Abstract
To resolve cellular heterogeneity, we developed a combinatorial indexing strategy to profile the transcriptomes of single cells or nuclei (sci-RNA-seq: Single cell Combinatorial Indexing RNA sequencing). We applied sci-RNA-seq to profile nearly 50,000 cells from the nematode Caenorhabditis elegans at the L2 stage, which is over 50-fold “shotgun cellular coverage” of its somatic cell composition. From these data, we define consensus expression profiles for 27 cell types, and recover rare neuronal cell types corresponding to as few as one or two cells in the L2 worm. We integrate these profiles with whole animal ChIP sequencing data to deconvolve the cell type specific effects of transcription factors. These data generated by sci-RNA-seq constitute a powerful resource for nematode biology, and foreshadow similar atlases for other organisms.
Individual cells are the natural unit of form and function in biological systems. However, conventional methods for profiling the molecular content of biological samples mask cellular heterogeneity, likely present even in ostensibly homogenous tissues (1). Recently, profiling the transcriptome of individual cells has emerged as a powerful strategy for resolving such heterogeneity. The expression levels of mRNA species are linked to cellular function, and therefore can be used to classify cell types (2–10) and to order cell states (11). Although methods for single cell RNA-seq have proliferated, they rely on the isolation of individual cells within physical compartments (12–20). Consequently, preparing single cell RNA-seq libraries with these methods can be expensive, the cost scaling linearly with the numbers of cells processed (21, 22).
We recently developed combinatorial indexing, a method using split-pool barcoding of nucleic acids to uniquely label a large number of single molecules or single cells. Single molecule combinatorial indexing can be used for haplotype-resolved genome sequencing and de novo genome assembly (23, 24), while single cell combinatorial indexing (“sci”) can be used to profile chromatin accessibility (sci-ATAC-seq) (25), genome sequence (sci-DNA-seq) (26), genome-wide chromosome conformation (sci-Hi-C) (27), and DNA methylation (sci-MET) (28) in large numbers of single cells.
Here we developed a combinatorial indexing method to uniquely label the transcriptomes of large numbers of single cells or nuclei (sci-RNA-seq). We then applied sci-RNA-seq to deeply profile single cell transcriptomes in the nematode C. elegans at the L2 stage. C. elegans is the only multicellular organism for which all cells and cell types are defined, as is its entire developmental lineage (29, 30). However, despite its modest cell count (e.g. 762 somatic cells per L2 larva), our knowledge of the molecular state of each cell and cell type remains fragmentary. We therefore saw an opportunity to generate a powerful resource for nematode biologists as well as for the single cell genomics community.
Overview of sci-RNA-seq
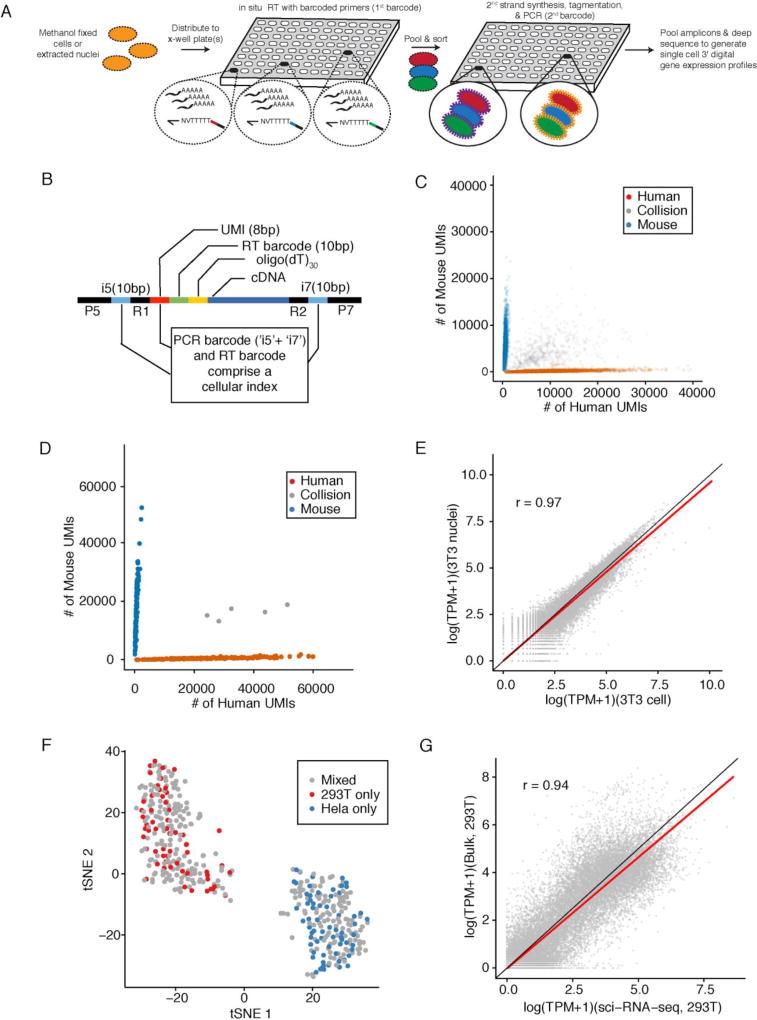
In its current form, sci-RNA-seq relies on the following steps (Fig. 1A): 1) Cells are fixed and permeabilized with methanol (alternatively, cells are lysed and nuclei recovered), and then split across 96- or 384-well plates. 2) A first molecular index is introduced to the mRNA of cells within each well with in situ reverse transcription (RT) incorporating a barcode-bearing, well-specific polyT primer containing unique molecular identifiers (UMI). 3) All cells are pooled and redistributed by fluorescence activated cell sorting (FACS) to 96- or 384-well plates in limiting numbers (e.g. 10–100 per well). Cells are gated on the basis of DAPI (4',6-diamidino-2-phenylindole) staining to discriminate single cells from doublets during sorting. 4) Second strand synthesis, transposition with Tn5 transposase, lysis, and PCR amplification are performed. The PCR primers target the barcoded polyT primer on one end, and the Tn5 adaptor insertion on the other end, such that resulting PCR amplicons preferentially capture the 3’ ends of transcripts. These primers introduce a second barcode, specific to each well of the PCR plate. 5) Amplicons are pooled and subjected to massively parallel sequencing, resulting in 3’-tag digital gene expression profiles, with each read associated with two barcodes corresponding to the first and second rounds of cellular indexing (Fig. 1B). In a variant of the method described below, we introduce a third round of cellular indexing during Tn5 transposition of double-stranded cDNA.
Fig. 1. sci-RNA-seq enables multiplex single cell transcriptome profiling.
(A) Schematic of sci-RNA-seq workflow. (B) Schematic of sci-RNA-seq library amplicons. Index2 and read1 covers the i5 index, UMI and RT barcode. Index1 and read2 covers the i7 index and cDNA fragment. (C) Scatter plot of unique human and mouse UMI counts from 384 × 384 sci-RNA-seq. Blue: inferred mouse cells (n = 5953). Red: inferred human cells (n = 3967). Grey: collisions (n = 884). (D) Scatter plot of unique human and mouse cell UMI counts from 96 × 96 sci-RNA-seq with optimized protocol. Blue: inferred mouse cells (n = 129). Red: inferred human cells (n = 160). Grey: collisions (n = 5). In (C) and (D), only cells originating from wells containing mixed human and mouse cells are shown. (E) Correlation between gene expression measurements in aggregated sci-RNA-seq profiles of NIH/3T3 cells (n = 238) vs. nuclei (n = 124). (F) tSNE plot of cells originating in wells containing HEK293T (red) (n = 60), HeLa S3 (blue) (n = 69) or a mixture (grey) (n = 321). (G) Correlation between gene expression measurements from aggregated sci-RNA-seq data vs. bulk RNA-seq data from a related protocol (33). (E) and (G) include linear regression (red) and y=x (black) lines.
The majority of cells pass through a unique combination of wells, resulting in a unique combination of barcodes for each cell that tags its transcripts. The rate of two or more cells receiving the same combination of barcodes can be tuned by adjusting how many cells are distributed to the second set of wells (25). Increasing the number of barcodes used during each round of indexing leads boosts the number of cells that can be profiled while reducing the effective cost per cell (fig. S1). Additional levels of indexing can potentially offer even greater complexity and lower costs. Multiple samples (e.g. different cell populations, tissues, individuals, time-points, perturbations, replicates, etc.) can be concurrently processed within one experiment, using different subsets of wells for each sample during the first round of indexing.
Scalability of sci-RNA-seq
We tested 262 sci-RNA-seq conditions with mammalian cells, optimizing the protocol and reaction conditions. We demonstrate scalability with 384 × 384 well sci-RNA-seq. During the first round of indexing, half of 384 wells contained pure populations of either human (HEK293T or HeLa S3) or mouse (NIH/3T3) cells, and the other half mixed human and mouse cells (Table S1). After barcoded RT, cells were pooled and then sorted to a new 384 well plate for the second round of barcoding and deep sequencing of pooled PCR amplicons. We recovered 15,997 single cell transcriptomes and readily assigned cells as human or mouse (Fig. 1C).
Optimization of sci-RNA-seq and application to nuclei
We performed optimized 96 × 96 well sci-RNA-seq on five cell populations, each present in distinct subsets of wells during the first round of barcoding (Table S1): HEK293T cells (8 wells); HeLa S3 cells (8 wells); an intraspecies mixture of HEK293T and HeLa S3 cells (32 wells); and interspecies mixtures of HEK293T and NIH/3T3 cells (24 wells) or nuclei (24 wells). We deeply sequenced the resulting library (~250,000 reads per cell; ~210,000 reads per nucleus; ~88% duplication rate), profiling 744 single cell and 175 single nucleus transcriptomes.
Transcriptomes in the 24 wells containing an interspecies mixture of human and mouse cells overwhelmingly mapped to the genome of one species or the other (289 of 294 cells), with only 5 ‘collisions’ (where collisions likely represent coincidental passage through the same wells by two or more cells) (Fig. 1D). Excluding collisions, we observed an average of 24,454 UMIs (5,604 genes) per human cell and 17,665 UMIs (4,065 genes) per mouse cell, with 1.9% and 3.3% of reads per cell mapping to the incorrect species.
Transcriptomes originating in the 24 wells containing an interspecies mixture of human and mouse nuclei also overwhelmingly mapped to the genome of one species or the other (172 of 175 nuclei), with only 3 collisions (fig. S2A). Excluding collisions, we observed an average of 32,951 UMIs (5,737 genes) per human nucleus and 20,123 UMIs (4,107 genes) per mouse nucleus (fig. S2B–C), with 2.2% and 1.9% of reads per cell mapping to the incorrect species. The greater UMI counts in nuclei are potentially due to the higher amounts of mRNA in cells resulting in a reduced RT efficiency per molecule. Consistent with this, optimizing the number of cells per RT reaction increased UMI counts per cell (31).
Estimates of gene expression from the aggregated transcriptomes of nuclei versus cells were well correlated (Pearson: 0.96 for HEK293T, 0.97 for NIH/3T3; Fig. 1E, fig. S2D). From cells, 81% of reads mapped to the expected strand of genic regions (47% exonic, 34% intronic), and 19% to intergenic regions or the unexpected strand of genic regions. From nuclei, 84% of reads mapped to the expected strand of genic regions (35% exonic, 49% intronic) and 16% to intergenic regions or the unexpected strand of genic regions, similar to previous studies (32). Whereas exonic reads show an expected enrichment at the 3’ ends of gene bodies, intronic reads do not, and may be the result of poly(dT) priming from poly(dA) tracts in heterogeneous nuclear RNA (fig. S3).
Transcriptomes originating in the 48 wells containing pure or an intraspecies mixture of HEK293T and HeLa S3 cells were readily separated into two clusters by t-stochastic neighbor embedding (t-SNE) (Figs. 1F and S4). Estimates of gene expression from the aggregated transcriptomes of all identified HEK293T cells versus a related bulk RNA-seq workflow (Tn5-RNA-seq (33)) without methanol fixation were well correlated (Pearson: 0.94, Fig. 1G).
Robustness of sci-RNA-seq
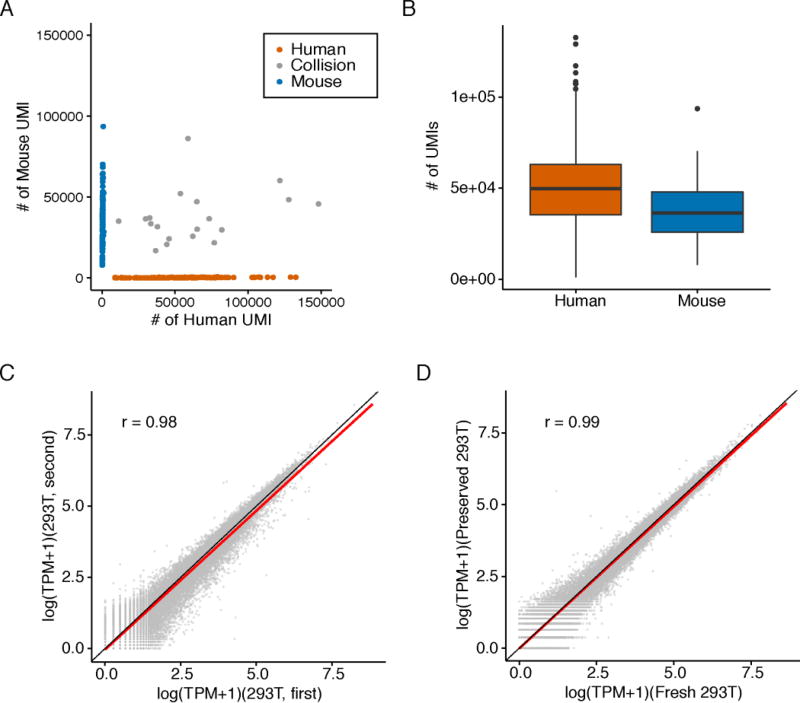
After optimizing the number of cells per RT reaction, we fixed a mixture of HEK293T and NIH/3T3 cells, and performed 16 × 84 well sci-RNA-seq (Table S1) (31). We recovered 185 human cells and 109 mouse cells with 22 collisions (Fig. 2A). At ~240,000 reads per cell (73% duplication rate), we observed an average of 49,043 UMIs (7,563 genes) per human cell and 36,737 UMIs (6,263 genes) per mouse cell (Fig. 2B, fig. S5A), with 0.9% and 1.2% of reads per cell mapping to the incorrect species. Although this and the previous experiment were performed two months apart on independently grown and fixed cells, the aggregated transcriptomes were well correlated (Pearson: 0.98 for HEK293T, 0.98 for NIH/3T3; Figs. 2C and S5B).
Fig. 2. sci-RNA-seq shows robust gene expression measurements.
(A) Scatter plot of unique human and mouse UMI counts from a 16 × 84 sci-RNA-seq experiment on mixed HEK293T and NIH/3T3 cells (Table S1). Blue: inferred mouse cells (n = 109). Red: inferred human cells (n = 168). Grey: collisions (n = 19). (B) Boxplots showing number of UMIs detected per cell. (C) Correlation between gene expression measurements in aggregated sci-RNA-seq profiles from experiments performed two months apart on independently grown and fixed cells. (D) Correlation between gene expression measurements in aggregated sci-RNA-seq profiles of fixed-fresh vs. fixed-frozen cells. (C) and (D) include linear regression (red) and y=x (black) lines.
We stored a portion of the methanol-fixed mixture of HEK293T and NIH/3T3 cells at −80C for 4 days and repeated sci-RNA-seq (Table S1). At ~200,000 reads per cell (73% duplication rate), we observed an average of 30,024 UMIs (5,965 genes) per human cell and 21,393 UMIs (4,503 genes) per mouse cell, with comparable purity (fig. S5C). The aggregated transcriptomes of the fixed-fresh vs. fixed-frozen cells were well correlated (Pearson: 0.99 for HEK293T cells, 0.98 for NIH/3T3 cells; Figs. 2D and S5D).
sci-RNA-seq with three levels of indexing
Two-level combinatorial indexing enables routine profiling of ~104 single cells per experiment. We tested an additional level of indexing during Tn5 transposition of double-stranded cDNA (25). We performed 16 × 6 × 16 well sci-RNA-seq on mixed HEK293T and NIH/3T3 cells after methanol fixation. After RT with 16 barcodes and second strand synthesis, cells were pooled and distributed to 6 wells for tagmentation with indexed Tn5 (6 barcodes), then pooled again and sorted to 16 wells for PCR with indexed primers. At ~20,000 reads per cell (51% duplication rate), we recovered 119 human and 62 mouse cells with 5 collisions (fig. S6A). The aggregated transcriptomes of three-level vs. two-level sci-RNA-seq were well correlated (Pearson: 0.96 for HEK293T, 0.94 for NIH/3T3; fig. S6B–C). Downsampling to 15,000 reads per cell, three-level indexing recovered fewer UMIs per cell than two-level indexing (3-level: on average, 6,033 for HEK293T, 3,640 for NIH/3T3; 2-level: 9,942 for HEK293T, 8,611 for NIH/3T3; fig. S6D–G), possibly due to lower efficiency of indexed vs. unindexed Tn5. This limitation notwithstanding, three-level combinatorial indexing has the potential to enable routine profiling of >106 single cells per experiment (fig. S6H; (31)).
Single cell RNA profiling of C. elegans
We next applied sci-RNA-seq to C. elegans. Of note, the cells in C. elegans larvae are much smaller, more variably sized, and have lower mRNA content than the mammalian cell lines on which we optimized the protocol. We pooled ~150,000 larvae synchronized at the L2 stage and dissociated them into single-cell suspensions. We then performed in situ RT across six 96-well plates (576 first-round barcodes), each well containing ~1,000 C. elegans cells and also ~1,000 human cells (HEK293T) as internal controls. After pooling all cells, we sorted the mixture of C. elegans and HEK293T cells to 10 new 96-well plates for PCR barcoding (960 second-round barcodes), gating on DNA content to distinguish between C. elegans and HEK293T cells. This sorting resulted in 96% of wells harboring only C. elegans cells (140 each), and 4% of wells harboring a mix of C. elegans and HEK293T cells (140 C. elegans and 10 HEK293T each).
This experiment yielded 42,035 C. elegans single-cell transcriptomes (UMI counts per cell for protein-coding genes ≥ 100). 94% of reads mapped to the expected strand of genic regions (92% exonic, 2% intronic). At a sequencing depth of ~20,000 reads per cell and a duplication rate of 80%, we identified a median of 575 UMIs mapping to protein-coding genes per cell (mean 1,121 UMIs and 431 genes per cell) (fig. S7A). Importantly, control wells containing both C. elegans and HEK293T cells demonstrated clear separation between species (fig. S7B), with 3.1% and 0.2% of reads per cell mapping to the incorrect species, respectively.
Identifying cell types
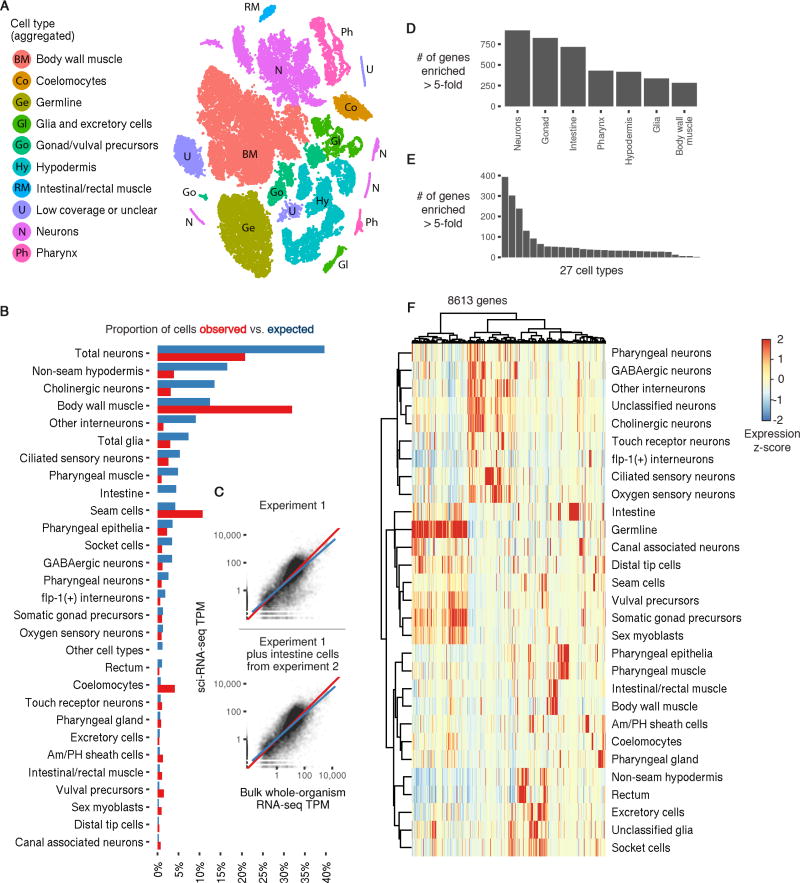
Semi-supervised clustering analysis segregated the cells into 29 distinct groups, the largest containing 13,205 (31.4%) and the smallest only 131 (0.3%) cells (Fig. 3A). Somatic cell types comprised 37,734 cells. We identified genes that were expressed specifically in a single cluster, and by comparing those genes to expression patterns reported in the literature, assigned the clusters to cell types (figs. S15–S23). Twenty-six cell types were represented in the 29 clusters: 19 represented exactly one literature-defined cell type, 7 contained multiple distinct cell types, 2 contained cells of a specific cell type but had abnormally low UMI counts, and 1 could not be readily assigned. Neurons, which were present in 7 clusters in the global analysis, were independently reclustered, initially revealing 10 major neuronal subtypes.
Fig. 3. A single sci-RNA-seq experiment highlights the single cell transcriptomes comprising the C. elegans larva.
(A) t-SNE visualization of the high-level cell types identified. (B) Bar plot showing the proportion of somatic cells profiled in the first sci-RNA-seq C. elegans experiment that could be identified as belonging to each cell type (red) compared to the proportion of cells from that type present in an L2 C. elegans individual (blue). (C) Scatter plots showing the log-scaled transcripts per million (TPM) of genes in the aggregation of all sci-RNA-seq reads (x axis) or in bulk RNA-seq (y axis; geometric mean of 3 experiments). Top plot includes only the first sci-RNA-seq experiment. Bottom plot also includes intestine cells from the second sci-RNA-seq experiment. (D) Number of genes that are enriched at least 5-fold in a specific tissue relative to the 2nd-highest-expressing tissue, excluding genes for which the differential expression between the 1st and 2nd-highest expressing tissues is not significant (q-value > 0.05). (E) Same as (D) except comparing cell types instead of tissues. (F) Heatmap showing the relative expression of genes in consensus transcriptomes for each cell type estimated by sci-RNA-seq. Genes are included if they have a size-factor-normalized mean expression of >0.05 in at least one cell type (8,613 genes in total). The raw expression data (UMI count matrix) is log-transformed, column centered and scaled (using the R function scale), and the resulting values are clamped to the interval [−2, 2].
Intestine cells were not represented in any cluster. Intestine cells comprise 2.5% of the somatic cells but are polyploid in C. elegans larvae (34) and also autofluorescent in the DAPI channel used to measure DNA content (35). We speculated they may have been excluded by how we gated on DNA content. We therefore performed a second 384 × 144 well C. elegans experiment, collecting all cells including polyploid cells on the basis of DAPI fluorescence (96 wells), or gating to enrich for polyploid cells (48 wells). Intestine cells were present (as compared with their absence in the previous experiment) and 2-fold enriched in wells gated for polyploidy. This experiment yielded 7,325 cells (UMI counts per cell for protein-coding genes ≥ 200), of which 6,335 were somatic and 511 intestine cells (fig. S8A).
Gene expression patterns in hypodermal cells suggested that the worm cells from the second C. elegans experiment were more tightly synchronized, overlapping but not identical in developmental timing to the first experiment (fig. S8B–F). C. elegans larvae feature pervasive oscillations in gene expression within each larval stage (36), making it difficult to distinguish biological variation from batch effects. However, the aggregated transcriptomes of human HEK293T cells from these same experiments were well correlated (Pearson: 0.97) and not readily separated by tSNE (fig. S9). This suggests that the variation observed is primarily due to differences in the developmental timing or preparation of the C. elegans larvae and cells, rather than technical variation in the sci-RNA-seq protocol. Regardless of its source, to minimize confounding by this variation, we only included the intestine cells from the second C. elegans experiment in subsequent analyses, with all other cell types being represented by the first experiment only.
The global and neuron-specific clustering analyses from the first C. elegans experiment, supplemented with intestine cells from the second experiment, allowed us to construct aggregate expression profiles for 27 cell types (Tables S2–S4; a 28th cell type, dopaminergic neurons, is excluded due to small cell numbers). These profiles are available online via GExplore (http://genome.sfu.ca/gexplore/gexplore_search_tissues.html; fig. S14). Comparing the observed proportions of each cell type to their known frequencies in L2 larvae showed that sci-RNA-seq captured many cell types at or near expected frequencies (Fig. 3B; 15/28 types had abundance ≥ 50%, and 27/28 had abundance ≥ 20%, of expectation).
Transcriptional programs can be readily distinguished within single cell transcriptome datasets at shallow sequencing depths (37). Thus, despite being able to distinguish many distinct cell types in the worm, our molecular definition for each would be incomplete. However, we observed that half of all C. elegans protein-coding genes were expressed in at least 100 cells in the full dataset, and 66% of protein-coding genes in at least 20 cells. This compares favorably with the estimates of expressed genes at the L2 stage from whole animal RNA-seq (69%) (38). The “whole worm” expression profile derived by aggregating all sci-RNA-seq reads correlated well with whole animal bulk RNA-seq (38) for L2 C. elegans (Fig. 3C; Spearman: 0.796 with cells from the first experiment only, 0.824 including intestine cells from the second experiment). Furthermore, 3,925 genes were enriched in a single tissue (differential expression at least five-fold greater than the 2nd-highest expressing tissue; Fig. 3D, Table S6), and 1,939 genes were enriched for expression in a single cell type (Fig. 3E, Table S7). Thus, despite the fact that sci-RNA-seq captures a minority of transcripts in each cell, our ‘oversampling’ of the cellular composition of the organism enables us to construct representative expression profiles for individual cell types (Fig. 3F).
Neuronal cell types
Because the transcripts of tissue or cell type clusters suggested subclasses within groups (Fig. 4A), we examined expression within several tissues in more detail. We confirmed and extended findings that anterior and posterior body wall muscle have distinct expression patterns (fig. S10A–B, Table S9, (39)), and also observed distinct expression patterns for posterior vs. other intestine cells (fig. S10C–D, Table S10) and amphid vs. phasmid sheath cells (fig. S10E–F, Table S11). But gene expression patterns were particularly diverse in neuronal cell types.
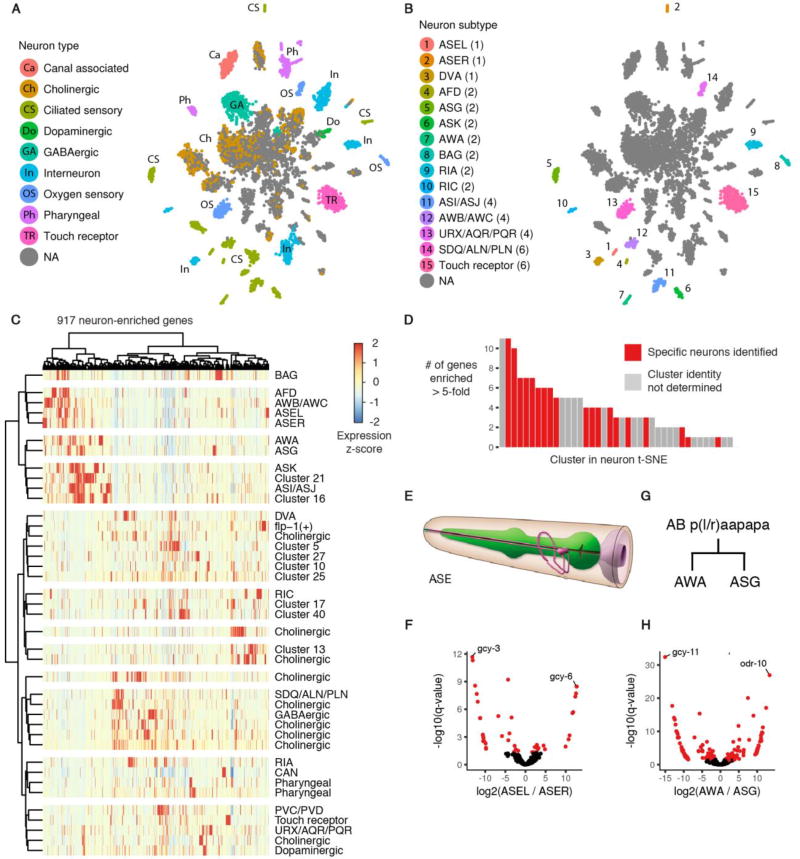
Fig. 4. sci-RNA-seq reveals the transcriptomes of fine-grained anatomical classes of C. elegans neurons.
(A) t-SNE visualization of high-level neuronal subtypes. Cells identified as neurons from the t-SNE clustering shown in Fig. 3A were re-clustered with t-SNE. (B) Clusters in the neuron t-SNE that can be identified as corresponding to one, two, or four specific neurons in an individual C. elegans larva. The number of neurons of each type are shown in parentheses. (C) Heatmap showing the relative expression of neuron-enriched genes across 40 neuron clusters identified by t-SNE and density peak clustering. Genes are included if their expression in the aggregate transcriptome of all neurons in our data is >5-fold higher than their expression in any other tissue, excluding cases where the differential expression is not significant (q-value > 0.05). (D) Distribution for each neuron cluster of the number of genes that are expressed >5-fold higher in that cluster than in the 2nd-highest expressing neuron cluster (q-value for differential expression < 0.05). (E) Cartoon illustrating the position of the left and right ASE neurons (pink) relative to the pharynx (green); reproduced with permission from www.wormatlas.org (60). (F) Volcano plot showing differentially expressed genes between the left and right ASE neurons. Points in red correspond to genes that are differentially expressed (q-value < 0.05) with a > 3-fold difference between the higher- and lower-expressing neuron(s). (G) The left AWA and ASG neurons arise from the embryonic cell AB plaapapa; the right AWA and ASG neurons arise from AB praapapa. (H) Volcano plot showing differentially expressed genes between the AWA and ASG neurons.
By morphological criteria, the 302 neurons of worm are classified into 118 distinct types (40) and from the database of reporter transgene expression patterns, most of these are postulated to have unique molecular signatures (41). Our initial re-clustering of neuronal cells divided them into 10 broad classes (Fig. 4A). Most classes of neurons were represented by several small but highly distinct clusters in the t-SNE plot. Further analysis of cluster-specific gene expression showed that many clusters corresponded to highly specific subsets of neurons in the L2 worm (Fig. 4B, Table S7). Three clusters corresponded to sets of four neurons in an individual worm, 8 clusters corresponded to a single pair of neurons (AFD, ASG, ASK, AWA, BAG, CAN, RIA, and RIC), and 3 clusters corresponded to exactly one neuron (ASEL, ASER, and DVA). Hierarchical clustering analysis showed that of the most of 917 genes highly enriched in neurons, compared to other tissues, were expressed in only a minority of neuronal clusters (Fig. 4C). 73% of neuron-enriched genes had no more than 10 neuron clusters (out of 40 total) in which they were expressed at ≥ 10% of the level of the highest-expressed cluster. 155 genes were highly enriched in a single neuron cluster relative to all others (Fig. 4D, Table S8).
Expression of marker genes, such as gcy-3 and gcy-6, were key in identifying two neuronal clusters as left ASE (ASEL) and right ASE (ASER) gustatory neurons, respectively (Fig. 4E). These neurons have asymmetry in gene expression (42), and we observe 44 genes to be differentially expressed (Fig. 4F, Table S12, fold difference > 3, FDR < 5%). mRNA from these neurons has previously been profiled with co-immunoprecipitation of RNA and a transgenic poly(A)-binding protein expressed specifically in ASEL or ASER, followed by microarray analysis (43). The differentially expressed genes we observe are consistent with this study (fig. S11), highlighting the ability of sci-RNA-seq to facilitate the analysis of cell types as rare as a single cell per individual.
Two neuronal clusters correspond to sister cells, the AWA and ASG neurons, (Fig. 4G), which arise from the same parental cell in the last round of C. elegans embryonic cell divisions. Their differentiation has previously been used as a model for the study of the regulation of cell fate decisions (44). In our data, 136 genes were differentially expressed between these two cell types (Fig. 4H, Table S13, fold difference > 3, FDR < 5%). The divergent transcriptomes of the AWA and ASG neurons, along with the left and right ASE neurons, highlight the potential of cells that are extremely closely related in morphology and developmental lineage to feature distinct programs of gene regulation.
Integration with transcription factor binding sites
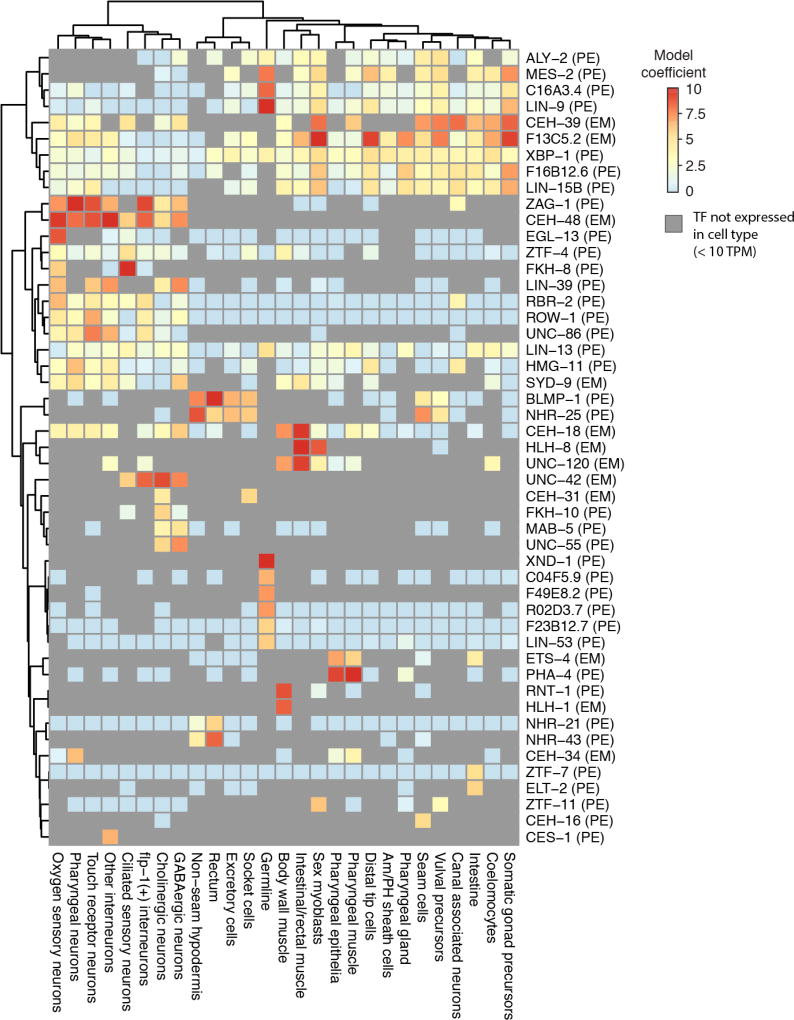
We hypothesized that correlating transcription factor (TF) binding patterns—profiled in ChIP-seq experiments from the modENCODE (45) and modERN (46) consortia—with cell type gene expression profiles could give insights into the regulatory programs underlying the gene expression profiles. For each of 27 cell types, we constructed regularized regression models to predict each gene’s expression as a function of the TF ChIP peaks present in its promoter (Fig. 5). We restricted a cell type’s model to those TFs that were detectably expressed within it (>10 transcripts per million (TPM)), increasing the proportion of TF-to-cell-type associations that are likely to reflect causal gene regulation. Our regression analysis predicted gene expression by selecting numerous regulators critical for development or proper function-specific cell types, including hlh-1 and unc-120 in body wall muscle (47), pha-4 in pharyngeal cell types (48), hlh-8 (CeTwist) in sex myoblasts (49), blmp-1 and nhr-25 in hypodermis (50, 51), elt-2 in the intestine (52), and xnd-1 in the germline (53, 54).
Fig. 5. Cell type specific expression profiles from sci-RNA-seq enable the deconvolution of whole-animal transcription factor ChIP-seq data.
For each of 27 cell types, a regularized regression model was fit to predict log-transformed gene expression levels in that cell type on the basis of ChIP-seq peaks in gene promoters (31). The ChIP-seq data was generated by the modENCODE (61) and modERN consortia (46), profiling transcription factor binding in whole C. elegans animals. “EM” next to a TF label indicates the ChIP-seq data for the TF is from an embryonic stage, while “PE” indicates the data is from a post-embryonic stage. Colors in the heatmap show the extent to which having a ChIP-seq peak for a given TF in a gene promoter correlates with increased expression in a given cell type. Peaks in “HOT regions” (31) are excluded. Grey cells in the heatmap correspond to cases where a TF is not expressed in a cell type (< 10 TPM), in which case ChIP-seq data for that TF is not considered by the regression model.
The regression identified several putative novel regulators of cell-type specific expression. For example, fkh-8, which is expressed specifically in ciliated sensory neurons (our data and reporter construct from (55)) was predictive of their gene expression program (fig. S12). The uncharacterized TF F49E8.2 is expressed specifically in the germline and associated with germline gene expression (fig. S12). F49E8.2 is an ortholog of the human gene “E2F-associated phosphoprotein” (EAPP) (56), and F49E8.2 ChIP-seq peaks co-localize with germline-specific EFL-1 peaks (ortholog of E2F, data from (57)) more often than could be expected due to chance (fig. S13ab, χ2-test, p = 2.8 × 10−21), suggesting that these proteins may physically interact. The hypodermis-associated TFs blmp-1 and nhr-25 were also associated with gene expression in socket cells, excretory cells, and rectal cells. nhr-25 is expressed 4.5-fold higher in socket cells than in seam cells (560 vs. 124 TPM) and 8.7-fold more than in the non-seam hypodermis (560 vs. 64 TPM), suggesting a role in glial development.
Discussion
Our method for single cell RNA-seq combinatorial indexing of cells or nuclei can be applied to profile the transcriptomes of tens-of-thousands of single cells per experiment through a library construction completed by a single individual in two days at a cost of $0.03-$0.20 per cell. sci-RNA-seq is compatible with cell fixation, which can minimize perturbations to cell state or RNA integrity before or during processing and facilitates the concurrent processing of multiple samples within a single experiment, potentially reducing batch effects relative to platforms requiring serial processing, an area of concern for the single cell RNA-seq field (58). Given that the second barcode is introduced after flow sorting, it is also possible to associate wells on the PCR plate with FACS-defined subpopulations. sci-RNA-seq is also compatible nuclei, which may be important for tissues for which unbiased cell disaggregation protocols are not well established (possibly most tissues). Lastly, sci-RNA-seq is scalable. We demonstrate up to 576 × 960 indexing, which enabled the generation of ~4 × 104 single cell transcriptomes in one experiment. However, processing of more cells with sub-linear cost scaling is possible by using more barcoded RT and PCR primers (e.g. 1,536 × 1,536 combinatorial indexing) and/or introducing additional rounds of indexing. With 384 × 384 × 384 combinatorial indexing, one can hypothetically profile the transcriptomes of over 10 million cells per experiment.
With sci-RNA-seq we generated a catalog of single cell transcriptomes with over 50-fold “shotgun cellular coverage” of the L2 C. elegans soma. We detect 18 non-neuronal cell types and a multitude of neuronal cell types, which we grouped into either 10 broad classes or 40 fine-grained clusters from an unsupervised analysis, highlighting the potential of an organism’s gene regulatory programs to be enacted at a fine-grained level. We anticipate these data will be a rich resource for nematode biology – a starting point for an atlas that leverages Sulston’s lineage map to define the molecular state of every cell throughout the life cycle of C. elegans. Furthermore, as illustrated by our experience with intestinal cells, the greater knowledge of “ground truth” for C. elegans may further the refinement of experimental and computational methods for recovering and distinguishing cell types and states.
sci-RNA-seq expands the repertoire of single cell molecular phenotypes that can be resolved by combinatorial indexing (25–28). Provided that multiple aspects of cellular biology can be concurrently barcoded, combinatorial indexing may also facilitate the scalable generation of ‘joint’ single cell molecular profiles (e.g. RNA-seq and ATAC-seq from each of many single cells). We also envision that large-scale, integrated profiling of the molecular states and lineage histories (59) of single cells in other organisms will begin to give shape to “global views” of their developmental biology.
Supplementary Material
Acknowledgments
The raw data have been deposited with the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) under accession code GSE98561 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=wduvwooqvrynngt&acc=GSE98561). These data are also made available as gene-by-cell matrices, along with a “vignette” in the form of a Jupyter notebook that shows examples of how to work with the data (http://waterston.gs.washington.edu/sci_RNA_seq_gene_count_data). We thank members of the Shendure, Trapnell and Waterston labs for helpful discussions and feedback, particularly A. Hill, V. Agarwal, M. Gasperinin, L. Starita, Y. Yin and B. Martin; S. Zimmerman and C. Berg for helpful technique suggestions; the modERN consortium for allowing us to use their ChIP-seq data; D. Prunkard, and L. Gitari in the Pathology Flow Cytometry Core Facility for their exceptional assistance in flow sorting; J. Rose, D. Maly, L. VandenBosch, and T. Reh lab for sharing the NIH/3T3 cell line; H. Hutter for adding our tissue-specific expression profiles on gExplore. HeLa S3 cells were used as part of this study. H. Lacks, and the HeLa cell line that was established from her tumor cells in 1951, have made significant contributions to scientific progress and advances in human health. We are grateful to H. Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. This work was funded by grants from the NIH (DP1HG007811 and R01HG006283 to JS; U41HG007355 and R01GM072675 to RHW, DP2 HD088158 to CT) and the Paul G. Allen Family Foundation (to JS), W. M. Keck Foundation (to CT and JS), Dale. F. Frey Award for Breakthrough Scientists (to CT) and Alfred P. Sloan Foundation Research Fellowship (to CT) and by the William Gates III Endowed Chair in Biomedical Sciences (RHW). DAC was supported in part by T32HL007828 from the National Heart, Lung, and Blood Institute. JS is an Investigator of the Howard Hughes Medical Institute.
REFERENCES AND NOTES
- 1.Trapnell C. Defining cell types and states with single-cell genomics. Genome Res. 2015;25:1491–1498. doi: 10.1101/gr.190595.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wills QF, et al. Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat. Biotechnol. 2013;31:748–752. doi: 10.1038/nbt.2642. [DOI] [PubMed] [Google Scholar]
- 5.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 8.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng W, et al. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 16.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 17.Grindberg RV, et al. RNA-sequencing from single nuclei. Proc. Natl. Acad. Sci. U. S. A. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christina Fan H, Fu GK, Fodor SPA. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 19.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol. Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Res. 2016;5 doi: 10.12688/f1000research.7223.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adey A, et al. In vitro, long-range sequence information for de novo genome assembly via transposase contiguity. Genome Res. 2014;24:2041–2049. doi: 10.1101/gr.178319.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amini S, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Genet. 2014;46:1343–1349. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cusanovich DA, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vitak SA, et al. Sequencing thousands of single-cell genomes with combinatorial indexing. Nat. Methods. 2017;14:302–308. doi: 10.1038/nmeth.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramani V, et al. Massively multiplex single-cell Hi-C. Nat. Methods. 2017;14:263–266. doi: 10.1038/nmeth.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulqueen RM, et al. Scalable and efficient single-cell DNA methylation sequencing by combinatorial indexing. 2017 doi: 10.1101/157230. [DOI] [Google Scholar]
- 29.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 30.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 31.Supplemental online materials.
- 32.Grindberg RV, et al. RNA-sequencing from single nuclei. Proc. Natl. Acad. Sci. U. S. A. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gertz J, et al. Transposase mediated construction of RNA-seq libraries. Genome Res. 2012;22:134–141. doi: 10.1101/gr.127373.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev. Biol. 1985;107:128–133. doi: 10.1016/0012-1606(85)90381-1. [DOI] [PubMed] [Google Scholar]
- 35.Clokey GV, Jacobson LA. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech. Ageing Dev. 1986;35:79–94. doi: 10.1016/0047-6374(86)90068-0. [DOI] [PubMed] [Google Scholar]
- 36.Hendriks G-J, Gaidatzis D, Aeschimann F, Großhans H. Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell. 2014;53:380–392. doi: 10.1016/j.molcel.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 37.Heimberg G, Bhatnagar R, El-Samad H, Thomson M. Low Dimensionality in Gene Expression Data Enables the Accurate Extraction of Transcriptional Programs from Shallow Sequencing. Cell Syst. 2016;2:239–250. doi: 10.1016/j.cels.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boeck ME, et al. The time-resolved transcriptome of C. elegans. Genome Res. 2016;26:1441–1450. doi: 10.1101/gr.202663.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruksana R, et al. Tissue expression of four troponin I genes and their molecular interactions with two troponin C isoforms in Caenorhabditis elegans. Genes Cells. 2005;10:261–276. doi: 10.1111/j.1365-2443.2005.00829.x. [DOI] [PubMed] [Google Scholar]
- 40.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 41.Hobert O, Glenwinkel L, White J. Revisiting Neuronal Cell Type Classification in Caenorhabditis elegans. Curr. Biol. 2016;26:R1197–R1203. doi: 10.1016/j.cub.2016.10.027. [DOI] [PubMed] [Google Scholar]
- 42.Hobert O, Johnston RJ, Chang S. Left–right asymmetry in the nervous system: the Caenorhabditis elegans model. Nat. Rev. Neurosci. 2002;3:629–640. doi: 10.1038/nrn897. [DOI] [PubMed] [Google Scholar]
- 43.Takayama J, Faumont S, Kunitomo H, Lockery SR, Iino Y. Single-cell transcriptional analysis of taste sensory neuron pair in Caenorhabditis elegans. Nucleic Acids Res. 2010;38:131–142. doi: 10.1093/nar/gkp868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarafi-Reinach TR, Melkman T, Hobert O, Sengupta P. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans. Development. 2001;128:3269–3281. doi: 10.1242/dev.128.17.3269. [DOI] [PubMed] [Google Scholar]
- 45.Araya CL, et al. Corrigendum: Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature. 2015;528:152. doi: 10.1038/nature16075. [DOI] [PubMed] [Google Scholar]
- 46.modERN consortia. ENCODE. (available at http://encodeproject.org/)
- 47.Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 2006;20:3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- 49.Harfe BD, et al. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998;12:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horn M, et al. DRE-1/FBXO11-dependent degradation of BLMP-1/BLIMP-1 governs C. elegans developmental timing and maturation. Dev. Cell. 2014;28:697–710. doi: 10.1016/j.devcel.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev. Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- 52.Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- 53.Wagner CR, Kuervers L, Baillie DL, Yanowitz JL. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature. 2010;467:839–843. doi: 10.1038/nature09429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mainpal R, Nance J, Yanowitz JL. A germ cell determinant reveals parallel pathways for germ line development in Caenorhabditis elegans. Development. 2015;142:3571–3582. doi: 10.1242/dev.125732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hope IA, Mounsey A, Bauer P, Aslam S. The forkhead gene family of Caenorhabditis elegans. Gene. 2003;304:43–55. doi: 10.1016/s0378-1119(02)01175-7. [DOI] [PubMed] [Google Scholar]
- 56.Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kudron M, et al. Tissue-specific direct targets of Caenorhabditis elegans Rb/E2F dictate distinct somatic and germline programs. Genome Biol. 2013;14:R5. doi: 10.1186/gb-2013-14-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tung P-Y, et al. Batch effects and the effective design of single-cell gene expression studies. Sci. Rep. 2017;7:39921. doi: 10.1038/srep39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna A, et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 2016;353:aaf7907. doi: 10.1126/science.aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Z A, Hall WormAtlas. 2017 (available at http://www.wormatlas.org/neurons/Individual%20Neurons/ASEframeset.html)
- 61.Araya CL, et al. Corrigendum: Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature. 2015 doi: 10.1038/nature16075. [DOI] [PubMed] [Google Scholar]
- 62.Zhang S, Banerjee D, Kuhn JR. Isolation and culture of larval cells from C. elegans. PLoS One. 2011;6:e19505. doi: 10.1371/journal.pone.0019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tange O. Others, Gnu parallel-the command-line power tool. The USENIX Magazine. 2011;36:42–47. [Google Scholar]
- 65.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014:btu638. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adey A, et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiu X, et al. Reversed graph embedding resolves complex single-cell developmental trajectories. bioRxiv. 2017:110668. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Habib N, et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodriguez A, Laio A. Clustering by fast search and find of density peaks. Science. 2014;344:1492–1496. doi: 10.1126/science.1242072. [DOI] [PubMed] [Google Scholar]
- 71.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9:432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strona G, Nappo D, Boccacci F, Fattorini S, San-Miguel-Ayanz J. A fast and unbiased procedure to randomize ecological binary matrices with fixed row and column totals. Nat. Commun. 2014;5:4114. doi: 10.1038/ncomms5114. [DOI] [PubMed] [Google Scholar]
- 74.Johnstone IL, Barry JD. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 1996;15:3633–3639. [PMC free article] [PubMed] [Google Scholar]
- 75.Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harterink M, et al. Neuroblast migration along the anteroposterior axis of C. elegans is controlled by opposing gradients of Wnts and a secreted Frizzled-related protein. Development. 2011;138:2915–2924. doi: 10.1242/dev.064733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nehrke K, Melvin JE. The NHX family of Na+-H+ exchangers in Caenorhabditis elegans. J. Biol. Chem. 2002;277:29036–29044. doi: 10.1074/jbc.M203200200. [DOI] [PubMed] [Google Scholar]
- 78.Murray JI, et al. Multidimensional regulation of gene expression in the C. elegans embryo. Genome Res. 2012;22:1282–1294. doi: 10.1101/gr.131920.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bacaj T, Tevlin M, Lu Y, Shaham S. Glia Are Essential for Sensory Organ Function in C. elegans. Science. 2008;322:744–747. doi: 10.1126/science.1163074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perens EA, Shaham S. C. elegans daf-6 encodes a patched-related protein required for lumen formation. Dev. Cell. 2005;8:893–906. doi: 10.1016/j.devcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 81.Harrison MM, Ceol CJ, Lu X, Horvitz HR. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16782–16787. doi: 10.1073/pnas.0608461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tabuchi TM, et al. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 2011;7:e1002074. doi: 10.1371/journal.pgen.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Latorre I, et al. The DREAM complex promotes gene body H2A.Z for target repression. Genes Dev. 2015;29:495–500. doi: 10.1101/gad.255810.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle. WormBook. 2006:1–16. doi: 10.1895/wormbook.1.81.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beg AA, Jorgensen EM. EXP-1 is an excitatory GABA-gated cation channel. Nat. Neurosci. 2003;6:1145–1152. doi: 10.1038/nn1136. [DOI] [PubMed] [Google Scholar]
- 86.Tilleman L, et al. An N-myristoylated globin with a redox-sensing function that regulates the defecation cycle in Caenorhabditis elegans. PLoS One. 2012;7:e48768. doi: 10.1371/journal.pone.0048768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghai V, Smit RB, Gaudet J. Transcriptional regulation of HLH-6-independent and subtype-specific genes expressed in the Caenorhabditis elegans pharyngeal glands. Mech. Dev. 2012;129:284–297. doi: 10.1016/j.mod.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 88.Ardizzi JP, Epstein HF. Immunochemical localization of myosin heavy chain isoforms and paramyosin in developmentally and structurally diverse muscle cell types of the nematode Caenorhabditis elegans. J. Cell Biol. 1987;105:2763–2770. doi: 10.1083/jcb.105.6.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Labouesse M. Epithelial junctions and attachments. WormBook. 2006:1–21. doi: 10.1895/wormbook.1.56.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Möhrlen F, Hutter H, Zwilling R. The astacin protein family in Caenorhabditis elegans. Eur. J. Biochem. 2003;270:4909–4920. doi: 10.1046/j.1432-1033.2003.03891.x. [DOI] [PubMed] [Google Scholar]
- 91.Hao L, Johnsen R, Lauter G, Baillie D, Bürglin TR. Comprehensive analysis of gene expression patterns of hedgehog-related genes. BMC Genomics. 2006;7:280. doi: 10.1186/1471-2164-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drace K, McLaughlin S, Darby C. Caenorhabditis elegans BAH-1 is a DUF23 protein expressed in seam cells and required for microbial biofilm binding to the cuticle. PLoS One. 2009;4:e6741. doi: 10.1371/journal.pone.0006741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aspöck G, Kagoshima H, Niklaus G, Bürglin TR. Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res. 1999;9:909–923. doi: 10.1101/gr.9.10.909. [DOI] [PubMed] [Google Scholar]
- 94.Page AP, Johnstone IL. The cuticle. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hong L, et al. MUP-4 is a novel transmembrane protein with functions in epithelial cell adhesion in Caenorhabditis elegans. J. Cell Biol. 2001;154:403–414. doi: 10.1083/jcb.200007075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jacob TC, Kaplan JM. The EGL-21 carboxypeptidase E facilitates acetylcholine release at Caenorhabditis elegans neuromuscular junctions. J. Neurosci. 2003;23:2122–2130. doi: 10.1523/JNEUROSCI.23-06-02122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kass J, Jacob TC, Kim P, Kaplan JM. The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J. Neurosci. 2001;21:9265–9272. doi: 10.1523/JNEUROSCI.21-23-09265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zahn TR, Macmorris MA, Dong W, Day R, Hutton JC. IDA-1, a Caenorhabditis elegans homolog of the diabetic autoantigens IA-2 and phogrin, is expressed in peptidergic neurons in the worm. J. Comp. Neurol. 2001;429:127–143. doi: 10.1002/1096-9861(20000101)429:1<127::aid-cne10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 99.Sieburth D, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 100.Korswagen HC, van der Linden AM, Plasterk RH. G protein hyperactivation of the Caenorhabditis elegans adenylyl cyclase SGS-1 induces neuronal degeneration. EMBO J. 1998;17:5059–5065. doi: 10.1093/emboj/17.17.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Combes D, Fedon Y, Toutant J-P, Arpagaus M. Multiple ace genes encoding acetylcholinesterases of Caenorhabditis elegans have distinct tissue expression. Eur. J. Neurosci. 2003;18:497–512. doi: 10.1046/j.1460-9568.2003.02749.x. [DOI] [PubMed] [Google Scholar]
- 102.Yu RY, Nguyen CQ, Hall DH, Chow KL. Expression of ram-5 in the structural cell is required for sensory ray morphogenesis in Caenorhabditis elegans male tail. EMBO J. 2000;19:3542–3555. doi: 10.1093/emboj/19.14.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yoshida A, et al. A glial K+/Cl− cotransporter modifies temperature-evoked dynamics in Caenorhabditis elegans sensory neurons. Genes Brain Behav. 2015 doi: 10.1111/gbb.12260. (available at http://onlinelibrary.wiley.com/doi/10.1111/gbb.12260/pdf) [DOI] [PubMed]
- 104.Karabinos A, Schulze E, Schünemann J, Parry DAD, Weber K. In vivo and in vitro evidence that the four essential intermediate filament (IF) proteins A1, A2, A3 and B1 of the nematode Caenorhabditis elegans form an obligate heteropolymeric IF system. J. Mol. Biol. 2003;333:307–319. doi: 10.1016/j.jmb.2003.08.041. [DOI] [PubMed] [Google Scholar]
- 105.Gruidl ME, et al. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13837–13842. doi: 10.1073/pnas.93.24.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kawasaki I, et al. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 2004;167:645–661. doi: 10.1534/genetics.103.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cram EJ, Shang H, Schwarzbauer JE. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J. Cell Sci. 2006;119:4811–4818. doi: 10.1242/jcs.03274. [DOI] [PubMed] [Google Scholar]
- 108.Kang SH, Kramer JM. Nidogen is nonessential and not required for normal type IV collagen localization in Caenorhabditis elegans. Mol. Biol. Cell. 2000;11:3911–3923. doi: 10.1091/mbc.11.11.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilkinson HA, Greenwald I. Spatial and temporal patterns of lin-12 expression during C. elegans hermaphrodite development. Genetics. 1995;141:513–526. doi: 10.1093/genetics/141.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnson RP, Kang SH, Kramer JM. C. elegans dystroglycan DGN-1 functions in epithelia and neurons, but not muscle, and independently of dystrophin. Development. 2006;133:1911–1921. doi: 10.1242/dev.02363. [DOI] [PubMed] [Google Scholar]
- 111.Starich TA, Hall DH, Greenstein D. Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics. 2014;198:1127–1153. doi: 10.1534/genetics.114.168815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ackley BD, et al. The NC1/endostatin domain of Caenorhabditis elegans type XVIII collagen affects cell migration and axon guidance. J. Cell Biol. 2001;152:1219–1232. doi: 10.1083/jcb.152.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kostas SA, Fire A. The T-box factor MLS-1 acts as a molecular switch during specification of nonstriated muscle in C. elegans. Genes Dev. 2002;16:257–269. doi: 10.1101/gad.923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Komatsu H, et al. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 2008;6:e196. doi: 10.1371/journal.pbio.0060196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Whitfield CW, Bénard C, Barnes T, Hekimi S, Kim SK. Basolateral localization of the Caenorhabditis elegans epidermal growth factor receptor in epithelial cells by the PDZ protein LIN-10. Mol. Biol. Cell. 1999;10:2087–2100. doi: 10.1091/mbc.10.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tcherepanova I, Bhattacharyya L, Rubin CS, Freedman JH. Aspartic proteases from the nematode Caenorhabditis elegans. Structural organization and developmental and cell-specific expression of asp-1. J. Biol. Chem. 2000;275:26359–26369. doi: 10.1074/jbc.M000956200. [DOI] [PubMed] [Google Scholar]
- 117.McGhee JD, et al. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 2007;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 118.Patton A, et al. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr. Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 119.Zhang Y, et al. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- 120.Kawano T, et al. C. elegans mig-6 encodes papilin isoforms that affect distinct aspects of DTC migration, and interacts genetically with mig-17 and collagen IV. Development. 2009;136:1433–1442. doi: 10.1242/dev.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J. Comp. Neurol. 2004;475:540–550. doi: 10.1002/cne.20189. [DOI] [PubMed] [Google Scholar]
- 122.Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J. Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Suo S, Kimura Y, Van Tol HHM. Starvation induces cAMP response element-binding protein-dependent gene expression through octopamine-Gq signaling in Caenorhabditis elegans. J. Neurosci. 2006;26:10082–10090. doi: 10.1523/JNEUROSCI.0819-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Janssen T, et al. Discovery of a cholecystokinin-gastrin-like signaling system in nematodes. Endocrinology. 2008;149:2826–2839. doi: 10.1210/en.2007-1772. [DOI] [PubMed] [Google Scholar]
- 125.Rand JB. Acetylcholine. WormBook. 2007:1–21. doi: 10.1895/wormbook.1.131.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jorgensen EM. GABA. WormBook. 2005:1–13. doi: 10.1895/wormbook.1.14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nass R, et al. A genetic screen in Caenorhabditis elegans for dopamine neuron insensitivity to 6-hydroxydopamine identifies dopamine transporter mutants impacting transporter biosynthesis and trafficking. J. Neurochem. 2005;94:774–785. doi: 10.1111/j.1471-4159.2005.03205.x. [DOI] [PubMed] [Google Scholar]
- 128.Suo S, Sasagawa N, Ishiura S. Cloning and characterization of a Caenorhabditis elegans D2-like dopamine receptor. J. Neurochem. 2003;86:869–878. doi: 10.1046/j.1471-4159.2003.01896.x. [DOI] [PubMed] [Google Scholar]
- 129.Oishi A, et al. FLR-2, the glycoprotein hormone alpha subunit, is involved in the neural control of intestinal functions in Caenorhabditis elegans. Genes Cells. 2009;14:1141–1154. doi: 10.1111/j.1365-2443.2009.01341.x. [DOI] [PubMed] [Google Scholar]
- 130.Hobson RJ, et al. SER-7, a Caenorhabditis elegans 5-HT7-like receptor, is essential for the 5-HT stimulation of pharyngeal pumping and egg laying. Genetics. 2006;172:159–169. doi: 10.1534/genetics.105.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Furuya M, Qadota H, Chisholm AD, Sugimoto A. The C. elegans eyes absent ortholog EYA-1 is required for tissue differentiation and plays partially redundant roles with PAX-6. Dev. Biol. 2005;286:452–463. doi: 10.1016/j.ydbio.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 132.Sengupta P, Chou JH, Bargmann CI. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- 133.Ortiz CO, et al. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics. 2006;173:131–149. doi: 10.1534/genetics.106.055749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lindemans M, et al. A neuromedin-pyrokinin-like neuropeptide signaling system in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2009;379:760–764. doi: 10.1016/j.bbrc.2008.12.121. [DOI] [PubMed] [Google Scholar]
- 135.Inada H, et al. Identification of guanylyl cyclases that function in thermosensory neurons of Caenorhabditis elegans. Genetics. 2006;172:2239–2252. doi: 10.1534/genetics.105.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yamada K, et al. Olfactory plasticity is regulated by pheromonal signaling in Caenorhabditis elegans. Science. 2010;329:1647–1650. doi: 10.1126/science.1192020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aurelio O, Hall DH, Hobert O. Immunoglobulin-domain proteins required for maintenance of ventral nerve cord organization. Science. 2002;295:686–690. doi: 10.1126/science.1066642. [DOI] [PubMed] [Google Scholar]
- 138.Cornils A, Gloeck M, Chen Z, Zhang Y, Alcedo J. Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development. 2011;138:1183–1193. doi: 10.1242/dev.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Birnby DA, et al. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in caenorhabditis elegans. Genetics. 2000;155:85–104. doi: 10.1093/genetics/155.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Etchberger JF, et al. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gray JM, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 144.Emtage L, Gu G, Hartwieg E, Chalfie M. Extracellular proteins organize the mechanosensory channel complex in C. elegans touch receptor neurons. Neuron. 2004;44:795–807. doi: 10.1016/j.neuron.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 145.Wang X, et al. The C. elegans L1CAM homologue LAD-2 functions as a coreceptor in MAB-20/Sema2–mediated axon guidance. J. Cell Biol. 2008;180:233–246. doi: 10.1083/jcb.200704178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.