Abstract
Breast cancer (BC) is the most common cancer and the second cause of death in women worldwide. Therapeutic options are increasing, but the response to treatments is not always efficient and the risk of recurrence covers decades. In this perspective, the need to have a proper follow-up for the therapeutic responses and for anticipating recurrence it is urgent in the clinical setting. Liquid biopsy provides the basic principle for a non-invasive method for the routinely monitoring of BC. However, due to the heterogeneity of tumors during onset and progression, the search for tumor DNA mutations of targeted genes in plasma/serum is a limiting factor. A possible approach overtaking this problem comes from the measurement of cell-free DNA integrity, which is an independent factor from the mutational status and theoretically is representative of all tumors. This review summarizes the state-of-the-art of cell-free DNA integrity researches in BC, the controversies and the future perspective.
Keywords: cfDNA integrity, Liquid biopsy, Breast cancer, ALU sequences, LINE-1 sequences
Core tip: Despite the potentiality of cell-free DNA integrity as a useful tool for the monitoring of Breast Cancer (BC), evinced in some clinical studies, the scientific community has not reached agreeable conclusions to translate the results from the bench-to-the-bedside yet. The main controversy regards the targets’ choice and the size of circulating cell-free tumor DNA fragments. This work underlines the utility of cell-free DNA Integrity evaluation for BC follow-up and at the same time highlights the common concepts explaining the different results in line of future directions.
INTRODUCTION
Breast cancer (BC) is still the most common cancer and the second cause of cancer-related death in women worldwide[1]. A timely knowledge of its occurrence, responsiveness to therapies and recurrence is becoming of paramount importance for clinicians to adopt specific and more efficient approaches with regards to any single patient’s health assistance. In clinical routine, the evaluation of serum markers as CEA or CA15-3 is still used for BC follow-up, but with a low specificity and sensibility[2-5]. Up to now, one of the most promising frontiers in this field is the liquid biopsy. Recently, the meta-analysis on the clinical utility of circulating tumor cells (CTC) in early BC or in metastatic BC (MBC) provides a solid rationale for their use in oncological settings[6-8]. However, their routinely use is still compromised by the relatively high cost of the technique.
Circulating cell-free DNA and qPCR measurement
From the blood circulation, it is possible to derive CTC, exosomes or cell-free nucleic acids (Figure 1). Cell-free DNA (cfDNA), consists of DNA fragments released after cell death processes from both tumor and normal cells. The circulating tumor DNA (ctDNA) can be differentiated from the rest of the cfDNA by looking at tumor-specific DNA changes, including mutations, gene amplifications, rearrangements and methylations[9] proving it as a valid non-invasive biomarker to monitor tumor growth, spread, clonal evolution and response to therapies[10]. This can be achieved either by a qualitative way (i.e., type of mutations) or quantitative way (i.e., copy number evaluation of mutated genes). However, the known mutations that can be used in liquid biopsy represent a limited percentage of patients. As an example, the most studied PI3KCA mutations all together have been found in about 30%-40% of BC patients[11].
Figure 1.
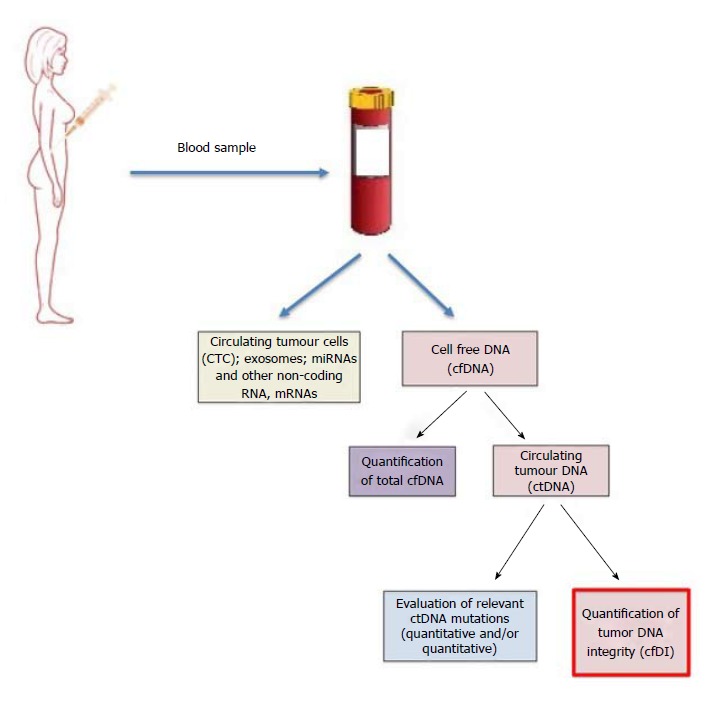
Diagram summarizing the possibility to monitor breast cancer from the blood circulating DNA.
Here, both low-cost and easy-to-be-perform methods that are not bound to one or few specific genetic mutations to predict occurrence and monitor disease progression in BC patients will be described in line of what is currently known in literature.
Briefly, real-time polymerase chain reaction-or quantitative PCR (qPCR) is a powerful advancement of PCR technology that enables the measurement of the starting amount of nucleic acids in the reaction without the need for post-PCR gel analysis. This is achieved by the possibility to detect in a real-time manner the amplification process by fluorescence and to measure the amplification products of samples at exponential phases. Through this technology the expression of a target is measured by fluorescent probes or DNA-labelling dyes. Of note, the qPCR dyes do not discriminate between specific or non- specific amplicon products, thus there is a need for an accurate testing of the annealing conditions and buffer reagents to guarantee specificity of the reaction. The quantification of an unknown sample can be absolute by using an internal amplification standard curve obtained with known DNA quantities or it can be relative by comparison of the difference in cycle threshold values (Ct) of a unknown sample with respect to reference (mainly expressed as ∆∆Ct values)[12,13]. Finally, to improve the accuracy of measurements, qPCR offers, together with the basic reagents, a passive fluorescein or ROX dyes to remove well-factors. The fluorescein acts as a passive reference dye, providing sufficient background fluorescence before the amplification reaction occurs, removing in this way the well factors- such as pipetting inaccuracies and fluorescence fluctuations-from the plate with the test samples.
Quantification of total circulating cell-free DNA
Some studies have focused on the quantification of total cfDNA levels using GAPDH, Beta-globin, Beta2-Microglobulin, hTERT or LINE-1 as potential target genes, making the higher levels of cfDNA as a way to distinguish benign from malignant BC[14-18]. Also SYBR Green’s fluorescence to measure total serum cfDNA has been investigated[19]. However, in our opinion, it is worth to consider how the total cfDNA levels are susceptible to increase also by the presence of other pathological conditions (e.g., infection, inflammation, etc.), thus influencing the results.
Quantification of cell-free DNA integrity
The detection of ctDNA levels using cell-free DNA integrity (cfDI) measurement, as ratio between longer and shorter DNA fragments, is more specific than total serum cfDNA and has been explored in BC by qPCR by many authors using SYBRGreen fluorescent dye (Table 1). In principle, normal cells, undergoing apoptosis, release DNA fragments of about 200 bp as the result of enzymatic cleavage of nucleosome units; whereas, tumor cells undergo many different death processes, including necrosis and autophagy, and they can release DNA fragments of different sizes[20,21]. Umetani et al[22], using ALU targets proposed cfDI for the first time as a valuable tool to identify primary BC, showing it could be suitable to define lymph node metastasis in a group of 83 patients compared to 51 healthy controls. They measured in serum shorter fragments of 115 bp that were considered as derived from apoptotic normal cells and larger ones of 247 bp as ctDNA, derived from necrosis/autophagy of cancer cells. The cfDI value calculated as the ratio quantity of longer over shorter fragments, ALU247/ALU115, was found to be higher in BC patients with high grade cancer compared to healthy controls. Accordingly to Umetani et al[22], Agostini et al[23] using the same ALU247 bp and ALU115bp targets demonstrated in plasma that cfDI value was twice higher in BC patients (n = 39) vs healthy controls (n = 49). Subsequently, Stötzer et al[24] proved in plasma that the ratio ALU247/115 were higher in patients with locally confined BC and MBC (n = 47) than benign BC (n = 12) (P < 0.001) but not vs healthy controls (n = 28). Moreover, this group evidenced that ALU concentrations alone were very interesting as markers for locally confined BC, while the use of cfDI was limited by the elevated levels found in some healthy controls. However, Iqbal et al[25] enrolling a larger number of women (148 patients vs 51 healthy controls) confirmed that the cfDI value, represented as ALU247/115 ratio, was significantly higher in serum of patients compared to healthy controls. Moreover, through a multivariate analysis, they showed a correlation between the cfDI value and the tumor size to predict the overall survival (OS) at 5 years and disease-free survival (DFS) at 4 years. Madhavan et al[21] also considered cfDI as a useful biomarker for BC in the largest patients’ cohort (82 BC and 201 MBC) by using different primer set for ALU sequences and introducing LINE-1 as another DNA repetitive element target. They quantified ALU 260 bp and LINE-1 266 bp amplicons vs ALU 111 bp and LINE-197 bp amplicons, respectively. They showed, differently than the other groups, cfDI value was lower in BC patients vs healthy control and positively correlated with a decrease in progression-free survival (PFS) (P = 0.0025 for ALU) and OS (P < 0.0001 for both ALU and LINE-1). Similarly, using the same ALU260/111 and LINE-1 266/197 ratios, Cheng et al[26] showed that cfDI was significantly lower in recurrent BC (n = 37) vs non-recurrent BC (n = 175) (P < 0.001 for both ALU and LINE-1 cfDI values) but they did not provide as an extra measure healthy controls. Interestingly, this latter research group showed that a higher risk of developing recurrence could be predicted by the reduction of cfDI value (P = 0.020 for ALU and P = 0.019 for LINE-1 cfDI values, respectively). Finally, it should be mentioned that Cheng et al[27] recently observed that higher cfDI values for both ALU and LINE-1 targets in MBC patients correlated with longer PFS and OS. However, Kamel et al[20] measuring the 400 bp and 100 bp amplicons of the Beta-actin from the DNA derived from plasma of 95 BC and 95 benign lesions vs 70 healthy controls estimated a cfDI- as difference between longer and shorter fragments- accordingly to Umetani et al[22] and the other authors[23-25], while yet differently from Madhavan et al[21]. In fact cfDI was found significantly higher in BC samples compared to those of benign and healthy subjects (P < 0.001). Moreover, they related those higher values to TNM stage, suggesting a cut-off to identify the more aggressive BC[20]. In agreement with Kamel et al[20], Maltoni et al[28] recently showed that tumour cells released longer DNA fragments than normal cells in the bloodstream. They quantified large fragments of 295 bp, 264 bp, 266 bp, 274 bp and short amplicons of 126, 128, 129, 129 bp from HER2, MYC, BCAS1 and PIK3CA, respectively, from the serum of healthy females (n = 10), non-recurrent BC (n = 58) and recurrent BC (n = 21). They estimated cfDI as the ratio between longer and shorter amplicons of these genes and demonstrated that BCAS1, MYC and PIK3CA long/short amplicons were significantly higher in patients compared to healthy controls (P = 0.002, P = 0.030 and P = 0.004, respectively). On the other hand, there was no significant difference for long/short amplicons of HER2[27].
Table 1.
cfDI evaluation for the monitoring of breast cancer
| Targets, length of the amplicons and primers’ sequences | Patients with primary BC | Results | Ref. |
| ALU, 115 bp | Healthy females (n = 51) and BC patients (n = 83) DNA from serum | The ratio ALU247/115 was higher in 51 patients with stage II (P = 0.005), stage III (P < 0.0001), stage IV (0.002) compared to healthy controls but not in 32 patients with stage 0 or I | Umetani et al[22], 2006 |
| FW: 5’-CCTGAGGTCAGGAGTTCGAG-3’ | |||
| RV: 5’-CCCGAGTAGCTGGGATTACA-3’ | |||
| ALU, 247 bp | |||
| FW: 5’-GTGGCTCACGCCTGTAATC-3’ | |||
| RV: 5’-CAGGCTGGAGTGCAGTGG-3’ | |||
| ALU, 115 bp | Healthy females (n = 49) and BC patients (n = 39) DNA from plasma | In the group of patients the ratio ALU247/115 was twice higher (P < 0.0001) than in the group of healthy controls | Agostini et al[23], 2012 |
| FW: 5’-CCTGAGGTCAGGAGTTCGAG-3’ | |||
| RV: 5’-CCCGAGTAGCTGGGATTACA-3’ | |||
| ALU, 247 bp | |||
| FW: 5’-GTGGCTCACGCCTGTAATC-3’ | |||
| RV: 5’-CAGGCTGGAGTGCAGTGG-3’ | |||
| ALU, 115 bp | Healthy females (n = 28), benign breast disease patients (n = 12), locally confined BC patients (n = 65) and MBC patients (n = 47) DNA from plasma | The ratio ALU247/115 was higher in patients with locally confined BC and metastatic BC than in benign BC (P < 0.001), but not vs healthy controls | Stötzer et al[24], 2014 |
| FW: 5’-CCTGAGGTCAGGAGTTCGAG-3’ | |||
| RV: 5’-CCCGAGTAGCTGGGATTACA-3’ | |||
| ALU, 247 bp | |||
| FW: 5’-GTGGCTCACGCCTGTAATC-3’ | |||
| RV: 5’-CAGGCTGGAGTGCAGTGG-3’ | |||
| ALU, 111 bp | Healthy females (n = 100), primary BC patients (n = 82) and MBC patients (n = 201) DNA from plasma | Both the ratios ALU 260/111 and LINE-1 266/97 were lower in primary BC patients (ALU: P = 0.046; LINE-1 P = 0.041) In MBC patients the lower values of cfDI were related to both a decrease in PFS (P = 0.0025 for ALU) and OS (P < 0.0001 for both ALU and LINE-1 fragments) | Madhavan et al[21], 2014 |
| FW: 5’-CTGGCCAACATGGTGAAAC-3’ | |||
| RV: 5’-AGCGATTCTCCTGCCTCAG-3’ | |||
| ALU, 260 bp | |||
| FW: 5’-ACGCCTGTAATCCCAGCA-3’ | |||
| RV: 5’-CGGAGTCTCGCTCTGTCG-3’ | |||
| LINE-1, 97 bp | |||
| FW: 5’-TGGCACATATACACCATGGAA-3’ | |||
| RV: 5’TGAGAATGATGGTTTCCAATTTC-3’ | |||
| LINE-1, 266 bp | |||
| FW: 5’-ACTTGGAACCAACCCAAATG-3’ | |||
| RV: 5’-CACCACAGTCCCCAGAGTG-3’ | |||
| ALU, 115 bp | Healthy females (n = 51) and BC patients (n = 148) DNA from serum | The ratio ALU 247/115 was significantly higher in patients compared to controls (P < 0.001) | Iqbal et al[25], 2015 |
| FW: 5’-CCTGAGGTCAGGAGTTCGAG-3’ | |||
| RV: 5’-CCCGAGTAGCTGGGATTACA-3’ | |||
| ALU, 247 bp | |||
| FW: 5’-GTGGCTCACGCCTGTAATC-3’ | |||
| RV: 5’-CAGGCTGGAGTGCAGTGG-3’ | |||
| Beta-actin, 100 bp | Healthy females (n = 70), benign lesions (n = 95) and BC patients (n = 95) DNA from plasma | cfDI value calculated as difference between 400 bp and 100 bp fragments Higher cfDI values were obtained in BC compared to benign lesions and healthy subjects (P < 0.001) | Kamel et al[20], 2016 |
| FW: 5’-GCACCACACCTTCTACAATGA-3’ | |||
| RV: 5’-GTCATCTTCTCGCGGTTGGC-3’ | |||
| Beta-actin, 400 bp | |||
| FW: 5-GCACCACACCTTCTACAATGA-3’ | |||
| (common primer) | |||
| RV: 5’-TGTCACGCACGATTTCCC-3’ | |||
| HER2, 126 bp | Healthy females (n = 10), BC patients (n = 79) DNA from serum | The ratios BCAS1 266/129, MYC 264/128, PIK3CA 274/129 were significantly higher in patients compared to controls (P = 0.002, P = 0.030 and P = 0.004, respectively) No significant values for HER2 targets | Maltoni et al[28], 2017 |
| FW-5-CCAGGGTGTTCCTCAGTTGT-3’ | |||
| RV-5- -GGAGTTCCTGCAGAGGACAG-3’ | |||
| HER2, 295 bp | |||
| FW-5’-CCAGGGTGTTCCTCAGTTGT-3’ | |||
| RV-5’-TCAGTATGGCCTCACCCTTC-3’ | |||
| MYC, 128 bp | |||
| FW-5-GGCATTTAAATTTCGGCTCA-3’ | |||
| RV-5-AAAAGCCAAATGCCAACTT-3’ | |||
| MYC, 264 bp | |||
| FW-5’-TGGAGTAGGGACCGCATATC-3’ | |||
| RV-5’-ACCCAACACCACGTCCTAAC-3’ | |||
| BCAS1, 129 bp | |||
| FW-5-GGGTCAGAGCTTCCTGTGAG-3’ | |||
| RV-5-TATCATGCCTTGGAGAACCA-3’ | |||
| BCAS1, 266 bp | |||
| FW-5’-GGGTCAGAGCTTCCTGTGAG-3’ | |||
| RV-5’-CGTTGTCCTGAAACAGAGCA-3’ | |||
| PIK3CA, 129 bp | |||
| FW-5’CTCCACGACCATCATCATCAGGT-3’ | |||
| RV-5’-TGGTTATTAATGAGCCTCACGG-3’ | |||
| PIK3CA, 274 bp | |||
| FW-5’-CTC CACGAC CAT CATCAGGT-3’ | |||
| RV-5’-CGAAGGTCACAAAGTCGTCT-3’ | |||
| ALU, 111 bp | Non-recurrent BC patients (n = 175) vs recurrent-BC patients (n = 37) No healthy females reported DNA from plasma | Both the ratios ALU260/111 and LINE1-266/97 were significantly lower during follow-up in recurrent BC vs non recurrent BC (P < 0.001 for both ALU and LINE-1 cfDI), Moreover, BC patients with a lower cfDI had higher risk of developing recurrence compared to patients with higher cfDI (P = 0.020 for ALU cfDI and P = 0.019 for LINE-1 cfDI, respectively) | Cheng et al[26], 2017 |
| FW: 5’-CTGGCCAACATGGTGAAAC-3’ | |||
| RV: 5’-AGCGATTCTCCTGCCTCAG-3’ | |||
| ALU, 260 bp | |||
| FW: 5’-ACGCCTGTAATCCCAGCA-3’ | |||
| RV: 5’-CGGAGTCTCGCTCTGTCG-3’ | |||
| LINE-1, 97 bp | |||
| FW: 5’-TGGCACATATACACCATGGAA-3’ | |||
| RV: 5’-TGAGAATGATGGTTTCCAATTTC-3’ | |||
| LINE-1, 266 bp | |||
| FW: 5’-ACTTGGAACCAACCCAAATG-3’ | |||
| RV: 5’-CACCACAGTCCCCAGAGTG-3’ | |||
| ALU, 111 bp | MBC patients (total n = 268) No healthy females DNA from plasma | Both the ratios ALU260/111 and LINE1-266/97 significantly increased in 268 MBC patients treated with one cycle of chemotherapy (MBCLB) compared to MBC at baseline (MBC1C) (P = 0.00017 for ALU -0.053 vs 0.063- and P = 0.0016 for LINE-1-0.45 vs 0.49) Moreover, in both MBCBL and MBC1C patients with a higher cfDI (for both ALU and LINE-1) correlated with a higher PFS and OS vs lower cfDI MBC patients | Cheng et al[27], 2018 |
| FW: 5’-CTGGCCAACATGGTGAAAC-3’ | |||
| RV: 5’-AGCGATTCTCCTGCCTCAG-3’ | |||
| ALU, 260 bp | |||
| FW: 5’-ACGCCTGTAATCCCAGCA-3’ | |||
| RV: 5’-CGGAGTCTCGCTCTGTCG-3’ | |||
| LINE-1, 97 bp | |||
| FW: 5’-TGGCACATATACACCATGGAA-3’ | |||
| RV: 5’TGAGAATGATGGTTTCCAATTTC-3’ | |||
| LINE-1, 266 bp | |||
| FW: 5’-ACTTGGAACCAACCCAAATG-3’ | |||
| RV: 5’-CACCACAGTCCCCAGAGTG-3’ |
BC: Breast cancer; cfDNA: Cell-free DNA; cfDI: Cell-free DNA integrity; ctDNA: Circulating tumour DNA; DFS: Disease free survival; MBC: Metastatic breast cancer; PFS: Progression-free survival; OS: Overall survival; qPCR: Quantitative real-time PCR; ddPCR: Droplet digital PCR.
DISCUSSION
The overall literature on cfDI is intriguing as it has an extraordinary potential for the monitoring of BC, but it remains to be clarified what is the expected value of cfDI: some authors claimed that ctDNA is made of longer amplicons than normal cfDNA, explaining why the cfDI increased in BC[20,22-25,27], whereas other research groups, using different primers, claimed the exact opposite[21,26].
Most of the authors, in their measurement of cfDI through the ALU sequences, decided to use a standard DNA curve, as for Umetani et al[22], to derive quantifications of their DNA[21-25,27], and used the fluorescein or ROX passive reference dyes to improve the quality of their results[23,25]. Additionally, the specificities of the amplification reactions for the different couple of primers described in the papers have been controlled by means of denaturation curves or gel electrophoresis. This implies that the different results by qPCR hardly can be attributable to the laboratory’s methodology, although we cannot completely exclude some variability in sample collection in the studies here described. Of note, differently than the other groups, Stötzer et al[24] have adopted a slightly different protocol for ALU amplifications by introducing UDP-DNA glycosidase.
Higher cfDI values in BC vs healthy controls were found in larger patients’ cohorts derived from independent clinical settings and by using more different targets compared to studies claiming lower cfDI values in the tumor (Figure 2). Of note, higher cfDI in tumor than healthy controls were found in those studies that have analyzed mainly BCs, which did not reach the metastatic setting[22,23,25], whereas lower cfDI than healthy controls were reported in a study using the largest MBC patients’ cohort up-to-date[21]. It is interesting to note that Umetani et al[22] proposed an increased cfDI value to predict local micrometastasis and recently Cheng et al[28] observed that cfDI value particularly decreased in BC patients with visceral metastasis. Thus we suggest that cfDI value can increase at initial stages of the BC and decrease in MBC. Surely, the most promising targets for the measurement of cfDI are represented by repetitive elements such as ALU and LINE-1 sequences, accounting for nearly 10% and 17% of the total genome, respectively. It is worth nothing that reproducible results were obtained when independent groups used the same ALU primer pairs, either those demonstrating higher cfDI[22-25] and those demonstrating lower cfDI in BC[21,26]. In our opinion, the methods of DNA extractions merely could have influenced the results. Interestingly, by looking with BLASTN genomic RefSeqGene Human at the target sites of ALU primers’ pairs used by the research groups obtaining divergent results, we observed different target sites for ALU247/115 pairs compared to the ALU260/111 ones. We cannot exclude that this could contribute to the opposite cfDI values obtained by the different research groups comparing BC vs healthy controls. Moreover, we would like to point out that the qPCR methodology by SYBR Green is not very sensitive in quantifying very small DNA fragments in diluted solutions[29], as it could be in liquid biopsy, and that the variability of amplification efficiency of a sample can be overtaken by many replicates and independent experiments, that are hard to performed with samples derived from liquid biopsy. In this respect, the determination of cfDI in liquid biopsy samples would benefit by more sensitive and accurate technologies such as digital droplet PCR (ddPCR).
Figure 2.
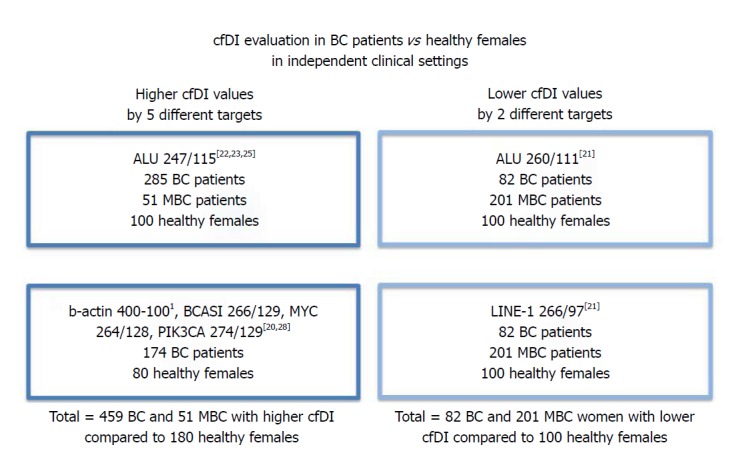
Summary of the literature data on cfDI determination in primary breast cancer vs healthy females. 1Note that cfDI by β-actin was evaluated as difference between large and short amplicons and not as ratio longer to shorter amplicons. BC: Breast cancer; MBC: Metastatic breast cancer.
In conclusion, monitoring primary and MBC through a non-invasive analysis such as that of circulating DNA remains one of the most interesting goals to achieve. Surely, the mutations in liquid biopsy are of paramount importance for targeted therapies and for monitoring response to treatment. However, the most interesting benefit-to-cost analysis for the follow-up of BC and its recurrence seems to be the evaluation of circulating cfDI. Future investigations for cfDI by ddPCR are warranted for the (1) testing for the choice of best targets; (2) clarification of the clinical significance of larger and shorter DNA fragments origin in serum/plasma; and (3) a better understanding of the potential clinical impact of cfDI in anticipating recurrence and responsiveness to therapies for all patients, independently from the mutational signature of BC.
Footnotes
Conflict-of-interest statement: No conflicts of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: Italy
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Peer-review started: November 25, 2017
First decision: January 15, 2018
Article in press: March 14, 2018
P- Reviewer: Hosseini M, Kanat O S- Editor: Cui LJ L- Editor: A E- Editor: Wang CH
Contributor Information
Navid Sobhani, Department of Medical, Surgical and Health Sciences, University of Trieste, Cattinara Academic Hospital, Trieste 34149, Italy.
Daniele Generali, Department of Medical, Surgical and Health Sciences, University of Trieste, Cattinara Academic Hospital, Trieste 34149, Italy.
Fabrizio Zanconati, Department of Medical, Surgical and Health Sciences, University of Trieste, Cattinara Academic Hospital, Trieste 34149, Italy.
Marina Bortul, Department of Medical, Surgical and Health Sciences, University of Trieste, Cattinara Academic Hospital, Trieste 34149, Italy.
Bruna Scaggiante, Department of Life Sciences, University of Trieste, Trieste 34127, Italy.
References
- 1.Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965. [DOI] [PubMed] [Google Scholar]
- 2.Duffy MJ, Evoy D, McDermott EW. CA 15-3: uses and limitation as a biomarker for breast cancer. Clin Chim Acta. 2010;411:1869–1874. doi: 10.1016/j.cca.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 3.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC Jr; American Society of Clinical Oncology. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 4.Lauro S, Trasatti L, Bordin F, Lanzetta G, Bria E, Gelibter A, Reale MG, Vecchione A. Comparison of CEA, MCA, CA 15-3 and CA 27-29 in follow-up and monitoring therapeutic response in breast cancer patients. Anticancer Res. 1999;19:3511–3515. [PubMed] [Google Scholar]
- 5.Shao Y, Sun X, He Y, Liu C, Liu H. Elevated Levels of Serum Tumor Markers CEA and CA15-3 Are Prognostic Parameters for Different Molecular Subtypes of Breast Cancer. PLoS One. 2015;10:e0133830. doi: 10.1371/journal.pone.0133830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bidard FC, Peeters DJ, Fehm T, Nolé F, Gisbert-Criado R, Mavroudis D, Grisanti S, Generali D, Garcia-Saenz JA, Stebbing J, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15:406–414. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 7.Bidard FC, Michiels S, Mueller V, Riethdorf S, Esserman L, Lucci A, Naume B, Horiguchi J, Gisbert-Criado R, Sleijfer S, et al. Abstract S3-01: IMENEO: International MEta-analysis of circulating tumor cell detection in early breast cancer patients treated by NEOadjuvant chemotherapy. Cancer Res. 2017;77:S3–S1. doi: 10.1158/1538-7445.SABCS16-S3-01. [DOI] [Google Scholar]
- 8.Bonora M, Wieckowsk MR, Chinopoulos C, Kepp O, Kroemer G, Galluzzi L, Pinton P. Molecular mechanisms of cell death: central implication of ATP synthase in mitochondrial permeability transition. Oncogene. 2015;34:1608. doi: 10.1038/bjc.2012.137. [DOI] [PubMed] [Google Scholar]
- 9.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 10.De Mattos-Arruda L, Caldas C. Cell-free circulating tumour DNA as a liquid biopsy in breast cancer. Mol Oncol. 2016;10:464–474. doi: 10.1016/j.molonc.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobhani N, Roviello G, Corona SP, Scaltriti M, Ianza A, Bortul M, Zanconati F, Generali D. The prognostic value of PI3K mutational status in breast cancer: a meta-analysis. J Cell Biochem. 2018 doi: 10.1002/jcb.26687. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Gal S, Fidler C, Lo YM, Taylor M, Han C, Moore J, Harris AL, Wainscoat JS. Quantitation of circulating DNA in the serum of breast cancer patients by real-time PCR. Br J Cancer. 2004;90:1211–1215. doi: 10.1038/sj.bjc.6601609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Tarhouny S, Seefeld M, Fan AX, Hahn S, Holzgreve W, Zhong XY. Comparison of serum VEGF and its soluble receptor sVEGFR1 with serum cell-free DNA in patients with breast tumor. Cytokine. 2008;44:65–69. doi: 10.1016/j.cyto.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Bechmann T, Andersen RF, Pallisgaard N, Madsen JS, Maae E, Jakobsen EH, Bak Jylling AM, Steffensen KD, Jakobsen A. Plasma HER2 amplification in cell-free DNA during neoadjuvant chemotherapy in breast cancer. J Cancer Res Clin Oncol. 2013;139:995–1003. doi: 10.1007/s00432-013-1413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler C, Radpour R, Barekati Z, Asadollahi R, Bitzer J, Wight E, Bürki N, Diesch C, Holzgreve W, Zhong XY. Levels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumors. Mol Cancer. 2009;8:105. doi: 10.1186/1476-4598-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang ZH, Li LH, Hua D. Quantitative analysis of plasma circulating DNA at diagnosis and during follow-up of breast cancer patients. Cancer Lett. 2006;243:64–70. doi: 10.1016/j.canlet.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 18.Sunami E, Vu AT, Nguyen SL, Giuliano AE, Hoon DS. Quantification of LINE1 in circulating DNA as a molecular biomarker of breast cancer. Ann N Y Acad Sci. 2008;1137:171–174. doi: 10.1196/annals.1448.011. [DOI] [PubMed] [Google Scholar]
- 19.Catarino R, Ferreira MM, Rodrigues H, Coelho A, Nogal A, Sousa A, Medeiros R. Quantification of free circulating tumor DNA as a diagnostic marker for breast cancer. DNA Cell Biol. 2008;27:415–421. doi: 10.1089/dna.2008.0744. [DOI] [PubMed] [Google Scholar]
- 20.Kamel AM, Teama S, Fawzy A, El Deftar M. Plasma DNA integrity index as a potential molecular diagnostic marker for breast cancer. Tumour Biol. 2016;37:7565–7572. doi: 10.1007/s13277-015-4624-3. [DOI] [PubMed] [Google Scholar]
- 21.Madhavan D, Wallwiener M, Bents K, Zucknick M, Nees J, Schott S, Cuk K, Riethdorf S, Trumpp A, Pantel K, et al. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat. 2014;146:163–174. doi: 10.1007/s10549-014-2946-2. [DOI] [PubMed] [Google Scholar]
- 22.Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, Martino S, Hoon DS. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24:4270–4276. doi: 10.1200/JCO.2006.05.9493. [DOI] [PubMed] [Google Scholar]
- 23.Agostini M, Enzo MV, Bedin C, Belardinelli V, Goldin E, Del Bianco P, Maschietto E, D’Angelo E, Izzi L, Saccani A, et al. Circulating cell-free DNA: a promising marker of regional lymphonode metastasis in breast cancer patients. Cancer Biomark. 2012;11:89–98. doi: 10.3233/CBM-2012-0263. [DOI] [PubMed] [Google Scholar]
- 24.Stötzer OJ, Lehner J, Fersching-Gierlich D, Nagel D, Holdenrieder S. Diagnostic relevance of plasma DNA and DNA integrity for breast cancer. Tumour Biol. 2014;35:1183–1191. doi: 10.1007/s13277-013-1158-4. [DOI] [PubMed] [Google Scholar]
- 25.Iqbal S, Vishnubhatla S, Raina V, Sharma S, Gogia A, Deo SS, Mathur S, Shukla NK. Circulating cell-free DNA and its integrity as a prognostic marker for breast cancer. Springerplus. 2015;4:265. doi: 10.1186/s40064-015-1071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng J, Cuk K, Heil J, Golatta M, Schott S, Sohn C, Schneeweiss A, Burwinkel B, Surowy H. Cell-free circulating DNA integrity is an independent predictor of impending breast cancer recurrence. Oncotarget. 2017;8:54537–54547. doi: 10.18632/oncotarget.17384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng J, Holland-Letz T, Wallwiener M, Surowy H, Cuk K, Schott S, Trumpp A, Pantel K, Sohn C, Schneeweiss A, et al. Circulating free DNA integrity and concentration as independent prognostic markers in metastatic breast cancer. Breast Cancer Res Treat. 2018;169:69–82. doi: 10.1007/s10549-018-4666-5. [DOI] [PubMed] [Google Scholar]
- 28.Maltoni R, Casadio V, Ravaioli S, Foca F, Tumedei MM, Salvi S, Martignano F, Calistri D, Rocca A, Schirone A, et al. Cell-free DNA detected by "liquid biopsy" as a potential prognostic biomarker in early breast cancer. Oncotarget. 2017;8:16642–16649. doi: 10.18632/oncotarget.15120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sedlackova T, Repiska G, Celec P, Szemes T, Minarik G. Fragmentation of DNA affects the accuracy of the DNA quantitation by the commonly used methods. Biol Proced Online. 2013;15:5. doi: 10.1186/1480-9222-15-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


