHI medium at 37°C with shaking at 220 rpm overnight. Then, 1% volumes of the culture were respectively transferred into 0.2% D-allose and D-glucose LB medium at 37°C with shaking at 220 rpm until the OD600 value reached 0.6. The bacteria were then harvested by centrifugation and total RNA was extracted using the Trizol method (Rio et al., 2010). The extracted RNA was used for RNA sequencing with BGISEQ-500, according to the sequencing procedure used by the Beijing Genomics Institute (BGI, China). After filtering out low quantity sequences, clean reads were generated and mapped to the reference genome using HISAT and Bowtie2 tools (Langmead et al., 2009; Kim et al., 2015). The Poisson distribution method was used to analyze differentially expressed genes (DEGs). Clustering analysis of DEGs was performed with cluster and java Treeview software (Michael Eisen, Stanford University, Stanford, CA). Gene Ontology (GO) (http://www.geneontology.org/) annotation was performed for the screened DEGs. Six pairs of specific primers were designed to verify DEGs based on the results of the transcriptome analysis (Supplementary Table 2). DNAs of above 278 L. monocytogenes strains were isolated by boiling, detected by PCR using the specific primers, and confirmed by agarose gel electrophoresis.
Plasmids construction and electro-transformation
To construct the D-allose related gene expression vectors, the DNA fragments, being obtained by PCR using the specific primers (Table 2), were purified from agarose gel electrophoresis using a DNA Gel Extraction kit (Takara, Shiga, Japan). The purified products were cloned into the SalI and BamHI sites of vector pIMK2 to form plasmids pAL1 to pAL7 (Figure 1) via In-Fusion or T4 ligase Cloning. Different lineage strains (Table 1) were used to prepare competent cells (Camilli et al., 1990). Plasmid pAL1 was electrotransformed into competent cells of the strains of all lineages except lineage II, and plasmids pAL2, pAL3, pAL4, pAL5, pAL6, and pAL7 were respectively introduced into competent cells of L. monocytogenes strain ICDC-LM188 (Table 1). The electroporation conditions were 2.5 KV, 400 Ω, 25 μF. Afterwards, the strains were grown on BHI plates containing 50 μg/ml kanamycin in a 37°C incubator for 24 h. The clones were detected by PCR using the specific primers (Supplementary Table 2).
Table 2.
Primers used to construct the expression vectors.
| Plasmids | Primers (5′-3′) |
|---|---|
| pAL1 | AL1 P1 F |
| CGCGGATCCTTAAATTGAGTCTTTTAAAGATAATTCAACGGGAACAATCTCTG | |
| AL1 P1 R | |
| CGCGTCGACTCAGTAGTTCAAATCATTTCCATTAG | |
| pAL2 | AL2 P1 F |
| CCATGGAAAAGGATCCTTAAATTGAGTCTTTTAAAGATAATTCAACGGGAACAATCTCTG | |
| AL2 P1 R | |
| AGTTGCTAGGTAACGATGTCTTAATTTTCGC | |
| AL2 P2 F | |
| GCGAAAATTAAGACATCGTTACCTAGCAACT | |
| AL2 P2 R | |
| ACCCCCTCGAGTCGACTTAAAGTACTGGTGTTAAAAGCGTAT | |
| pAL3 | AL3 P1 F |
| CCATGGAAAAGGATCCTTAAATTGAGTCTTTTAAAGATAATTCAACGGGAACAATCTCTG | |
| AL3 P1 R | |
| TTGCAAATTAAACATTTACGATTTCTGCTTGCCATTCTAAAGTACGTATAAGGT | |
| AL3 P2 F | |
| AAGCAGAAATCGTAAATGTTTAATTTGCAAAAAGGTTTTCCAG | |
| AL3 P2 R | |
| ACCCCCTCGAGTCGACTCAGTAGTTCAAATCATTTCCATTAG | |
| pAL4 | AL4 P1 F |
| CCATGGAAAAGGATCCTTAAATTGAGTCTTTTAAAGATAATTCAACGGGAACAATCTCTG | |
| AL4 P1 R | |
| TTTTTGATAATCCATTTAATCATTTTCATCTTCAATTCTAGCAATCATTTCGACGCG | |
| AL4 P2 F | |
| GATGAAAATGATTAAATGGATTATCAAAAACTAGCTAAAGAG | |
| AL4 P2 R | |
| ACCCCCTCGAGTCGACTCAGTAGTTCAAATCATTTCCATTAG | |
| pAL5 | AL5 P1 F |
| CCATGGAAAAGGATCCTTAAATTGAGTCTTTTAAAGATAATTCAACGGGAACAATCTCTG | |
| AL5 P1 R | |
| TACTTCTTGCTCCATTCATCGTAATTCCTCCATCTTTTCTTG | |
| AL5 P2 F | |
| GAGGAATTACGATGAATGGAGGCAAGAAGTAACACGACTAAT | |
| AL5 P2 R | |
| ACCCCCTCGAGTCGACTCAGTAGTTCAAATCATTTCCATTAG | |
| pAL6 | AL6 P1 F |
| CCATGGAAAAGGATCCTTAAATTGAGTCTTTTAAAGATAATTCAACGGGAACAATCTCTG | |
| AL6 P1 R | |
| AATAGCAATTTTCATATGAAAAAAACATCCATAAAAGATATCGC | |
| AL6 P2 F | |
| GGATGTTTTTTTCATATGAAAATTGCTATTGGAAATGATCATGTTGG | |
| AL6 P2 R | |
| ACCCCCTCGAGTCGACTCAGTAGTTCAAATCATTTCCATTAG | |
| pAL7 | AL7 F |
| CGCGGATCCATGAGCAAAAAACTGATTTGTCCTTCTAT | |
| AL7R | |
| CGCGTCGACTCAGTAGTTCAAATCATTTCCATTAG |
Figure 1.
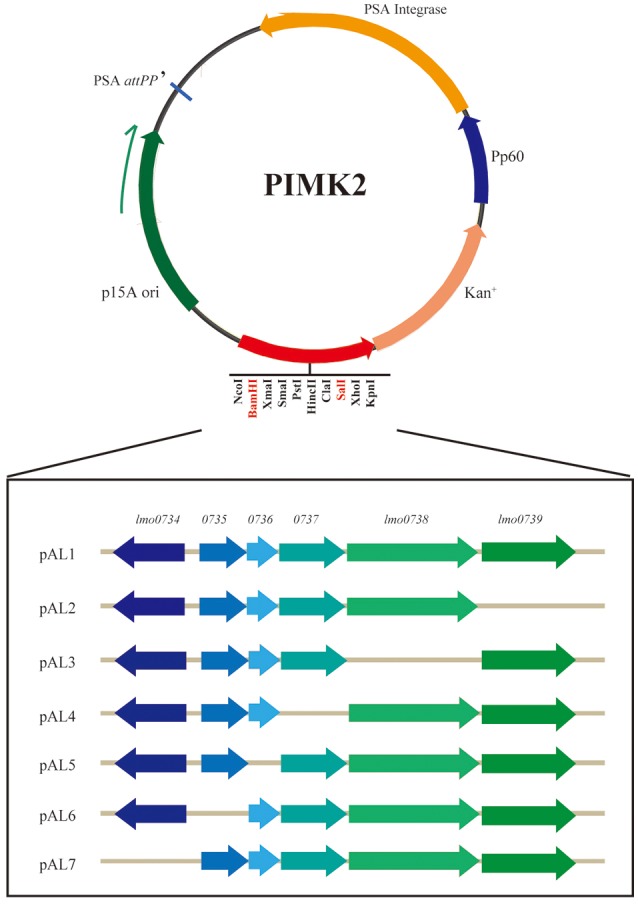
Schematic diagram of the expression vector construction. The gene sequences were cloned into restriction endonuclease recognition sites BamHI and SalI. Each recombinant plasmid lacked one gene in the cluster. The direction of transcription of each is marked by an angled arrow.
Growth phenotype and curve analysis
The positive clones were incubated in BHI medium at 37°C with shaking at 220 rpm overnight, 1% volumes of cultures were transferred into BHI medium with shaking at 37°C until the OD600 value reached 0.6. Afterwards, 1% volumes of inoculum were cultured in 0.2% D-allose MWB medium with shaking 220 rpm at 37°C for 24 h. The cells growth status was monitored (Zwietering et al., 1990; Markkula et al., 2012). Biolog phenotype microarrays (Biolog, USA) were used to analyze the utilization of D-allose by L. monocytogenes strain ICDC-LM188 and its recombinant strains (Miller and Rhoden, 1991). Based on the manufacturer's protocol for Listeria, the Biolog 96-well micro-plates PM1 and PM2a were used to test carbohydrate metabolism; plate PM2a contains D-allose. Clones were inoculated into 20 ml 1 × Buffer IF-0a, and the turbidity of the suspension was checked until 81% light transmittance was achieved. Then, 1.76 ml of the cell suspension was transferred into 22.24 ml PM1 and PM2a inoculating fluid, and 100 μl/well of the cell suspension was inoculated in 96-well micro-plates, which were incubated in an OmniLog instrument at 37°C for 24 h. Growth curve analysis of the recombinant strains was carried out according to a previous description (Pontinen et al., 2017) using the Bioscreen C microbiology reader (Growth Curves Ltd., Helsinki, Finland.) at 37°C, 0.2% D-allose medium was used as the broth.
Results
D-allose utilization and growth assay
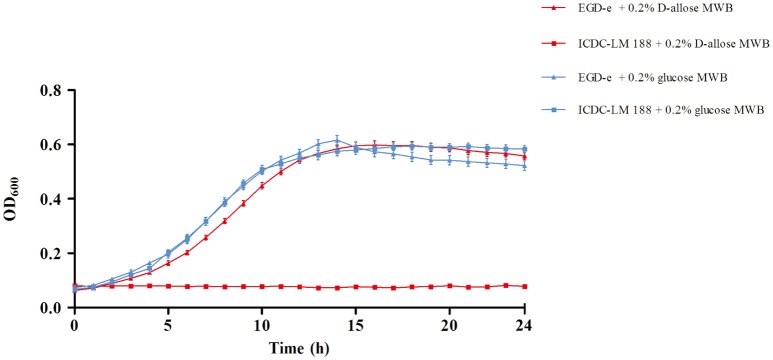
In our previous study, D-allose was discovered to be a carbon source for L. monocytogenes growth, and L. monocytogenes could grow in the novel Listeria Allose Enrichment Broth (LAEB). In addition to D-allose, tryptone, peptone, and Lab-lemco powder could contributed a small amount of carbon source and energy in the LAEB medium (Liu et al., 2017). In this study, to identify D-allose utilization in the growth of L. monocytogenes, MWB medium was used (Premaratne et al., 1991). D-glucose was replaced by D-allose in MWB medium as the sole carbon and energy source for growth. L. monocytogenes strains EGD-e and ICDC-LM188 were used as experimental references, because EGD-e could grow well in LAEB medium, while ICDC-LM188 could not (Liu et al., 2017). Growth rates were calculated (Zwietering et al., 1990; Markkula et al., 2012) and were similar between EGD-e and ICDC-LM 188 in 0.2% D-glucose MWB medium. Moreover, there was no obvious difference in the growth of EGD-e between D-glucose and D-allose medium (Figure 2). By contrast, no growth of ICDC-LM 188 was observed in the 0.2% D-allose MWB medium (Figure 2). Thus, ICDC-LM188 could not utilize D-allose as carbon and energy source.
Figure 2.
Growth curve of L. monocytogenes EGD-e and ICDC-LM188. The red line shows growth in 0.2% D-allose modified Welshimer's broth (MWB) medium, the blue line shows growth in 0.2% D-glucose MWB medium. Triangles represent EGD-e and squares represent ICDC-LM188.
In addition, 278 experimental strains were cultured as above to verify their D-allose utilization. The result showed that 171 strains belonging to lineage II could grow in D-allose MWB medium; however, the other 107 strains, including 91 strains of lineage I and 16 strains of lineages III and IV, could not (Table 3). The result was analyzed using Fisher's test, which revealed that there was a correlation (Pearson r = 0.7071, P < 0.001) between the lineages and D-allose utilization.
Table 3.
D-allose utilization in L. monocytogenes strains.
| Growth in D-allose medium | ||||
|---|---|---|---|---|
| + | − | Total | ||
| I | 0 | 91 | 91 | |
| Lineage | II | 171 | 0 | 171 |
| III | 0 | 16 | 16 | |
| Total | 171 | 107 | 278 | |
Transcriptome analysis and D-allose metabolism related gene detection
Using RNA-Seq technology, more than 23 million clean reads were acquired after filtering out low quality reads. The transcriptomes were successfully sequenced and deposited in GenBank (Accession No. SRR6281666 and SRR6281667). According to the quality control and expression level calculated using the RSEM algorithm and the fragments per kilobase of transcript per million mapped reads (FPKM) method (Li and Dewey, 2011), the differential expression of 15 genes were upregulated and 13 genes were downregulated in D-allose utilizing group compared with the D-glucose utilizing group. Moreover, pathway enrichment analysis of the DEGs was performed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and the functions of the DEGs were classified as nucleotide metabolism and carbohydrate metabolism in the secondary pathway of KEGG (Supplementary Table 3).
Subsequently, whole-genome alignment of different lineage strains was conducted and a gene cassette (lmo0734 to 0739) of strain EGD-e was observed to be absent in L. monocytogenes lineage I, III, and IV strains (Figure 3A). This specific gene cassette was highly expressed under D-allose utilization conditions (Figure 3B). DNA was extracted from 278 experimental strains, including different lineages, to verify the presence of the genes in the different lineages. All strains of lineage II contained the gene cassette; however, it was not present in any of the other lineages. Notably, only the strains of lineage II could utilize D-allose as a carbon source for growth. These results were analyzed using Fisher's test, which showed that there was a correlation between the six genes and D-allose utilization in L. monocytogenes (Pearson r = 0.7071, P < 0.001).
Figure 3.
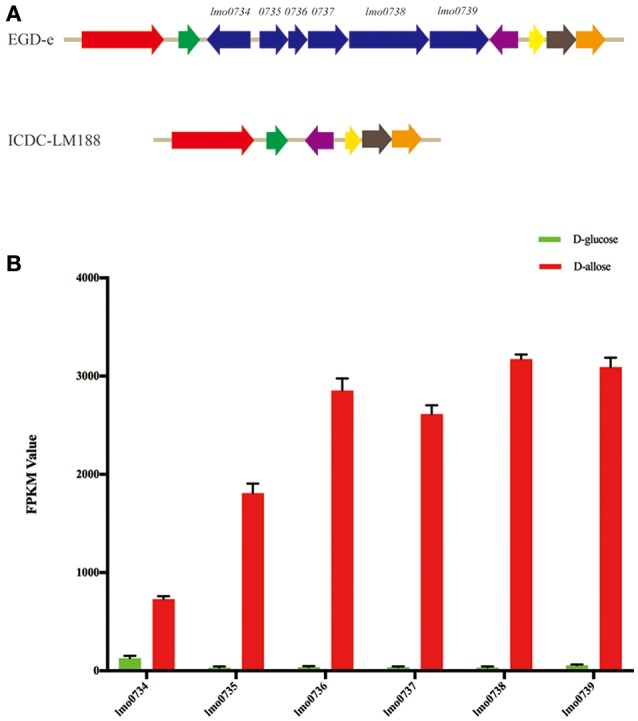
Genome analysis and gene expression levels. (A) The cluster of whole-genome alignment between L. monocytogenes EGD-e and ICDC-LM188. The same color angled arrows represent the same genes. Six genes of EGD-e were found to be absent in ICDC-LM188 (between the green and purple angled arrows). (B) Comparison of the expression level comparisons of five different genes between D-glucose and D-allose (P < 0.001). FPKM, fragments per kilobase of transcript per million mapped reads.
Growth and biolog characterization of recombinant strains
D-allose utilization of the recombinant strains was verified using a growth assay (Table 1). D-allose supported the growth in different lineage strains containing pAL1 (containing all six genes; Supplementary Figure 1A). This revealed that the specific gene cassette was involved in D-allose metabolism in different lineage strains of L. monocytogenes. To further identify the key genes in the D-allose metabolism of L. monocytogenes, plasmids pAL2, pAL3, pAL4, pAL5, pAL6, and pAL7 were constructed according to the schematics shown in Figure 1, and were transferred separately into strain ICDC-LM188. Strains RS802, RS803, and RS804 grew on D-allose medium; however, strains RS805, RS806, and RS807 did not (Supplementary Figure 1B). This revealed that genes lmo0734, lmo0735, and lmo0736 were essential for D-allose metabolism of L. monocytogenes.
The recombinant strains RS801, RS802, RS803, and RS804 showed different growth rates in MWB medium with 0.2% D-allose (Figure 4). The growth curves were analyzed in Graphpad Prism, the specific growth rates were calculated (Table 4). According to t-test analysis (Zwietering et al., 1990; Pontinen et al., 2017), the growth rates of RS801 and RS802 were similar, while RS803 and RS804 were similar. Moreover, the growth rates of RS801 and RS802 were higher than those of RS803 and RS804 (P < 0.01).
Figure 4.
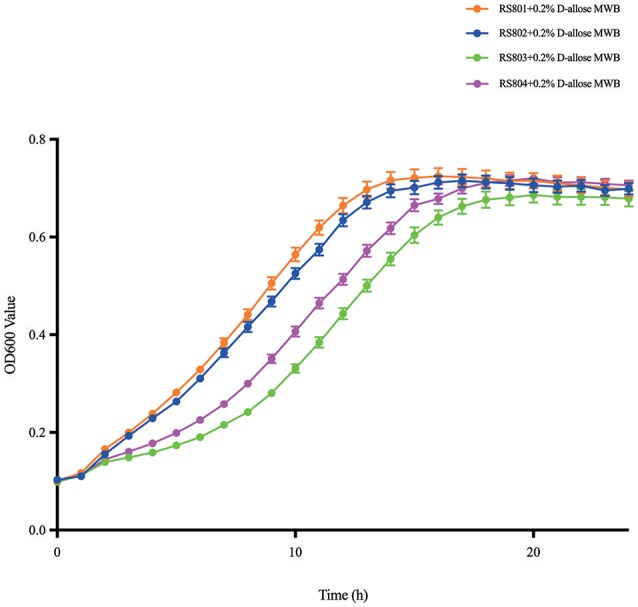
Growth curve of RS801, RS802, RS803, and RS804. Graph showing growth in 0.2% D-allose modified Welshimer's broth (MWB). Orange line: RS801; blue line: RS802; green line: RS803; purple line: RS804.
Table 4.
Growth rates of recombinant strains in D-allose.
| Strains | Growth rate ± SD (OD600 units/h) |
|---|---|
| RS801 | 0.1504 ± 0.006 |
| RS802 | 0.1486 ± 0.005 |
| RS803 | 0.1055 ± 0.005 |
| RS804 | 0.1082 ± 0.004 |
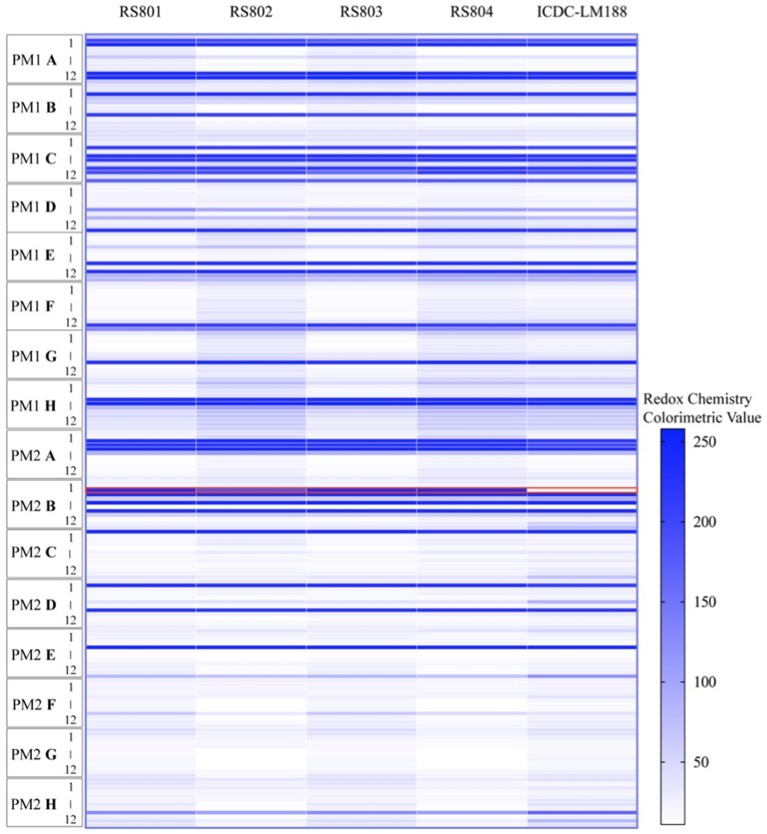
Biolog phenotype microarrays showed that recombinant strains RS801, RS802, RS803, and RS804 could grow on D-allose basal broth while ICDC-LM188 could not (Figure 5). In addition to metabolism of D-allose, the recombinant strains and ICDC-LM188 showed similar utilization of other carbohydrates. This revealed that the six genes (lmo0734-0739) represented a specific cassette involved in the carbohydrate metabolism of D-allose.
Figure 5.
Phenotype microarray analysis of the utilization of carbohydrate by ICDC-LM188, RS801, RS802, RS803, and RS804. All plates were incubated at 37°C for 24 h. PM represent a PM plate, A–H and 1–12, respectively, represent the column and row of the PM plate. Each horizontal line in the heat map shows a type of carbohydrate metabolism. The red border marking the line represents the utilization of D-allose.
Discussion
L. monocytogenes has been described as a resistant and ubiquitous bacterium that can survive in harsh conditions. Under nutrient-limited conditions, some subgroup lineage II strains can grow faster than some subgroups of lineage I strains (Tang et al., 2015), and most strains of lineage II were reported to be isolated more frequently from food samples than from patients (Chenal-Francisque et al., 2011; Montero et al., 2015). Some lineage II strains have been reported to form biofilms to adapt to harsh environments (Wong, 1998; Møretrø and Langsrud, 2004). In the previous study of LAEB, we found that some L. monocytogenes strains could utilize D-allose for growth (Liu et al., 2017). In the present study, we further confirmed that only strains of lineage II could utilize D-allose as a carbon source, and the other lineages could not, including 91 lineage I strains and 17 lineage III strains. This suggested the presence of special carbohydrate metabolisms in different lineage strains, which might play important roles in the growth and isolation of L. monocytogenes in different environments. The hypothesis needs to be studied further in the future.
In our study, a specific gene cassette associated with D-allose was present in lineage II strains, which allowed them to utilize D-allose. The cassette has been reported to be unique to lineage II and atypical 4b, but is absent in other lineages (Doumith et al., 2004; Milillo et al., 2009; Lee et al., 2012). In particular, lmo0737 is regarded as a specific gene to identify lineage II (Ward et al., 2008). Additionally, the present of the specific gene cassette has been reported to have no impact on invasion and growth in nutrient-deprived conditions in knockout mutants of EGD-e (Milillo et al., 2009). Nonetheless, in this study, we verified that the gene cassette presents in lineage II strains and is related to D-allose metabolism.
Gene lmo0734 is defined as a Lac I regulator in genome of EGD-e, and it is a member of the D-allose metabolism operon of L. monocytogenes. Transcriptional fusions in the als operon were Lac+ in the original background in D-allose metabolism of E. coli K12 (Kim et al., 1997). Genes lmo0735 and lmo0736 are regarded to have alsE and alsI functions. The three genes have been reported to be essential in D-allose metabolism of L. monocytogenes. In previous studies, alsE was required in D-allose metabolism of E. coli K12 and A. aerogenes, while alsI proved to be essential in A. aerogenes and dispensable in E. coli K12 (Gibbins and Simpson, 1964; Kim et al., 1997; Poulsen et al., 1999). Gene lmo0737 was annotated as a hypothetical protein belonging to a member of TIM phosphate binding superfamily. Phosphorylase kinase is one of the TIM family (Nagano et al., 1999). D-allose is phosphorylated to D-allulose by the product of alsK, a phosphorylase kinase. Gene lmo0737 might be the equivalent to alsK, being responsible for phosphorylation for D-allose. Gene lmo0738 is annotated as encoding a PTS beta-glucoside transporter subunit IIABC in EGD-e, which has the same function as the products of alsA, B, and C, namely transferring D-allose across the membrane. Genes lmo0737 and lmo0738 have been reported to be dispensable in D-allose metabolism in other bacteria. Nonetheless, absence of lmo0737 and lmo0738 decreased the growth rate in D-allose MWB medium. The requirement for lmo0734 to lmo0736 and the dispensability of lmo0738 to lmo0739 suggested that lmo0734 to lmo0736 are irreplaceable, while other genes might compensate for the activities of lmo0737 and lmo0738. Interestingly, ribose has been reported as an analog of D-allose, and D-allose can be metabolized via some proteins in the D-ribose metabolic pathway (Kim et al., 1997). Gene lmo0739 encodes 6-phospho-beta-glucosidase in L. monocytogenes. Currently, there are no studies to indicate 6-phospho-beta-glucosidase plays a role in D-allose metabolism. In addition, lmo0739 is absent in genome of Listeria ivanovii PAM55, which can utilize D-allose (Liu et al., 2017).
In the transcriptome analysis, there were other DEGs in addition to genes lmo0734 to lmo0739. Among the downregulated genes, genes lmo0096 and lmo0097 mainly participate in mannose metabolism. D-allose metabolism is a part of mannose metabolism (KEGG Pathway map00051). D-allose metabolism might have an affect on mannose metabolism. Genes lmo2761 to lmo2765 have functions in D-cellobiose metabolism, which is associated with D-glucose (KEGG Pathway map00050), which is isomer of D-allose. However, a clear relationship between D-allose and D-cellobiose metabolism has not been demonstrated. As for the correlation between these genes and D-allose metabolism, it need to be studied in future.
Conclusion
Our previous study demonstrated that the utilization of D-allose in a novel enrichment broth could increase the isolation rate of L. monocytogenes. The present study firstly proposes D-allose metabolism in L. monocytogenes and identified the key genes involved in this metabolism. We determined the distribution of these genes in different lineage strains. We provide preliminarily evidence for the utilization of D-allose in the novel enrichment broth for L. monocytogenes lineage II strains. Our results will serve as a reference for the optimization of novel broth (LAEB) to increase the isolation of other lineages strains in the future and for further research into the carbohydrate metabolism of L. monocytogenes.
Author contributions
LZ: Designed the project, analyzed data, and wrote manuscript; DL, LL, and YaW: Analyzed data; YiW and CY: Carried out the experiments.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Mega Project of Research on the Prevention and Control of HIV/AIDS, Viral Hepatitis Infectious Diseases, Ministry of Science and Technology of China [grant number 2013ZX10004-101 to CY]; the State Key Laboratory of Infectious Disease Prevention and Control [grant number 2015SKLID507 to CY]; and the National Institute for Communicable Disease Control and Prevention, China [grant number 2016ZZKTB09 to CY].
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00621/full#supplementary-material
References
- Camilli A., Portnoy A., Youngman P. (1990). Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172, 3738–3744. 10.1128/jb.172.7.3738-3744.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenal-Francisque V., Lopez J., Cantinelli T., Caro V., Tran C., Leclercq A., et al. (2011). Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 17, 1110–1112. 10.3201/eid/1706.101778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M., Cazalet C., Simoes N., Frangeul L., Jacquet C., Kunst F., et al. (2004). New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72, 1072–1083. 10.1128/IAI.72.2.1072-1083.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbins L. N., Simpson F. J. (1964). The incorporation of D-allose into the glycolytic pathway by Aerobacter aerogenes. Can. J. Microbio. 10, 829–836. 10.1139/m64-108 [DOI] [PubMed] [Google Scholar]
- Kathariou S. (2002). Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65, 1811–1829. 10.4315/0362-028X-65.11.181 [DOI] [PubMed] [Google Scholar]
- Kim C., Song S., Park C. (1997). The D-allose operon of Escherichia coli K-12. J. Bacteriol. 179, 7631–7637. 10.1128/jb.179.24.7631-7637.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P., Chow M. Y., Loessner M. J., Portnoy D. A., Calendar R. (2002). Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184, 4177–4186. 10.1128/JB.184.15.4177-4186.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Ward T. J., Graves L. M., Wolf L. A., Sperry K., Siletzky R. M., et al. (2012). Atypical Listeria monocytogenes serotype 4b strains harboring a lineage II-specific gene cassette. Appl. Environ. Microbio. 78, 660–667. 10.1128/AEM.06378-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. (2008). Handbook of Listeria monocytogenes. Boca Raton, FL: Taylor & Francis CRC Press. [Google Scholar]
- Liu D., Wang Y., Wang Y., Zhang L., Luo L., Liu K., et al. (2017). Development of a novel Listeria enrichment broth for the isolation of pathogenic Listeria. J. Food Prot. 80, 1768–1776. 10.4315/0362-028X.JFP-16-529 [DOI] [PubMed] [Google Scholar]
- Low J. C., Donachie W. (1997). A review of Listeria monocytogenes and listeriosis. Vet. J. 153, 9–29. 10.1016/S1090-0233(97)80005-6 [DOI] [PubMed] [Google Scholar]
- Luo L., Zhang Z., Wang H., Wang P., Lan R., Deng J., et al. (2017). A 12-month longitudinal study of Listeria monocytogenes contamination and persistence in pork retail markets in China. Food Control 76, 66–73. 10.1016/j.foodcont.2016.12.037 [DOI] [Google Scholar]
- Markkula A., Lindstrom M., Johansson P., Bjorkroth J., Korkeala H. (2012). Roles of four putative DEAD-box RNA helicase genes in growth of Listeria monocytogenes EGD-e under heat, pH, osmotic, ethanol, and oxidative stress conditions. Appl. Environ. Microbiol. 78, 6875–6882. 10.1128/AEM.01526-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead P. S., Slutsker L., Dietz V., Mccaig L. F., Bresee J. S., Shapiro C., et al. (1999). Food-related illness and death in the United States. Emerg. Infect. Dis. 5, 607–625. 10.3201/eid0506.990624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milillo S. R., Badamo J. M., Wiedmann M. (2009). Contributions to selected phenotypic characteristics of large species-and lineage-specific genomic regions in Listeria monocytogenes. Food Microbiol. 26, 212–223. 10.1016/j.fm.2008.08.010 [DOI] [PubMed] [Google Scholar]
- Miller J. M., Rhoden D. L. (1991). Preliminary evaluation of Biolog, a carbon source utilization method for bacterial identification. J. Clin. Microbiol. 29, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero D., Bodero M., Riveros G., Lapierre L., Gaggero A., Vidal R. M., et al. (2015). Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 6:384. 10.3389/fmicb.2015.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møretrø T., Langsrud S. (2004). Listeria monocytogenes: biofilm formation and persistence in food-processing environments. Biofilms 1, 107–121. 10.1017/S1479050504001322 [DOI] [Google Scholar]
- Murata A., Sekiya K., Watanabe Y., Yamaguchi F., Hatano N., Izumori K., et al. (2003). A novel inhibitory effect of D-allose on production of reactive oxygen species from neutrophils. J. Biosci. Bioeng. 96, 89–91. 10.1016/S1389-1723(03)90104-6 [DOI] [PubMed] [Google Scholar]
- Nagano N., Hutchinson E. G., Thornton J. M. (1999). Barrel structures in proteins: automatic identification and classification including a sequence analysis of TIM barrels. Protein Sci. 8, 2072–2084. 10.1110/ps.8.10.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi R. H., den Bakker H. C., Wiedmann M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. 10.1016/j.ijmm.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Pine L., Malcolm G. B., Brooks J. B., Daneshvar M. I. (1989). Physiological studies on the growth and utilization of sugars by Listeria species. Can. J. Microbiol. 35, 245–254. 10.1139/m89-037 [DOI] [PubMed] [Google Scholar]
- Pontinen A., Lindstrom M., Skurnik M., Korkeala H. (2017). Screening of the two-component-system histidine kinases of Listeria monocytogenes EGD-e. LiaS is needed for growth under heat, acid, alkali, osmotic, ethanol and oxidative stresses. Food Microbiol. 65, 36–43. 10.1016/j.fm.2017.01.018 [DOI] [PubMed] [Google Scholar]
- Poulsen T. S., Chang Y. Y., Hovejensen B. (1999). d-Allose Catabolism of Escherichia coli: involvement of alsI and regulation of als regulon expression by allose and ribose. J. Bacteriol. 181, 7126–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premaratne R. J., Lin W. J., Johnson E. A. (1991). Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57, 3046–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio D. C., Ares M., Hannon G. J., Nilsen T. W. (2010). Purification of RNA using TRIzol (TRI Reagent). Cold Spring Harb. Protoc. 2010:pdb.prot5439. 10.1101/pdb.prot5439 [DOI] [PubMed] [Google Scholar]
- Roberts A., Nightingale K., Jeffers G., Fortes E., Kongo J. M., Wiedmann M. (2006). Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiol. 152(Pt 3), 685–693. 10.1099/mic.0.28503-0 [DOI] [PubMed] [Google Scholar]
- Siddiqi R., Khan M. A. (1982). Vitamin and nitrogen base requirements for Listeria monocytogenes and haemolysin production. Zentralblatt Für Bakteriologie Mikrobiologie Und Hygiene.abt.originale.a Medizinische Mikrobiologie Infektionskrankheiten Und Parasitologie 253, 225–235. [PubMed] [Google Scholar]
- Tang S., Wiedmann M., Gardner A. L., Brown A. M., Boor K. J., Bergholz T. M. (2015). Clonal clustering using 10-gene multilocus sequence typing reveals an association between genotype and Listeria monocytogenes maximum growth rate in defined medium. Foodborne Pathog. Dis. 12, 972–982. 10.1089/fpd.2015.2019 [DOI] [PubMed] [Google Scholar]
- Ward T. J., Ducey T. F., Usgaard T., Dunn K. A., Bielawski J. P. (2008). Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 74, 7629–7642. 10.1128/AEM.01127-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C. L. (1998). Biofilms in food processing environments. J. Dairy Sci. 81, 2765–2770. 10.3168/jds.S0022-0302(98)75834-5 [DOI] [PubMed] [Google Scholar]
- Zhang W. (2007). Listeria, Listeriosis, and Food Safety, 3rd Edn. Boca Raton, FL: CRC Press. [Google Scholar]
- Zilelidou E. A., Rychli K., Manthou E., Ciolacu L., Wagner M., Skandamis P. N. (2015). Highly invasive Listeria monocytogenes strains have growth and invasion advantages in strain competition. PLoS ONE 10:e0141617. 10.1371/journal.pone.0141617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilelidou E., Karmiri C. V., Zoumpopoulou A. G., Mavrogonatou B. E., Kletsas C. D., Tsakalidou C. E., et al. (2016). Listeria monocytogenes strains underrepresented during selective enrichment with an ISO method might dominate during passage through simulated gastric fluid and in vitro infection of Caco-2 cells. Appl. Environ. Microbiol. 82, 6846–6858. 10.1128/AEM.02120-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwietering M. H., Jongenburger I., Rombouts F. M., Van't Riet K. (1990). Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56, 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.