Summary
The vertebrate extinction rate over the past century is approximately 22 – 100 times greater than background extinction rates [1] and large mammals are particularly at risk [2, 3]. Quaternary megafaunal extinctions have been attributed to climate change [4], overexploitation [5] or a combination of the two [6]. Rhinoceroses (Family: Rhinocerotidae) have a rich fossil history replete with iconic examples of climate-induced extinctions [7], but current pressures threaten to eliminate this group entirely. The Sumatran Rhinoceros (Dicerorhinus sumatrensis) is among the most imperiled mammals on earth. The 2011 population was estimated at ≤ 216 wild individuals [8] and currently the species is extirpated, or nearly so, throughout the majority of its former range [8–12]. Understanding demographic history is important in placing current population status into a broader ecological and evolutionary context. Analysis of the Sumatran Rhinoceros genome reveals extreme changes in effective population size throughout the Pleistocene. Population expansion during the early to middle Pleistocene was followed by decline. Ecological niche modeling indicated that changing climate likely played a role in the decline of Sumatran Rhinoceros as less suitable habitat on an emergent Sundaland corridor isolated Sumatran Rhinoceros populations. By the end of the Pleistocene the Sundaland corridor was submerged, populations were fragmented and consequently reduced to low Holocene levels from which they would never recover. Past events denuded the Sumatran Rhinoceros of genetic diversity either through population decline or fragmentation or some combination of the two and likely made the species even more susceptible to later exploitation and habitat loss.
Keywords: Demography, Ecological niche modeling, Evolution, Pairwise sequentially Markovian coalescent, Pleistocene, Rhinoceros, Sundaland, Whole genome sequencing
eTOC Blurb
Mays et al. report the first genome sequence for the Sumatran Rhinoceros. Genomic analysis reveals a fluctuating population history, ending at low levels by the end of the Pleistocene. Ecological niche models suggest that changing climate during the Pleistocene influenced habitat availability and likely led to declining or fragmented populations.
Results and Discussion
Genomic coalescent analyses allow for hypothesis testing regarding demographic history, an approach that is particularly useful when studying recently extinct or highly endangered species where sampling is often extremely limited [13]. Studies have shown that currently imperiled or recently extinct species tend to have experienced long-term population decline [14, 15] or have relatively low effective population size (Ne) caused by dramatic population fluctuation [16]. It is of biological and conservation importance to examine the driving forces behind these historical changes in populations. Climate is likely a causal factor in shaping population dynamics of many species [6, 17]. Populations denuded of genetic diversity by past climate fluctuations are especially vulnerable to current exploitation and habitat degradation [16]. To address questions at the intersection of climate and population change we coupled a demographic analysis using a Pairwise Sequential Markovian Coalescent (PSMC) method based on whole genome sequencing with Ecological Niche Models (ENMs) to elucidate the demographic history of the Sumatran Rhinoceros as it relates to past climate change (see STAR Methods).
Our study reports the first draft genome assembly for the Sumatran Rhinoceros. Jellyfish 2.2.3 [18] supported a genome size of 2.53 Gb sequenced at a peak coverage of 46×. Our estimated genome size is broadly congruent with other estimates of genome size in the Perissodactyla (http://www.genomesize.com) [19]. Heterozygosity was low (approximately 1.3 single nucleotide polymorphism (SNP) sites per 1,000 bp of autosomal sequence) and comparable to that found in whole genome studies in recently extinct mammals [17, 20] and approaching that of inbred domestic species such as the Horse (Equus caballus) [21].
Prior studies place the Sumatran Rhinoceros within the dicerorhine Eurasian rhinoceroses with close evolutionary affiliations with the Wooly Rhinoceroses (Coelodonta spp.) and Stephanorhinus spp. [7, 22, 23]. Fossils from Myanmar attributed to Dicerorhinus have been dated to the mid to late Pliocene [24] and from Guangxi, China to the early Pleistocene [25]. Earlier fossils attributed to Dicerorhinus likely belong to other dicerorhine genera such as Stephanorhinus [23]. Fossil evidence therefore suggests that Dicerorhinus originated in Northern Indochina and South China during the middle to late Pliocene with at least one lineage eventually expanding southward into Indochina and Sundaland during a period when the landmasses in the region were emergent and in their present-day configurations [26]. After the Pliocene the region was periodically submerged, isolating terrestrial biotas [27]. PSMC analysis of the Sumatran Rhinoceros genome complements this fossil record with a demographic history derived from genomic data.
The PSMC analyses revealed the population dynamics of the Sumatran Rhinoceros from approximately 7 Ma to 1 ka (Figure 1, Figure S1, Table 1). PSMC analyses based on all scaffolds and autosomal scaffolds returned similar results and therefore we only reported the results for the latter. Sumatran Rhinoceros populations likely experienced substantial population fluctuations since the beginning of the Pleistocene (2.58 Ma). The degree and timing of these fluctuations depended on estimates of substitution rate and generation time, but the trend in Pleistocene population change was similar across separate analyses. Applying a substitution rate of 2.34 × 10−8 substitutions/site/generation [28] and a generation time of 12 years [29] we estimated a peak Ne (rounded to the nearest 100 individuals) of 57,800 occurring approximately 950 ka and a minimal Ne of 700 occurring approximately 9 ka and a net drop in Ne of 31,200 across the Pleistocene (Figure 1, Table 1). Separate PSMC analyses based on upper and lower estimates of substitution rate from the literature [13, 30, 31] revealed a peak Ne (41,000 – 112,800) sometime during the early to mid Pleistocene and minimal Ne (500 – 1,300) by the end of the Pleistocene (Figure S1, Table 1). Population decline characterized Sumatran Rhinoceros populations throughout most of the middle to late Pleistocene (Figure 1, Figure S1, Table 1).
Figure 1. Demographic history of the Sumatran Rhinoceros.
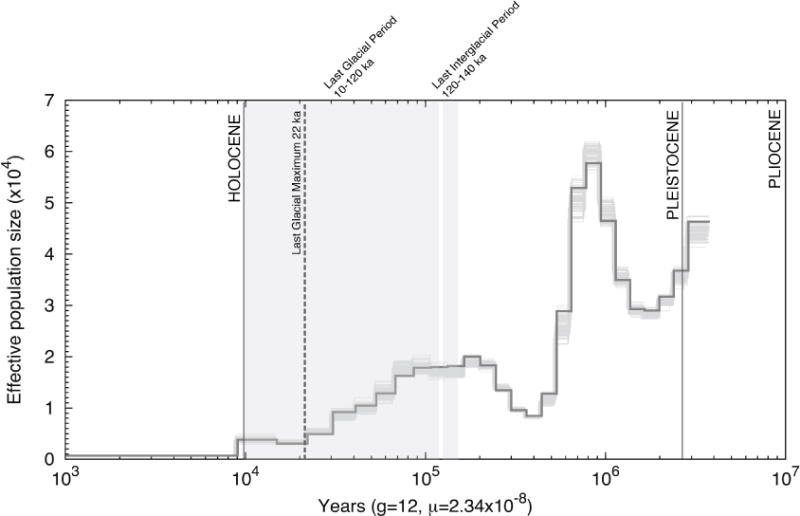
The PSMC analysis is applied to the genomic sequences of the Sumatran Rhinoceros converted to demographic units (individuals and years) assuming a generation time of g = 12 years and a substitution rate of μ = 1.95 × 10−9 substitutions/site/year (2.34 × 10−8 substitutions/site/generation). The x-axis indicates time before present in years on a log scale and the y-axis indicates effective population size. The bold grey curve shows the estimate based on original data, and the light grey curves show the estimates for 100 bootstrapped sequences. The two gray shaded areas indicate the last glacial period (LGP) and the last interglacial period (LIG) and the dashed line demarcates the approximate time of the last glacial maximum (LGM, see also Figure S1).
Table 1.
Effective population size over time.
| Substitutions/site/generation | Minimum Ne (time of min Ne in ka) | Maximum Ne (time of max Ne in ka) | Ne at 12ka | Ne at 2.58Ma | Net change in Ne during the Pleistocene |
|---|---|---|---|---|---|
| 1.2 × 10−8 | 1,300 (17) | 112,800 (1,800) | 1,300 | 55,300 | −54,000 |
| 2.34 × 10−8 | 700 (9) | 57,800 (950) | 3,600 | 34,800 | −31,200 |
| 3.3 × 10−8 | 500 (6.5) | 41,000 (650) | 2,300 | 30,800 | −28,500 |
Effective population size (Ne) variation across three PSMC analyses using different estimates of the per generation substitution rate and a generation time of g = 12. All population sizes are rounded to the nearest 100 individuals (see also Figure S1).
An increase in Ne occurring during the early to middle Pleistocene is indicative of a demographic expansion that likely co-occurred with a range expansion of Sumatran Rhinoceros from an ancestral, more northerly Asian distribution into Southeast Asia and Sundaland. The expansion of Sumatran Rhinoceros across an exposed Sundaland would correspond to similar expansions of continental mammals into the region. By the middle Pleistocene continental fauna replaced many island taxa that evolved in isolation during the early Pleistocene [32] and PSMC analyses suggest that the Sumatran Rhinoceros was also part of this early to middle Pleistocene invasion of Sundaland. Following this early to middle Pleistocene demographic expansion were dramatic population fluctuations throughout the remainder of the Pleistocene often occurring in association with climate and/or sea level changes. Population fluctuations might explain relatively low and long-term decline in Ne of the Sumatran Rhinoceros from middle to late Pleistocene [16].
The duration of the last glacial period (LGP, ca. 10–120 ka) [27] and the transition between the Pleistocene and the Holocene coincides with dramatic population changes in many species. Genomic analyses reveal abrupt declines in Ne associated with the end of the LGP for many north temperate and arctic megafauna [17, 31, 33, 34], or steady declines throughout the LGP [35]. Genomic studies of other species, including sub-tropical and tropical species, also suggest declines in Ne during the LGP for crocodilians [36], birds [15, 16] and mammals [13, 14]. Nadachowska-Bryska et al. [15] found the LGP coincided with significant declines in Ne for 22 of 38 avian species studied. The LGP was likewise a period of population decline for the Sumatran Rhinoceros ending at their current and minimal Ne by the Pleistocene-Holocene boundary.
Comparisons among studies of demographic changes based on PSMC are fraught with assumptions. While the shape of the Ne curve remains consistent, magnitude and timing of changes in Ne are biased by both substitution rate and generation time [15]. Substitution rates used in the analyses are estimates derived from studies of other large mammals [13, 28, 30, 31] and represent a source of variation in the PSMC analyses in estimating the timing and magnitude of the Ne curve.
PSMC analyses reveal a low recent estimate of Ne for the Sumatran Rhinoceros that has remained low since the end of the LGP (Figure 1, Figure S2, Table 1). Population declines due to recent human exploitation and habitat loss are likely acting on a population denuded of genetic diversity during the Pleistocene. However, PSMC is a poor indicator of very recent Ne given the comparatively small sample size associated with very recent coalescent events [13]. Future studies using coalescent approaches that incorporate variation across multiple genomes [37] would aid in corroborating these patterns. However, given the paucity of wild rhinoceros samples in general and the deliberate inbred nature of the captive Sumatran Rhinoceros population, obtaining multiple genetically independent samples for sequencing in this species is challenging.
ENMs suggest that past climate change may have contributed significantly to the population dynamics of Sumatran Rhinoceros. Predicted present-day distributions of Sumatran Rhinoceros are similar between the ‘All occurrences’ (D. sumatrensis and Rhinoceros spp., Figure 2A) and ‘SR occurrences’ (D. sumatrensis, Figure 2D) data sets, and are in general agreement with their current distribution [11, 38]. Predicted present-day distribution of the subspecies D. s. sumatrensis (DSS occurrences, Figure 2G) is restricted to Sumatra and Malay Peninsula, and does not extend to other areas within the Sundaland region (e.g. Borneo, Java). This pattern is consistent with the known distribution of this subspecies and suggests that climatic conditions alone may be sufficient to limit range expansion of D. s. sumatrensis.
Figure 2. Predicted distributions of Sumatran Rhinoceros.
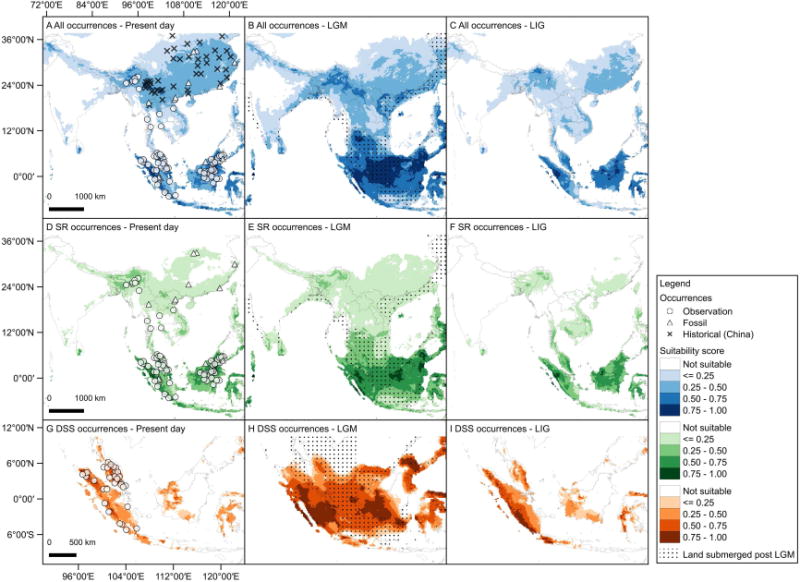
All occurrences (top panel) include Dicerorhinus sumatrensis and Rhinoceros spp.; SR occurrences (middle panel) include D. sumatrensis; DSS occurrences (bottom panel) include SR occurrences from Sumatra and Peninsula Malay (D. s. sumatrensis). Occurrences for Rhinoceros spp. are denoted with an × while known Sumatran Rhinoceros occurrences are denoted with open circles. Likely historical occurrences of Sumatran Rhinoceros are denoted by triangles. A grid is overlaid on the maps in the second column to denote emergent land during the last glacial maximum (LGM). The areas with suitability scores lower than the minimum training presence threshold are considered ‘not suitable.’ The land submerged post LGM are the areas ca. 120 m below sea level on the bathymetric map (see also Figure S2, Figure S3, Table S2 and Table S3).
All three ENMs for Sumatran Rhinoceros (all occurrences, SR occurrences, DSS occurrences) revealed significant changes in predicted distributions associated with Pleistocene climate change from the last interglacial (LIG) [39] through the last glacial maximum (LGM) [27, 40] to present day (Figure 2). The central Sundaland corridor was submerged at the end of the LGP creating an western refugium in Sumatra and an eastern refugium in Borneo [41]. Predicted distributions are similar between LIG (Figure 2C, 2F, and 2I) and present day (Figure 2A, 2D, and 2G), both of which are smaller and more fragmented than that during LGM (Figure 2B, 2E, and 2H). Predicted present-day distributions fall predominantly within tropical and subtropical moist broadleaf forest for all three ENMs (Table S2). Predicted LGM distributions were concentrated in the Sundaland region (Figure 2B, 2E, and 2H), and the highest proportion of LGM distributions were associated with tropical grassland followed by monsoon and dry forest and tropical forest. However, for the DSS model 32% of predicted LGM distribution fell within tropical forest indicating that for this subspecies tropical forest closely rivals tropical grassland as the vegetation found in the most suitable climate niche during the LGM (Table S2). If forest cover restricts the ecological niche, at least for the subspecies D. s. sumatrensis, their LGM distribution would have been greatly reduced and become highly fragmented (Figure 2, Table S2). For instance, removing the ‘tropical grassland’ in central Sundaland reduced predicted LGM distributions by 21–34% (Figure S2, Table S2). The rise in sea level, particularly in the Sundaland region [41], also reduced the predicted distributions for Sumatran Rhinoceros from the LGM to present day by 25 – 39% (Figure 2, Table S2).
Among the dicerorhine rhinoceroses only the Sumatran Rhinoceros is known as a tropical forest species with the rest being primarily or exclusively open woodland, grassland, and savannah species with more temperate distributions [7, 22, 23]. Modern Sumatran Rhinoceros typically have a preference for secondary forest and in some locales are associated with riparian, disturbed and even edge habitat [12, 42]. Given the close evolutionary relationships between Sumatran Rhinoceros and more temperate, grassland, and open forest species, the ancestral preferred habitat for ancestral Sumatran Rhinoceros when it expanded into Southeast Asia during the early Pleistocene may have been more open with populations adapting to more forested habitats over time.
A broad north-south savannah corridor may have extended through Sundaland during the late Pleistocene [43–46] (Figure S2). This belt of open vegetation running through central Sundaland between what are now the islands of Sumatra and Borneo has been under some debate [44, 47]. However, limited migration during the LGP between west (Sumatra) and east Sundaland (Borneo) has been suggested for mammals [48], snakes and frogs [49] and rainforest termites [44]. Divergence among these taxa within Sundaland is likely due to vicariance events that predate the Pleistocene indicating the Sundaland corridor acted as a barrier to dispersal for many taxa. The Sundaland savannah corridor may have been a dynamic, mosaic landscape comprised of both open and closed vegetation habitats [45, 46]. Whether such mosaic landscape was part of the niche for any species in the genus Dicerorhinus, Sumatran Rhinoceros sensu lato or the Sumatran/Malay Peninsula subspecies (D. s. sumatrensis) during LGP is unclear.
Given the strong favoring of tropical and subtropical moist broadleaf forest in all three present-day ENMs and known habitat preferences [12, 42] favorable climate may not have been associated with favorable vegetation during the LGM. In addition, PSMC analyses revealed demographic decline throughout the LGP suggesting the central Sundaland corridor may have functioned as a “soft” barrier to dispersal for Sumatran Rhinoceros populations in Sumatra/Malay Peninsula and Borneo that would in effect promote population divergence [50]. Contraction of lowland and upland tropical forest during the LGP has resulted in the current refugial state of these habitats and likely contributed to population bottlenecks in many Sundaland species [51]. The concordance between the contractions of predicted distributions and genetic evidence of a declining population throughout the LGP suggests a role for climate in the reduction of Sumatran Rhinoceros populations to levels by the end of the Pleistocene from which they would never recover.
Distinguishing population declines from population structuring is difficult using PSMC [33]. Sumatran Rhinoceros has been historically divided into three subspecies: a historically extinct D. s. lasiotis occurring in Northern Indochina, South China, Myanmar and far eastern India, D. s. sumatrensis on the Malay Peninsula and Sumatra and D. s. harrisoni on the island of Borneo [42, 50, 52]. The latter two subspecies are likely the descendants of populations trapped in refugia either during the LGP when a drier central Sundaland corridor acted as a barrier to dispersal, by the end of the LGP or during earlier interglacial periods when the corridor was submerged. D. s. lasiotis however may have been isolated from other populations since the LIG when large portions of Indochina were unsuitable in terms of climatic conditions (Figure 2C and 2F). The ENM analysis restricted to occurrences of D. s. sumatrensis (the subspecies from which our genome data was derived) is the model showing the most dramatic contraction of predicted distribution due to the inundation of the Sundaland corridor. Therefore, the conclusion that climate played a role in population decline is at least strongly suggested for D. s. sumatrensis if not for the entire species.
Climate however is not the only potential cause of extinctions and population declines at the Pleistocene-Holocene boundary. Depredation and habitat changes by expanding Homo sapiens populations are implicated in the extinctions of many megafaunal species [5, 53]. Excavations at the Niah cave site on the island of Borneo reveals that forest was cleared by humans for cultivation during the Holocene [54] and that humans hunted local animals, including the Sumatran Rhinoceros, as early as the late Pleistocene [55]. Hunting by Pleistocene humans in Southeast Asia has been implicated in the extirpation of Orangutan (Pongo spp.) from parts of its range and the extinction of Stegodon and Giant Pangolin (Manis palaeojavanica)[56]. It is likely that recent human exploitation and habitat loss have been acting on Sumatran Rhinoceros populations already denuded of genetic diversity since the Pleistocene and have thus accelerated their extinction trajectory.
Coupling analyses from genome data and ENM is a powerful tool in elucidating the patterns and process associated with past demographic changes in populations. For critically endangered species, this approach may provide a more objective ecological and evolutionary context for designing conservation strategies. We hope our genome sequence may serve as a reference for broader population genomics in this imperiled species.
Star ★ Methods
KEY RESOURCES TABLE
See ‘Star Methods: Key Resources Table’.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Dicerorhinus sumatrensis sumatrensis | Cincinnati Museum Center | CMC M4249 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Critical Commercial Assays | ||
| TruSeq® DNA PCR-Free LT Library Preparation Kit (24 samples) | Illumina, Inc. | 20015962 |
| Nextera Mate Pair Library Prep Kit (12 indexes, 48 gel-free samples or 12 gel-plus samples) | Illumina, Inc. | FC-132-1001 |
| HiSeq PE Rapid Cluster Kit v2 | Illumina, Inc. | PE-402-4002 |
| HiSeq Rapid SBS Kit v2 | Illumina, Inc. | FC-402-4023 |
| Deposited Data | ||
| Whole genome shotgun sequence assembly | This paper | NCBI: PEKH00000000 |
| Raw whole genome sequencing reads | This paper | NCBI: PRJNA415733 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| Trimmomatic 0.33 | [57] | http://www.usadellab.org/cms/?page=trimmomatic |
| kmergenie | [58] | http://kmergenie.bx.psu.edu |
| Jellyfish 2.2.3 | [18] | http://www.genome.umd.edu/jellyfish.html |
| DISCOVAR de novo | [59] | https://software.broadinstitute.org/software/discovar/blog/ |
| SOAP de novo 2.04 | [60] | http://soap.genomics.org.cn/soapdenovo.html |
| Extreme Science and Engineering Discovery Environment (XSEDE) | [61] | https://www.xsede.org |
| Google Earth | Google Inc. | https://www.google.com/earth/ |
| Burrows Wheeler Aligner Program (BWA) 0.715 | [69] | http://bio-bwa.sourceforge.net |
| Basic Local Alignment Search Tool (BLAST) 2.5.0 | [70] | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| SAM Tools 1.3.1 | [71] | http://www.htslib.org |
| PICARD 2.4.0 | Broad Institute | https://github.com/broadinstitute/picard |
| BAM Tools 1.3.1 | [72] | https://github.com/pezmaster31/bamtools |
| Genome Analysis Toolkit (GATK) 3.6 | [81] | https://software.broadinstitute.org/gatk/ |
| Pairwise Sequentially Markovian Coalescent (PSMC) 0.6.5 | [13] | https://github.com/lh3/psmc |
| MAXENT 3.3.3 | [75] | https://biodiversityinformatics.amnh.org/open_source/maxent/ |
| Other | ||
Contact for reagent resources and sharing
Further information and requests for protocols and datasets should be directed to and will be fulfilled by the Lead Contact, Herman L. Mays Jr. (maysh@marshall.edu)
Experimental model and subject details
Tissue was collected from a captive, wild-caught male Sumatran Rhinoceros collected in Indonesia in Retak Mudik, Sub-District of Ipuh, District of Bengkulu Utara, and Province of Bengkulu on the island of Sumatra and exported to the Cincinnati Zoo and Botanical Garden on April 10, 1991. This specimen (named “Ipuh”) was euthanized due to deteriorating health on February 18, 2013 and tissue samples from skeletal muscle, heart and liver were collected during the necropsy and separate samples of each tissue type were stored in ethanol or RNAlater kept at −80°C. Genomic DNA was isolated from each tissue type using standard phenol-chloroform-isoamyl alcohol extraction methods. Tissues and specimen voucher material (mounted skin and complete disarticulated skeleton) were deposited at the Cincinnati Museum Center (CMC M4249).
Method details
Genome sequencing
Whole genome, shotgun sequencing was performed on an Illumina HiSeq 1500 at the Marshall University Genomics Core Facility. One paired-end library and eight mate pair libraries were prepared from purified genomic DNA and sequenced. We prepared the paired end library using Illumina TruSeq® DNA PCR-Free LT Library Preparation Kit from genomic DNA according to the manufacturer’s instructions; average insert size for this library was 462 base pairs (bp). These libraries were sequenced in three separate 2 × 250 bp paired-end HiSeq1500 Rapid Runs. Gel-free and gel-plus mate pair libraries were prepared using the Nextera Mate Pair Library Prep Kit according to the manufacturer’s instructions. Gel-plus libraries were prepared from DNA fragments in three size ranges: 4-6kb, 6-9kb and 9-12kb. Adaptor enrichment (library amplification) was 10 cycles of PCR for gel-free libraries and 15 cycles of PCR for gel-plus libraries. Two replicates were generated for each gel-free and gel-plus mate pair library, resulting in 8 libraries in total. Average library insert sizes for gel-free and gel-plus libraries ranged from 345 to 515 bp and from 240 to 363 bp, respectively. Mate pair libraries were sequenced in a 2 × 150 bp paired-end Rapid Run mode. Illumina HiSeq sequencing used the HiSeq PE Rapid Cluster Kit v2 and HiSeq Rapid SBS Kit v2 sequencing kits.
Genome assembly
Trimming of sequencing reads was done using Trimmomatic 0.33 [57] and K-mer estimation was performed using kmergenie [58]. Genome size and coverage was estimated from trimmed fastq files by 25-mers in Jellyfish 2.2.3 [18]. De novo genome assembly from the Illumina libraries was conducted via a pipeline combining DISCOVAR de novo [59] and SOAPdenovo2 2.04 [60]. Contigs were generated by passing the paired-end reads through DISCOVAR de novo, running on a 12 TB node on the Bridges computing cluster at Pittsburgh Supercomputing Center via a startup allocation from the Extreme Science and Engineering Discovery Environment (XSEDE)[61]. Resulting contigs were combined with the mate pair libraries and assembled into scaffolds using the “scaff” command from SOAPdenovo2. After preprocessing, 570,526,774 paired-end DNA sequencing reads were used to assemble contigs with DISCOVAR de novo. The resulting contigs, with an N50 of 80,701 bp, were combined with reads from mate pair libraries and assembled into scaffolds using SOAPdenovo2. This process generated 1.1 million scaffolds, 4,588 of which were greater than 100 kb, spanning a total of 2.96 Gb with an N50 of 0.6 Mb.
Occurrence data for ecological niche modeling
We built ecological niche models (ENMs) for Sumatran Rhinoceros at a resolution of 10 arc-minutes (ca. 18.5 km × 18.5 km at the equator) given the relatively low resolution of the occurrence data (e.g. only 26% of the 19 occurrences reported in Meijaard [9] had an accuracy of < 20 km). Sumatran Rhinoceros tend to have large home ranges with low population densities (home range: ca. 10–30 km2; population density: ca. 0.02–0.04 km2) [62] and as such our comparatively coarse spatial resolution is likely ecologically relevant.
Occurrences were obtained from the literature [9–11, 38, 63–68] and georeferenced in GoogleEarth©. We established three occurrence data sets. An all occurrences data set (132 occurrences) included Sumatran Rhinoceros (D. sumatrensis) and putative Rhinoceros spp.; the SR occurrences data set (91 occurrences) included occurrences from all recognized subspecies of the Sumatran Rhinoceros (SR); and a DSS occurrences data set (30 occurrences) included SR occurrences from Sumatra and the Malay Peninsula, which are assigned to the subspecies D. s. sumatrensis (DSS) [52]. Although the historical geographic range of Sumatran Rhinoceros is indeterminate, partly due to their sympatric distribution with Rhinoceros spp. (R. unicornis, R. sondaicus), modern observations, fossil records and historical documents indicate that they once occurred in Bhutan and northeastern India, through southern China, Myanmar, Thailand, Cambodia, Lao PDR, Vietnam and the Malay Peninsula, and the islands of Sumatra and Borneo in Indonesia [11, 38, 68]. Therefore, we set the spatial extent of the ENMs to include all known occurrences of Sumatran Rhinoceros and sympatric Rhinoceros spp., an area ranging from 71° to 124° E and 11° S to 38° N (herein ‘South Asia’). However, for DSS occurrences, we reduced the spatial extent to the Sundaland region, ranging from 90° to 124° E and 11° S to 11° N (i.e. the northern boundary set at Isthmus of Kra). It is necessary to reduce the study area for DSS occurrences because they are spatially clustered, which may lead to model overfitting when pseudo-absence data are randomly drawn from a large study area. For statistical analysis of these models see section below.
Quantification and statistical analysis
Demographic analysis using PSMC
The Burrows-Wheeler Aligner program (BWA 0.7.15) [69] was used to map raw sequencing reads against the de novo assembled genome containing all scaffolds or scaffolds excluding those that are X-chromosome-linked (i.e. autosomal scaffolds). The BWA-mem algorithm was used with default parameters. We searched X-chromosome-linked scaffolds from the assembled genome by blasting all scaffolds against the X-chromosomes of human (Homo sapiens; GCA_000001405.25), mouse (Mus musculus; GCA_000001635.7) and horse (Equus caballus; GCA_000002305.1), respectively, using BLAST+ 2.5.0 [70]. We assumed the blasted scaffolds that were shared among the three independent analyses as X-chromosome-linked scaffolds in the Sumatran Rhinoceros genome. The BLAST+ parameters were set as: −evalue = 1e-10; −word_size = 15; −max_target_seqs = 1000. We then excluded X-chromosome-linked scaffolds from the assembled genome to test for their effect on the genome-based estimates of demographic history.
SAMtools 1.3.1 [71] was used to sort and merge reads from different sequencing lanes. The program Picard 2.4.0 (https://broadinstitute.github.io/picard/) was used to remove duplicate reads from the BWA mapped records. Sequencing depth was estimated using BamTools 1.3.1 [72]. The Genome Analysis Toolkit (GATK 3.6) [73] was used for local realignment and base quality recalibration to the mapped records before calling consensus sequences. Recalibration based on a concordant SNP dataset was done with SAMtools “mpileup” and GATK “UnifiedGeontyper” programs.
We applied the SAMtools package to produce diploid consensus sequences containing heterozygous (i.e., single-nucleotide polymorphism, SNP) sites for the BWA aligned records using the “mpileup”, “bcftools” and “vcfutils.pl” programs. Several filters and options were added to keep only those consensus sequences with high confidence: (1) the option “–C50” was used to lower mapping quality for reads containing excessive mismatches; (2) the minimum mapping quality for an alignment to be included (-q) was set to 25; (3) sites with sequencing depths (-d) smaller than a third and (-D) larger than twice of the average depth of the aligned genome were excluded from the consensus sequence assignment, and (4) the sequences with consensus quality lower than 20 were filtered out. The first three filters were performed when using SAMtools for consensus sequence calling, and the fourth one was performed using the “fq2psmcfa” program in the PSMC package. We calculated the percentage of SNP sites of the consensus sequences.
We used the Pairwise Sequentially Markovian Coalescent (PSMC 0.6.5)[13] model to infer the effective population sizes (Ne) of the Sumatran Rhinoceros over time based on the genome sequences with SNP sites. The program “fq2psmcfa” provided by the PSMC package was used to divide the consensus sequences to 100-bp bins as input files for PSMC analysis. The minimal consensus quality of sequence for considering the fq2psmcfa conversion was set to 20. We set N (the number of iterations) = 25, t (Tmax) = 15 and p (atomic time interval) = 4+25*2+4+6.
We used a substitution rate based on comparisons between cattle, dog and human genomes of 1.95 × 10−9 substitutions/site/year [28]. In addition, we report supplementary PSMC analyses based on two other substitution rates from studies of human and horses (Equus spp.) genomes, which were 1.0 × 10−9 substitutions per site per year [13, 31], and that of the Przewalski’s Horse (Equus przewalskii) genome, which was 2.75 × 10−9 substitutions per site per year [30], to define potential bounds for population size and the timing of demographic changes. Other estimates of substitution rates averaged across mammalian orders fall within this range (2.22 × 10−9 substitutions/site/year) [74]. We estimated a generation time of 12 years based on doubling the average maximum age at sexual maturity (6.5 years for males and 5.5 years for females) [29]. Thus the substitution rates of 1.2 × 10−8, 2.34 × 10−8, and 3.3 × 10−8 substitutions/site/generation were used to convert the PSMC output to scales in years and individuals. Bootstrap tests with 100 replicates were performed by splitting the converted PSMC input sequences to shorter segments using the program “splitfa” in the PSMC package, and then randomly sampling the segments using the “-b” option for PSMC analyses.
Ecological niche modeling
We constructed ENMs in Maxent 3.3.3 [75] with bioclimate variables from Worldclim [76] as predictors. We retained the bioclimate variables that are not highly correlated with one another (|r|≥0.8) for the given study area (i.e. South Asia, Sundaland) and have a non-zero permutation importance to model fit (for the lists of bioclimate variables used in the ENMs; Table S1). The ENMs built under current climates were projected to paleoclimates during the last interglacial period (LIG; ca. 120 – 140 ka) [39] and the last glacial maximum (LGM; ca. 22 ka) [40]. The multivariate similarity surface (MESS) was used to detect areas with novel paleoclimate conditions (i.e. climate conditions that fall outside of the training range) [77]. The MESS results indicated that most of the study area did not present novel paleoclimate conditions (Figure S3). To produce predicted distributions, we applied the minimum training presence threshold (i.e. the areas with suitability scores lower than the threshold values are considered ‘not suitable’). The area under the receiver operating characteristic curve (AUC) of present-day ENMs ranged from 0.82 to 0.91. The partial receiver operating characteristic curves were estimated at omission rate of 0%, 1% and 5%, with bootstrapped mean AUC ratios > 1 (p < 0.001 based on 1,000 replicates) for all present-day ENMs across the three occurrence data sets [78], suggesting appropriate model fit.
Sumatran Rhinoceros occur in dense forests such as rainforests, secondary forests and closed-canopy woodlands [38], which could further limit their distribution. However, adding vegetation type as a predictor to ENMs is difficult in our case because paleo-vegetation data is lacking for LIG and difficult to reconcile between LGM and modern vegetation data. As an alternative, we calculated the proportion of present-day suitable areas that falls within each biome type [79] and the proportion of LGM suitable areas that falls within each vegetation type [80].
Data and software availability
The genome sequence assembly has been deposited at DNA DataBank of Japan (DDBJ), the European Nucleotide Archive (ENA), and GenBank at the National Center for Biotechnology Information (NCBI) under the accession PEKH00000000. The version described in this paper is version PEKH01000000. Raw sequencing reads were deposited in the Sequence Read Archive at the NCBI and accessed via accession number PRJNA415733.
Supplementary Material
Highlights.
This study reports the first whole genome sequence for the Sumatran Rhinoceros.
Sumatran Rhinoceros underwent large population fluctuations during the Pleistocene.
Pleistocene climate change dramatically influenced available habitat.
Changes in population may have been due to population decline and/or fragmentation.
Acknowledgments
Thanks to John Goering and Robert Lindner Jr for their generous support. Thanks to Glenn Storrs, Jay Kalagayan and Elizabeth Peirce of Cincinnati Museum Center for facilitating the acquisition of donor support. Thanks to David Might, David Noem, Loree Celebreeze, Chris Moran, Dorinda Whitsett and Cameron Mays for preparing the voucher specimen and Peijia Tsai for aid in the PSMC analyses. Thanks to Haowen Tong of the Chinese Academy of Sciences for verifying fossil occurrences in China. Robin O’Keefe provided helpful comments on the manuscript. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) supported by National Science Foundation grant number ACI-1053575. DNA sequencing was performed at the Marshall University Genomics Core Facility. The Marshall University Genomics Core Facility is supported in part by NIH/NIGMS P20GM103434, which funds the IDeA WV-INBRE program. This paper is dedicated to the staff of the Cincinnati Zoo and Botanical Garden who cared for Ipuh during his 22 years in Cincinnati. The authors have no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
H.L.M. conceived the study, contributed to lab work and genome analysis, and in drafting the manuscript.
C-M. H. contributed to the PSMC analysis and in drafting the manuscript.
P-J. S. contributed to the ENM analysis and in drafting the manuscript.
J. D. contributed to genome assembly and analysis and in drafting the manuscript.
M. J. contributed to genome assembly and analysis and genome data archival.
S-F. Y. contributed to the PSMC analysis.
T. L. R. contributed to acquiring the samples, the ENM analysis and in drafting the manuscript.
D. A. O. contributed to acquiring the samples and in drafting the manuscript.
J. F. contributed to the lab work associated with genome sequencing.
S. R. contributed to genome assembly and analysis.
D. A. P. contributed to the lab work associated with genome sequencing.
All authors reviewed the manuscript.
References
- 1.Ceballos G, Ehrlich PR, Barnosky AD, Garcia A, Pringle RM, Palmer TM. Accelerated modern human-induced species losses: Entering the sixth mass extinction. Sci Adv. 2015;1:1400253. doi: 10.1126/sciadv.1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardillo M, Mace GM, Jones KE, Bielby J, Bininda-Emonds OR, Sechrest W, Orme CD, Purvis A. Multiple causes of high extinction risk in large mammal species. Science. 2005;309:1239–1241. doi: 10.1126/science.1116030. [DOI] [PubMed] [Google Scholar]
- 3.Ripple WJ, Newsome TM, Wolf C, Dirzo R, Everatt KT, Galetti M, Hayward MW, Kerley GI, Levi T, Lindsey PA, et al. Collapse of the world’s largest herbivores. Sci Adv. 2015;1:1400103. doi: 10.1126/sciadv.1400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper A, Turney C, Hughen KA, Brook BW, McDonald HG, Bradshaw CJ. Abrupt warming events drove Late Pleistocene Holarctic megafaunal turnover. Science. 2015;349:602–606. doi: 10.1126/science.aac4315. [DOI] [PubMed] [Google Scholar]
- 5.Alroy J. A multispecies overkill simulation of the end-Pleistocene megafaunal mass extinction. Science. 2001;292:1893–1896. doi: 10.1126/science.1059342. [DOI] [PubMed] [Google Scholar]
- 6.Lorenzen ED, Nogues-Bravo D, Orlando L, Weinstock J, Binladen J, Marske KA, Ugan A, Borregaard MK, Gilbert MTP, Nielsen R, et al. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature. 2011;479:359–364. doi: 10.1038/nature10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng T, Wang X, Fortelius M, Li Q, Wang Y, Tseng ZJ, Takeuchi GT, Saylor JE, Saila LK, Xie G. Out of Tibet: Pliocene woolly rhino suggests high-plateau origin of ice age megaherbivores. Science. 2011;333:1285–1288. doi: 10.1126/science.1206594. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad Zafir AW, Payne J, Mohamed A, Lau CF, Sharma DSK, Alfred R, Williams AC, Nathan S, Ramono WS, Clements GR. Now or never: what will it take to save the Sumatran rhinoceros Dicerorhinus sumatrensis from extinction? Oryx. 2011;45:225–233. [Google Scholar]
- 9.Meijaard E. The Sumatran Rhinoceros in Kalimantan, Indonesia: Its possible distribution and conservation prospects. Pachyderm. 1996;21:15–23. [Google Scholar]
- 10.Choudhury A. The status of the Sumatran rhinoceros in northeastern India. Oryx. 1997;31:151–152. [Google Scholar]
- 11.Antoine PO. Pleistocene and Holocene rhinocerotids (Mammalia, Perissodactyla) from the Indochinese Peninsula. C R Palevol. 2012;11:159–168. [Google Scholar]
- 12.Pusparini W, Sievert PR, Fuller TK, Randhir TO, Andayani N. Rhinos in the parks: An island-wide survey of the last wild population of the Sumatran rhinoceros. PLoS ONE. 2015;10:0136643. doi: 10.1371/journal.pone.0136643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao S, Zheng P, Dong S, Zhan X, Wu Q, Guo X, Hu Y, He W, Zhang S, Fan W, et al. Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat Genet. 2013;45:67–71. doi: 10.1038/ng.2494. [DOI] [PubMed] [Google Scholar]
- 15.Nadachowska-Brzyska K, Li C, Smeds L, Zhang G, Ellegren H. Temporal dynamics of avian populations during Pleistocene revealed by whole-genome sequences. Curr Biol. 2015;25:1375–1380. doi: 10.1016/j.cub.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung CM, Shaner PJ, Zink RM, Liu WC, Chu TC, Huang WS, Li SH. Drastic population fluctuations explain the rapid extinction of the passenger pigeon. Proc Natl Acad Sci USA. 2014;111:10636–10641. doi: 10.1073/pnas.1401526111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palkopoulou E, Mallick S, Skoglund P, Enk J, Rohland N, Li H, Omrak A, Vartanyan S, Poinar H, Gotherstrom A, et al. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr Biol. 2015;25:1395–1400. doi: 10.1016/j.cub.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishan A, Dandekar P, Nathan N, Hamelik R, Miller C, Shaw J. DNA index, genome size, and electronic nuclear volume of vertebrates from the Miami Metro Zoo. Cytometry A. 2005;65:26–34. doi: 10.1002/cyto.a.20130. [DOI] [PubMed] [Google Scholar]
- 20.Rogers RL, Slatkin M. Excess of genomic defects in a woolly mammoth on Wrangel island. PLoS Genet. 2017;13:1006601. doi: 10.1371/journal.pgen.1006601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orlando L, Leonard JA, Thenot A, Laudet V, Guerin C, Hänni C. Ancient DNA analysis reveals woolly rhino evolutionary relationships. Mol Phylogenet Evol. 2003;28:485–499. doi: 10.1016/s1055-7903(03)00023-x. [DOI] [PubMed] [Google Scholar]
- 23.Tong HW. Evolution of the non-Coelodonta dicerorhine lineage in China. C R Palevol. 2012;11:555–562. [Google Scholar]
- 24.Maung-Thein ZM, Takai M, Tsubamoto T, Egi N, Thaung H, Nishimura T, Maung M, Zaw W. A review of fossil rhinoceroses from the Neogene of Myanmar with description of new specimens from the Irrawaddy Sediments. J Asian Earth Sci. 2010;37:154–165. [Google Scholar]
- 25.Tong HW, Guérin C. Early Pleistocene Dicerorhinus sumatrensis remains from the Liucheng Gigantopithecus Cave, Guangxi, China. Geobios. 2009;42:525–539. [Google Scholar]
- 26.Hall R. Plate tectonics of Cenozoic SE Asia and the distribution of land and sea. In: Hall R, Holloway JD, editors. Biogeography and Geological Evolution of SE Asia. The Netherlands: Backhuys Publishers; 1998. pp. 99–131. [Google Scholar]
- 27.Rohling EJ, Fenton M, Jorissen FJ, Bertrand P, Ganssen G, Caulet JP. Magnitudes of sea-level lowstands of the past 500,000 years. Nature. 1998;394:162–165. [Google Scholar]
- 28.Liu GE, Matukumalli LK, Sonstegard TS, Shade LL, Van Tassell CP. Genomic divergences among cattle, dog and human estimated from large-scale alignments of genomic sequences. BMC Genomics. 2006;7:140. doi: 10.1186/1471-2164-7-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth TL, Reinhart PR, Romo JS, Candra D, Suhaery A, Stoops MA. Sexual maturation in the Sumatran rhinoceros (Dicerorhinus sumatrensis) Zoo Biol. 2013;32:549–555. doi: 10.1002/zoo.21089. [DOI] [PubMed] [Google Scholar]
- 30.Goto H, Ryder OA, Fisher AR, Schultz B, Kosakovsky Pond SL, Nekrutenko A, Makova KD. A massively parallel sequencing approach uncovers ancient origins and high genetic variability of endangered Przewalski’s horses. Genome Biol Evol. 2011;3:1096–1106. doi: 10.1093/gbe/evr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 32.van den Berg GD, de Vos J, Sondaar PY. The Late Quaternary palaeogeography of mammal evolution in the Indonesian Archipelago. Palaeogeogr Palaeoclimatol Palaeoecol. 2001;171:382–408. [Google Scholar]
- 33.Gautier M, Moazami-Goudarzi K, Leveziel H, Parinello H, Grohs C, Rialle S, Kowalczyk R, Flori L. Deciphering the wisent demographic and adaptive histories from individual whole-genome sequences. Mol Biol Evol. 2016;33:2801–2814. doi: 10.1093/molbev/msw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Kim HL, Burhans RC, Drautz DI, Wittekindt NE, et al. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci USA. 2012;109:E2382–2390. doi: 10.1073/pnas.1210506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens HJ, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393–398. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G, Vandewege MW, St John JA, Capella-Gutierrez S, Castoe TA, et al. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science. 2014;346:1254449. doi: 10.1126/science.1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheehan S, Harris K, Song YS. Estimating variable effective population sizes from multiple genomes: a sequentially Markov conditional sampling distribution approach. Genetics. 2013;194:647–662. doi: 10.1534/genetics.112.149096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foose TJ, van Strien NJ, editors. Asian rhinos: status survey and conservation action plan. Gland, Switzerland, and Cambridge, U.K: IUCN; 1997. [Google Scholar]
- 39.Otto-Bliesner BL, Marshall SJ, Overpeck JT, Miller GH, Hu A. Simulating Arctic climate warmth and icefield retreat in the last interglaciation. Science. 2006;311:1751–1753. doi: 10.1126/science.1120808. [DOI] [PubMed] [Google Scholar]
- 40.Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM. The last glacial maximum. Science. 2009;325:710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- 41.Sathiamurthy E, Voris HK. Nat Hist J. S2. Chulalongkorn Univ; 2006. Maps of Holocene sea level transgression and submerged lakes on the Sunda Shelf; pp. 1–43. [Google Scholar]
- 42.Dinerstein E. Family: Rhinocerotidae (Rhinoceroses) In: Wilson DE, Mittermeier RA, editors. Handbook of Mammals of the World Vol 2 Hoofed Mammals. Vol. 2. Barcelona: Lynx Edicions; 2011. pp. 144–181. [Google Scholar]
- 43.Heaney LR. A synopsis of climatic and vegetational change in Southeast Asia. Clim Change. 1991;19:53–61. [Google Scholar]
- 44.Gathorne-Hardy FJ, Syaukani Davies RG, Eggleton P, Jones DT. Quaternary rainforest refugia in south-east Asia: using termites (Isoptera) as indicators. Biol J Linn Soc. 2002;75:453–466. [Google Scholar]
- 45.Meijaard E. Ph D. The Australian National University; 2004. Solving mammalian riddles: A reconstruction of the Tertiary and Quaternary distribution of mammals and their palaeoenvironments in island South-East Asia. [Google Scholar]
- 46.Bird MI, Taylor D, Hunt C. Palaeoenvironments of insular Southeast Asia during the Last Glacial Period: a savanna corridor in Sundaland? Quat Sci Rev. 2005;24:2228–2242. [Google Scholar]
- 47.Wurster CM, Bird MI, Bull ID, Creed F, Bryant C, Dungait JA, Paz V. Forest contraction in north equatorial Southeast Asia during the Last Glacial Period. Proc Natl Acad Sci USA. 2010;107:15508–15511. doi: 10.1073/pnas.1005507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandon-Jones D. The Asian Colobinae (Mammalia: Cercopithecidae) as indicators of Quaternary climatic change. Biol J Linn Soc. 1996;59:327–350. [Google Scholar]
- 49.Inger R, Voris H. The biogeographical relations of the frogs and snakes of Sundaland. J Biogeogr. 2001;28:863–891. [Google Scholar]
- 50.Morales JC, Andau PM, Supriatna J, Zainuddin ZZ, Melnick D. Mitochondrial DNA variability and conservation genetics of the Sumatran Rhinoceros. Conserv Genet. 1997;11:539–543. [Google Scholar]
- 51.Cannon CH, Morley RJ, Bush AB. The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc Natl Acad Sci USA. 2009;106:11188–11193. doi: 10.1073/pnas.0809865106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson DE, Reeder DM. Mammal species of the world: a taxonomic and geographic reference. Johns Hopkins University Press; 2005. [Google Scholar]
- 53.Miller G, Fogel M, Magee J, Gagan M, Clarke S, Johnson B. Ecosystem collapse in Pleistocene Australia and a human role in megafaunal extinction. Science. 2005;309:287–290. doi: 10.1126/science.1111288. [DOI] [PubMed] [Google Scholar]
- 54.Hunt CO, Rushworth G. Cultivation and human impact at 6000 cal yr B.P. in tropical lowland forest at Niah, Sarawak, Malaysian Borneo. Quat Res. 2017;64:460–468. [Google Scholar]
- 55.Piper PJ, Rabett RJ. Hunting in a tropical rainforest: evidence from the Terminal Pleistocene at Lobang Hangus, Niah Caves, Sarawak. Int J Osteoarchaeol. 2009;19:551–565. [Google Scholar]
- 56.Corlett RT. The impact of hunting on the mammalian fauna of tropical Asian forests. Biotropica. 2007;39:292–303. [Google Scholar]
- 57.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chikhi R, Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2014;30:31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 59.Love RR, Weisenfeld NI, Jaffe DB, Besansky NJ, Neafsey DE. Evaluation of DISCOVAR de novo using a mosquito sample for cost-effective short-read genome assembly. BMC Genomics. 2016;17:187. doi: 10.1186/s12864-016-2531-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD, et al. XSEDE: Accelerating Scientific Discovery. Comput Sci Eng. 2014;16:62–74. [Google Scholar]
- 62.Laurie A. Behavioural ecology of the greater one-horned rhinoceros (Rhinoceros unicornis) J Zool. 1982;196:307–341. [Google Scholar]
- 63.Wang Z, Zhao W, Sun G. The eco-environmental model of rhinoceros extinction in China. Pol Ecol Stud. 1993;19:29–34. [Google Scholar]
- 64.Wang Z, Xu F, Sun G. A preliminary analysis of the relationship between the extinction of rhinoceros and human population pressure in China. Acta Ecol Sin. 1997;17:640–644. [Google Scholar]
- 65.Tong H. Les rhinoceros des sites a fossiles humains de Chine. L’Anthropologie. 2000;104:523–529. [Google Scholar]
- 66.Xu ZF. The effects of paying tribute to the imperial court in the history of rhinoceros extinction and elephant endangerment in southern Yunnan. Chinese Biodiversity. 2000;8:112–119. [Google Scholar]
- 67.Tong H. Age profiles of rhino fauna from the Middle Pleistocene Nanjing man site, south China explained by the rhino specimens of living species. Int J Osteoarchaeol. 2001;11:231–237. [Google Scholar]
- 68.Rookmaker K. Distribution and extinction of the rhinoceros in China: review of recent Chinese publications. Pachyderm. 2006;40:102–106. [Google Scholar]
- 69.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barnett DW, Garrison EK, Quinlan AR, Stromberg MP, Marth GT. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci USA. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Phillips S, Anderson R, Schapire R. Maximum entropy modeling of species geographic distributions. Ecol Modell. 2006;190:231–259. [Google Scholar]
- 76.Hijmans R, Cameron S, Parra J, Jones P, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- 77.Elith J, Kearney M, Phillips S. The art of modelling range-shifting species. Methods Ecol Evol. 2010;1:330–342. [Google Scholar]
- 78.Narayani B. Tool for Partial-ROC version 1.0. Biodiversity Institute; Lawrence, KS: 2008. [Google Scholar]
- 79.Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GVN, Underwood EC, D’Amico JA, Itoua I, Strand HE, Morrison JC, et al. Terrestrial ecoregions of the world: A new map of life on Earth: A new global map of terrestrial ecoregions provides an innovative tool for conserving biodiversity. BioScience. 2001;51:933–938. [Google Scholar]
- 80.Ray N, Adams JM. A GIS-based vegetation map of the world at the last glacial maximum (25,000–15000 BP) Internet Archaeol. 2001;11 http://dx.doi.org/10.11141/ia.11111.11142. [Google Scholar]
- 81.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


