This article reports the crystal structure of the apo form and the AMPPNP- and Mn2+- bound forms of the essential helicase of ZIKV refined to 1.78 and 1.3 Å resolution, respectively.
Keywords: Zika virus, helicase, crystal structure, ligands
Abstract
Recent studies suggest a link between infection by Zika virus (ZIKV) and the development of neurological complications. The lack of ZIKV-specific therapeutics has alarmed healthcare professionals worldwide. Here, crystal structures of apo and AMPPNP- and Mn2+-bound forms of the essential helicase of ZIKV refined to 1.78 and 1.3 Å resolution, respectively, are reported. The structures reveal a conserved trimodular topology of the helicase. ATP and Mn2+ are tethered between two RecA-like domains by conserved hydrogen-bonding interactions. The binding of ligands induces the movement of backbone Cα and side-chain atoms. Numerous solvent molecules are observed in the vicinity of the AMPPNP, suggesting a role in catalysis. These high-resolution structures could be useful for the design of inhibitors targeting the helicase of ZIKV for the treatment of infections caused by ZIKV.
1. Introduction
Recent outbreaks of Zika virus (ZIKV) have revealed several disconcerting facts about the infection, which was previously considered to be mild and self-limiting. A link has now been established between ZIKV infection and the development of microcephaly in newborns (Heukelbach & Werneck, 2016 ▸; Russo et al., 2017 ▸; Calvet et al., 2016 ▸). In addition, higher incidences of another neurological complication, Guillain–Barré syndrome, have been noted in areas where ZIKV is endemic (Siu et al., 2016 ▸; Miller et al., 2017 ▸; Krauer et al., 2017 ▸; Dub & Fontanet, 2017 ▸). Both of these neurological abnormalities are associated with high mortality rates. Of particular concern is the rapid spread of the virus via transmission from mosquitoes to humans and via human-to-human transmission. More than 70 countries across different continents have reported cases of ZIKV (Maurer-Stroh et al., 2016 ▸). Central and South American countries have a significant disease burden, with estimates of more than a million cases of ZIKV infection (Korzeniewski et al., 2016 ▸). Currently, there are no vaccines or ZIKV-specific antiviral drugs available to stall the spread of the virus (Korzeniewski et al., 2016 ▸; Yun & Lee, 2017 ▸; Russo et al., 2017 ▸). It is therefore imperative to gain an understanding of the viral protein functions that aid the virus in its propagation, so that they can be targeted by inhibitors to prevent the spread of the virus.
ZIKV belongs to the Flaviviridae family and is grouped into the Flavivirus genus (Xin et al., 2017 ▸; Bollati et al., 2010 ▸). The genome of ZIKV consists of a single positive-stranded RNA (Yun & Lee, 2017 ▸; Marano et al., 2016 ▸). Similar to many other flaviviruses, ZIKV is primarily transmitted to humans via mosquitoes. Mosquito bites deposit the virus in the epidermis and dermis, where the virus gains entry into keratinocytes and fibroblasts (Routhu & Byrareddy, 2017 ▸; Aziz et al., 2017 ▸). Inside the cells, a single polypeptide chain is translated from the viral genome. This polypeptide is processed by host and viral proteases to produce structural proteins including E, C and prM/M as well as seven other nonstructural proteins (Sironi et al., 2016 ▸; Cunha et al., 2016 ▸). These proteins carry out the replication of the genome and other tasks that are necessary for the assembly of progeny. ZIKV is postulated to involve cellular processes such as autophagy and apoptosis during its life cycle to subvert the host defence machinery (Cugola et al., 2016 ▸). In addition to epidermal keratinocytes and cutaneous fibroblasts, ZIKV is also known to infect dendritic and neuronal cells such as astrocytes, as well as microglial cells, using TAM proteins (AXL and Tyro3) as receptors (Meertens et al., 2017 ▸). The ability of ZIKV to use a wide range of receptors belonging to the C-type lectin and the TIM and TAM classes aids the virus in infecting a large variety of host cells (Hamel et al., 2015 ▸).
During the replication of the viral genome, single-stranded RNA is made available to the RNA-dependent RNA polymerase (RdRp) by an endogenous helicase activity. The NS3 protein of ZIKV harbours the helicase activity. Similar to other flaviviruses, this protein is multifunctional and has a serine protease domain that is covalently linked to the helicase. The N-terminal residues 1–180 make up the protease (NS3pro), while the remaining amino acids 181–618 fold into the helicase. Since both of the functions that are performed by NS3 are crucial for propagation of the virus, the protein is conceivably a target for the development of antiviral drugs. The structure of the helicase of ZIKV is known in apo (Tian, Ji, Yang, Xie et al., 2016 ▸), pyrophosphate-bound (Jain et al., 2016 ▸), ATP- and Mn2+-bound (Tian, Ji, Yang, Zhang et al., 2016 ▸) and RNA-bound (Tian, Ji, Yang, Xie et al., 2016 ▸) forms. In addition, structures of helicases from other flaviviruses such as Dengue virus (DV; Luo et al., 2008 ▸), Yellow fever virus (YFV; Wu et al., 2005 ▸) and Japanese encephalitis virus (JEV; Yamashita et al., 2008 ▸), as well as hepaciviruses including Hepatitis C virus (HCV; Gu & Rice, 2010 ▸), have previously been described. The structures reveal a highly conserved trimodular architecture for the helicase. The locations of the ATP-, divalent metal ion- and nucleic acid-binding sites are also very similar in these structures, suggesting a highly conserved mechanism for unwinding nucleic acids. Here, we describe crystal structures of the apo as well as AMPPNP- and Mn2+-bound forms of the helicase of ZIKV refined to 1.78 and 1.30 Å resolution, respectively. These structures represent a marked improvement in resolution over previously determined structures of the helicase from ZIKV and could aid in the development of much-needed therapeutic interventions targeting ZIKV.
2. Materials and methods
2.1. Protein preparation
The helicase domain of NS3 (henceforth referred to as NS3H) from ZIKV (residues 176–617) was amplified by PCR using forward (5′-CGCGGATCCATGGTTGAATGTTTCGAACCCTCGATGCTGAAGAAGAAGCAG-3′) and reverse (5′-CCGCTCGAGCTATCTTTTTCCAGCGGCGAATTCTTTGAACGACTTC-3′) primers. The PCR product was digested and ligated into a pET-28a plasmid, where a thrombin cleavage site was inserted between a hexahistidine tag and the N-terminus of the NS3H sequence. The construct was transformed into Escherichia coli BL21(DE3) competent cells (TIANGEN). The transformed E. coli cells were cultured in 2 l flasks in the presence of 100 µg ml−1 ampicillin to an OD600 nm of 0.8 and were then induced at 298 K with IPTG at a concentration of 0.2 mM for 18 h.
At the end of the incubation time, the cells were harvested by centrifugation. The harvested cells were resuspended in lysis buffer (20 mM HEPES–NaOH pH 7.0, 150 mM NaCl, 4 mM MnCl2) and the suspension was homogenized using an ultrahigh-pressure cell disrupter (JNBIO) at 277 K. After high-speed centrifugation to remove cell debris, the supernatant containing soluble NS3H protein was mixed with Ni–NTA agarose beads equilibrated with the lysis buffer and loaded into a 15 ml disposable polypropylene column. The column was washed with ten column volumes of wash buffer consisting of 20 mM HEPES–NaOH pH 7.0, 150 mM NaCl, 4 mM MnCl2, 20 mM imidazole; 200 µl 2 mg ml−1 thrombin protease was then added to the column and the column was incubated overnight at 277 K. The flowthrough from the column containing tagless NS3H was collected and buffer-exchanged into buffer A consisting of 100 mM HEPES–NaOH pH 7.0, 150 mM NaCl, 2 mM DTT using 10 kDa cutoff centrifugal concentrators. The protein was further purified using a prepacked 5 ml HiTrap Mono Q column. Protein bound to the column was eluted using a linear gradient of NaCl (0–500 mM) in buffer B (100 mM HEPES–NaOH pH 7.0, 2 mM DTT, 500 mM NaCl). Fractions containing the protein were pooled, concentrated to 1 ml and injected onto a Superdex G75 gel-filtration column equilibrated with 20 mM HEPES–NaOH buffer pH 7.0, 150 mM NaCl, 10 mM MnCl2, 2 mM DTT. The protein eluted in a single peak. Fractions corresponding to the peak were pooled and concentrated to 7 mg ml−1 before screening for crystallization conditions.
2.2. Crystallization, data collection and structure determination
Crystallization was performed at 291 K using the hanging-drop vapour-diffusion technique. Commercially available sparse-matrix screens were used for screening. Drops consisting of 1 µl protein solution were mixed with an equal volume of reservoir solution and equilibrated against 200 µl reservoir solution. Crystals appeared in a large number of conditions of the PEG/Ion screen (Hampton Research) within 24 h. After optimization, crystals that were grown in a solution consisting of 8% Tacsimate pH 6.0, 20% PEG 3350 were of sufficient quality for data collection. To determine the structure of the NS3H–AMPPNP–Mn2+ complex, we first dissolved AMPPNP powder in lysis buffer (20 mM HEPES–NaOH pH 7.0, 150 mM NaCl, 10 mM MnCl2). The final concentration of AMPPNP in this buffer was 10 mM. Crystals were soaked in this solution for 2 min prior to flash-cooling in liquid nitrogen. Initial X-ray diffraction studies indicated that there was no need for additional cryoprotection components. Crystals were cooled in a stream of nitrogen at 100 K during X-ray diffraction data collection.
The structure of the apo form of ZIKV NS3H was solved by molecular replacement (MR) with Phaser from the CCP4 suite (Winn et al., 2011 ▸) using the DV RNA helicase structure (PDB entry 2bmf; Xu et al., 2005 ▸) as a search template. The model obtained from MR was manually improved with Coot (Emsley & Cowtan, 2004 ▸) and subsequently subjected to refinement by PHENIX (Adams et al., 2010 ▸). The structure of the NS3H–AMPPNP–Mn2+ complex was solved by MR using the structure of the apo form of NS3H as a search template. The final refinement statistics are summarized in Table 1 ▸. Structural figures were produced using PyMOL (DeLano, 2002 ▸).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| ZIKV NS3H | ||
|---|---|---|
| Apo (PDB entry 5jps) | AMPPNP/Mn2+ (PDB entry 5y4z) | |
| Unit-cell parameters | ||
| a (Å) | 53.55 | 53.75 |
| b (Å) | 69.04 | 68.78 |
| c (Å) | 57.15 | 56.97 |
| α, β, γ (°) | 90.0, 91.9, 90.0 | 90.0, 92.5, 90.0 |
| Space group | P21 | P21 |
| Wavelength (Å) | 0.9785 | 0.9785 |
| Resolution (Å) | 50.00–1.79 (1.82–1.79) | 50.00–1.30 (1.32–1.30) |
| Total No. of reflections | 263403 (12750) | 418035 (134) |
| No. of unique reflections | 39152 (1903) | 75138 (112) |
| Completeness (%) | 99.2 (97.7) | 73.5 (2.2) |
| Average I/σ(I) | 23.9 (3.9) | 26.1 (0.5) |
| R merge † (%) | 5.1 (34.5) | 8.2 (74.5) |
| No. of reflections used [σ(F) > 0] | 37162 | 75065 |
| R work ‡ (%) | 18.0 | 19.3 |
| R free ‡ (%) | 21.5 | 21.5 |
| R.m.s.d., bond distances (Å) | 0.020 | 0.007 |
| R.m.s.d., bond angles (°) | 1.625 | 1.198 |
| Average B value (Å2) | 21.6 | 22.3 |
| No. of protein atoms | 3406 | 3471 |
| No. of ligand atoms | 0 | 32 |
| No. of solvent atoms | 398 | 426 |
| Ramachandran plot | ||
| Favoured regions (%) | 96.46 | 95.43 |
| Generously allowed regions (%) | 3.30 | 4.34 |
| Disallowed regions (%) | 0.24 | 0.23 |
R
merge = 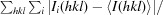
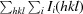 , where 〈I(hkl)〉 is the mean of the observations I
i(hkl) of reflection hkl.
, where 〈I(hkl)〉 is the mean of the observations I
i(hkl) of reflection hkl.
R
work = 
 ; R
free is the R factor for a preselected subset (5%) of reflections that were not included in refinement.
; R
free is the R factor for a preselected subset (5%) of reflections that were not included in refinement.
3. Results
3.1. Overall structure of the helicase from Zika virus
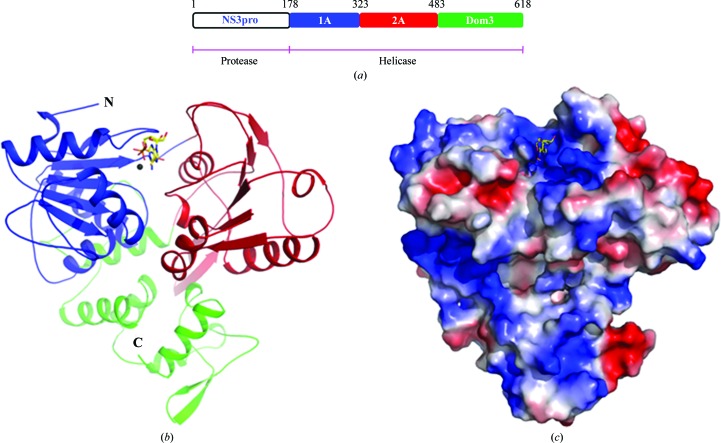
The NS3 protein of ZIKV is 618 residues in length (Fig. 1 ▸ a). Residues 178–618 make up the helicase. The structure of the region stretching from residues 178 to 618, encompassing the helicase of NS3, was solved by molecular replacement using the structure of the helicase from DV (PDB entry 2bmf) as the search template. Electron density for amino acids Thr247–His253 and His196–Gly200 was not observed in the structure of the apoenzyme. However, both these regions of the protein are well ordered in the structure of the helicase bound to the ATP analogue AMPPNP and Mn2+ (Fig. 1 ▸ b). The structures of the apo form and the AMPPNP- and Mn2+-bound form of NS3H from ZIKV were refined to 1.78 Å (R work = 18.04%; R free = 21.46%) and 1.3 Å (R work = 19.16%; R free = 21.58%) resolution, respectively. The final models of both structures exhibit good stereochemistry, the statistics for which are listed in Table 1 ▸.
Figure 1.
Overall structure of the helicase from ZIKV. (a) Diagrammatic representation of the domain boundaries of the NS3 protein from ZIKV. Crystal structures of the region encompassing residues 178–618 are reported in this study. (b) Cartoon representation of the structure of the helicase from ZIKV bound to AMPPNP and Mn2+. The convention used for colouring the domains of the structure is shown in (a). AMPPNP is shown as sticks, while the Mn2+ ion is shown as a sphere. The N- and C-termini of the protein are marked. (c) Surface electrostatic potential representation of the structure of the helicase from ZIKV. Blue represents positive potential and red negative potential.
The helicase of ZIKV is made up of three domains (Fig. 1 ▸). In the current structure, Val176–Gly483 make up two α/β domains that are similar in topology. Both of these domains contain a central, curved β-sheet that is flanked by α-helices on either side. While five β-strands arranged in parallel make up the β-sheet of domain 1, the β-sheet of domain 2 is made up of six β-strands that are positioned parallel with respect to each other. These two domains are reminiscent of the RecA-like domains 1A and 2A that are found in typical helicases; for example, the NS3 helicases from DV, JEV, YFV and HCV. In contrast to the two compact RecA-like domains, domain 3 consists of loosely packed helices and a lone peripheral antiparallel β-hairpin that is solvent-exposed. Interestingly, a similar β-hairpin protrudes out of RecA-like domain 2A using two long loops and is seen to be buried in a hydrophobic pocket located on the surface of domain 3. Just like domain 2A, domain 1A also interacts with domain 3. Two α-helices and a long loop of domain 3 make contact with domain 1A. Thus, the three domains of NS3 can be envisioned as occupying the vertices of a triangle. When viewed from the top, there are troughs with positively charged surfaces running through inter-domain interfaces, presumably for binding ligands such as RNA and ATP as well as for facilitating movement of the domains during the processive unwinding of nucleic acids (Fig. 1 ▸ c).
Helicases belonging to the SF-II family are characterized by the presence of seven conserved sequence motifs. In NS3H from ZIKV, all seven motifs are located on RecA-like domains 1A and 2A. Specifically, motifs I, Ia and II are located on domain 1A, while motifs IV–VI are located on domain 2A. Motif III is located on the loop connecting domain 1A to domain 2A.
3.2. Structure of Zika virus helicase bound to the ATP analogue AMPPNP and Mn2+
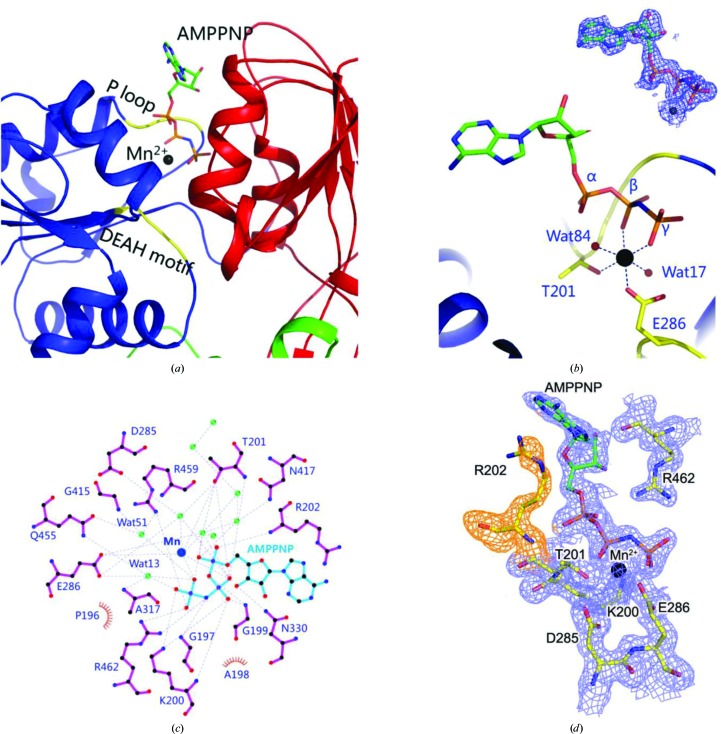
To investigate the mode of binding of ATP and Mn2+ and the structural changes induced by the binding of these ligands, we solved the structure of the helicase in complex with the ATP analogue AMPPNP and Mn2+. The ligands bind in a cavity formed between the two RecA-like domains (domains 1A and 2A; Figs. 1 ▸ b and 2 ▸ a). Although we used MnCl2 in the buffer, there is a possibility that some of the sites are occupied by Mg2+ instead of Mn2+. The only contact of the adenosine ring of AMPPNP with the protein is its stacking with the side chain of Arg202. The three N atoms, NH1, NH2 and NE, of the side chain of Arg202 are less than 3.5 Å away from the aromatic ring of the adenosine moiety. The ribose ring is localized in space by a 2.93 Å hydrogen bond between the O3 atom and the ND2 atom of Asn330. The Walker A motif or the P-loop comprising of the 197GAGKT201 residues is observed to tether the phosphates firmly (Fig. 2 ▸ a). The backbone amide N atoms of this motif are engaged in hydrogen-bonding interactions with the O atoms of the α- and β-phosphates of ATP. In addition, the amine group of Lys200 forms hydrogen bonds to the O atoms of the β-phosphate as well as the γ-phosphate.
Figure 2.
Mode of binding of the ATP analogue AMPPNP and Mn2+ by the ZIKV helicase. (a) AMPPNP and Mn2+ bind between the two RecA-like domains (coloured blue and red as in Fig. 1 ▸ a). The conserved P-loop and the DEAH motif are highlighted in yellow. AMPPNP is shown in stick representation, while Mn2+ is shown as a sphere. (b) Octahedral coordination of Mn2+. Hydrogen bonds are shown as dashed lines. AMPPNP is shown in stick representation, while the Mn2+ ion and solvent molecules are shown as spheres. (c) LigPlot representation of the interactions of AMPPNP and Mn2+ with solvent molecules and the amino acids of the protein. Hydrogen bonds are shown as dashed lines. Green spheres represent waters. Hydrophilic amino acids and AMPPNP are shown in stick representation. The position of Mn2+ is marked as a blue sphere. Hydrophobic amino acids are shown with a semicircle. (d) 2F o − F c electron density for AMPPNP, Mn2+ and surrounding residues contoured at 1σ.
ZIKV helicase is a DEAH-box-type helicase. A Walker B motif comprising of residues 285DEAH288 engages the γ-phosphate and the manganese ion (Fig. 2 ▸ a). The octahedral coordination of Mn2+ involves the formation of hydrogen bonds to Thr201, Glu286, the β-phosphate, the γ-phosphate and two solvent molecules (Figs. 2 ▸ b and 2 ▸ c). Arg459 and Arg462 from motif VI are in the vicinity of the phosphates. Both of these residues donate a hydrogen bond to the O atom of the γ-phosphate. Lastly, some of the contacts of the protein with ATP and Mn2+ are mediated by water molecules (Fig. 2 ▸ c). Thus, ATP and Mn2+ are bound tightly by numerous hydrogen-bonding interactions (Fig. 2 ▸ c). The electron density for the ligands and surrounding residues was clear (Fig. 2 ▸ d). The interactions of the ligands with the surrounding residues help to position the ligands optimally for catalysis. Currently, there are no data on the specificity of flavivirus helicases for nucleotides.
3.3. Homologous structures
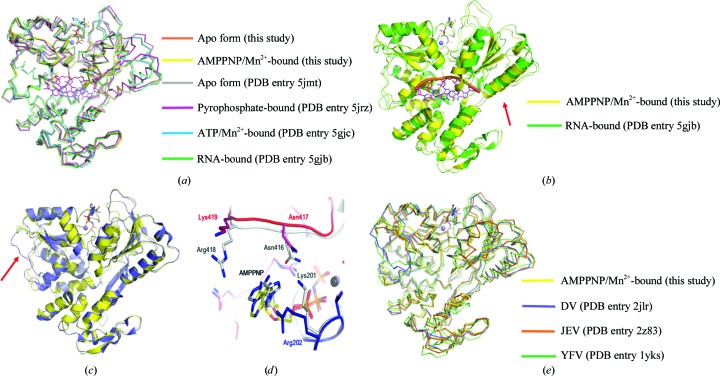
Structures of NS3H in its apo form (PDB entry 5jmt; Tian, Ji, Yang, Xie et al., 2016 ▸), bound to ATP as well as Mn2+ (PDB entry 5gjc; Tian, Ji, Yang, Zhang et al., 2016 ▸), bound to RNA (PDB entry 5gjb; Tian, Ji, Yang, Zhang et al., 2016 ▸) and bound to pyrophosphate (PDB entry 5jrz; Jain et al., 2016 ▸) have previously been reported. The resolution of our structure of NS3H bound to AMPPNP and Mn2+ is the highest to be reported for NS3H from ZIKV to date. This resolution permits interpretation of the locations of water molecules with greater confidence. This is important because solvent molecules play an essential role in catalysis by NS3H.
Superimposition of the apo structure of ZIKV NS3H (PDB entry 5jmt) and the ATP/Mn2+-bound structure (PDB entry 5gjc) on our structures reveals an r.m.s.d. of less than 0.5 Å between overlapping Cα atoms over the entire length of the helicase (Fig. 3 ▸ a). However, when the pyrophosphate-bound form of the helicase (PDB entry 5jrz) is superimposed with our structures, the r.m.s.d. increases to 1.3 Å (Fig. 3 ▸ a). The r.m.s.d. further increases to 1.5 Å when the RNA-bound structure of NS3H (PDB entry 5gjb) is superimposed on our structures. The largest deviation between the structures is observed in the positions of two large helices of RecA-like domain 2A in the RNA-bound structure (Fig. 3 ▸ b). In the RNA-bound structure of NS3H these helices have moved by about 4 Å from the positions observed in the other structures. Such a structural change is conceivable in order to make way for the binding of RNA. The overall topologies of all of the structures of NS3H bound to different ligands are very similar (Fig. 3 ▸ a).
Figure 3.
Comparison of the conformations of ZIKV NS3H. (a) Superimposition of the structures of ZIKV NS3H. The structures are shown as ribbons. RNA, AMPPNP and ATP are shown in stick representation; the divalent cation is shown as a sphere. (b) Superimposition of a cartoon representation of the RNA-bound form of ZIKV NS3H (green) on a cartoon representation of the AMPPNP- and Mn2+-bound form of ZIKV NS3H (yellow). Red arrows indicate the helices of Rec-like domain 2A that move upon RNA binding. (c) Superimposition of a cartoon representation of the helicase from DV (slate) on a cartoon representation of the AMPPN- and Mn2+-bound form of ZIKV NS3H (yellow). The red arrow indicates the position of the loop in the DV helicase that needs to move during RNA binding. (d) Superimposition studies indicate that the mode of tethering of the adenosine moiety of AMPPNP by the DV (grey) and ZIKV helicases is different. Amino acids and AMPPNP are shown in stick representation. (e) Superimposition of the structures of helicases from ZIKV, DV, JEV and YFV. The structures are shown as ribbons. AMPPNP and ATP are shown in stick representation; the divalent cation is shown as a sphere.
A search for structural homologues from other viruses identified the structure of the helicase from DV (Luo et al., 2008 ▸) as the highest structural match (PDB entry 2jlr; Z-score of 58, r.m.s.d. of 1.3 Å for 433 matching Cα positions; Fig. 3 ▸ c). Comparison of AMPPNP- and divalent cation-bound structures of the helicases from the two sources reveals a major difference in the Leu236–Val258 region of RecA-like domain 1A (Fig. 3 ▸ c). This region forms a long loop in the ZIKV helicase. In the structure of the DV helicase this region contains a β-strand. More importantly, the loop is displaced by ∼18 Å from its position in the ZIKV helicase. Furthermore, the distance between the two RecA-like domains of the DV helicase is almost 2 Å less when compared with the distance between the RecA-like domains of the helicase from ZIKV. There is also some difference in how the two helicases engage the adenosine ring of the ATP analogue (Fig. 3 ▸ d). While the side chain of Arg202 of the ZIKV helicase stacks against the heterocyclic ring of adenosine, the amine group of an equivalent lysine residue, Lys201, found in the DV helicase forms two ionic interaction: 3.3 Å with the N6 atom and 3.6 Å with the amine N7 atom of the adenosine moiety. Furthermore, the side chains of Asn416 and Arg418 of the DV helicase are within van der Waals radii of the adenosine ring. The equivalent residues of the ZIKV helicase do not interact with the adenosine ring of the ATP analogue. Thus, although the overall structures of the helicases from ZIKV and DV are very similar, alternative solutions exist to accomplish similar tasks as exemplified by the binding of adenosine by the helicase. In addition to the helicase from DV, the overall structures of the helicases from JEV (Wu et al., 2005 ▸) and YFV (Yamashita et al., 2008 ▸) are also very similar in topology to the ZIKV helicase (Fig. 3 ▸ e).
3.4. Conformational changes upon the binding of ATP and Mn2+
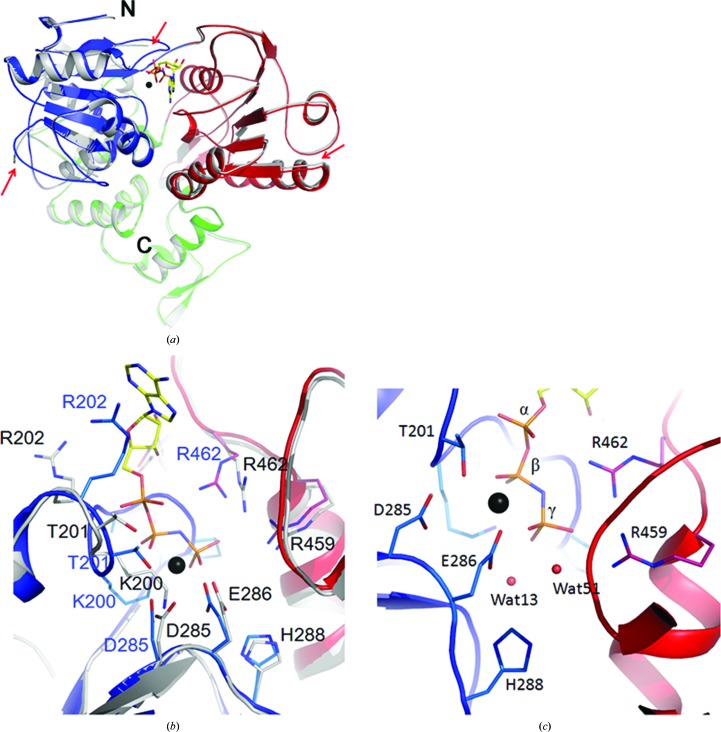
Helicases are molecular motors that undergo conformational changes to accomplish their functions. The binding and hydrolysis of ATP induces movement of the helicase along the RNA (Matlock et al., 2010 ▸; Frick, 2007 ▸; Appleby et al., 2011 ▸; Luo et al., 2008 ▸; Enemark & Joshua-Tor, 2008 ▸). To assess the conformational changes induced by ligand binding, we compared the structure of the apo form of the helicase with the structure of the AMPPNP- and Mn2+-bound helicase (Fig. 4 ▸ a). There are three obvious changes in the positions of the backbone Cα atoms between the two structures. The first, and perhaps the most significant, change observed is the ordering of the Leu236–Val258 loop in the ligand-bound structure. This region of RecA-like domain 1A forms the sixth β-strand in NS3 helicases from flaviviruses such as DV, YFV and JEV (Luo et al., 2008 ▸; Yamashita et al., 2008 ▸; Wu et al., 2005 ▸). In our structure of the apo form of ZIKV NS3H most of this region is disordered. However, in the AMPPNP- and Mn2+-bound structure this region forms a well ordered loop (Fig. 4 ▸ a). Interestingly, this same loop is ordered in another structure of the apo form of NS3H (PDB entry 5jmt), while it is disordered in the ATP/Mn2+-bound form of the helicase (PDB entry 5gjc). This Leu236–Val258 loop is solvent-exposed and is in proximity to an α-helix that is equivalent in position to the spring α-helix of HCV.
Figure 4.
Structural manoeuvres of ZIKV NS3 during the unwinding of nucleic acids. (a) Superimposition of the structure of the apo form of NS3H (grey) on the AMPPNP/Mn2+-bound form of the helicase. The structures are shown as cartoons. Red arrows indicate the differences in the two structures. (b) Movement of the residues from their positions in the apo form of ZIKV helicase (grey sticks) to a new position (sticks coloured based on the convention used in Fig. 1 ▸; blue, domain 1A; red, domain 2A) upon the binding of AMPPNP and Mn2+ is shown. (c) The positions of two solvent molecules, Wat13 and Wat51, either of which could play the role of catalytic nucleophile, are shown. The phosphate groups of AMPPNP and the residues surrounding them are shown in stick representation. In all of the panels, AMPPNP is shown in stick representation; Mn2+ is shown as a black sphere.
The second difference involves the ordering of the Walker A motif or the P-loop. In the apo form of the structure, the loop harbouring the Walker A motif (His196–Gly200) is disordered. The same region is ordered in the presence of AMPPNP and Mn2+ (Fig. 4 ▸ a). The third difference is found in the position of RecA-like domain 2A. This domain has moved inwards towards the ligands by ∼1 Å. In addition to the shift in the positions of the Cα atoms, there is some movement of the side chains upon the binding of ligands (Fig. 4 ▸ b). In particular, the guanidium N atoms of Arg202 have moved by 6.3 Å from their positions in the apo form of the protein to stack with the adenosine moiety. In addition, the side chains of the DEAH motif have moved closer to the ligands (Fig. 4 ▸ b). Although His288 of this motif has moved inwards from its position in the structure of the apo form of the protein, the N atoms of His288 are still not within hydrogen-bonding distance of Glu286 (4.0 Å) or the two putative candidates for performing the role of nucleophile: Wat13 (4.6 Å) and Wat51 (4.0 Å) (Fig. 4 ▸ c). It is possible that binding of the substrate RNA would facilitate further movement, enabling these residues to facilitate catalysis. Other notable movements include a 3.2 Å shift of the guanidium NH2 atom of Arg462 to form a hydrogen bond to the α-phosphate and a 2.8 Å shift of the side chain of Thr201 to form a coordinate bond to Mn2+ (Fig. 4 ▸ b).
3.5. Crystal contacts of the Leu236–Val258 loop
The position of the Leu236–Val258 loop is significantly different in the structures of NS3H from ZIKV. Importantly, this loop is close to the RNA-binding site. A number of weak hydrophobic interactions seem to localize the Leu236–Val258 loop in space in the RNA-bound structure of ZIKV NS3H (PDB entry 5gjb). Specifically, the aromatic ring of Tyr243 is observed to interact with the side chains of Leu236, Val241, Val248, Val250 and Leu260 that surround it. In addition, the side chain of Met244 interacts with the side chains of Arg242 and Arg269, while the hydroxyl group of Try243 interacts with the Cβ atom of Glu233. Three residues of the loop that are in proximity to the RNA, Thr245, Thr246 and Ala247, are observed to interact with the guanidium N atoms of Arg226. Several other weak interactions between residues of the loop and water-mediated contacts help in the folding and localization of the loop away from the RNA-binding channel. Similar interactions of the loop are observed in the structures of pyrophosphate-bound (PDB entry 5jrz), AMPPNP/Mn2+-bound (this study) and the apo form (this study) of ZIKV NS3H. In another structure of the apo form of NS3H of ZIKV reported previously (PDB entry 5jmt), the region comprising of Ala247–Glu256 has shifted by 2–3 Å from the position observed in the abovementioned structures. The most significant difference in the position of the loop and its crystal contacts is observed in the structures of the homologous helicases from DV (PDB entry 2jlr) and YFV (PDB entry 1yks). The tyrosine equivalent to Tyr243 of NS3H is in proximity to the RNA-binding site, with its side chain stacked against the side chain of Arg225. In addition, the tyrosine interacts with the side chain of Cys261. Furthermore, residues Gln243–Asp250 of the loop encircle the side chain of Arg268. The histidine residue in the loop, His252, interacts with the side chain of Arg274. These crystal contacts seem to position the loop in a conformation that blocks the RNA-binding channel at one end. Interestingly, in the structure of the ATP/Mn2+-bound form of NS3H from ZIKV (PDB entry 5gjc) and the structure of the homologous helicase from JEV (PDB entry 2z83) the same loop appears to occupy an intermediate position; however, the loop is partially disordered in these structures. Thus, the Leu236–Val258 loop of NS3H from ZIKV is probably flexible, and the contacts formed as a result of crystallization might be responsible for the differences in the position of the loop observed in the structures of NS3H from ZIKV and its homologues.
4. Discussion
The crystal structure of the helicase from ZIKV reveals a topology that is conserved across flaviviruses (Tian, Ji, Yang, Zhang et al., 2016 ▸; Tian, Ji, Yang, Xie et al., 2016 ▸; Cao et al., 2016 ▸; Jain et al., 2016 ▸). In addition, the mode of binding of ATP and Mn2+ by the helicase is shared between viruses belonging to the Flavivirus genus. Conceivably, the mechanism of hydrolysis of ATP that drives the unwinding of RNA is also shared. The mode of binding of the ATP analogue AMPPNP as well as Mn2+ to the ZIKV helicase provides a number of mechanistic insights into the reaction catalyzed by the helicase. There are two solvent molecules in the active site in the vicinity of the γ-P atom of the ATP analogue (Fig. 4 ▸ c). Either of these solvent molecules could play the role of the nucleophile. The positions of both of these waters are conserved in the structure of the helicase from DV (Luo et al., 2008 ▸). While Wat13 is 3.5 Å away from the P centre, Wat51 is 3.7 Å away from the P centre. There is a possibility that both of these solvent molecules could move closer to the P centre upon the binding of RNA. Glu286 (Glu285 in DV helicase) is hydrogen-bonded to both of these solvent molecules. The solvent molecule is activated for nucleophilic attack upon abstraction of a proton. Gln455 (Gln456 in DV helicase) probably assists in the activation by polarizing the water further. Moreover, His288 is within van der Waals distance of Glu286, which probably enhances the activation of the glutamic acid. Alternatively, His288 could activate Wat13 or Wat51 directly. However, in the current structures His288 is not within hydrogen-bonding distance of these solvent molecules. It could probably move closer to the solvent molecules upon the binding of RNA. Gln455 is likely to assist in proper positioning of the nucleophile by forming a hydrogen bond. Positively charged residues such as Lys200, Arg459 and Arg462 as well as the manganese ion probably help in the redistribution of the charges during catalysis. The presence of arginines near the P centre also raises the possibility of substrate-assisted catalysis, as observed in Ras-like GTPases, where the γ-phosphate serves as a base and the side chain of arginine stabilizes the pentacoordinated transition state.
Considering the fact that ATP hydrolysis drives unwinding of the RNA, the conformational changes observed between the structures of the apo form and the AMPPNP- and Mn2+-bound form of the helicase of ZIKV were surprisingly not very large. Similar observations have been made for the DV and HCV helicases (Luo et al., 2008 ▸; Gu & Rice, 2010 ▸). In fact, the binding of RNA to the helicase, and not ATP, induced a much larger movement of the domains of the helicase of DV.
The available structures of NS3H from ZIKV bound to different ligands including RNA, ATP, AMPPNP, Mn2+ and pyrophosphate help in interpreting the structural movements that would be required or induced upon binding RNA (Fig. 3 ▸ a). The largest manoeuvre that would be required for binding RNA is the movement of the Leu236–Val258 loop. The loop blocks the path of RNA in the structure of NS3H from ZIKV bound to ATP and Mn2+ (PDB entry 5gjc) as well as in the AMPPNP- and Mn2+-bound structure of the DV helicase (PDB entry 2jlr; Fig. 3 ▸ c). Interestingly, the conformation of this loop region in the apo, pyrophosphate-bound and AMPPNP/Mn2+-bound forms of NS3H is similar to that observed in the RNA-bound form, suggesting that this loop is flexible and needs to be localized during RNA binding. RNA binds at the junction where the two RecA-like domains meet domain 3. Two large helices of RecA-like domain 2A move to make way for the RNA (Fig. 3 ▸ b). The gap here is large and domain 3 would close in to seal the gap after the binding of RNA. RecA-like domain 2A would close in on domain 1A upon binding of all the ligands and this would propel the helicase towards a closed conformation. In this conformation, the side chains of the active site are in an optimal position to hydrolyze ATP. The steps after hydrolysis cannot be interpreted from the currently available structural data, but would require the release of ADP and Pi, which would bring the two RecA-like domains back to the positions observed in the apo form of the helicase bound to RNA. During this movement, the backtracking of RNA is prevented by an unknown mechanism. In the next round, the binding of ATP would move the RecA-like domain 2A closer to domain 1A again, which results in threading of the RNA through the channel, and the cycle is repeated to produce single-stranded RNA devoid of secondary structure.
5. Conclusion
In conclusion, the high-resolution crystal structures of the essential helicase from ZIKV in its apo and AMPPNP- and Mn2+-bound forms reveal a conserved topology that is shared with other flaviviruses, such as DV, JEV and YFV, which cause infections in humans. Our studies reveal that the binding of ATP results in the ordering of disordered regions and induces conformational changes in the helicase. The structure of the AMPPNP- and Mn2+-bound helicase provides information about the amino acids of ZIKV that are required for the hydrolysis of ATP that drives the unwinding of RNA. These results provide a framework for the design of inhibitors targeting the helicase to prevent the spread of ZIKV.
Supplementary Material
PDB reference: Zika virus helicase, apo, 5jps
PDB reference: complex with AMPPNP and Mn2+, 5y4z
Acknowledgments
We thank Dr Lijie Wu and the staff of beamline BL19U at Shanghai Synchrotron Radiation Facility (SSRF) and of the Photon Factory (Japan) for their generous help in diffraction data collection and determination of the crystal structure. LL, JW, ZJ and SN designed the research, performed the experiments, analyzed the data and wrote the paper. LL conceived and supervised the project, designed the research, analyzed the data and wrote the paper. The authors declare that they have no competing interests.
Funding Statement
This work was funded by National Natural Science Foundation of China grants 31000332, 31100208, 31100877, 31170782, and 31110103915.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Appleby, T. C., Anderson, R., Fedorova, O., Pyle, A. M., Wang, R., Liu, X., Brendza, K. M. & Somoza, J. R. (2011). J. Mol. Biol. 405, 1139–1153. [DOI] [PMC free article] [PubMed]
- Aziz, H., Zia, A., Anwer, A., Aziz, M., Fatima, S. & Faheem, M. (2017). J. Med. Virol. 89, 943–951. [DOI] [PubMed]
- Bollati, M. et al. (2010). Antiviral Res. 87, 125–148. [DOI] [PMC free article] [PubMed]
- Calvet, G. et al. (2016). Lancet Infect. Dis. 16, 653–660.
- Cao, X., Li, Y., Jin, X., Li, Y., Guo, F. & Jin, T. (2016). Nucleic Acids Res. 44, 10505–10514. [DOI] [PMC free article] [PubMed]
- Cugola, F. R. et al. (2016). Nature (London), 534, 267–271.
- Cunha, M. S. et al. (2016). Genome Announc. 4, e00032-16.
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Dub, T. & Fontanet, A. (2017). Rev. Neurol. (Paris), 173, 361–363. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Enemark, E. J. & Joshua-Tor, L. (2008). Curr. Opin. Struct. Biol. 18, 243–257. [DOI] [PMC free article] [PubMed]
- Frick, D. N. (2007). Curr. Issues Mol. Biol. 9, 1–20. [PMC free article] [PubMed]
- Gu, M. & Rice, C. M. (2010). Proc. Natl Acad. Sci. USA, 107, 521–528. [DOI] [PMC free article] [PubMed]
- Hamel, R. et al. (2015). J. Virol. 89, 8880–8896. [DOI] [PMC free article] [PubMed]
- Heukelbach, J. & Werneck, G. L. (2016). Lancet, 388, 846–847. [DOI] [PubMed]
- Jain, R., Coloma, J., García-Sastre, A. & Aggarwal, A. K. (2016). Nature Struct. Mol. Biol. 23, 752–754. [DOI] [PMC free article] [PubMed]
- Korzeniewski, K., Juszczak, D. & Zwolińska, E. (2016). Int. Marit. Health, 67, 31–37. [DOI] [PubMed]
- Krauer, F., Riesen, M., Reveiz, L., Oladapo, O. T., Martínez-Vega, R., Porgo, T. V., Haefliger, A., Broutet, N. J., Low, N. & WHO Zika Causality Working Group (2017). PLoS Med. 14, e1002203. [DOI] [PMC free article] [PubMed]
- Luo, D., Xu, T., Watson, R. P., Scherer-Becker, D., Sampath, A., Jahnke, W., Yeong, S. S., Wang, C. H., Lim, S. P., Strongin, A., Vasudevan, S. G. & Lescar, J. (2008). EMBO J. 27, 3209–3219. [DOI] [PMC free article] [PubMed]
- Marano, G., Pupella, S., Vaglio, S., Liumbruno, G. M. & Grazzini, G. (2016). Blood Transfus. 14, 95–100. [DOI] [PMC free article] [PubMed]
- Matlock, D. L., Yeruva, L., Byrd, A. K., Mackintosh, S. G., Langston, C., Brown, C., Cameron, C. E., Fischer, C. J. & Raney, K. D. (2010). Biochemistry, 49, 2097–2109. [DOI] [PMC free article] [PubMed]
- Maurer-Stroh, S., Mak, T.-M., Ng, Y.-K., Phuah, S.-P., Huber, R. G., Marzinek, J. K., Holdbrook, D. A., Lee, R. T. C., Cui, L. & Lin, R. T. P. (2016). Euro Surveill. 21, 30347. [DOI] [PMC free article] [PubMed]
- Meertens, L. et al. (2017). Cell. Rep. 18, 324–333. [DOI] [PubMed]
- Miller, E., Becker, Z., Shalev, D., Lee, C. T., Cioroiu, C. & Thakur, K. (2017). J. Neurol. Sci. 375, 367–370. [DOI] [PubMed]
- Routhu, N. K. & Byrareddy, S. N. (2017). J. Neuroimmune Pharmacol. 12, 219–232. [DOI] [PMC free article] [PubMed]
- Russo, F. B., Jungmann, P. & Beltrao-Braga, P. C. B. (2017). Cell. Microbiol. 19, e12744. [DOI] [PubMed]
- Sironi, M., Forni, D., Clerici, M. & Cagliani, R. (2016). PLoS Negl. Trop. Dis. 10, e0004978. [DOI] [PMC free article] [PubMed]
- Siu, R., Bukhari, W., Todd, A., Gunn, W., Huang, Q. S. & Timmings, P. (2016). Neurology, 87, 1623–1624. [DOI] [PubMed]
- Tian, H., Ji, X., Yang, X., Xie, W., Yang, K., Chen, C., Wu, C., Chi, H., Mu, Z., Wang, Z. & Yang, H. (2016). Protein Cell, 7, 450–454. [DOI] [PMC free article] [PubMed]
- Tian, H., Ji, X., Yang, X., Zhang, Z., Lu, Z., Yang, K., Chen, C., Zhao, Q., Chi, H., Mu, Z., Xie, W., Wang, Z., Lou, H., Yang, H. & Rao, Z. (2016). Protein Cell, 7, 562–570. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wu, J., Bera, A. K., Kuhn, R. J. & Smith, J. L. (2005). J. Virol. 79, 10268–10277. [DOI] [PMC free article] [PubMed]
- Xin, Q.-L., Deng, C.-L., Chen, X., Wang, J., Wang, S.-B., Wang, W., Deng, F., Zhang, B., Xiao, G. & Zhang, L.-K. (2017). J. Virol. 91, e00554-17. [DOI] [PMC free article] [PubMed]
- Xu, T., Sampath, A., Chao, A., Wen, D., Nanao, M., Chene, P., Vasudevan, S. G. & Lescar, J. (2005). J. Virol. 79, 10278–10288. [DOI] [PMC free article] [PubMed]
- Yamashita, T., Unno, H., Mori, Y., Tani, H., Moriishi, K., Takamizawa, A., Agoh, M., Tsukihara, T. & Matsuura, Y. (2008). Virology, 373, 426–436. [DOI] [PubMed]
- Yun, S.-I. & Lee, Y.-M. (2017). J. Microbiol. 55, 204–219. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Zika virus helicase, apo, 5jps
PDB reference: complex with AMPPNP and Mn2+, 5y4z