Abstract
Geranylgeranyl diphosphate (GGPP) is the precursor for the biosynthesis of gibberellins, carotenoids, chlorophylls, isoprenoid quinones, and geranylgeranylated proteins in plants. There is a small gene family for GGPP synthases encoding five isozymes and one related protein in Arabidopsis, and all homologs have a putative localization signal to translocate into specific subcellular compartments. Using a synthetic green fluorescent protein (sGFP), we studied the subcellular localization of these GGPP synthases. When these fusion proteins were expressed by the cauliflower mosaic virus 35S promoter in Arabidopsis, GGPS1-sGFP and GGPS3-sGFP proteins were translocated into the chloroplast, GGPS2-sGFP and GGPS4-sGFP proteins were localized in the endoplasmic reticulum, and the GGPS6-sGFP protein was localized in the mitochondria. Both GGPS1 and GGPS3 proteins synthesized in vitro were taken up into isolated intact pea chloroplasts and processed to the mature form. RNA-blot and promoter-β-glucuronidase (GUS) analysis showed that these GGPP synthases genes are organ-specifically expressed in Arabidopsis. GGR and GGPS1 were ubiquitously expressed, while GGPS2, GGPS3, and GGPS4 were expressed specifically in the flower, root, and flower, respectively. These results suggest that each GGPP synthase gene is expressed in different tissues during plant development and GGPP is synthesized by the organelles themselves rather than being transported into the organelles. Therefore, we predict there will be specific pathways of GGPP production in each organelle.
A large variety of products are derived from isoprenoids in plants for their growth and response to environmental changes (Gray, 1987). Geranylgeranyl diphosphate (GGPP) is one of the key isoprenoids to be converted into compounds necessary for plant growth, such as gibberellins, carotenoids, chlorophylls, isoprenoid quinones, and geranylgeranylated small G proteins such as Rho, Rac, and Rab (Brown and Goldstein, 1993). Plant hormone gibberellins are necessary for seed germination and normal plant growth and are synthesized from a common precursor, ent-kaurene. This precursor is produced from GGPP with two-step cyclization via copalyl diphosphate. Carotenoids are synthesized from 2 molecules of GGPP by head-to-head condensation, and have an essential function for protection against potentially harmful photo-oxidative processes. Thus, GGPP synthase is a crucial branch point enzyme that is responsible for the various aspects of plant growth and development. The mechanism for the enzymatic reaction with condensation of isopentenyl diphosphate (IPP) and other substrates such as farnesyl diphosphate (FPP) to produce C20 GGPP is conserved among organisms from bacteria (Mende et al., 1997) to humans (Kainou et al., 1999).
Recently, two different IPP synthetic pathways have been found in plants. One is the mevalonate pathway that starts from condensation of acetyl-coenzyme A (acetyl-CoA), and the other is the glyceraldehyde phosphate/pyruvate pathway, the so called non-mevalonate pathway, in which pyruvate, not mevalonate, is a precursor of IPP. By labeling experiments with [13C]Glc and using a specific inhibitor for mevalonate biosynthesis, it has been shown that IPP is produced by the non-mevalonate pathway in plastids, whereas cytosolic and mitochondrial IPPs are derived from mevalonate (Schwender et al., 1996; Lichtenthaler et al., 1997; Disch et al., 1998). These findings imply that the enzymes involved in IPP synthesis are localized in different compartments in plants.
Prenyl diphosphate synthases, the enzymes utilizing IPP, are also thought to be distributed in three subcellular compartments: the cytosol, mitochondria, and plastids (Gray, 1987; Kleinig, 1989). In plant cells, isoprenoids such as carotenoids, chlorophylls, and ent-kaurene have been shown to be synthesized from GGPP in plastids (Bartley and Scolnik, 1995; Hedden and Kamiya, 1997), whereas sterols are formed from FPP and protein prenylation has been proposed to occur in the cytoplasm/endoplasmic reticulum (ER). In mitochondria, isoprenoid quinone is necessary for the respiratory chain reactions as an electron transporter to obtain energy (Trumpower, 1981), and an isoprenoid side chain of ubiquinone has been proposed to be synthesized from IPP with an allylic substrate FPP or GGPP (Okada et al., 1998). The Arabidopsis farnesyl diphosphate synthase FPS1L, one of the FPP synthase isoforms generated from the FPS1 gene, has been shown to be translocated into mitochondria (Cunillera et al., 1997), and the rice farnesyl diphosphate synthase FPPS1 is localized in the chloroplasts of mesophyll cells (Sanmiya et al., 1999). These data indicate that each subcellular compartment has its own pathway to produce isoprenoid compounds using different enzymes and IPP derived from either the mevalonate or non-mevalonate pathway.
There have been many reports of the isolation of GGPP synthase genes from plants. GGPP synthase genes have been characterized in Capsicum annuum (Kuntz et al., 1992; Badillo et al., 1995), Sinapis alba (Laferriere and Beyer, 1991), Lupinus albus (Aitken et al., 1995), Catharanthus roseus (Bantignies et al., 1995), and Arabidopsis. In Arabidopsis, two functionally active and five putative GGPP synthase gene sequences have been reported: GGR, GGPS1, GGPS2, GGPS3, GGPS4, GGPS5, and GGPS6 (Scolnik and Bartley, 1994, 1995, 1996; Zhu et al., 1997a, 1997b). However, GGPS2 and GGPS5 genes are considered to be identical because there is no difference at the nucleotide level between them, and the only difference is that the GGPS5 protein is missing six amino acids at its N terminus compared with the GGPS2 protein. Thus, at present, there are six GGPP synthases isozymes in Arabidopsis. These Arabidopsis GGPP synthases have putative localization signals in their N-terminal regions to transfer them into specific subcellular compartments, and the GGPS6 protein has already been shown to be translocated into mitochondria in tobacco Bright Yellow-2 (BY-2) cells (Zhu et al., 1997b).
While knowledge of the subcellular localization of GGPP synthase is crucially important for our understanding of compartmentalization of isoprenoid biosynthesis, a localization study for other homologs of GGPP synthase has not yet been reported. It is likely that synthesis of GGPP in Arabidopsis is regulated and compartmentalized in different organelles where different kinds of GGPP synthases occur. In this report, we attempt to understand the biological significance of each GGPP synthase homolog in Arabidopsis by investigating their localization using a synthetic green fluorescent protein (sGFP) (Chiu et al., 1996). In addition, the expression of these GGPP synthase genes at different developmental stages and in different organs was examined by RNA gel-blot and promoter-GUS analysis. Based on our results, Arabidopsis GGPP synthases can be classified into three groups: cytosolic/ER, plastidic, and mitochondrial enzymes.
MATERIALS AND METHODS
Materials
Arabidopsis ecotype Columbia (Col-1) was used throughout the study. Plants were grown under continuous light at 22°C on Murashige and Skoog (MS) medium (GIBCO-BRL, Cleveland). Roots, rosette leaves, cauline leaves, stems, and flowers were harvested separately for RNA preparation. Plants grown on soil for 4 weeks under the same conditions as above were used for vacuum infiltration.
Construction of Plasmids
For functional assay, four sets of oligonucleotide primers (for the GGR gene, sense, 5′-ATGGATCCGATGTTGTTTAGTGGTTC-3; antisense, 5′-CAAGCGAAGAAGC- TCTGG-3′; for the GGPS1 gene, sense, 5′-GGTGAGAATTTCAGATTTCAG-3′; antisense, 5′-CCGGATACGATTACACCAACAAAC-3′; for the GGPS3 gene, sense, 5′-AATCTAGACATGGCTACTACTGTTC-3′; antisense, 5′-TCAGTTGTGTCTGAAAGC-3′; for the GGPS4 gene, sense, 5′-ATGGATCCAATGGAAGCTCAAAATATC-3′; antisense, 5′-TCTAGACAATTTTCAGTGGTTTCTGTTGGC-3′) were designed from the sequence of each GGPP synthase gene to amplify the open reading frame (ORF) of these genes.
A PCR and a reverse transcriptase (RT)-PCR were performed in 50 μL of reaction mixture with 100 pmol of gene-specific primer sets and 0.1 μg of genomic DNA or 1 μg of total RNA extracted from frozen Arabidopsis plants. The amplified products were cloned into the pT7 blue-T vector (Novagen, Madison, WI), and sequenced. The resulting plasmids, pTG10, pTG11, pTG13, and pTG14, expressed the GGR, GGPS1, GGPS3, and GGPS4 protein, respectively, as LacZ-fused proteins. For higher expression of GGR and GGPS4 proteins, 1.2-kb BamHI-HindIII fragments of both GGR and GGPS4 genes prepared from pTG10 and pTG14 were cloned into the same site of the pQE31 vector (Qiagen, Valencia, CA) to yield pQG10 and pQG14, respectively.
For the localization study we made four kinds of putative localization signal (LS)-sGFP fusion constructs. The oligonucleotide primers GG1s (5′-CCTCTAGAATGGCTTCAGTGACTCTA-3′) and GG1a (5′-AAGGATCCGAGAGGAACAGCTGAAT-3′), GG2s (5′-CCTCTAGAATGAAAAATGG- AACCACA-3′) and GG2a (5′-AAGGATCCGACTGGAATTGCTTCTT-3′), GG3s (5′-AATCTAGACATGGCTACTACTGTTC-3′) and GG3a (5′-GGATCCGAGCGGTACAGAA- ACGTCG-3′), GG4s (5′-AATCTAGAAATGGAAGCTCAAAATA-3′) and GG4a (5′-GGATCCAAGTGGAATTGCTTCATC-3′) were used to amplify each LS sequence by PCR. These amplified fragments (LS1, LS2, LS3, and LS4) were digested with XbaI and BamHI site and cloned into the same site of the pTH121 vector (Zhu et al., 1997b) to obtain LS-sGFP fused gene fragments. XbaI-SacI LS-sGFP fragments were then prepared from pTH121 harboring distinct LS fragments and transferred to the same site of pBI101 (CLONTECH Laboratories, Palo Alto, CA). The resulting plasmids, pBIG117, pBIG118, pBIG119, and pBIG120, were used to make transgenic Arabidopsis in addition to pBIG121 and pBIC121, both of which were made previously (Zhu et al., 1997b).
For histochemical analysis, we made five kinds of promoter-GUS constructs. 5′-Untranslated regions of each GGPP synthase gene were amplified by PCR with oligonucleotide primers of UTR1s (5′-AGAAGCTTACAAGTTG-3′) and UTR1a (5′-TCTAGAGGCGATTTCTGAAATC-3′), UTR2s (5′-AAGCTTAATGACGATAGGTTTG-3′ and UTR2a (5′-TCTAGATTTTCATCAAAATCAAATC-3′), UTR3s (5′-CAATTATCGCTCGGTTCACATG-3′) and UTR3a (5′-GGCCTATCCACCGTCTCTAATTG-3′), UTR4s (5′-ATTGTTCAGCCTTCTACATCC-3′ and UTR4a (5′-TTGATCAACA- ATATAGGTGATTG-3′), and UTR6s (5′-AAGCTTTCTAACTAACCAC-3′) and UTR6a (5′-TCTAGAGTAAACTTTATTGAG-3′) designed from genomic sequence information on the database.
These amplified fragments (UTR1, UTR2, UTR3, UTR4, and UTR6) were cloned into the pT7 blue-T vector, and these sequences were confirmed. HindIII-XbaI-digested fragments of UTR1 and UTR6 or the XbaI-digested fragment of UTR2 were cloned into the same site of pHM1 vector, which has the GUS gene downstream of the cloning site to yield pHG1, pHG6, and pHG2, respectively. UTR3 and UTR4 fragments were cut out using SalI and BamHI sites or SalI and SmaI sites, respectively. Then these fragments were both cloned into the corresponding site of the pBI101 vector to yield pHG3 and pHG4, respectively.
In Vitro Enzyme Assay for GGPP Synthase
Escherichia coli M15 cells (Qiagen) transformed with pQG10, pTG11, pTG13, and pQG14 were inoculated in Luria-Bertani medium containing 100 μg/mL ampicillin, then cultured for 18 h at 37°C after induction with 1 mm isopropylthio-β-galactoside. The E. coli cells harvested by centrifugation were washed and resuspended with buffer A (100 mm potassium phosphate, pH 7.4, 1 mm EDTA, and 1 mm β-mercaptoethanol), and then disrupted by a sonicator with three times 30-s burst and 30-s interval. Sonicated cells were centrifuged at 10,000g for 20 min, and then supernatants were collected to use in in vitro enzyme reactions. Crude enzymes were incubated with [14C]IPP and FPP in buffer A containing 5 mm MgCl2, and elongated isoprenoid products were extracted by n-butanol and examined on thin-layer chromatography after hydrolysis of the extracted products, as described previously (Okada et al., 1998).
Visualization of sGFP
Five-day-old Arabidopsis seedlings transformed with LS-sGFP fusion fragments were examined for visualization of both the sGFP signal and chlorophyll autofluorescence using an microscope (model BX60F5, Olympus Optical, Tokyo) equipped with epifluorescence. To observe sGFP fluorescence, an excitation filter (BP470–490, Olympus), dichroic mirror (DM505, Olympus), and emission filter (BA515–550, Olympus) were used. To observe chlorophyll autofluorescence from the chloroplast, an excitation filter (BP520–550, Olympus), dichroic mirror (DM565, Olympus), and emission filter (BA580IF, Olympus) were used. Plant materials from at least 20 independently obtained T2 seedlings were put on the glass slide and covered with a coverslip in 50 mm potassium phosphate buffer. Hypocotyl epidermis and guard cells of stoma in true leaves of transgenic plants were used for observation of these fluorescent signals.
Histochemical Staining with X-Gluc
Plant tissues were vacuum-infiltrated with 1 mm X-Gluc solution (Jefferson, 1987), which consists of 50 mm sodium phosphate and 0.06% (v/v) Triton X-100. The infiltrated tissues were incubated overnight at 37°C. Colored pigments including chlorophylls were removed by 70% (v/v) ethanol at room temperature. Plant samples were viewed under a dissecting microscope (MZAPO, Leica Microsystems, Wetzlar, Germany). For observation of thin-sectioned tissues, the stained plants were fixed with 1% (v/v) glutaraldehyde in 0.1 m phosphate buffer, pH 7.0, for 12 h. After dehydration of the tissues by a series of ethanol solutions, tissues were embedded in Paraplast (Sigma-Aldrich, St. Louis), and 5-μm sections were cut using a microtome. Sections were viewed under an microscope (BX60F5, Olympus).
RNA Extraction and Gel-Blot Analysis
Total RNAs were isolated from different tissues including roots, rosette leaves, cauline leaves, stems, and flower clusters of 28-d-old Arabidopsis. Plant tissues frozen in liquid N2 were homogenized using a mortar and pestle, and RNA was extracted using TRIZOL reagent (GIBCO-BRL) according to the manufacturer's instructions. Total RNAs (10 μg) were separated on a 1.2% (w/v) agarose gel containing 2.2 m formaldehyde and 0.5 mm ethidium bromide, and the RNAs were transferred to a Hybond+ membrane (Amersham-Pharmacia Biotech, Uppsala) (Sambrook et al., 1989). Equal loading of total RNA on the membrane was confirmed with a UV transmitter (Stratagene, La Jolla, CA). Six separated membranes fixed by a UV closs-linker (Stratagene) were hybridized independently with 0.8 to 1.0 kb of random-primed 32P-labeled DNA fragments derived from plasmids pTG10, pTG11, pTG13, pTG14, pATEN2 (Zhu et al., 1997a), and pGGPS6-1 (Zhu et al., 1997b). The hybridizations were carried out under high-stringency conditions at 65°C to avoid cross-hybridization between six GGPP synthase genes. Membranes were washed twice under high-stringency conditions of 1× SSC and 0.1% (w/v) SDS at room temperature, followed by 0.1× SSC and 0.1% (w/v) SDS at 65°C for 20 min. The membranes were then exposed to the image plate and quantitated by the image analyzer (model BAS2000, Fuji Photo Film, Tokyo).
In Vitro Translation and Protein Uptake into Chloroplast
For preparing the intact pea chloroplasts from young leaves of 10-d-old pea seedlings, we used the homogenization protocol described by Albrecht and Sandmann (1994). Isolated intact chloroplasts were washed once with the intake buffer (50 mm HEPES-KOH, pH 7.5, 330 mm sorbitol) and used for uptake experiments. The in vitro-translated proteins were synthesized from plasmids pTG11 and pTG13, both of which have GGPS1 and GGPS3 cDNA under the T7 promoter, respectively, using the TNT coupled reticulocyte system (Promega, Madison, WI) according to the manufacturer's manual. Radio-labeled GGPS1 and GGPS3 proteins (30 μL) were mixed and incubated for 1 h at room temperature with 160 μL of the intact chloroplasts in the uptake reaction mixture consisting of 10 μL of 250 mm Met, 25 μL of 60 mm Mg-ATP (pH 7.0), and 30 μL of 2× intake buffer. Chloroplasts were collected by centrifugation with 1 mL of 40% (v/v) Percoll, and resuspended in 500 μL of intake buffer to use for protease treatment. Protease treatment was carried out by adding 50 μL of 2 mg/mL thermolysin (Sigma) in 50 mm CaCl2 solution and incubating for 30 min on ice. For the control experiment, 50 μL of 50 mm CaCl2 was added instead. Chloroplast pellets were collected by centrifugation with 1 mL of 40% (v/v) Percoll and examined on SDS-PAGE after washing with intake buffer containing 1 mm EDTA. The signals of imported proteins were detected by exposing to the imaging plate with the image analyzer (BAS2000, Fuji).
RESULTS
Enzymatic Activity of Arabidopsis GGPP Synthases
Although the sequences of six genes (GGR, GGPS1, GGPS2, GGPS3, GGPS4, and GGPS6) for GGPP synthase of Arabidopsis have been deposited in the database (Fig. 1), only two isozymes, GGPS5 and GGPS6, were shown to have the enzymatic activity of GGPP synthase (Zhu et al., 1997a, 1997b). To identify the activity for other putative GGPP synthase homologs, we amplified GGR, GGPS1, GGPS3, and GGPS4 genes by PCR using gene-specific oligonucleotide primers based on the sequences of GGPP synthase genes. cDNAs for GGR, GGPS1, and GGPS3 were successfully obtained by RT-PCR using total RNA prepared from 28-d-old wild-type plants. Although cDNA for GGPS4 could not be amplified by RT-PCR, genomic DNA of GGPS4 was amplified and sequenced to examine the existence of an intron sequence, but no intron sequence was found in the GGPS4 gene. The ORF of GGPS4 consists of 373 amino acids. Comparison of the sequences between PCR-amplified genomic DNA and RT-PCR-amplified cDNA revealed that the GGR gene had a 145-bp intron sequence at position 383 to 528, and the GGPS3 gene had a 108-bp intron sequence at position 631 to 739, whereas the GGPS1 gene did not have any intron sequence.
Figure 1.
Multiple alignment of GGPP synthases from Arabidopsis and a photosynthetic bacterium. Deduced amino acid sequences of five Arabidopsis GGPP synthases (ATGGPS1, ATGGPS2, ATGGPS3, ATGGPS4, and ATGGPS6) and one related protein (ATGGR) and a Rhodobacter capsulatus (RCCRTE) GGPP synthase were compared with each other. Amino acids conserved in more than three enzymes are indicated in white on black type. Numbered lines show the seven domains that are highly conserved among prenyl diphosphate synthases with two Asp-rich motifs for substrates binding (domains II and VI). The box at the N-terminal region indicates putative localization signals of Arabidopsis GGPP synthases. Accession numbers of corresponding genes are follows: ATGGR, L40577; ATGGPS1, L25813; ATGGPS2, U44876; ATGGPS3, AB023038; ATGGPS4, AC006135; ATGGPS6, AB000835; and RCCRTE, Z11165.
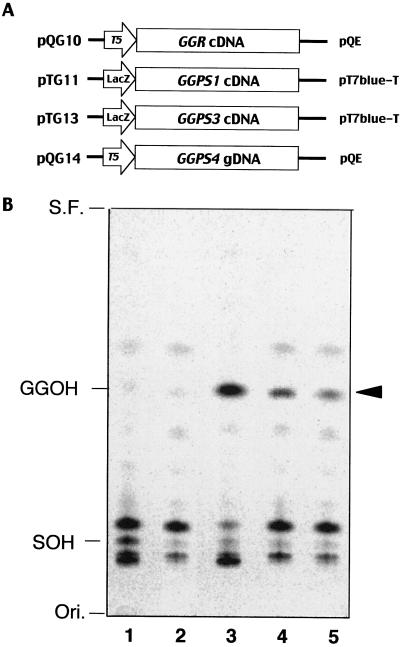
We constructed four plasmids for expressing the different GGPP synthase ORFs (GGR, GGPS1, GGPS3, and GGPS4) of Arabidopsis (Fig. 2A) and expressed them in E. coli. Crude extracts from the E. coli culture expressing exogenous GGPP synthases were used for enzyme reaction with [14C]IPP and FPP as substrates in 50 mm potassium phosphate buffer containing 5 mm magnesium. Elongated isoprenoid products were isolated by n-butanol and hydrolyzed to the alcohol form of isoprenoid by acid phosphatase, then separated by thin-layer chromatography. While octaprenol and undecaprenol, both of which are derived from host enzymes, were detected from an E. coli M15 harboring vector alone (Fig. 2B, lane 1), products that co-migrated with authentic geranylgeraniol visualized by iodine vapor were detected in addition to octaprenol and undecaprenol from E. coli M15 harboring the plasmid pTG11, pTG13, and pQG14 (Fig. 2B, lanes 3–5). However, we could not detect any GGPP synthase activity from E. coli expressing pQG10 (Fig. 2B, lane 2) even though highly expressed GGR protein was detected by western blotting (data not shown). These results indicated that all of these genes encoded functional GGPP synthases except the GGR protein, which is not necessarily an active GGPP synthase and might need an extra component for its enzymatic function.
Figure 2.
In vitro functional assay of Arabidopsis GGPP synthase. A, Four kinds of plasmids with different GGPP synthase genes (GGR, GGPS1, GGPS3, and GGPS4) were constructed. cDNA for GGR, GGPS1, and GGPS3 and genomic DNA for GGPS4 were used for plasmid constructions. B, Crude enzymes from E. coli culture expressing these genes were incubated with [14C]IPP and FPP. Reaction products were extracted by n-butanol, hydrolyzed to the alcohol form of isoprenoids, then separated on reverse-phase thin-layer chromatography. Arrowhead indicates detected geranylgeraniol synthesized by exogenously expressed GGPP synthases of Arabidopsis. Authentic geranylgeraniol (GGOH) and solanesol (SOH) visualized temporary by iodine vapor are indicated on the left. Ori., Origin; S.F., solvent front; lane 1, M15/pQE31(negative control); lane 2, M15/pQG10; lane 3, M15/pTG11; lane 4, M15/pTG13; and lane 5, M15/pQG14.
Differential Subcellular Localization of GGPP Synthase Isozymes
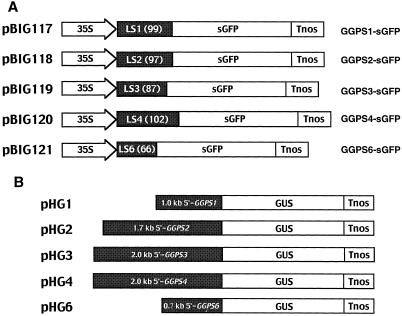
It is generally accepted that GGPP synthase is present in plastids, since GGPP synthase from Capsicum annuum has been located in the chromoplast of ripening fruit (Kuntz et al., 1992). However, we previously showed that the GGPS6 protein is imported into mitochondria in tobacco BY-2 cells (Zhu et al., 1997b) using the sGFP protein as a marker (Chiu et al., 1996). This result leads us to the idea that every GGPP synthase homolog of Arabidopsis has the possibility of being translocated into subcellular locations other than the plastid. To examine the localization of GGPP synthases with confirmed enzymatic activity, we made five kinds of sGFP constructs, which were fused with a putative localization signal consisting of 66 to 102 N-terminal amino acids of five functionally active GGPP synthases of Arabidopsis (Fig. 3A). These plasmids were introduced into Arabidopsis with Agrobacterium tumefaciens-mediated transformation using the vacuum infiltration method (Bechtold et al., 1993), and then sGFP signals of transgenic plants were observed using a fluorescence microscope.
Figure 3.
Plasmids for sGFP fusion and promoter-GUS constructs. A, Five sGFP fusion plasmids with different sorts of putative localization signals (LS1, LS2, LS3, LS4, and LS6) fused to sGFP were constructed on pBI121. The numbers in the LS regions indicate amino acids lengths of signal peptide. 35S, Cauliflower mosaic virus 35S promoter; Tnos, nopaline synthase gene terminator. B, Five GGPP synthases genes promoters-GUS fusion plasmids were made. These plasmids had untranslated promoter regions of GGPS1 (0.9 kb), GGPS2 (1.7 kb), GGPS3 (2.0 kb), GGPS4 (2.0 kb), and GGPS6 (0.7 kb), which are transcriptionally fused by the GUS gene.
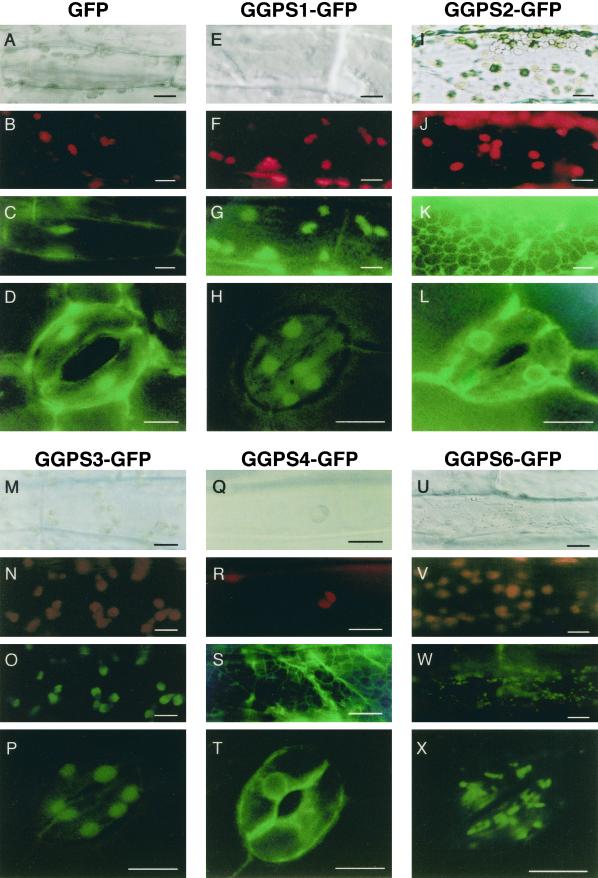
In the control plant expressing sGFP alone, the fluorescence of sGFP was detected in the cellular membrane and nucleus under blue light (Fig. 4, C and D), and chloroplasts were seen as red dots due to the autofluorescence of chlorophylls under green light (Fig. 4B). It has been said that sGFP itself tends to associate with the membrane and to accumulate in the nucleus due to the potential sequences promoting translocation to the nucleus in sGFP (Baulcombe et al., 1995). When the GGPS1-sGFP protein or the GGPS3-sGFP protein was expressed, relatively large fluorescent granules were seen in the chloroplasts of the hypocotyl epidermal cells (Fig. 4, F, G, N, and O). Green fluorescence was also seen from chloroplasts of guard cells of the true leaf (Fig. 4, H and P). In root tissues, relatively large fluorescent granules were also observed (data not shown). Because root tissues have etioplasts rather than chloroplasts, the GGPS1-sGFP and the GGPS3-sGFP proteins will be translocated into etioplasts in root tissues.
Figure 4.
Subcellular localization of GGPS-sGFP fused proteins in Arabidopsis. The cells from hypocotyls of 3-d-old plants and true leaves of 10-d-old plant of stable T2 transgenic Arabidopsis were used for the observation of GGPS-sGFP fused proteins by fluorescence microscope. A, E, I, M, Q, and U, Normal microscopic pictures under natural light. B, F, J, N, R, and V, Red chlorophyll autofluorescence from chloroplasts. Red granular signals of chloroplasts were seen. C, G, K, O, S, and W, Fluorescence images of sGFP from hypocotyl epidermal cells. D, H, L, P, T, and X, Fluorescence images of sGFP from guard cells of stoma. sGFP itself tends to localize in the nucleus (C and D). GGPS1-sGFP and GGPS3-sGFP proteins are localized in chloroplasts (G and H or O and P). GGPS2-sGFP and GGPS4-sGFP proteins are localized in the ER and reticulated patterns of the signal were seen (K and L or S and T). GGPS6-sGFP protein is localized in small particles of the mitochondria (W and X). The three pictures at the top were taken in identical regions of the cells of hypocotyl epidermis. Bars = 10 μm.
The GGPS1-sGFP protein was also found to be translocated into plastid-like organelles, probably proplastids, in BY-2 cells (data not shown). These results indicated that the GGPS1 and GGPS3 proteins may be translocated into chloroplasts at the upper part of the plant and into etioplasts in root tissues. In the GGPS2-sGFP and GGPS4-sGFP proteins, green fluorescence was observed in the mesh of net forms that are the reticulated ER membrane in hypocotyl epidermal cells (Fig. 4, K and S). In guard cells of true leaves, green fluorescence was observed mainly around the nucleus, which is likely the ER membrane (Fig. 4, L and T). These results indicated that both GGPS2 and GGPS4 proteins are localized in the ER membrane. In the case of the GGPS6-sGFP protein, which was previously shown to be imported into mitochondria in tobacco BY-2 cells, small green fluorescent granules were detected in hypocotyl epidermal cells (Fig. 4W), and these mitochondria-located signals were apparently different from those of chloroplasts (Fig. 4V). In guard cells, green fluorescence was also seen as dots smaller than those of chloroplasts (Fig. 4X), supporting the idea that the GGPS6 protein is imported into the mitochondria of Arabidopsis. These findings suggest that each GGPP synthase is translocated into a certain organelle, and GGPP is synthesized in organelles themselves rather than being transported into the organelles from other compartments.
Import of GGPS1 and GGPS3 Proteins into Pea Chloroplasts
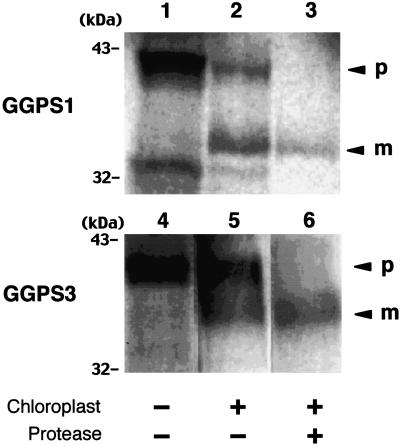
To determine whether both GGPS1 and GGPS3 full-length proteins (both of which are most likely to be chloroplast proteins) are really taken up into purified pea chloroplasts or not, in vitro-translated GGPS1 and GGPS3 proteins labeled with [35S]Met were mixed with purified intact pea chloroplasts. As shown in Figure 5, the precursor forms of both the GGPS1 and the GGPS3 proteins were detected on the autoradiogram as bands of about 41 and 38 kD, respectively (Fig. 5, lanes 1 and 4). When these proteins were mixed with pea chloroplasts, both the precursor and the mature forms of these proteins were detected (Fig. 5, lanes 2 and 5), whereas only the mature form of these proteins of 34 kD (GGPS1) and 36 kD (GGPS3) were detected after protease treatment (Fig. 5, lanes 3 and 6). These results indicated that both the GGPS1 and GGPS3 proteins were successfully translocated into pea chloroplasts and then processed to the mature form of the enzyme. These results are consistent with the results from sGFP experiments showing chloroplast-localized GGPS1-GFP- and GGPS3-GFP-fused proteins.
Figure 5.
In vitro import analysis of GGPS1 and GGPS3 proteins into pea chloroplast. Labeled proteins were made using rabbit reticulolysate and [35S]Met by in vitro translation. Lanes 1 to 3, GGPS1 protein; lanes 4 to 6, GGPS3 protein. Lanes 1 and 4 show the total in vitro-translated proteins of GGPS1 and GGPS3, respectively. Lanes 2 and 5 and lanes 3 and 6 show the labeled proteins after uptake by isolated pea chloroplasts with (lanes 3 and 6) or without (lanes 2 and 5) protease treatment. Arrowheads indicate the precursor (p) and mature (m) forms of those proteins. Molecular masses of the protein markers (kD) are indicated on the left.
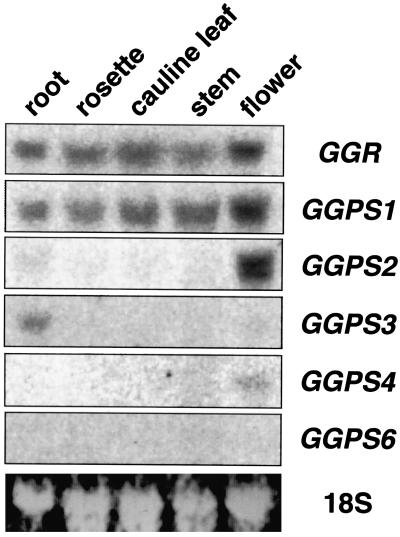
Tissue Distribution of GGPP Synthase Gene Transcripts
To examine the expression of five GGPP synthase genes and one related gene, northern-blot analysis was carried out with the cDNAs as probes using a number of tissues (including roots, rosette leaves, cauline leaves, mature stems, and flower clusters) of 28-d-old Arabidopsis plants. We checked the cross-hybridization of each probe to other GGPP synthase genes by Southern hybridization. No signals other than those from each GGPP synthase gene were detected under the same conditions in northern-blot analysis. As shown in Figure 6, we found that the GGR mRNA and GGPS1 mRNA were ubiquitously expressed in all organs we examined, and that the GGPS3 mRNA was mainly expressed in roots, whereas the GGPS2 mRNA was detected mainly in flower clusters. The GGPS4 mRNA was faintly detected in flower clusters and the GGPS6 mRNAs were barely detected, indicating that expression levels of these two genes are quite low. These results suggest that the expression of the GGPS1, GGPS2, and GGPS3 genes, but not the GGPS4 and GGPS6 genes, are regulated at the transcriptional level in an organ-specific manner.
Figure 6.
Expression patterns of five GGPP synthase genes and one related gene of Arabidopsis. Total RNAs were prepared from separated parts of 3-week-old Arabidopsis including roots, rosette leaves, cauline leaves, elongated stems, and flower clusters. Each 10 μg of total RNA was loaded on formaldehyde-agarose gel containing ethidium bromide. The blotted membranes were hybridized with radiolabeled DNA probes of GGR, GGPS1, GGPS2, GGPS3, GGPS4, and GGPS6. Bottom lane shows 18S rRNA on the nylon membrane visualized by the UV transmitter.
Histochemical Analysis Using Promoter-GUS Constructs
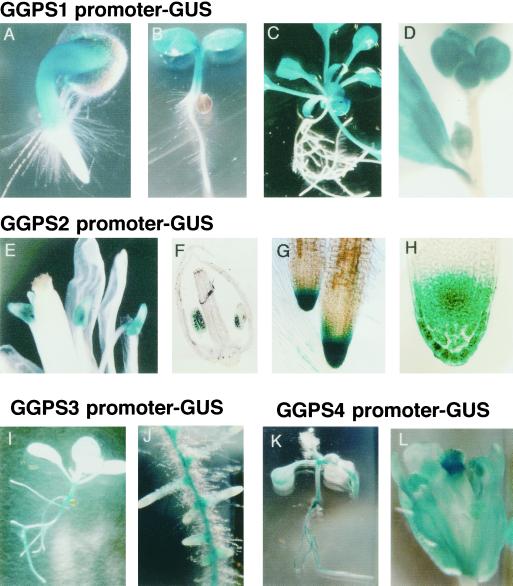
By histochemical analysis using promoter-GUS fusion gene constructs along with northern analysis, tissue- and developmentally specific expression of GGPP synthase genes were observed. We made five constructs that have 0.7 to 2.0 kb of the 5′-untranslated region of these genes fused with the GUS gene (Fig. 3B), and stable T2 transgenic Arabidopsis were used for the histochemical analysis.
GGPS1 promoter activity was detected strongly in upper plant parts such as hypocotyl and cotyledons of 2-d-old seedlings (Fig. 7A), and this promoter activity was still active in cotyledons and at rosette leaves in 6-d-old seedlings and 12-d-old rosette plants (Fig. 7, B and C). In 28-d-old adult plants, this promoter activity was always detected in rosette and cauline leaves and was also detected in flower buds (Fig. 7D). There was almost no promoter activity in the root, while GGPS1 mRNA was detected in roots by northern analysis. This discrepancy may have been caused by the presence of basal parts of the hypocotyl in the root samples used for RNA samples. GGPS2 promoter activity was detected in flowers, especially in anthers at the flowering stage (Fig. 7, E and F), and was also detected around the root tip, especially in the root cap (Fig. 7, G and H), at all developmental stages. GGPS3 promoter activity was detected slightly in whole parts of the plant, but relatively high activity was detected in the basal part of the hypocotyl (Fig. 7I) and the vascular tissues of roots of 12-d-old plants (Fig. 7J). GGPS4 promoter activity was detectable in the vascular tissues in the root, hypocotyl, and veins in 6-d-old plants (Fig. 7K), and was strongly detected in flowers, especially evident in the stigmas of 28-d-old plants (Fig. 7L). GGPS6 promoter activity was almost undetectable, as in northern-blot analysis. These results indicate that each GGPP synthase gene has a specific place to be expressed in the plant organs, even if the subcellular localization of the proteins is exactly the same, such as the GGPS1 and GGPS3 proteins or the GGPS2 and GGPS4 proteins (Fig. 4).
Figure 7.
Histochemical localization of different GGPP synthase gene promoter activities during development of transgenic Arabidopsis. GGPS1 promoter-GUS: A, 2-d-old seedling; B, 6-d-old seedling; C, 12-d-old rosette plant; D, flower cluster of 28-d-old plant. GGPS2 promoter-GUS: E, Flowers after opening; F, 5-μm section of flower; G, root tip of 12-d-old rosette plant; H, 5-μm section of root tip. GGPS3 promoter-GUS: I, 6-d-old seedling; J, root. GGPS4 promoter-GUS: K, 6-d-old seedling; L, flower of 28-d-old plant.
DISCUSSION
Amino acid sequence comparison between five GGPP synthases with one related protein from Arabidopsis and one from a bacterium revealed that all of them have seven highly conserved regions implicated in similar enzymatic functions (Fig. 1). These conserved domains, especially domain II and domain VI, are well conserved among organisms and have been shown to be the substrates binding sites (Joly and Edwards, 1993). Because of their high amino acid similarity among GGPP synthase and some other prenyl diphosphate synthases ranging from GPP synthase to decaprenyl diphosphate synthase, it is impossible to define the enzymatic function of polyprenyl diphosphate synthase from information about their primary structure alone—it is important to also detect enzymatic activity of the expressed proteins.
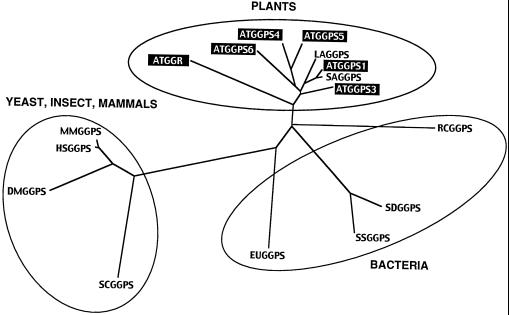
Our in vitro enzyme assay in this study and in the previous study showed that five GGPP synthases encoded by the GGPS1, GGPS2, GGPS3, GGPS4, and GGPS6 genes have detectable activity, but not the one encoded by the GGR. However, the GGR gene is unlikely to be a pseudogene, because GGR mRNA was detected in whole parts of the adult plant when examined by northern analysis. Scolnik and Bartley (1995) also reported that the GGR gene was constitutively expressed in seedlings grown in light, dark, and light with norflurazon. GGR may not encode a GGPP synthase, because multiple alignment of these GGPP synthases showed that the GGR protein has some different features compared with other Arabidopsis homologs: (a) The third Asp at the DDxxD motif in domain VI is not conserved in the GGR protein. (b) The GGR protein lacks several amino acids between domain IV and V, and just after domain VI. (c) The C-terminal region of the GGR protein is redundant compared with other homologs, e.g. Rhodobacter capsulatus GGPP synthase. In the phylogenetic tree drawn by the CLUSTAL W program (Thompson et al., 1994), the GGR protein was classified slightly far from other GGPP synthases of the plant group, including five genes of Arabidopsis (Fig. 8). Furthermore, the large subunit of GPP synthase from peppermint has considerable similarity to Arabidopsis GGR protein (Burke et al., 1999), suggesting that the GGR gene would encode the GPP synthase large subunit rather than GGPP synthase.
Figure 8.
Phylogeny of GGPP synthases from plants, bacteria, yeast, insect, and mammals. A phylogenetic tree was drawn using the CLUSTAL W program. Whole amino acids including N-terminal localization signals of GGPP synthase from plant species were used to calculate. AT, Arabidopsis; LA, Lupinus albus, accession number U15778; SA, Sinapis alba, accession number X98795; RC, Rhodobacter capsulatus, accession number Z11165; SD, Sulfolobus acidocaldarius, accession number D28748; SS, Sulfolobus solfataricus, accession number Y08257; EU, Erwinia uredovora, accession number D90087; SC, Saccharomyces cerevisiae, accession number U31632; DM, Drosophila melanogaster, accession number AF049659; MM, Mus musculus, accession number AB016044; and HS, Homo sapiens, accession number AB016043.
Five Arabidopsis GGPP synthases and their related proteins were found to have the redundant N-terminal peptides that may be used as a translocation signal into specific subcellular locations. Localization of GGPP synthases using the PSORT program showed that the GGR protein is in the cytoplasm, both GGPS1 and GGPS3 proteins are translocated into chloroplasts, GGPS2 and GGPS4 proteins are localized in the vacuole and ER, respectively, and the GGPS6 protein is imported into mitochondria. Plastid localization of a GGPP synthase has been reported in C. annuum, and this protein was detected in chloroplasts and chromoplasts using an immunocytochemical method (Kuntz et al., 1992).
Our localization studies in Arabidopsis GGPP synthases using the cauliflower mosaic virus 35S promoter-driven sGFP showed that the GGPS1-sGFP and GGPS3-sGFP proteins were translocated into chloroplasts in the upper part of the plant, and into leukoplasts (etioplasts) in the root tissues. A protein uptake experiment using intact pea chloroplasts also showed that both of these proteins were translocated into chloroplasts and processed to the mature form. These results indicate that the GGPS1 and GGPS3 proteins supply GGPP for the biosynthesis of chlorophyll, carotenoids, and ent-kaurene, precursors of gibberellin, in the plastids. Although GGPS3 mRNA could not be detected from leaves, in vitro-synthesized GGPS3 protein and GGPS3-sGFP-fused protein were clearly detected to translocate into chloroplast. This inconsistency probably occurred because of the use of a strong expression system with the cauliflower mosaic virus 35S promoter or the T7 promoter to express the protein; in nature, the GGPS3 protein would be translocated into the etioplast but not the chloroplast.
The GGPS4 protein was localized in the ER, and this result is consistent with that prediction by the PSORT program. While the GGPS2 protein was predicted by the PSORT program to be translocated into the vacuole, the GGPS2-sGFP protein and the GGPS4-sGFP protein were actually found in the ER. Since the ER plays a central part in lipid and protein biosynthesis, and almost all proteins that are secreted to the cell exterior are initially derived from the ER lumen, the GGPS2 and GGPS4 proteins might be targeted to the ER and then transferred to the cell surface via the Golgi apparatus. After translocation or during transportation, the GGPS2 and GGPS4 proteins could function for geranylgeranylation of proteins such as Rab proteins, which have been shown to be related to the ER/Golgi protein transport system (Mizoguchi et al., 1990).
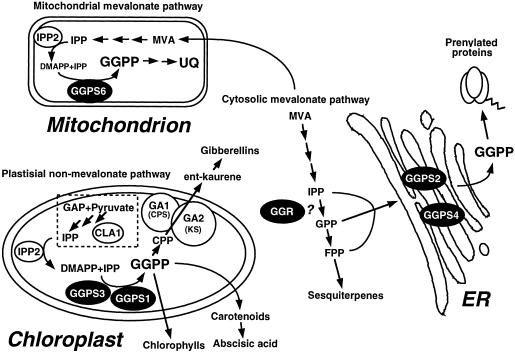
It has been reported that the ER/Golgi system is a major site of plastoquinone biosynthesis in spinach leaves (Osowska-Rogers et al., 1994). In this respect, the GGPS2 and GGPS4 proteins may be involved in the production of the side chain of plastoquinone. The GGPS6-sGFP protein was translocated into the mitochondria in Arabidopsis, and this result was in good agreement with the result we previously reported using tobacco BY-2 cells (Zhu et al., 1997b). GGPS6 protein could supply GGPP to be the precursor of the side chain of ubiquinone in the mitochondria. Based on our localization study, five GGPP synthases of Arabidopsis can be classified into three groups consisting of the cytosolic/ER type, the plastidial type, and the mitochondrial type (Fig. 9). The GGPS1 and GGPS3 proteins belong to the plastidial type of GGPP synthase, the GGPS2 and GGPS4 proteins to the cytosolic/ER type, and the GGPS6 protein to the mitochondrial type. The cytosolic/ER and mitochondrial type GGPP synthases, GGPS2, GGPS4, and GGPS6, are thought to use IPP, which is produced through mevalonate. In contrast, the plastidial type enzymes, GGPS1 and GGPS3, are thought to use IPP derived from the non-mevalonate pathway in plastids, since β-carotene, prenyl chains of chlorophylls, and plastoquinone have been shown to be synthesized from pyruvate and glyceraldehyde phosphate in labeling experiments with 13C-Glc (Schwender et al., 1996; Lichtenthaler et al., 1997).
Figure 9.
Schematic mode of localizations and possible functions of five GGPP synthases in Arabidopsis. The GGPS1 and GGPS3 proteins are translocated into chloroplasts for the biosynthesis of gibberellins, carotenoids, abscisic acids, and chlorophylls. The GGPS2 and GGPS4 proteins are localized on the ER membrane, and the GGPP produced by these enzymes is used for protein prenylation. The GGPS6 protein is translocated into the mitochondria, and the product could be used for ubiquinone biosynthesis. The GGR protein, for which enzymatic activity could not be detected in vitro, might stay in the cytosol to produce GPP. The differentiated subcellular localization of GGPP synthases makes it possible to have GGPP pools inside plant cells. CLA1, Putative transketolase; CPP, copalyl diphosphate; DMAPP, dimethylallyl diphosphate; GA1, CPP synthase; GA2, ent-kaurene synthase; GAP, glyceraldehyde-3-phosphate; IPP2, IPP isomerase; and MVA, mevalonate.
Histochemical analysis using GUS promoter stains showed the tissue-specific expression of each GGPP synthase gene. GGPS1, a plastidial type, was strongly expressed in aerial parts of the plant, especially in rapidly growing tissues such as shoot apices, cotyledons, and vascular tissues of the hypocotyl. This expression almost overlapped with the Arabidopsis GA1 gene that encodes ent-kaurene synthase, and it is thought that the GGPS1 promoter is active in tissues with developing chloroplasts, where the biosynthesis of gibberellins is initiated. Because the GGPS1 protein seems to have a higher activity than those of the other isozymes, the GGPS1 gene must encode a major type of GGPP synthase in Arabidopsis. Transcripts of the GGPS3 gene, the other type of gene for plastidial GGPP synthase, were mainly detected in roots by northern analysis, and GUS expression was also strongly observed in root vascular tissues.
The results from this study indicate that the GGPS3 gene must be mainly expressed in the roots, where the translated GGPS3 protein could be imported into the etioplasts, a possible subcellular location for GA biosynthesis in root tissue. GA biosynthesis in root tissue is still unclear, but evidence that a low level of GA is necessary for promoting root growth has been reported (Tanimoto, 1991). GUS activity for the GGPS2 gene promoter was observed in the root cap and an anther, and northern analysis showed that GGPS2 mRNA was predominantly expressed in flower clusters and only slightly in the root. In contrast, GGPS4 mRNA was detected by northern analysis at a low level in flowers, but promoter activity was observed in vascular tissues of roots and in the stigma, stamen, and sepals of flowers. Overnight GUS staining with X-Gluc might have brought more sensitive detection of the promoter activity than northern analysis. We speculate that the GGPS2 protein was only produced at the anther and root cap, and the GGPS4 protein in the vascular tissues of roots and the pistil. It could be that these two enzymes have an important function for geranylgeranylation of the protein other than terpene biosynthesis. GGPS6 mRNA was not detected by northern analysis or indicated by GUS activity. The GGPS6 protein, as well as the Arabidopsis FPS1 protein, can be expected to contribute to the biosynthesis of isoprenoid quinone. Because this promoter activity was very weak, the GGPS6 gene might be expressed in very specific conditions such as strong environmental stress.
In Arabidopsis five isozymes of GGPP synthase are differentially expressed in a variety of organs. Each of them is differentially expressed at the subcellular level, where they have distinctive biological roles. Recently, GGPP synthase was shown to regulate cell cycle progression in human lymphocytes (Tatsuno et al., 1997), and geranylgeraniol, the alcohol form of GGPP, was found to induce apoptosis of tumor cells (Ohizumi et al., 1995). Some of the Arabidopsis GGPP synthases might have a physiological function rather than the well-known role of being responsible for terpene biosynthesis. Further study by screening of knockout plants for these genes would be helpful to clarify the physiological functions of each GGPP synthase isozyme in Arabidopsis.
ACKNOWLEDGMENTS
We thank Dr. Y. Niwa (University of Shizuoka, Japan) for providing the plasmid 35SO-sGFP (S65T), and Drs. S. Mita and K. Nakamura (Nagoya University, Japan) for providing the plasmid pABH-Hm1. We are grateful to Prof. Richard Kendrick (Frontier Research Program, RIKEN), Drs. Xiangjia Min (RIKEN), Hiroshi Kawaide (RIKEN), and Shinjiro Yamaguchi (Duke University, Durham, NC) for their comments on this manuscript, and Yukiji Tachiyama (RIKEN) for her technical assistance with DNA sequencing.
Footnotes
This work was supported by the Frontier Research Program, The Institute of Physical and Chemical Research (RIKEN).
LITERATURE CITED
- Aitken SM, Attucci S, Ibrahim RK, Gulick PJ. A cDNA encoding geranylgeranyl pyrophosphate synthase from white lupin. Plant Physiol. 1995;108:837–838. doi: 10.1104/pp.108.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Sandmann G. Light-stimulated carotenoid biosynthesis during transformation of maize etioplast is regulated by increased activity of isopentenyl pyrophosphate isomerase. Plant Physiol. 1994;105:529–534. doi: 10.1104/pp.105.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badillo A, Steppuhn J, Deruere J, Camara B, Kuntz M. Structure of a functional geranylgeranyl pyrophosphate synthase gene from Capsicum annuum. Plant Mol Biol. 1995;27:425–428. doi: 10.1007/BF00020196. [DOI] [PubMed] [Google Scholar]
- Bantignies B, Liboz T, Ambid C. Nucleotide sequence of a Catharanthus roseusgeranylgeranyl pyrophosphate synthase gene. Plant Physiol. 1995;110:336. [Google Scholar]
- Bartley GE, Scolnik PA. Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell. 1995;7:1027–1038. doi: 10.1105/tpc.7.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC, Chapman S, Santa Cruz S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thalianaplants. CR Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- Brown MS, Goldstein JL. Protein prenylation: mad bet for Rab. Nature. 1993;366:14–15. doi: 10.1038/366014a0. [DOI] [PubMed] [Google Scholar]
- Burke CC, Wildung MR, Croteau RC. Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc Natl Acad Sci USA. 1999;96:13062–13067. doi: 10.1073/pnas.96.23.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirao T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Cunillera N, Boronat A, Ferrer A. The Arabidopsis thalianaFPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J Biol Chem. 1997;272:15381–15388. doi: 10.1074/jbc.272.24.15381. [DOI] [PubMed] [Google Scholar]
- Disch A, Hemmerlin A, Bach TJ, Rohmer M. Mevalonate-derived isopentenyl diphosphate is the biosynthetic precursor of ubiquinone prenyl side chain in tobacco BY-2 cells. Biochem J. 1998;331:615–621. doi: 10.1042/bj3310615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JC. Control of isoprenoid biosynthesis in higher plants. Adv Bot Res. 1987;14:25–91. [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Joly A, Edwards PA. Effect of site-directed mutagenesis of conserved aspartate and arginine residues upon farnesyl diphosphate synthase activity. J Biol Chem. 1993;268:26983–26989. [PubMed] [Google Scholar]
- Kainou T, Kawamura K, Tanaka K, Matsuda H, Kawamukai M. Identification of the GGPS1genes encoding geranylgeranyl diphosphate synthases from mouse and human. Biochim Biophys Acta. 1999;1437:333–340. doi: 10.1016/s1388-1981(99)00028-1. [DOI] [PubMed] [Google Scholar]
- Kleinig H. The role of plastids in isoprenoid biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:39–59. [Google Scholar]
- Kuntz M, Romer S, Suire C, Hugueney P, Weil JH, Schantz R, Camara B. Identification of a cDNA for the plastid-located geranylgeranyl pyrophosphate synthase from Capsicum annuum: correlative increase in enzyme activity and transcript level during fruit ripening. Plant J. 1992;2:25–34. doi: 10.1111/j.1365-313x.1992.00025.x. [DOI] [PubMed] [Google Scholar]
- Laferriere A, Beyer P. Purification of geranylgeranyl diphosphate synthase from Sinapis albaetioplasts. Biochim Biophys Acta. 1991;1077:167–172. [Google Scholar]
- Lichtenthaler HK, Schwender J, Disch A, Rohmer M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 1997;400:271–274. doi: 10.1016/s0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- Mende K, Homann V, Tudzynski B. The geranylgeranyl diphosphate synthase gene of Gibberella fujikuroi: isolation and expression. Mol Gen Genet. 1997;255:96–105. doi: 10.1007/s004380050477. [DOI] [PubMed] [Google Scholar]
- Mizoguchi A, Kim S, Ueda T, Kikuchi A, Yorifuji H, Hirokawa N, Takai Y. Localization and subcellular distribution of smg p25A, a ras p21-like GTP-binding protein, in rat brain. J Biol Chem. 1990;265:11872–11879. [PubMed] [Google Scholar]
- Ohizumi H, Masuda Y, Nakajo S, Sakai I, Ohsawa S, Nakaya K. Geranylgeraniol is a potent inducer of apoptosis in tumor cells. J Biochem. 1995;117:11–13. doi: 10.1093/oxfordjournals.jbchem.a124695. [DOI] [PubMed] [Google Scholar]
- Okada K, Kainou T, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M. Molecular cloning and mutational analysis of the ddsA gene encoding decaprenyl diphosphate synthase from Gluconobacter suboxydans. Eur J Biochem. 1998;255:52–59. doi: 10.1046/j.1432-1327.1998.2550052.x. [DOI] [PubMed] [Google Scholar]
- Osowska-Rogers S, Swiezewska E, Andersson B, Dallner G. The endoplasmic reticulum-Golgi system is a major site of plastoquinone synthesis in spinach leaves. Biochem Biophys Res Commun. 1994;205:714–721. doi: 10.1006/bbrc.1994.2724. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanmiya K, Ueno O, Matsuoka M, Yamamoto N. Localization of farnesyl diphosphate synthase in chloroplast. Plant Cell Physiol. 1999;40:348–354. doi: 10.1093/oxfordjournals.pcp.a029549. [DOI] [PubMed] [Google Scholar]
- Schwender J, Seemann M, Lichtenthaler HK, Rohmer M. Biosynthesis of isoprenoids (carotenoids, sterols, prenyl side-chains of chlorophylls and plastoquinone) via a novel pyruvate/glyceraldehyde 3-phosphate non-mevalonate pathway in the green alga Scenedesmus obliquus. Biochem J. 1996;316:73–80. doi: 10.1042/bj3160073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik PA, Bartley GE. Nucleotide sequence of an Arabidopsis cDNA for geranylgeranyl pyrophosphate synthase. Plant Physiol. 1994;104:1469–1470. doi: 10.1104/pp.104.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik PA, Bartley GE. Nucleotide sequence of a putative geranylgeranyl pyrophosphate synthase (GenBank accession no. L40577) from Arabidopsis (PGR 95-018) Plant Physiol. 1995;108:1343. doi: 10.1104/pp.104.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik PA, Bartley GE. Two more members of Arabidopsis geranylgeranyl pyrophosphate synthase family (PGR 96-014) Plant Physiol. 1996;110:1435. doi: 10.1104/pp.104.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto E. Gibberellin requirement for the normal growth of roots. In: Takahashi N, Phinney BO, MacMillan J, editors. Gibberellins. New York: Springer; 1991. pp. 229–240. [Google Scholar]
- Tatsuno I, Tanaka T, Oeda T, Yasuda T, Kitagawa M, Saito Y, Hirai A. Geranylgeranylpyrophosphate, a metabolite of mevalonate, regulates the cell cycle progression and DNA synthesis in human lymphocytes. Biochem Biophys Res Commun. 1997;241:376–382. doi: 10.1006/bbrc.1997.7825. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpower BL. New concepts on the role of ubiquinone in the mitochondrial respiratory chain. J Bioeng Biomembr. 1981;13:1–24. doi: 10.1007/BF00744743. [DOI] [PubMed] [Google Scholar]
- Zhu XF, Suzuki K, Okada K, Tanaka K, Nakagawa T, Kawamukai M, Matsuda H. Cloning and functional expression of a novel geranylgeranyl pyrophosphate synthase gene from Arabidopsis thaliana in Escherichia coli. Plant Cell Physiol. 1997a;38:357–361. doi: 10.1093/oxfordjournals.pcp.a029174. [DOI] [PubMed] [Google Scholar]
- Zhu XF, Suzuki K, Saito T, Okada K, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M. Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thalianais localized in mitochondria. Plant Mol Biol. 1997b;35:331–341. doi: 10.1023/a:1005898805326. [DOI] [PubMed] [Google Scholar]