Abstract
Introduction
Infectious bursal disease virus (IBDV) is a causative agent of immunosuppressive disorder resulting in significant losses to the world poultry industry. This study describes the molecular characterisation of an atypical IBDV from a field outbreak that occurred in vaccinated chicken flocks in Latvia in 2011.
Material and Methods
Ten bursae of Fabricius from each flock were collected for laboratory examination. Virus isolation was performed in embryonated eggs and CEF culture. The RT-PCR aimed at hypervariable domain of VP2 gene combined with sequencing was performed for detection and identification of IBDV.
Results
The molecular examinations confirmed the IBDV infection. The analysis of the amino acid sequence revealed that the strain possessed four amino acids at VP2 protein (222A, 256I, 294I, and 299S), indicating a genetic relatedness to a very virulent IBDV. However, some unique or rare amino acid substitutions (219L, 220F, 254D, 279N, and 280T) were also detected.
Conclusion
The obtained results demonstrate the occurrence of IBDV with a high mutation rate within the hypervariable domain of VP2 peptide, and highlight the necessity of implementation of IBDV surveillance in Eastern European poultry industry to determine whether this strain is an exception or a new wave of IBDV with new genetic features emerged in the field.
Keywords: chickens, infectious bursal disease virus, VP2, Latvia
Introduction
Infectious bursal disease virus (IBDV) is responsible for a highly contagious immunosuppressive disease in young chickens. The virus belongs to Birnaviridae family, Avibirnavirus genus, and has a bisegmented double-stranded RNA genome (4). The isolates have been classified into three different pathotypes: classical virulent (cIBDV), variant, and very virulent (vvIBDV) (21), but nowadays mainly the last one causes prolonged immunosuppression and involves broiler rearing problems, such as reduced feed conversion and lack of flock uniformity. Segments A and B of IBDV encode for five viral proteins (VP1 to VP5). The VP2 protein builds the capsid and contains conformation-dependent antigenic epitopes responsible for the induction of neutralising antibodies (3, 14). This protein also possesses a hypervariable region (hvVP2) which has higher mutation rates than other regions of IBDV (11). The sequence of hvVP2 may vary, but typically very virulent viruses have amino acids 222A, 256I, 294I, and 299S. Molecular detection and characterisation of IBDV is mainly based on the sequence of the hypervariable region of the VP2 peptide (8, 9, 17, 18). The basic tools for virus eradication include implementation of an appropriate vaccination programme and strict biosecurity. For this purpose, a constant virus monitoring should be implemented.
The aim of this study was genetic characterisation of a very virulent IBDV detected in vaccinated broiler flocks in Latvia.
Material and Methods
Virus samples
At the beginning of 2011 an increased mortality, lack of flock uniformity, and reduced feed conversion were observed in four broiler flocks in a farm in Latvia (Bauska area). The birds in each flock were reared in different houses, and at the sampling time they were 14 (two flocks), 37, and 39 days of age. All chickens, at two to three weeks of age, had been immunised with live vaccines containing an intermediate strain (Nobilis D78, MSD Animal Health, the Netherlands) administered in drinking water. In all diseased broilers the bursa of Fabricius was enlarged and congested in post-mortem examination. In total, 10 specimens of the bursa from each flock were collected for laboratory examination.
Sample preparation
The tissue samples were homogenised (20% w/v in PBS), centrifuged (3500 × g for 15 min), and the supernatants were used for further examinations.
Isolation of the virus
Isolation of the virus was carried out on 10-day-old SPF embryonated eggs (VALO-Biomedia, Germany) using chorioallantoic membrane route (CAM) according to the OIE Diagnostic Manual (20). The chorioallantoic membranes and embryos were homogenised and centrifuged as described above, and stored at temperature below –70°C. The SPF chicken embryo fibroblast (CEF) cell cultures were also used for the isolation of the virus. Several passages were performed with a bursa-derived (three passages) and the virus erlier propagated in embryonated eggs (four passages) samples. The cells were cultivated in Eagle’s medium (Sigma, USA) with the addition of 10% foetal calf serum (Gibco, UK) and 1 × antibiotic antimycotic solution (Sigma, USA).
Viral RNA extraction and RT-PCR amplification
Viral RNA was extracted from clarified supernatants of the bursa of Fabricius, embryos, and cell cultures using a commercial kit (RNeasy Mini Kit, Qiagen, Germany) according to the manufacturer’s protocol. The RT-PCR amplification of the partial sequence of VP2 gene was achieved using primers VP2bisF: 5’- ACCTTCCAAGGAAGCCTGAGTG -3’ and VP2bisR 5’- ATCAGCTCGAAGTTGCTCACC -3’ in order to generate an amplicon of 739 bp, from nucleotide position 513 to 1252 (numbering according to Bayliss et al. (1)). The reverse transcription (RT) and PCR reactions were performed using a commercial kit (OneStep RT-PCR Kit, Qiagen, Germany) in 25 μL of reaction mixture containing 1.5 μL of 10 μM of each primer, 1 μL of 10 μM dNTP, 1 μL of enzyme mix, 5 μL of both buffers, and 7.5 μL of water. The RT was performed at 50°C for 30 min. PCR was conducted in the following conditions: initial denaturation at 95°C for 15 min, followed by 35 cycles of amplification (denaturation at 94°C for 30 sec, annealing at 61°C for 30 sec, and elongation at 72°C for 1 min). Final extension was conducted at 72°C for 10 min. The product was visualised on 2% ethidium bromide stained agarose gel after electrophoretic separation. The RT-PCR products (cDNA) were sequenced in both directions using the same set of primers by a commercial service (Genomed, Poland).
Phylogenetic analysis of the VP2 partial sequence
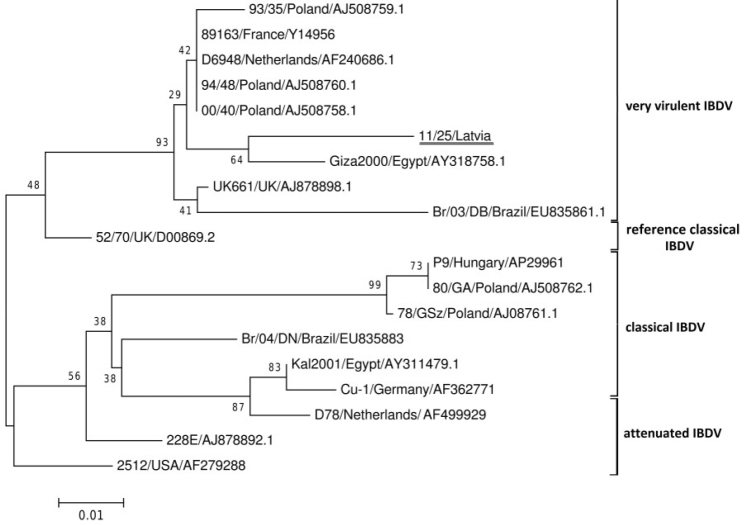
The analysis of the obtained sequences was performed using a MEGA v.6.0 software. The nucleotide and deduced amino acid sequences were aligned using ClustalW method. Phylogenetic trees were generated using the neighbour joining (NJ) method with 1000 bootstrap replicates. Nucleotide sequences of VP2 coding region of 18 IBDV strains of different pathotypes and origins were obtained from GenBank (accession numbers Fig. 1) and included in the analysis. The sequences of hypervariable domain of VP2 gene of the viruses detected in the bursae of Fabricius from examined chickens as well as those isolated from the embryos were the same. The nt sequence of a virus strain designated as 11/25 was submitted to GenBank with the accession number KF013951.
Fig. 1.
Phylogenetic tree based on alignment of the partial VP2 aa sequence (148 aa)
Results
The RNA extracted from the supernatants originating from two flocks as well as from the inoculated SPF embryonated eggs was positive in RT-PCR, indicating IBDV infection. Virus isolation from the embryonated eggs was successful in the first two passages, and typical changes in the form of congestions in the cerebral region and mottled necrosis in embryo liver were observed. The cytopathic effect of the virus in CEF cells was observed only in bursa-derived samples in the second and third passages. The sequence analysis of the virus isolated in CEF revealed high similarity to D78 strain.
Phylogenetic analysis revealed that the isolated 11/25 IBDV strain clustered with the very virulent group of IBDV (vvIBDV) (Fig. 1) and appeared the most closely related to vvIBDV isolated in Egypt in 2000 (accession number AY318758). The presence of four conserved amino acids (222A, 256I, 294I, and 299S) characteristic for vvIBDV was identified in the studied strain (Fig. 2). However, additional five amino acid alterations (219L, 220F, 254D, 279N, and 280T) were also detected. They were located within the major hydrophilic peak A (219L, 220F), the minor peak 1 (254D), and the minor peak 2 (279N, 280T).
Fig. 2.
Alignment of deduced amino acid (aa) sequence of the VP2 variable domain from aa position 216 to 353 of IBDV strains. Dots indicate aa positions identical to F52/70. Major hydrophilic peaks are boxed with red line, and minor hydrophilic peaks are boxed with green line
Discussion
Very virulent IBDV’s are highly pathogenic for young chickens and they are able to break through high levels of maternally derived antibodies (MDA). Despite the fact that the virus circulates in the field and causes losses in poultry production (personal information from practising veterinarians), there are no surveillance programmes devoted to IBDV detection and its genetic characterisation in Eastern Europe. Furthermore, knowledge about current epidemiological field situation is essential for better eradication of IBDV by implementation of adequate immunoprophylaxis. Our results indicate the presence of the virus with a high degree of genetic changes in this part of Europe, as compared to the previously published IBDV molecular characterisation (5). Moreover, to our knowledge, this is the first report of such amino acid profile detected in one strain. Some of these changes were previously reported in Egyptian IBDV strains (220F and 279N) (18), Polish and Hungarian strains isolated in the late 1970s, and more recently in Italian and Brazilian strains (254D and 279N) (5, 8, 17). The role of some of these substitutions is known. For example, the position 254 was found important for antigenicity in US variant strains (10). Moreover, Domańska et al. (5) demonstrated that their presence at position 254D/N modified reactivity of Polish and Hungarian early classical virulent strains with some neutralising monoclonal antibodies (Mab5). In turn, the significance of substitutions at positions 279 (D→N) and 284 (A→T) was proved to be crucial for adaptation of very virulent IBDV to replicate in cell cultures (15, 19). Four-fold passage of the 11/25 strain on CEF cells halted the growth of this isolate (confirmed by RT-PCR). This may probably be attributed to the lack of threonine (T) at position 284.
The effect of other substitutions found within the major hydrophilic peak A (219L, 220F) or minor hydrophilic peak 1 (280T) is not known, but since the hypervariable domain of VP2 represents the molecular basis of antigenic variation, the identified amino acid alterations could result in the viral escape from the neutralising effect of antibodies and might explain the failure of the applied vaccination schedule observed in this case. It is worth noting that with the exception of 279N, amino acid sequences of the D78 vaccine strains used in the studied flocks do not possess four other missense substitutions listed above, and therefore we reject the possibility that the obtained sequence of 11/25 strain is the result of detection and characterisation of mixed populations of viruses (i.e. vaccine and field strains) (Fig. 2).
The obtained results demonstrate the occurrence of IBDV with a high mutation rate within the hypervariable domain of VP2 peptide, and further investigations in Bauska region are needed to determine whether this strain is an exception or a new wave of IBDV with new genetic features emerged in the field. The presented results highlight the importance of the constant monitoring of the field situation, especially in the context of repeated vaccination breakdowns and continuous growth of chicken meat production in the region.
Footnotes
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: The research was financially supported by the National Veterinary Research Institute in Puawy, Poland.
Animal Rights Statement: None required.
References
- 1.Bayliss C.D., Spies U., Shaw K., Peters R.W., Papageorgiou A., Müller H., Boursenell M.E.. A comparison of the sequences of segment A of four infectious bursal disease virus strain and identification of a variable region in VP2. J Gen Virol. 1990;71:1303–1312. doi: 10.1099/0022-1317-71-6-1303. [DOI] [PubMed] [Google Scholar]
- 2.Brandt M., Yao K., Liu M., Heckert R.A., Vakharia V.N.. Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J Virol. 2001;75:11974–11982. doi: 10.1128/JVI.75.24.11974-11982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coulibaly F., Chevalier C., Gutsche I., Pous J., Navaza J., Bressanelli S., Delmas B., Rey F.A.. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell. 2005;120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Dobos P., Hill B.J., Hallett R., Kells D.T., Becht H., Teninges D.. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J Virol. 1979;32:593–605. doi: 10.1128/jvi.32.2.593-605.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domańska K., Mato T., Rivallan G., Śmietanka K., Minta Z., de Boisseson C., Toquin D., Lomniczi B., Palya V., Eterradossi N.. Antigenic and genetic diversity of early European isolates of infectious bursal disease virus prior to the emergence of the very virulent viruses: early European epidemiology of infectious bursal disease virus revisited? Arch Virol. 2004;149:465–480. doi: 10.1007/s00705-003-0230-6. [DOI] [PubMed] [Google Scholar]
- 6.Durairaj V., Sellers H.S., Linnemann E.G., Icard A.H., Mundt E.. Investigation of the antigenic evolution of field isolates using the revers genetic system of infectious bursal disease virus (IBDV) Arch Virol. 2011;156:1717–1728. doi: 10.1007/s00705-011-1040-x. [DOI] [PubMed] [Google Scholar]
- 7.Escaffre O., Le Nouën C., Amelot M., Ambroggio X., Ogden K.M., Guionie O., Toquin D., Müller H., Islam M.R., Eterradossi N.. Both genome segments contribute to the pathogenicity of very virulent infectious bursal disease virus. J Virol. 2013;87:2767–2780. doi: 10.1128/JVI.02360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandes M.J.B., Simoni I.C., Vogel M.G., Harakava R., Rivas E.B., Oliveira M.B.C., Kanashiro A.M.I., Tessari E.N.C., Gama N.M.S.Q., Arns C.W.. Molecular characterization of Brazilian infectious bursal disease virus isolated from 1997 to 2005. Avian Dis. 2009;53:449–454. doi: 10.1637/8531-112808-Reg.1. [DOI] [PubMed] [Google Scholar]
- 9.Islam M.T., Le T.H., Rahman M.M., Islam M.A.. Molecular characterization of two Bangladeshi infectious bursal disease virus isolates using the hypervariable sequence of VP2 as a genetic marker. J Vet Sci. 2012;13:405–412. doi: 10.4142/jvs.2012.13.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackwood D.J., Jackwood R.J., Sommer S.E.. Identification and comparison of point mutations associated in classic and variant infectious bursal disease viruses. Virus Res. 1997;49:131–137. doi: 10.1016/s0168-1702(97)01465-2. [DOI] [PubMed] [Google Scholar]
- 11.Jackwood D.J., Sommer-Wagner S.. Genetic characteristics of infectious bursal disease viruses from four continents. Virology. 2007;365:369–375. doi: 10.1016/j.virol.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 12.Jackwood D.J., Sreedevi B., LeFever L.J., Sommer-Wagner S.E.. Studies on naturally occurring infectious bursal disease viruses suggest that a single amino acid substitution at position 253 in VP2 increases pathogenicity. Virology. 2008;377:110–116. doi: 10.1016/j.virol.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 13.Le Nouën C., Rivallan G., Toquin D., Darlu P., Morin Y., Beven V., Boisseson C., Cazaban C., Comte S., Gardin Y., Eterradossi N.. Very virulent infectious bursal disease virus: reduced pathogenicity in a rare natural segment-B reasserted isolate. J Gen Virol. 2006;87:209–216. doi: 10.1099/vir.0.81184-0. [DOI] [PubMed] [Google Scholar]
- 14.Letzel T., Coulibaly F., Rey F.A., Delmas B., Jagt E., Van Loon A.A.M.W., Mundt E.. Molecular and structural bases for the antigenicity of VP2 of infectious bursal disease virus. J Virol. 2007;81:12827–12835. doi: 10.1128/JVI.01501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim B.L., Cao Y., Yu T., Mo W.C.. Adaptation of very virulent infectious bursal disease virus to chicken embryonic fibroblasts by site-directed mutagenesis of residues 279 and 284 of viral coat protein VP2. J Virol. 1999;73:2854–2862. doi: 10.1128/jvi.73.4.2854-2862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Z., Zhang L., Wang N., Chen Y., Gao L., Wang Y., Gao H., Gao Y., Li K., Qi X., Wang X.. Naturally occurring reassortant infectious bursal disease virus in northern China. Virus Res. 2015;203:92–95. doi: 10.1016/j.virusres.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Martin A.M., Fallacara F., Barbieri I., Tosi G., Rivallan G., Eterradossi N., Ceruti R., Cordioli P.. Genetic and antigenic characterization of infectious bursal disease virus isolated in Italy during the period 2002–2005. Avian Dis. 2007;51:863–872. doi: 10.1637/7904-020107-REGR.1. [DOI] [PubMed] [Google Scholar]
- 18.Metwally A.M., Yousif A.A., Shaheed I.B., Mohammed W.A., Samy A.M., Reda I.M.. Re-emergence of very virulent IBDV in Egypt. Int J Virol. 2009;5:1–17. [Google Scholar]
- 19.Noor M., Mahmud M.S., Ghose P.R., Roy U., Nooruzzaman M., Chowdhury E.H., Das P.M., Islam M.R., Muller H.. Further evidence for the association of distinct amino acid residues with in vitro and in vivo growth of infectious bursal disease virus. Arch Virol. 2014;159:701–709. doi: 10.1007/s00705-013-1885-2. [DOI] [PubMed] [Google Scholar]
- 20.OIE (World Organisation for Animal Health) Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris: 2016. Infectious bursal disease (Gumboro disease). Chapter 2.3.12; pp. 1–21. [Google Scholar]
- 21.Van den Berg T.P.. Acute infectious bursal disease in poultry: A review. Avian Pathol. 2000;29:175–194. doi: 10.1080/03079450050045431. [DOI] [PubMed] [Google Scholar]