The crystal structure of chorismate mutase from B. phymatum reveals the prototypical AroQγ topology, as observed in the homolog from M. tuberculosis.
Keywords: chorismate mutase, Burkholderia phymatum, structural genomics, Seattle Structural Genomics Center for Infectious Disease, isomerase, shikimate pathway
Abstract
The bacterium Burkholderia phymatum is a promiscuous symbiotic nitrogen-fixating bacterium that belongs to one of the largest groups of Betaproteobacteria. Other Burkholderia species are known to cause disease in plants and animals, and some are potential agents for biological warfare. Structural genomics efforts include characterizing the structures of enzymes from pathways that can be targeted for drug development. As part of these efforts, chorismate mutase from B. phymatum was produced and crystallized, and a 1.95 Å resolution structure is reported. This enzyme shares less than 33% sequence identity with other homologs of known structure. There are two classes of chorismate mutase: AroQ and AroH. The bacterial subclass AroQγ has reported roles in virulence. Chorismate mutase from B. phymatum has the prototypical AroQγ topology and retains the characteristic chorismate mutase active site. This suggests that substrate-based chorismate mutase inhibitors will not be specific and are likely to affect beneficial bacteria such as B. phymatum.
1. Introduction
Burkholderia are nonfermenting motile Gram-negative bacteria that are among the largest groups of species of Betaproteobacteria and include infective and symbiotic species (Yabuuchi et al., 1992 ▸; Sawana et al., 2014 ▸). B. glumae and B. gladioli cause infection in plants (Cui et al., 2016 ▸). B. phymatum was identified from root-nodule isolates from tropical legumes and is capable of symbiotic nitrogen fixation with the legumes Machaerium lunatum and Mimosa pudica (Vandamme et al., 2002 ▸). Owing to the importance of Burkholderia, there is a need to clarify the structures of enzymes that play important roles in the life cycles of these bacteria. Enzymes in the shikimate pathway are important for the biochemical synthesis of aromatic amino acids, folic acid, ubiquinone and other aromatic compounds (Mousdale & Coggins, 1991 ▸; Abell, 1999 ▸; Haslam, 1993 ▸; Coggins et al., 2003 ▸). Chorismate mutases catalyze the Claisen rearrangement of chorismate to prephenate, which is the first committed step in the shikimate pathway, and these enzymes are only found in fungi, bacteria and higher plants. Chorismate mutases are isomerases that are also known as hydroxyphenylpyruvate synthase and belong to two classes: AroQ and AroH. The AroH class are monofunctional enzymes that have a trimeric α/β-barrel topology, while the AroQ class are made up of a homodimer of three helical subunits and are sometimes bifunctional enzymes. The AroQ class is more common in nature than the AroH class, and members of the bacterial subclass AroQγ have reported roles in virulence and are considered to be drug targets, as they are absent in mammals (Haslam, 1993 ▸; Roberts et al., 1998 ▸). Structure-based therapeutic discovery efforts using chorismate mutase from Mycobacterium tuberculosis are under way and have been reviewed elsewhere (Khanapur et al., 2017 ▸). As part of structural genomics studies at the Seattle Structural Genomics Center for Infectious Disease, chorismate mutase from B. phymatum was produced and crystallized, and its high-resolution crystal structure was determined (Raymond et al., 2011 ▸; Myler et al., 2009 ▸; Baugh et al., 2013 ▸).
2. Materials and methods
2.1. Macromolecule production
Chorismate mutase from B. phymatum was cloned, expressed and purified by the Seattle Structural Genomics Center for Infectious Disease (SSGCID; Myler et al., 2009 ▸; Stacy et al., 2011 ▸) following standard protocols described previously (Bryan et al., 2011 ▸; Choi et al., 2011 ▸; Serbzhinskiy et al., 2015 ▸). Dr Mary Lidstrom of the University of Washington provided the genomic DNA from B. phymatum STM815 (Moulin et al., 2014 ▸). The mature protein (UniProt ID B2JYH9, GenBank ID ACC76687.1) encoding amino acids 18–182 was PCR-amplified from genomic DNA using the primers shown in Table 1 ▸. The resultant amplicon was cloned into the ligation-independent cloning (LIC; Aslanidis & de Jong, 1990 ▸) pET-14b-based expression vector pBG1861, which was kindly provided by Wesley Van Voorhis, University of Washington and encodes a noncleavable 6×His fusion tag (MAHHHHHHM-ORF). Plasmid DNA was transformed into chemically competent Escherichia coli BL21(DE3)-R3 Rosetta cells. The cells were expression-tested, and two litres of culture were grown using auto-induction medium (Studier, 2005 ▸) in a LEX Bioreactor (Epiphyte Three Inc.). The expression clone was assigned the SSGCID target identifier BuphA.00160.b.B2.
Table 1. Macromolecule-production information.
| Source organism | Paraburkholderia phymatum (strain DSM 17167/CIP 108236/LMG 21445/STM815) |
| DNA source | GenBank ID ACC76687.1 |
| Forward primer | CTCACCACCACCACCACCATATGGGAGCGCAGCAGGATGCG |
| Reverse primer | ATCCTATCTTACTCACTTAAGATTTGACACATATCCGTGCGAC |
| Cloning vector | pBG1861 |
| Expression vector | pBG1861 |
| Expression host | E. coli BL21(DE3)-R3 Rosetta |
| Complete amino-acid sequence of the construct produced | MAHHHHHHGAQQDAFVPLVRSMADRLNTADQVALSKWDTGQPVYDGQREAQVIANAATMASEYGLTAEDAINIFSDQVEANKEVQYALLNNWRRQGDAPATPRQSLAGVIRPILDKLQASIMQNLQSVAPLRSIADCHALVASAVGQVAEQASLDVLHRAALDRAVARICVKS |
BuphA.00160.b.B2 was purified by a two-step protocol consisting of immobilized metal-affinity chromatography (IMAC) followed by size-exclusion chromatography (SEC). All chromatography runs were performed on an ÄKTApurifier 10 (GE Healthcare) using automated IMAC and SEC programs as described previously (Bryan et al., 2011 ▸). Specifically, SEC was performed on a HiLoad 26/600 Superdex 75 column (GE Healthcare) using a mobile phase consisting of 300 mM NaCl, 20 mM HEPES, 5% glycerol, 1 mM TCEP pH 7.0. The peak fractions eluted as a single peak consistent with monomeric protein when denatured and run on a reduced SDS–PAGE gel; these fractions eluted with a projected molecular weight of 22 kDa, indicating that the protein could either be a monomer or a dimer in solution. The peak fractions were concentrated to 44.8 mg ml−1 using an Amicon Ultra 15 30 kDa molecular-weight cutoff concentrator (Millipore, Billerica, Massachusetts, USA). Aliquots of 200 µl were flash-frozen in liquid nitrogen and stored at −80°C until use for crystallization. Both the clone and the purified protein can be ordered at https://apps.sbri.org/SSGCIDTargetStatus/Target/BuphA.00160.b.
2.2. Crystallization
Established crystallization approaches at the SSGCID were followed. Briefly, recombinant BuphA.00160.b.B2 was diluted to 22.4 mg ml−1. Single crystals were obtained by vapor diffusion in sitting drops directly from condition D5 of the Microlytic MCSG1 screen, using ammonium formate and polyethylene glycol (PEG) 3350 as precipitants (Table 2 ▸). 0.4 µl protein solution and 0.4 µl precipitant solution were combined using a robot and the resulting 0.8 µl drop was equilibrated against a reservoir containing 80 µl precipitant solution.
Table 2. Crystallization.
| Method | Vapor diffusion, sitting drop |
| Plate type | Rigaku Reagents XJR |
| Temperature (K) | 290 |
| Protein concentration (mg ml−1) | 22.4 |
| Buffer composition of protein solution | 300 mM NaCl, 20 mM HEPES, 5% glycerol, 1 mM TCEP pH 7.0 |
| Composition of reservoir solution | 200 mM ammonium formate pH 6.6, 20% PEG 3350 |
| Protein:precipitant | 0.4 µl:0.4 µl |
| Volume of reservoir (µl) | 80 |
2.3. Data collection and processing
Data collection and processing were performed using established protocols at the SSGCID. Specifically, a single crystal was transferred into a cryosolution that consisted of 90% crystallization solution and 10% ethylene glycol, flash-cooled in liquid nitrogen and transferred into a puck for data collection on beamline 21-ID-F at the Advanced Photon Source (APS). Data were processed using XDS and XSCALE (Kabsch, 2010 ▸). Additional data-collection information is provided in Table 3 ▸. The raw images and detailed data-collection information are available for download (https://proteindiffraction.org/project/5ts9/).
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | Beamline 21-ID-F, APS |
| Wavelength (Å) | 0.97872 |
| Temperature (K) | 100 |
| Detector | Rayonix MX-300 CCD |
| Crystal-to-detector distance (mm) | 250 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 200 |
| Exposure time per image (s) | 1 |
| Space group | P21 |
| a, b, c (Å) | 62.59, 151.12, 73.08 |
| α, β, γ (°) | 90, 90.84, 90 |
| Mosaicity (°) | 0.206 |
| Resolution range (Å) | 50–1.95 (2.00–1.95) |
| Total No. of reflections | 417948 (30787) |
| No. of unique reflections | 98259 (7236) |
| Completeness (%) | 99.7 (99.70) |
| CC1/2 | 0.996 (0.808) |
| Multiplicity | 4.25 (4.25) |
| 〈I/σ(I)〉 | 11.7 (2.8) |
| R merge | 0.096 (0.538) |
| R r.i.m. † | 0.108 (0.595) |
| Overall B factor from Wilson plot (Å2) | 18.9 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution and refinement
The structure was solved by molecular replacement with MOLREP (Lebedev et al., 2008 ▸; Vagin & Teplyakov, 2010 ▸) using PDB entry 4oj7 (Seattle Structural Genomics Center for Infectious Disease, unpublished work) as the starting model. Model building and structure refinement were performed using the PHENIX package (Adams et al., 2010 ▸) and the resulting structure-refinement data are provided in Table 4 ▸.
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 47–1.95 (2.00–1.95) |
| Completeness (%) | 99.7 |
| σ Cutoff | F > 1.360σ(F) |
| No. of reflections, working set | 98234 (6842) |
| No. of reflections, test set | 2017 (132) |
| Final R cryst | 0.156 (0.199) |
| Final R free | 0.206 (0.260) |
| No. of non-H atoms | |
| Protein | 9644 |
| Ligand | 0 |
| Solvent | 1295 |
| Total | 10939 |
| R.m.s.d. | |
| Bonds (Å) | 0.009 |
| Angles (°) | 0.923 |
| Average B factors (Å2) | |
| Protein | 21.2 |
| Water | 30.7 |
| Ramachandran plot | |
| Most favored (%) | 99 |
| Allowed (%) | 1 |
3. Results and discussion
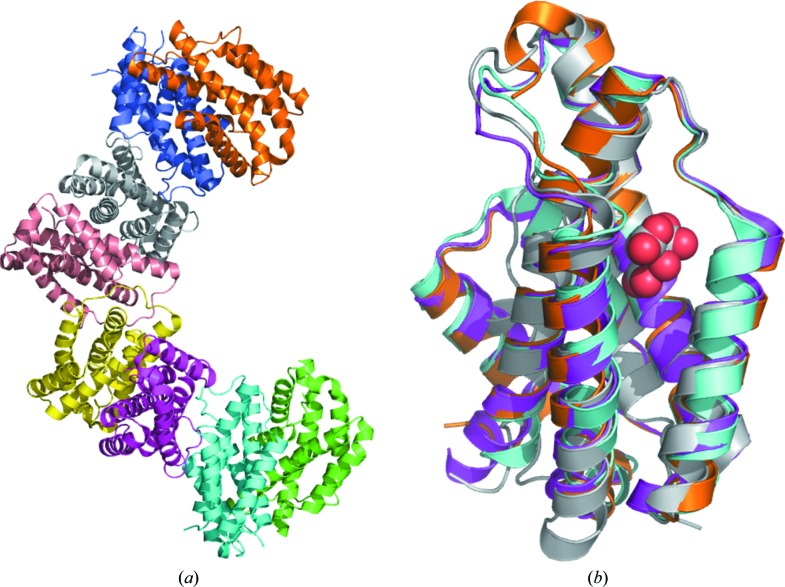
The structure of chorismate mutase from B. phymatum was solved in the monoclinic space group P21 as a tetramer of prototypical AroQ chorismate mutase homodimers (Fig. 1 ▸; Švarcová et al., 2015 ▸; Guo et al., 2003 ▸). All monomers have the AroQγ topology of three intertwined helices as observed in the secreted chorismate mutases from B. thailandensis (PDB entry 4oj7), Mycobacterium tuberculosis (PDB entry 2fp2; Ökvist et al., 2006 ▸) and Yersina pestis (PDB entry 2gbb; Kim et al., 2008 ▸) and are composed entirely of helices connected by short loops (Fig. 2 ▸).
Figure 1.
(a) Ribbon diagram of the tetramer of dimers for chorismate mutase from B. phymatum. (b) Superposed monomers of chorismate mutases with AroQγ topology from B. phymatum (PDB entry 5ts9; magenta), B. thailandensis (PDB entry 4oj7; cyan), M. tuberculosis (PDB entry 2fp2; gold) and Y. pestis (PDB entry 2gbb; gray with the citrate molecule shown as spheres).
Figure 2.
Structural and primary-sequence alignment of chorismate mutases from B. phymatum, B. thailandensis (PDB entry 4oj7), M. tuberculosis (PDB entry 2fp2), Y. pestis (PDB entry 2gbb) and E. coli (PDB entry 1ecm). The secondary-structure elements shown are α-helices (α), 310-helices (η), β-strands (β) and β-turns (TT). Identical residues are shown in white on a red background and conserved residues are shown in red. This figure was generated using ESPript (Gouet et al., 1999 ▸, 2003 ▸).
The structures most similar to BuphA.00160.b.B2 were identified by PDBeFold (http://www.ebi.ac.uk/msd-srv/ssm) analysis using the default threshold cutoffs of 70% for the percentages of the secondary structure of the target chain identified in the query protein and of the secondary structure of the query chain (Krissinel & Henrick, 2004 ▸). Out of the 132 724 entries in the PDB, there were only two unique matches identified at the default cutoff: the chorismate mutases from B. thailandensis (PDB entry 4oj7) and from M. tuberculosis (PDB entries 2fp2, 2f61 and 2ao2; Ökvist et al., 2006 ▸; Kim et al., 2006 ▸; Qamra et al., 2006 ▸). PDB entry 4oj7, the most similar structure, shares a sequence identity of 33% with an r.m.s.d. of 1.3 Å for 100% matched sequence identity. The percentage of matched sequence identity is the fraction of the secondary structure of the target chain that was identified in the query protein. The M. tuberculosis structures share 25% sequence identity; specifically, PDB entry 2fp2 has an r.m.s.d. of 1.14 Å for 86% matched sequence identity. Other chorismate mutases, for example the homolog from Y. pestis, share ∼20% sequence identity and did not meet the cutoff of PDBeFold. Overall, the tertiary structure of the B. phymatum enzyme is more similar to that of the homolog from B. thailandensis than to those from M. tuberculosis and Y. pestis (Figs. 1 ▸ b and 2 ▸).
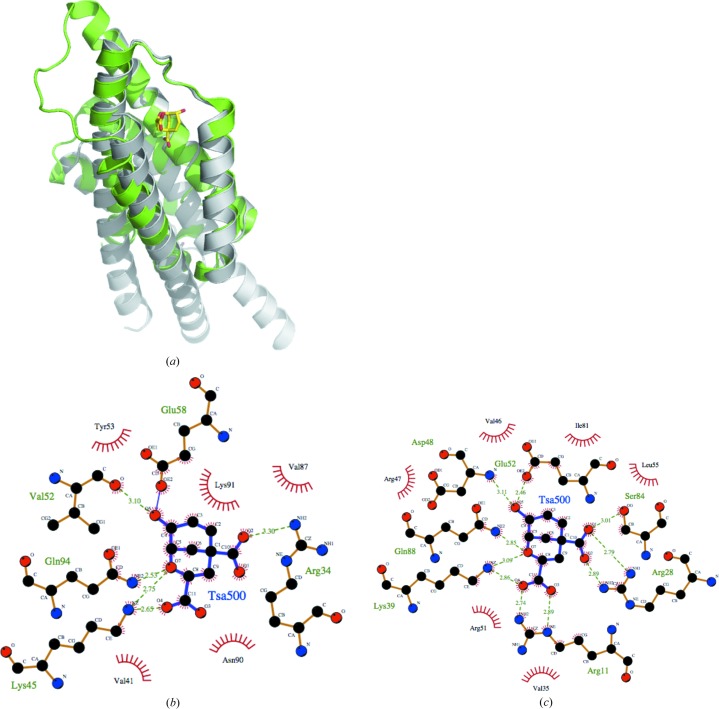
There are no known structures of any AroQγ chorismate mutase with chorismate or its analogs in the active site. However, there is a reported structure of an AroQα chorismate mutase from E. coli (PDB entry 1ecm) complexed with a transition-state analog (Lee et al., 1995 ▸). The overall topology of AroQα chorismate mutase is different from that of AroQγ, with longer helices (Fig. 3 ▸ a). While AroQα and AroQγ chorismate mutases have low sequence similarity (typically ∼20% sequence similarity), the amino-acid residues that make up the active site are largely conserved (Figs. 3 ▸ b and 3 ▸ c). This suggests that inhibitors designed to target the active sites of chorismate mutases from infective organisms may also affect those of beneficial ones.
Figure 3.
(a) Superposition of a monomer of chorismate mutase from E. coli (PDB entry 1ecm; gray) with that from B. phymatum (PDB entry 4oj7; green). The transition-state analog (TSA) in the active site of PDB entry 1ecm is shown as yellow sticks. LIGPLOT diagrams reveal conserved residues in the substrate-binding sites of the chorismate mutases from (b) B. phymatum and (c) E. coli interacting with the transition-state analog (TSA) derived from the E. coli structure.
4. Conclusions
The structure of chorismate mutase from B. phymatum reveals that it has a conserved AroQγ topology and a substrate-binding cavity with highly conserved residues. Inhibitors for this conserved cavity may not be selective between beneficial and infective organisms, suggesting that substrate-based chorismate mutase inhibition may not be the best approach for selective inhibitor design.
Supplementary Material
PDB reference: chorismate mutase from Burkholderia phymatum, 5ts9
Acknowledgments
The SSGCID consortium is directed by Dr Peter Myler (principal investigator) and comprises many different scientists working at multiple centers towards determining the three-dimensional structures of proteins from biodefense organisms and emerging infectious diseases. In particular, we would like to thank the SSGCID cloning, protein production and X-ray crystallography groups at the Center for Infectious Disease Research, the University of Washington and Beryllium Discovery Corporation.
Funding Statement
This work was funded by National Institute of Allergy and Infectious Diseases grants HHSN272200700057C and HHSN272201200025C.
References
- Abell, C. (1999). Comprehensive Natural Products Chemistry, edited by O. Meth-Cohn, D. Barton & K. Nakanishi, Vol. 1, pp. 573–607. Amsterdam: Elsevier.
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res. 18, 6069–6074. [DOI] [PMC free article] [PubMed]
- Baugh, L. et al. (2013). PLoS One, 8, e53851. [DOI] [PMC free article] [PubMed]
- Bryan, C. M., Bhandari, J., Napuli, A. J., Leibly, D. J., Choi, R., Kelley, A., Van Voorhis, W. C., Edwards, T. E. & Stewart, L. J. (2011). Acta Cryst. F67, 1010–1014. [DOI] [PMC free article] [PubMed]
- Choi, R., Kelley, A., Leibly, D., Nakazawa Hewitt, S., Napuli, A. & Van Voorhis, W. (2011). Acta Cryst. F67, 998–1005. [DOI] [PMC free article] [PubMed]
- Coggins, J. R., Abell, C., Evans, L. B., Frederickson, M., Robinson, D. A., Roszak, A. W. & Lapthorn, A. P. (2003). Biochem. Soc. Trans. 31, 548–552. [DOI] [PubMed]
- Cui, Z., Zhu, B., Xie, G., Li, B. & Huang, S. (2016). Rice Sci. 23, 111–118.
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res. 31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Guo, H., Cui, Q., Lipscomb, W. N. & Karplus, M. (2003). Angew. Chem. Int. Ed. 42, 1508–1511. [DOI] [PubMed]
- Haslam, E. (1993). Shikimic Acid: Metabolism and Metabolites. Chichester: John Wiley & Sons.
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Khanapur, M., Alvala, M., Prabhakar, M., Shiva Kumar, K., Edwin, R. K., Sri Saranya, P. S., Patel, R. K., Bulusu, G., Misra, P. & Pal, M. (2017). Bioorg. Med. Chem. 25, 1725–1736. [DOI] [PubMed]
- Kim, S.-K., Reddy, S. K., Nelson, B. C., Robinson, H., Reddy, P. T. & Ladner, J. E. (2008). FEBS J. 275, 4824–4835. [DOI] [PubMed]
- Kim, S.-K., Reddy, S. K., Nelson, B. C., Vasquez, G. B., Davis, A., Howard, A. J., Patterson, S., Gilliland, G. L., Ladner, J. E. & Reddy, P. T. (2006). J. Bacteriol. 188, 8638–8648. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Lebedev, A. A., Vagin, A. A. & Murshudov, G. N. (2008). Acta Cryst. D64, 33–39. [DOI] [PMC free article] [PubMed]
- Lee, A. Y., Karplus, P. A., Ganem, B. & Clardy, J. (1995). J. Am. Chem. Soc. 117, 3627–3628.
- Moulin, L. et al. (2014). Stand. Genomic Sci. 9, 763–774. [DOI] [PMC free article] [PubMed]
- Mousdale, D. M. & Coggins, J. R. (1991). Target Sites for Herbicide Action, edited by R. C. Kirkwood, pp. 29–56. New York: Plenum.
- Myler, P. J., Stacy, R., Stewart, L., Staker, B. L., Van Voorhis, W. C., Varani, G. & Buchko, G. W. (2009). Infect. Disord. Drug Targets, 9, 493–506. [DOI] [PMC free article] [PubMed]
- Ökvist, M., Dey, R., Sasso, S., Grahn, E., Kast, P. & Krengel, U. (2006). J. Mol. Biol. 357, 1483–1499. [DOI] [PubMed]
- Qamra, R., Prakash, P., Aruna, B., Hasnain, S. E. & Mande, S. C. (2006). Biochemistry, 45, 6997–7005. [DOI] [PubMed]
- Raymond, A., Haffner, T., Ng, N., Lorimer, D., Staker, B. & Stewart, L. (2011). Acta Cryst. F67, 992–997. [DOI] [PMC free article] [PubMed]
- Roberts, F., Roberts, C. W., Johnson, J. J., Kyle, D. E., Krell, T., Coggins, J. R., Coombs, G. H., Milhous, W. K., Tzipori, S., Ferguson, D. J., Chakrabarti, D. & McLeod, R. (1998). Nature (London), 393, 801–805. [DOI] [PubMed]
- Sawana, A., Adeolu, M. & Gupta, R. S. (2014). Front. Genet. 5, 429. [DOI] [PMC free article] [PubMed]
- Serbzhinskiy, D. A., Clifton, M. C., Sankaran, B., Staker, B. L., Edwards, T. E. & Myler, P. J. (2015). Acta Cryst. F71, 594–599. [DOI] [PMC free article] [PubMed]
- Stacy, R., Begley, D. W., Phan, I., Staker, B. L., Van Voorhis, W. C., Varani, G., Buchko, G. W., Stewart, L. J. & Myler, P. J. (2011). Acta Cryst. F67, 979–984. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Švarcová, M., Krátký, M. & Vinšova, J. (2015). Curr. Med. Chem. 22, 1383–1399. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vandamme, P., Goris, J., Chen, W. M., de Vos, P. & Willems, A. (2002). Syst. Appl. Microbiol. 25, 507–512. [DOI] [PubMed]
- Yabuuchi, E., Kosako, Y., Oyaizu, H., Yano, I., Hotta, H., Hashimoto, Y., Ezaki, T. & Arakawa, M. (1992). Microbiol. Immunol. 36, 1251–1275. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: chorismate mutase from Burkholderia phymatum, 5ts9