The putative large and small terminase genes of bacteriophage N4 that are essential for genome packaging were identified, cloned and expressed, and the proteins were purified. The purified wild-type large terminase exhibited basal and small terminase-stimulated ATPase activity and produced thin plate-like crystals under one condition.
Keywords: bacteriophage N4, genome packaging, dsDNA virus, large terminase, small terminase
Abstract
Genome packaging is a critical step in the assembly of dsDNA bacteriophages and is carried out by a powerful molecular motor known as the large terminase. To date, wild-type structures of only two large terminase proteins are available, and more structural information is needed to understand the genome-packaging mechanism. Towards this goal, the large and small terminase proteins from bacteriophage N4, which infects the Escherichia coli K12 strain, have been cloned, expressed and purified. The purified putative large terminase protein hydrolyzes ATP, and this is enhanced in the presence of the small terminase. The large terminase protein was crystallized using the sitting-drop vapour-diffusion method and the crystal diffracted to 2.8 Å resolution using a home X-ray source. Analysis of the X-ray diffraction data showed that the crystal belonged to space group P212121, with unit-cell parameters a = 53.7, b = 93.6, c = 124.9 Å, α = β = γ = 90°. The crystal had a solvent content of 50.2% and contained one molecule in the asymmetric unit.
1. Introduction
Genome packaging in dsDNA phages involves the translocation of a unit-length genome into the preformed capsid by genome-packaging machinery consisting of large and small terminases and a portal protein (Casjens, 2011 ▸; Feiss & Rao, 2012 ▸; Sun et al., 2010 ▸). The large terminase is a multifunctional protein with an N-terminal ATPase domain (Kanamaru et al., 2004 ▸; Mitchell et al., 2002 ▸) that drives the packaging motor and physically interacts with the portal protein (Sun et al., 2008 ▸). The C-terminal domain possesses the nuclease centre and is required for DNA cleavage during the packaging process (Alam et al., 2008 ▸; Smits et al., 2009 ▸; Kanamaru et al., 2004 ▸; Mitchell et al., 2002 ▸). Mutational and structural evidence suggest that the C-terminal domain is also responsible for the DNA-translocase activity (Sun et al., 2008 ▸; Kanamaru et al., 2004 ▸; Zhao et al., 2013 ▸). The small terminase binds to the packaging-initiation sites on the viral genome (Jackson et al., 1978 ▸; Catalano et al., 1995 ▸) and also regulates the activity of the large terminase (Baumann & Black, 2003 ▸; Leffers & Rao, 2000 ▸; Al-Zahrani et al., 2009 ▸). In terms of force generation, phage-packaging motors are reported to be some of the most powerful molecular motors (Smith et al., 2001 ▸; Fuller et al., 2007 ▸). Thus, structurally as well as functionally, phage genome-packaging motors are excellent prototypes to understand how the energy released from ATP hydrolysis is used for DNA translocation.
Crystallization of the full-length large terminase from many phages has proved to be unsuccessful. To date, the only full-length wild-type large terminase structures available are those from phage Sf6 (gp2; Zhao et al., 2013 ▸) and Geobacillus stearothermophilus phage D6E (Xu et al., 2017 ▸). The crystal structure of phage T4 large terminase (gp17) with a 33-amino-acid deletion at the C-terminal end and with a D255E/E256D mutation in its ATPase active centre has been reported (Sun et al., 2008 ▸). The crystallizations of phage P22 (Roy & Cingolani, 2012 ▸) and herpesvirus (Nadal et al., 2010 ▸) large terminases have been reported to be difficult. The structure of the mutant T4 large terminase revealed that the ATPase domain bears a nucleotide-binding fold consisting of six adjacent parallel β-strands belonging to the additional-strand catalytic glutamate (ASCE) superfamily, which is typical of many ATPase and translocation proteins. From the structural information, an electrostatic force-dependent DNA-translocation model was proposed (Sun et al., 2008 ▸). The crystal structure of the wild-type full-length large terminase from phage Sf6 (gp2) revealed an N-terminal RecA-like ATPase domain and a C-terminal RNase H-like nuclease domain connected by a linker region which is thought to be involved in the regulation of various functions of gp2 (Zhao et al., 2013 ▸). A DNA-translocation mechanism based on movement of the linker region has been proposed (Zhao et al., 2013 ▸).
Despite the available structural information, the mechanism of genome translocation by the large terminase and how it is coordinated with its ATPase activity is still largely unknown. Therefore, structural studies of large terminase proteins from different phages are essential. Here, we report the purification, ATPase activity and crystallization of the putative large terminase protein from bacteriophage N4, a lytic phage that infects the Escherichia coli K12 strain (Molina et al., 1965 ▸). Phage N4 possesses a linear dsDNA genome of 72 kbp and encodes 72 proteins (Zivin et al., 1980 ▸; Ohmori et al., 1988 ▸). A remarkable feature of phage N4 is that it packages 3–5 copies of the virion-encoded RNA polymerase (vRNAP) along with its genome, which is utilized for transcription of the early viral genes during infection (Rothman-Denes & Schito, 1974 ▸; Falco et al., 1977 ▸). The viral proteins that are responsible for genome packaging in phage N4 have not been characterized. It is not known whether the packaging of vRNAP is carried out by the genome-packaging machinery or requires a different translocating protein, or whether it is coupled to capsid assembly.
2. Materials and methods
2.1. Sequence analysis of phage N4 terminase
Phage N4 gp68, which was previously annotated as the large terminase (Kulikov et al., 2012 ▸; Chan et al., 2014 ▸), and gp69, which has the appropriate size and location in the genome as previously reported (Kulikov et al., 2012 ▸), were selected for sequence analysis. A multiple sequence alignment was generated using T-Coffee (https://www.ebi.ac.uk/tools/msa/tcoffee; Notredame et al., 2000 ▸) and the secondary structure was predicted using Jpred4 (http://www.compbio.dundee.ac.uk/jpred/; Drozdetskiy et al., 2015 ▸). The probability of coiled-coil motif (CCM) formation in gp69 was predicted using COILS (https://www.ch.embnet.org/software/COILS_form.html; Lupas et al., 1991 ▸).
2.2. Cloning, expression and purification
gp68 (gene product 68; YP_950546.1), a 60.4 kDa protein, was identified as the putative large terminase, with all of the critical motifs required for ATP binding and hydrolysis, while owing to its proximity to the large terminase gene and other sequence features, as detailed in §3, gp69 (YP_950547.1), a 25.7 kDa protein, was identified as the potential small terminase (Kulikov et al., 2012 ▸; Chan et al., 2014 ▸).
Phage N4 genomic DNA was used to amplify the gp68 and gp69 genes and the amplified genes were cloned in pET-28a at the XhoI and NheI (Thermo Fisher Scientific) restriction-enzyme sites, which added a tag of 2.3 kDa. Hence, the molecular weights of the expressed proteins were 62.7 and 28 kDa for gp68 and gp69, respectively. The obtained clones were further confirmed by sequencing. E. coli XL10 Gold competent cells were used to maintain the recombinant plasmid and E. coli BL21 (DE3) pLysS cells were used for overexpression and purification.
Overexpression of gp68 and gp69 was induced with 0.3 mM ispropyl β-d-1-thiogalactopyranoside at 30°C for 3 h. The E. coli cells were harvested by centrifugation (8500g for 10 min at 4°C) and the cell pellet was resuspended in 20 ml binding buffer (50 mM Tris–HCl pH 7.5, 20 mM imidazole, 5 mM MgCl2, 10% glycerol) with benzamidine (6.3 mg) and 400 µl 40 mM phenylmethylsulfonyl fluoride (PMSF). The cells were lysed by sonication and the lysate was centrifuged at 15 000g for 40 min. The supernatant obtained was filtered through a 0.22 µm filter and the protein was purified by three successive chromatographic steps: nickel-affinity, anion-exchange and size-exclusion chromatography. Briefly, the supernatant containing the soluble protein was loaded onto a 1 ml His-Trap column (GE Healthcare) equilibrated with 50 mM Tris–HCl buffer pH 7.5 containing 20 mM imidazole, 5 mM MgCl2 and 10% glycerol. Bound protein was eluted with a linear imidazole gradient (50–600 mM) using an ÄKTAprime plus (GE Healthcare). Based on SDS–PAGE analysis, eluted protein fractions containing the desired protein were pooled and loaded onto a 1 ml HiTrap Q HP column (GE Healthcare) pre-equilibrated with 50 mM Tris–HCl buffer pH 7.5 containing 5 mM NaCl, 5 mM MgCl2 and 10% glycerol. Bound protein was eluted using a linear NaCl gradient (20–500 mM). Fractions containing the desired protein were pooled, concentrated to 1 ml and loaded onto a Superdex 200 preparative-grade column connected to an ÄKTA FPLC system (GE Healthcare). The column was pre-equilibrated with 20 mM Tris–HCl buffer pH 7.5 containing 100 mM NaCl and 5% glycerol. The eluted protein was pooled and concentrated, and was either stored at −80°C or used immediately to set up crystallization trials. The yields of gp68 and gp69 were about 0.6 and 4 mg per litre of culture, respectively.
2.3. ATPase assay
ATPase assays were performed by a colorimetric method to detect and quantify the inorganic phosphate (Pi) released using malachite green dye (Lanzetta et al., 1979 ▸). The reaction mixture (20 µl) consisting of purified gp68 (2 µM) either alone or in the presence of gp69 (4 µM), 1 mM ATP in 20 mM Tris–HCl buffer pH 8.0, 100 mM NaCl and 5 mM MgCl2 was incubated for 30 min at 37°C. The reaction was stopped by adding EDTA to a final concentration of 50 mM. Malachite green dye was added to the reaction mixture and allowed to stand at room temperature for 20 min. The Pi released was quantified by measuring the optical density at 630 nm.
2.4. Crystallization
Purified gp68 was concentrated to 6 mg ml−1 and used for crystallization. The initial crystallization trials were performed with a Phoenix crystallization robot (Protein Crystallography Facility, IIT Bombay) using the JCSG Core I Suite (Qiagen) and the sitting-drop vapour-diffusion method at 22°C. A drop volume of 1 µl (0.5 µl reservoir solution and an equal volume of protein solution) was used. Conditions were further optimized using the hanging-drop vapour-diffusion method in a 48-well plate (NEST), equilibrating the drops against 75 µl reservoir solution. Each drop consisted of 2 µl gp68 protein solution and an equal volume of reservoir solution. For optimization, we tried concentrations of 2–40%(w/v) PEG 8000 and 20–150 mM HEPES. The best crystals were obtained in a condition consisting of 10%(w/v) PEG 8000, 0.1 M HEPES pH 7.5 using 3 µl gp68 protein solution and an equal volume of reservoir solution. A single cooled crystal was used for data collection. Additionally, crystals were harvested, soaked in 0.5 M potassium bromide for 30 s and used for data collection.
2.5. X-ray diffraction data collection and processing
X-ray diffraction data were collected from native and potassium bromide-soaked crystals using the home X-ray radiation source: a Rigaku MicroMax-007 HF generator equipped with an R-AXIS IV++ detector at the Protein Crystallography Facility, Centre for Research in Nanotechnology and Science (CRNTS), IIT Bombay. Data were collected at 100 K under a continuous flow of nitrogen. Various concentrations of glycerol (10, 20, 25, 30 and 35%) were tested as cryoprotectants. Diffraction data were obtained after cooling the crystal in a liquid-nitrogen cryostream (100 K) using a solution consisting of 10%(w/v) PEG 8000, 0.1 M HEPES pH 7.5, 35% glycerol as a cryoprotectant. Each data set was collected with a crystal rotation of 0.5° per frame at a wavelength of 1.5418 Å using a Cu Kα X-ray radiation source. The diffraction images were indexed and integrated using XDS (Kabsch, 2010 ▸). The data-collection statistics are presented in Table 1 ▸. Owing to low sequence identity to available crystal structures of homologous proteins, an initial phase could not be obtained using the molecular-replacement method. Structure-solution trials with the heavy-atom-derivatized crystals using the multiple isomorphous replacement (MIR) and multiple anomalous dispersion (MAD) methods are therefore in progress.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the outer resolution shell.
| Native crystal | Potassium bromide-soaked crystal | |
|---|---|---|
| Wavelength (Å) | 1.5418 | 1.5418 |
| Temperature (K) | 100 | 100 |
| Detector type | R-AXIS IV++ | R-AXIS IV++ |
| Crystal-to-detector distance (mm) | 200 | 200 |
| Oscillation range per image (°) | 0.5 | 0.5 |
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å, °) | a = 52.7, b = 93.6, c = 124.9, α = β = γ = 90 | a = 53.2, b = 94.2, c = 122.9, α = β = γ = 90 |
| Mosaicity (°) | 0.4 | 0.4 |
| Resolution (Å) | 40–2.8 (2.9–2.8) | 40–3.8 (3.9–3.8) |
| Total No. of reflections | 79218 (8071) | 40776 (2994) |
| No. of unique reflections | 15803 (1560) | 11741 (869) |
| Multiplicity | 5.0 (5.1) | 3.4 (3.4) |
| Completeness (%) | 99.7 (99.9) | 99.7 (99.8) |
| Average I/σ(I) | 11.0 (2.0) | 7.0 (2.0) |
| R meas (%) | 12.6 (86.3) | 23.7 (87.3) |
| CC1/2 | 1.0 (0.8) | 1.0 (0.5) |
| Overall B factor from Wilson plot (Å2) | 56.2 | 74.9 |
3. Results and discussion
3.1. Sequence analysis of phage N4 large and small terminase proteins
Although gp68 was suggested to be the large terminase in two previous studies (Kulikov et al., 2012 ▸; Chan et al., 2014 ▸), to date no sequence analysis or functional characterization has been performed to show that gp68 and gp69 are the large and small terminases, respectively, of phage N4. Hence, we performed a thorough sequence analysis of these two proteins, followed by cloning, expression and ATPase-activity measurements to ascertain their functions before setting up crystallization trials.
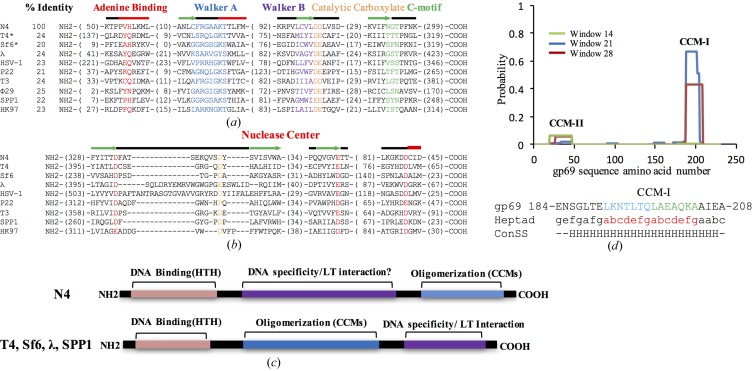
Although gp68 (YP_950546.1) showed a limited sequence identity of less than 24% to other well characterized large terminases, characteristic conservation of the ATPase signature (Walker A motif, Walker B motif and C-motif) at the N-terminus (Fig. 1 ▸ a) and a triad of metal (Mg2+) coordinating acidic residues that make up the nuclease catalytic centre in the C-terminal domain (nuclease centre; Fig. 1 ▸ b) helped in the selection of gp68 as the potential large terminase (Mitchell et al., 2002 ▸; Rao & Feiss, 2015 ▸; Alam et al., 2008 ▸).
Figure 1.
The protein encoded by the gp68 gene of phage N4 is a probable terminase. (a, b) Alignments of the N-terminal ATPase domain (a) and the C-terminal nuclease domain (b) of gp68 with other viral terminases. Motifs are shown in the following colours: adenine-binding motif, red; Walker A motif, blue; Walker B motif, purple; catalytic carboxylate, orange; coupling motif, green. The consensus secondary structure of the conserved region is also shown. In the alignment of the C-terminal region, the conserved aspartates and glutamates which form the probable catalytic triad (red) and a conserved fourth aspartate residue present in the loop (yellow) are shown with their predicted secondary structures. The alignment was manually edited from the T-Coffee output. The consensus secondary structure was obtained using JPred. Thick red line, α-helix; green arrow, β-sheet; thin black line, no predicted structure. Numbers in parentheses represent the numbers of residues not shown in the alignment. All sequences were obtained from GenBank. The accession numbers of the sequences used are as follows: N4, YP_950546.1; T4, NP_049776.1; Sf6, NP_958178.1; λ, P03708.1; HSV, P04295.2; P22, BAG12600.1; T3, P10310.1; φ29, P11014.1; SPP1, P54308.1; HK97, NP_037698.1. (c) Comparison of the domain organization in gp69 (top panel) and other well characterized small terminases (lower panel). HTH, helix–turn–helix motif; CCM, coiled-coil motif; LT, large terminase. (d) Probability plot for CCMs in the gp69 sequence from phage N4 as predicted by COILS. Probability scores are on a scale of 0–1 (where 1 is 100%) and are plotted against the amino-acid residue number. Peaks in the graph indicate probable coiled-coil regions. The default output of probabilities in scanning windows of 14, 21 and 28 residues are shown in green, blue and red, respectively. The heptad frame for CCM-I generated using COILS (abcdefg) and the consensus secondary structure (ConSS) predicted by JPred are shown below the CCM-I amino-acid sequence of gp69.
Small terminase proteins from different phages share very limited sequence homology. In most phages, this protein is encoded by a gene preceding the large terminase gene. However, in the case of phage N4 the gene preceding gp68 (gp67) appears to be a tail protein. However, gp69 (YP_950547.1), the gene succeeding gp68, showed the characteristic presence of the coiled-coil motif (CCM; Lupas, 1996 ▸), which is a highly conserved and common feature among most small terminase sequences (Kondabagil & Rao, 2006 ▸; Figs. 1 ▸ c and 1 ▸ d). The CCMs are required for oligomerization of the small terminase and ATPase stimulation of the large terminase (Kondabagil & Rao, 2006 ▸). A possible helix–turn–helix (HTH) motif involved in DNA binding was also predicted in the N-terminal region (Fig. 1 ▸ c), as seen in other small terminases (Pabo & Sauer, 1984 ▸; Kypr & Mrázek, 1986 ▸; Chai et al., 1992 ▸; Lin et al., 1997 ▸; Zhao et al., 2010 ▸). Because of the overall conservation of the sequence features, gp69 was selected for overexpression along with gp68.
3.2. Purification of gp68 and gp69
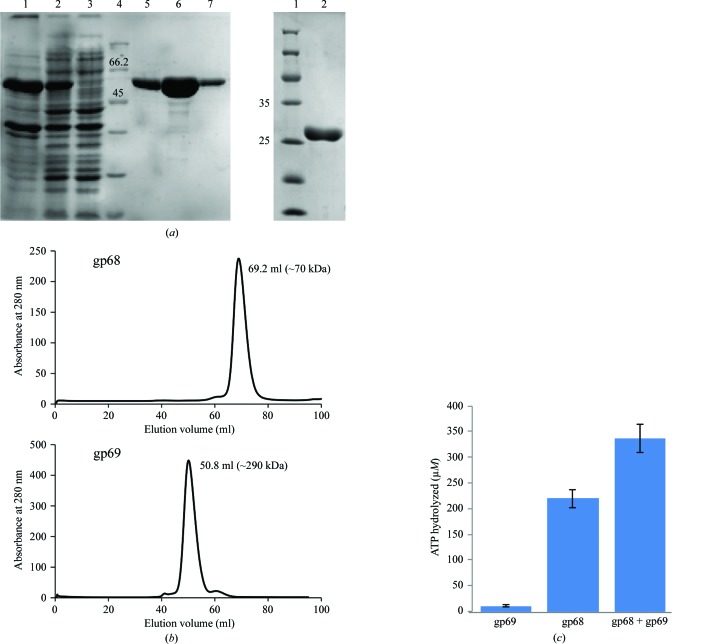
After three successive rounds of chromatography, near-homogenous preparations of both large and small terminases were obtained (Fig. 2 ▸ a). Purified gp68 eluted as a monomer during size-exclusion chromatography, while gp69 eluted as a higher molecular-weight fraction of about 290 kDa that roughly corresponded to a decamer (Fig. 2 ▸ b). Furthermore, MALDI analysis of the purified proteins showed that gp68 is a monomer, whereas gp69 showed the presence of various high-molecular-weight species including dimers and trimers (data not shown).
Figure 2.
Purification of phage N4 putative large and small terminases and the ATP-hydrolysis activity of the large terminase protein gp68. (a) Left: SDS polyacrylamide gel showing purified gp68 (62.7 kDa). Lane 1, pellet; lane 2, supernatant; lane 3, flowthrough; lane 4, protein marker (labelled in kDa); lane 5, eluate from Ni2+–NTA column; lane 6, eluate from Q HP column; lane 7, eluate from Superdex 200 16/300 pg column. Right: SDS polyacrylamide gel showing the purified gp69 (28 kDa). (b) Elution profile of gp68 and gp69 from a Superdex 200 16/300 pg column. The peak for gp68 at 69.2 ml corresponds to the monomeric form of gp68 (top profile) and the peak at 50.8 ml approximately represents the decameric form of gp69 (bottom profile) according to standard molecular-weight markers. (c) gp68 possesses ATP-hydrolysis activity that is further stimulated by the small terminase gp69. See §2 for details.
3.3. gp68 exhibits basal ATPase activity that is enhanced by about twofold in the presence of gp69
Purified gp68 readily hydrolyzes ATP, which was further stimulated in the presence of gp69 (Fig. 2 ▸ c). Unlike the T4 and SPP1 large terminases, which exhibited very little basal ATPase activity (Leffers & Rao, 2000 ▸; Baumann & Black, 2003 ▸; Gual et al., 2000 ▸), gp68 showed significant basal ATPase activity, as observed in the case of the phage λ large terminase gpA (Rubinchik et al., 1994 ▸; Catalano, 2000 ▸; Fig. 2 ▸ c). It was also observed that the maximum extent of stimulation of about twofold was achieved at a gp68:gp69 molar ratio of 1:2 and that a further increase in gp69 concentration (to up to eight times the molar ratio of gp68) did not result in any appreciable increase in the ATPase activity (data not shown). Furthermore, the extent of stimulation across different batches of purified proteins varied from 1.5-fold to twofold. An increase in the ATPase activity of gp68 by ∼1.5-fold is reported here (Fig. 2 ▸ c). Furthermore, gp69 did not show significant ATPase activity on its own. Interestingly, different small and large terminase systems exhibit different levels of stimulation. The extent of stimulation of the large terminase ATPase activity in the presence of the small terminase in phage N4 is comparable to that of phage SPP1 (∼3.5-fold; Gual et al., 2000 ▸) but differs from that of phage T4 (>50–100-fold; Leffers & Rao, 2000 ▸; Baumann & Black, 2003 ▸). The stoichiometry of the holoterminase complex in phage N4 has yet to be established. Studies have suggested that in the case of SPP1 and λ phages, the maximum ATPase stimulation was achieved at a small terminase to large terminase ratio of 2:1 (Camacho et al., 2003 ▸; Tomka & Catalano, 1993 ▸), while in the case of P22 it was 1:2 (McNulty et al., 2015 ▸) and in T4 the maximum stimulation required was 1:1 (one oligomer of small terminase comprising of about eight gp16 molecules to one subunit of large terminase; Kanamaru et al., 2004 ▸). These results, taken together with sequence analysis, indicate that gp68 and gp69 are the probable large and small terminase proteins of phage N4.
3.4. Crystallization and X-ray diffraction data collection
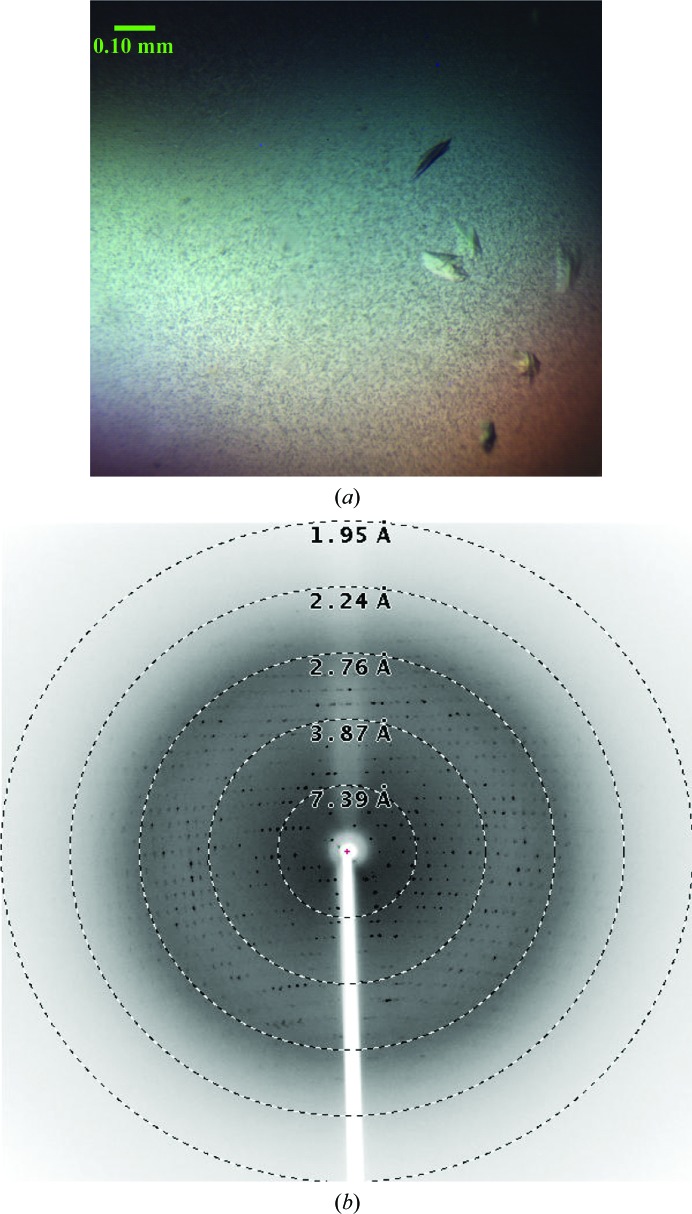
Because of the difficulties associated with the purification of gp68, only a small quantity of purified gp68 was obtained and the protein was concentrated to ∼6 mg ml−1 for crystallization. Only 288 crystallization drops could be set up using the 96 conditions of the JCSG Core I suite (Qiagen). After 5 d, very thin and small plate-like crystals were observed in a condition consisting of 10% PEG 8000, 0.1 M HEPES pH 7.5. Further optimization carried out using an increased drop volume produced plate-like crystals in the same condition [10%(w/v) PEG 8000, 0.1 M HEPES pH 7.5] that were suitable for testing using X-rays. The crystals were observed after 3 d of incubation (Fig. 3 ▸ a) in the crystallization drop. A few crystals were washed and dissolved in water. The solution containing the dissolved crystals was analyzed using SDS–PAGE and the result confirmed that the crystals were indeed of gp68 (data not shown). The appearance of very small needle-like crystals was observed in another condition. However, owing to the small size of the needles they could not be mounted for X-ray diffraction experiments, and further optimization of the crystallization condition did not yield any crystals.
Figure 3.
Crystals of the bacteriophage N4 large terminase gp68 and a representative diffraction pattern. (a) The crystals used for the study had dimensions of 0.1 × 0.05 × 0.05 mm. (b) A representative diffraction pattern from a single crystal of the bacteriophage N4 large terminase collected at 0.5° oscillation per frame using an R-AXIS IV++ detector. An exposure time of 10 min was used for data collection. Resolution shells are shown as dotted circles.
A plate-like crystal of gp68 diffracted to 2.5 Å resolution at our home X-ray source (Fig. 3 ▸ b); however, diffraction data with optimal quality could be processed to only 2.8 Å resolution. Data-processing statistics are shown in Table 1 ▸. The data suggested that the gp68 crystal belonged to space group P212121, with unit-cell parameters a = 53.7, b = 93.6, c = 124.9 Å, α = β = γ = 90°. The Matthews coefficient (V M; Matthews, 1968 ▸) was calculated to be 2.47 Å3 Da−1, which corresponded to one molecule of gp68 per asymmetric unit, with a solvent content of about 50.2%. Attempts to solve the structure by the molecular-replacement method failed because of the low sequence identity to the available templates in the PDB. Based on the signal-to-noise ratio (2.0) of the highest resolution shell and the Wilson B factor (56.2 Å2), it appears that these crystals would diffract to higher resolution on a synchrotron-radiation source.
We also collected another diffraction data set at 3.8 Å resolution from a crystal soaked in potassium bromide (Table 1 ▸). The diffraction data from the bromide-derivatized crystal were indexed in space group P212121, with unit-cell parameters a = 53.2, b = 94.2, c = 122.9 Å, α = β = γ = 90°. These derivative data will be used to find the positions of bromide ions using the single isomorphous replacement (SIR) method. If the positions of the bromide ions are identified, similar potassium bromide derivatives will be prepared for anomalous data collection to solve the structure using the multiple anomalous dispersion (MAD) method. Further analysis of this potassium bromide-derivative data, purification of protein and structure-solution trials using other heavy-atom-derivatized crystals are in progress.
4. Conclusion
Here, we report the purification, crystallization and X-ray crystallographic analysis of the bacteriophage N4 large terminase protein gp68 expressed in an E. coli expression host. Similar to other well studied phage large and small terminases, the purified phage N4 putative large terminase is a monomer, while the putative small terminase is an oligomer. The purified large terminase protein exhibited a basal ATPase activity that was stimulated by ∼1.5-fold in the presence of the small terminase, while the small terminase lacked ATPase activity on its own. Considering the unsuccessful crystallization attempts using other large terminase proteins and the availability of a very limited number of large terminase crystal structures, the successful crystallization of the wild-type large terminase from phage N4 is a minor but significant development towards obtaining its atomic structure, which will help in understanding the genome-packaging mechanism.
Acknowledgments
We acknowledge support from the Protein Crystallography Facility, Centre for Research in Nanotechnology and Science (CRNTS), Indian Institute of Technology Bombay.
Funding Statement
This work was funded by Council of Scientific and Industrial Research grant 37(1508)/11/EMR-II. Indian Institute of Technology Bombay grant P11IRCCSG004 to Kiran Kondabagil. Department of Biotechnology , Ministry of Science and Technology grant .
References
- Alam, T. I., Draper, B., Kondabagil, K., Rentas, F. J., Ghosh-Kumar, M., Sun, S., Rossmann, M. G. & Rao, V. B. (2008). Mol. Microbiol. 69, 1180–1190. [DOI] [PubMed]
- Al-Zahrani, A. S., Kondabagil, K., Gao, S., Kelly, N., Ghosh-Kumar, M. & Rao, V. B. (2009). J. Biol. Chem. 284, 24490–24500. [DOI] [PMC free article] [PubMed]
- Baumann, R. G. & Black, L. W. (2003). J. Biol. Chem. 278, 4618–4627. [DOI] [PubMed]
- Camacho, A. G., Gual, A., Lurz, R., Tavares, P. & Alonso, J. C. (2003). J. Biol. Chem. 278, 23251–23259. [DOI] [PubMed]
- Casjens, S. R. (2011). Nature Rev. Microbiol. 9, 647–657. [DOI] [PubMed]
- Catalano, C. E. (2000). Cell. Mol. Life Sci. 57, 128–148. [DOI] [PMC free article] [PubMed]
- Catalano, C. E., Cue, D. & Feiss, M. (1995). Mol. Microbiol. 16, 1075–1086. [DOI] [PubMed]
- Chai, S., Bravo, A., Lüder, G., Nedlin, A., Trautner, T. A. & Alonso, J. C. (1992). J. Mol. Biol. 224, 87–102. [DOI] [PubMed]
- Chan, J. Z. M., Millard, A. D., Mann, N. H. & Schäfer, H. (2014). Front. Microbiol. 5, 506. [DOI] [PMC free article] [PubMed]
- Drozdetskiy, A., Cole, C., Procter, J. & Barton, G. J. (2015). Nucleic Acids Res. 43, W389–W394. [DOI] [PMC free article] [PubMed]
- Falco, S. C., Vander Laan, K. & Rothman-Denes, L. B. (1977). Proc. Natl Acad. Sci. USA, 74, 520–523. [DOI] [PMC free article] [PubMed]
- Feiss, M. & Rao, V. B. (2012). Adv. Exp. Med. Biol. 726, 489–509. [DOI] [PubMed]
- Fuller, D. N., Raymer, D. M., Kottadiel, V. I., Rao, V. B. & Smith, D. E. (2007). Proc. Natl Acad. Sci. USA, 104, 16868–16873. [DOI] [PMC free article] [PubMed]
- Gual, A., Camacho, A. G. & Alonso, J. C. (2000). J. Biol. Chem. 275, 35311–35319. [DOI] [PubMed]
- Jackson, E. N., Jackson, D. A. & Deans, R. J. (1978). J. Mol. Biol. 118, 365–388. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kanamaru, S., Kondabagil, K., Rossmann, M. G. & Rao, V. B. (2004). J. Biol. Chem. 279, 40795–40801. [DOI] [PubMed]
- Kondabagil, K. R. & Rao, V. B. (2006). J. Mol. Biol. 358, 67–82. [DOI] [PubMed]
- Kulikov, E., Kropinski, A. M., Golomidova, A., Lingohr, E., Govorun, V., Serebryakova, M., Prokhorov, N., Letarova, M., Manykin, A., Strotskaya, A. & Letarov, A. (2012). Virology, 426, 93–99. [DOI] [PubMed]
- Kypr, J. & Mrázek, J. (1986). J. Mol. Biol. 191, 139–140. [DOI] [PubMed]
- Lanzetta, P. A., Alvarez, L. J., Reinach, P. S. & Candia, O. A. (1979). Anal. Biochem. 100, 95–97. [DOI] [PubMed]
- Leffers, G. & Rao, V. B. (2000). J. Biol. Chem. 275, 37127–37136. [DOI] [PubMed]
- Lin, H., Simon, M. N. & Black, L. W. (1997). J. Biol. Chem. 272, 3495–3501. [DOI] [PubMed]
- Lupas, A. (1996). Trends Biochem. Sci. 21, 375–382. [PubMed]
- Lupas, A., Van Dyke, M. & Stock, J. (1991). Science, 252, 1162–1164. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McNulty, R., Lokareddy, R. K., Roy, A., Yang, Y., Lander, G., Heck, A. J., Johnson, J. E. & Cingolani, G. (2015). J. Mol. Biol. 427, 3285–3299. [DOI] [PMC free article] [PubMed]
- Mitchell, M. S., Matsuzaki, S., Imai, S. & Rao, V. B. (2002). Nucleic Acids Res. 30, 4009–4021. [DOI] [PMC free article] [PubMed]
- Molina, A. M., Pesce, A. & Shito, G. C. (1965). Boll. Ist. Sieroter. Milan. 44, 329–337.
- Nadal, M., Mas, P. J., Mas, P. J., Blanco, A. G., Arnan, C., Solà, M., Hart, D. J. & Coll, M. (2010). Proc. Natl Acad. Sci. USA, 107, 16078–16083. [DOI] [PMC free article] [PubMed]
- Notredame, C., Higgins, D. G. & Heringa, J. (2000). J. Mol. Biol. 302, 205–217. [DOI] [PubMed]
- Ohmori, H., Haynes, L. L. & Rothman-Denes, L. B. (1988). J. Mol. Biol. 202, 1–10. [DOI] [PubMed]
- Pabo, C. O. & Sauer, R. T. (1984). Annu. Rev. Biochem. 53, 293–321. [DOI] [PubMed]
- Rao, V. B. & Feiss, M. (2015). Annu. Rev. Virol. 2, 351–378. [DOI] [PMC free article] [PubMed]
- Rothman-Denes, L. B. & Schito, G. C. (1974). Virology, 60, 65–72. [DOI] [PubMed]
- Roy, A. & Cingolani, G. (2012). J. Biol. Chem. 287, 28196–28205. [DOI] [PMC free article] [PubMed]
- Rubinchik, S., Parris, W. & Gold, M. (1994). J. Biol. Chem. 269, 13586–13593. [PubMed]
- Smith, D. E., Tans, S. J., Smith, S. B., Grimes, S., Anderson, D. L. & Bustamante, C. (2001). Nature (London), 413, 748–752. [DOI] [PubMed]
- Smits, C., Chechik, M., Kovalevskiy, O. V., Shevtsov, M. B., Foster, A. W., Alonso, J. C. & Antson, A. A. (2009). EMBO Rep. 10, 592–598. [DOI] [PMC free article] [PubMed]
- Sun, S., Kondabagil, K., Draper, B., Alam, T. I., Bowman, V. D., Zhang, Z., Hegde, S., Fokine, A., Rossmann, M. G. & Rao, V. B. (2008). Cell, 135, 1251–1262. [DOI] [PubMed]
- Sun, S., Rao, V. B. & Rossmann, M. G. (2010). Curr. Opin. Struct. Biol. 20, 114–120. [DOI] [PMC free article] [PubMed]
- Tomka, M. A. & Catalano, C. E. (1993). J. Biol. Chem. 268, 3056–3065. [PubMed]
- Xu, R.-G., Jenkins, H. T., Antson, A. A. & Greive, S. J. (2017). Nucleic Acids Res. 45, 13029–13042. [DOI] [PMC free article] [PubMed]
- Zhao, H., Christensen, T. E., Kamau, Y. N. & Tang, L. (2013). Proc. Natl Acad. Sci. USA, 110, 8075–8080. [DOI] [PMC free article] [PubMed]
- Zhao, H., Finch, C. J., Sequeira, R. D., Johnson, B. A., Johnson, J. E., Casjens, S. R. & Tang, L. (2010). Proc. Natl Acad. Sci. USA, 107, 1971–1976. [DOI] [PMC free article] [PubMed]
- Zivin, R., Malone, C. & Rothman-Denes, L. B. (1980). Virology, 104, 205–218. [DOI] [PubMed]