Abstract
Introduction
In 2014–2015, the epidemic of classical swine fever (CSF) occurred in many large-scale pig farms in different provinces of China, and a subgenotype 2.1d of CSF virus (CSFV) was newly identified.
Material and Methods
The phylogenetic relationship, genetic diversity, and epidemic status of the 2014–2015 CSFV isolates, 18 new CSFV isolates collected in 2015, and 43 other strains isolated in 2014–2015 were fully analysed, together with 163 CSFV reference isolates.
Results
Fifty-two 2014–2015 isolates belonged to subgenotype 2.1d and nine other isolates belonged to subgenotype 2.1b. The two subgenotype isolates showed unique molecular characteristics. Furthermore, the 2.1d isolates were found to possibly diverge from 2.1b isolates.
Conclusion
This study suggests that the Chinese CSFVs will remain pandemic.
Keywords: swine, classical swine fever virus, evolution, subgenotype 2.1d, China
Introduction
Classical swine fever (CSF), a highly contagious and often fatal disease of pigs, is listed by the Office International des Epizooties (OIE) and remains a significant economic problem of swine industry in numerous regions of the world (5, 18). The disease is caused by CSF virus (CSFV), a member of the Pestivirus genus within the Flaviviridae family. The genus also includes bovine viral diarrheoa virus types I and II (BVDV I and II) and border disease virus (BDV) (17). Pigs can be infected by other pestiviruses, but these viruses usually do not spread efficiently outside their typical ruminant hosts (20).
CSFV is a single positive-stranded, enveloped RNA virus. The genome is approximately 12.3 kb in length, comprising a single, long open reading frame (ORF) that encodes four structural proteins (C, Erns, E1, and E2) and eight non-structural proteins (Npro, P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B), flanked by two non-coding regions at the 5 untranslated region (5 UTR) and 3 UTR (16).
Based on the phylogenetic analysis of the sequences of 5 UTR, E2 envelope glycoprotein gene, and NS5B polymerase gene CSFVs were divided into three genotypes (1, 2, and 3) and 10 subgenotypes (1.1–1.3, 2.1–2.3, and 3.1–3.4) (20). Subgenotype 2.1 isolates were further classified into 2.1a and 2.1b and were reported to be epidemic in many countries (4). Recently, subgenotypes 2.1c and 1.4 were reported (10, 23). Four subgenotypes (1.1, 2.1, 2.2, and 2.3) of CSFV isolates have existed in mainland China and contributed to CSFV outbreaks (24, 27). Among these isolates, subgenotype 2.1 isolates, especially 2.1b, have long been the predominant strains (2, 24, 27).
In 2014–2015, CSF outbreaks appeared in many regions of China, especially in Shandong province. A subgenotype 2.1d was newly identified by our laboratory (30). Then other researchers reported similar isolates (8, 15). In the present study, 18 newly emerged CSFV isolates collected in 2015 are reported for the first time and the phylogenetic relationship, genetic diversity, and epidemic status of all new CSFV isolates collected in 2014–2015 have been fully analysed.
Material and Methods
Sample collection
From August 2014 to July 2015, more than 50 clinical samples were collected, including lungs, spleen, kidneys, and serum samples from suspected CSFV-infected pigs on different large-scale pig farms distributed over six provinces (Shandong, Jilin, Heilongjiang, Jiangsu, Hebei, and Inner Mongolia). The samples were homogenised in Dulbecco's modified Eagle's medium (DMEM, Gibco, USA) using a TissuLyser II (Qiagen, Germany) for RNA extraction.
Genome sequencing
CSFV-positive samples were selected for E2 gene sequencing. The primers used to amplify the E2 gene (E2-forward: GTAAATATGTGTGTGTTAGACCAGA, E2-reverse: GTGTGGGTAATTGAGTTCCCTATCA), methods of RNA extraction, RT-PCR, and genome sequencing were described previously (30).
Phylogenetic analysis
Based on the E2 full-length sequences of 61 new CSFV isolates collected in 2014–2015 and 163 reference CSFV isolates worldwide, phylogenetic trees were constructed using the neighbour-joining method with 1000 bootstraps in MEGA 6.06 software (26). MUSCLE in MEGA software generated multiple sequence alignments (25).
Nucleotide (nt) and amino acid (aa) analysis of E2.
The nt and aa sequence homologies between the 61 new CSFV isolates and 11 representative CSFV isolates, including Shimen (1.1), Brescia90 (1.2), CSF0306 (1.3), CSF0705 (1.4), Paderborn (2.1a), HEBZ (2.1b), GDPY.2008 (2.1c), Zj0801 (2.1d), LAL-290 (2.2), Alfort (2.3), and P97 (3.4), were assessed using the Clustal W method of Lasergene (Version 7.1) (DNASTAR Inc., USA). To explore the genetic variation characteristic of the new isolates, the aa sequences of E2 of 38 new 2014–2015 CSFV isolates were fully analysed, together with 27 representative CSFV isolates from China and other countries.
Results
Detection of suspected CSFV samples
More than 40 samples collected from 25 different large-scale pig farms between August 2014 and July 2015 were identified as CSFV-positive by RT-PCR. The E2 genes of 36 positive samples were sequenced. In total, 18 strains isolated in 2014 were reported previously (30). The other 18 strains isolated in 2015 were reported for the first time. In addition, the E2 gene sequences of other 25 strains isolated in 2014–2015 were obtained from NCBI. The detailed information on these isolates was shown in (Table 1).
Table 1.
Characteristics of new CSFV isolates collected in 2014–2015
| No | Virus strain | Collection date | Collection area | Accession no |
|---|---|---|---|---|
| 1 | SDJNa-14 c | 2014.04 | Shandong | KT953589 |
| 2 | SDQU1408a | 2014.08 | Shandong | Unsubmitted |
| 3 | HLJJY1409a | 2014.09 | Heilongjiang | Unsubmitted |
| 4 | HLJTB1409a | 2014.09 | Heilongjiang | Unsubmitted |
| 5 | JLMC1409a | 2014.09 | Jilin | Unsubmitted |
| 6 | SDWH11409a | 2014.09 | Shandong | Unsubmitted |
| 7 | SDWH21409a | 2014.09 | Shandong | Unsubmitted |
| 8 | SDWH31409a | 2014.09 | Shandong | Unsubmitted |
| 9 | HLJLYG1410a | 2014.10 | Heilongjiang | Unsubmitted |
| 10 | SDJN(D)1410a | 2014.10 | Shandong | Unsubmitted |
| 11 | SDLS(C)1410a | 2014.10 | Shandong | Unsubmitted |
| 12 | SDLW1410 a | 2014.10 | Shandong | Unsubmitted |
| 13 | SDSG(A)1410a | 2014.10 | Shandong | Unsubmitted |
| 14 | SDSG(B)1410a | 2014.10 | Shandong | Unsubmitted |
| 15 | SDWH(F)1410a | 2014.10 | Shandong | Unsubmitted |
| 16 | SDZC1411 a | 2014.11 | Shandong | Unsubmitted |
| 17 | HLJSH1412a | 2014.12 | Heilongjiang | Unsubmitted |
| 18 | JSZL a | 2014.12 | Jiangsu | KT119352 |
| 19 | SDLY-14c | 2014.12 | Shangdong | KT953604 |
| 20 | SDJN(E)1412a | 2014.12 | Shandong | Unsubmitted |
| 21 | SDHS9150129b | 2015.01 | Shandong | Unsubmitted |
| 22 | SDHS1015012b | 2015.01 | Shandong | Unsubmitted |
| 23 | JL2015 b | 2015.02 | Jilin | Unsubmitted |
| 24 | NMG2015 b | 2015.02 | Inner Mongolia | Unsubmitted |
| 25 | HB150309b | 2015.03 | Hebei | Unsubmitted |
| 26 | SD19-15 c | 2015.03 | Shangdong | KT953603 |
| 27 | SDJNi2-15 c | 2015.03 | Shangdong | KT953588 |
| 28 | SDJNi3-15 c | 2015.03 | Shangdong | KT953596 |
| 29 | SDLK150320b | 2015.03 | Shandong | Unsubmitted |
| 30 | SDLW2-15 c | 2015.03 | Shangdong | KT953597 |
| 31 | SDQZ150319b | 2015.03 | Shandong | Unsubmitted |
| 32 | SDZB2-15 c | 2015.03 | Shangdong | KT953590 |
| 33 | JL150418b | 2015.04 | Jilin | Unsubmitted |
| 34 | NK150425b | 2015.04 | Heilongjiang | Unsubmitted |
| 35 | SDJNi1-15 c | 2015.04 | Shangdong | KT953587 |
| 36 | SDJNi4-15 c | 2015.04 | Shangdong | KT953598 |
| 37 | SDJNi5-15 c | 2015.04 | Shangdong | KT953600 |
| 38 | SDJNi6-15 c | 2015.04 | Shangdong | KT953601 |
| 39 | SDLW1-15 c | 2015.04 | Shangdong | KT953593 |
| 40 | SDLY-15 c | 2015.04 | Shangdong | KT953594 |
| 41 | SDMZ1-15 c | 2015.04 | Shangdong | KT953599 |
| 42 | SDQZ150414b | 2015.04 | Shandong | Unsubmitted |
| 43 | SDTA2-15 c | 2015.04 | Shangdong | KT953591 |
| 44 | SDTA3-15 c | 2015.04 | Shangdong | KT953595 |
| 45 | SDWK-15 c | 2015.04 | Shangdong | KT953602 |
| 46 | SDZC150416b | 2015.04 | Shandong | Unsubmitted |
| 47 | HB150528b | 2015.05 | Hebei | Unsubmitted |
| 48 | HeN1505 c | 2015.05 | Henan | KU556758 |
| 49 | SDZC150514b | 2015.05 | Shandong | Unsubmitted |
| 50 | SDZC150526b | 2015.05 | Shandong | Unsubmitted |
| 51 | HLJSH150609b | 2015.06 | Heilongjiang | Unsubmitted |
| 52 | SDHZ-15 c | 2015.06 | Shangdong | KT953607 |
| 53 | SDMZ2-15 c | 2015.06 | Shangdong | KT953606 |
| 54 | SDSK-15 c | 2015.06 | Shangdong | KT953608 |
| 55 | SDTA4-15 c | 2015.06 | Shangdong | KT953610 |
| 56 | SDXLS-15 c | 2015.06 | Shangdong | KT953609 |
| 57 | SDXT-15 c | 2015.06 | Shangdong | KT953611 |
| 58 | SDZC150601b | 2015.06 | Shandong | Unsubmitted |
| 59 | SDZB-15 c | 2015.06 | Shangdong | KT953605 |
| 60 | HLJHEB15071b | 2015.07 | Heilongjiang | Unsubmitted |
| 61 | HLJSH150702 b | 2015.07 | Heilongjiang | Unsubmitted |
The isolates reported previously by other laboratories
The isolates reported previously by our laboratory
The isolates first reported in this study
Phylogenetic analysis of 2014–2015 isolates
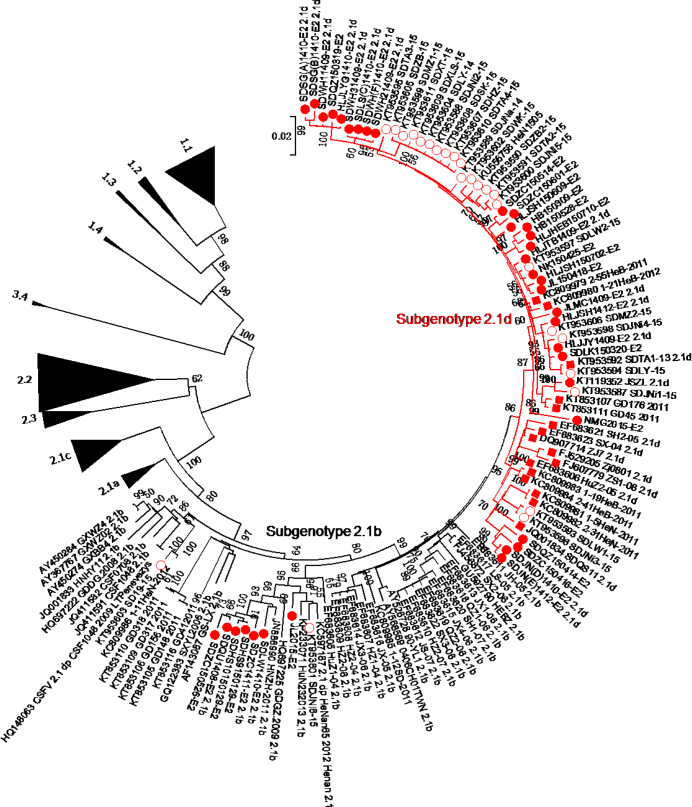
A total of 224 full-length E2 gene sequences, including 61 isolates identified in 2014–2015, formed the phylogenetic tree (Fig. 1). CSFV isolates were divided into genotypes 1, 2, and 3. Genotype 1 and 2 isolates were further divided into subgenotypes 1.1-1.4 and 2.1-2.3, respectively. In addition, subgenotype 2.1 isolates were further subdivided into 2.1a, 2.1b, 2.1c, and a new group 2.1d.
Fig. 1.
Phylogenetic analysis of the 61 new isolates collected in 2014–2015 and other 163 reference CSFV isolates based on the full-length E2 gene sequences. For the new 2014–2015 isolates, 37 strains (●) were isolated by our laboratory and 25 strains (○) were isolated by others. Red lines indicate the subgenotype 2.1d, including 52 strains (● or ○) isolated in 2014–2015 and 16 strains (■) isolated between 2004 and 2013
Of 61 isolates identified between 2014 and 2015, 52 isolates (JLMC1409, HLJTB1409, SDWH11409, SDWH21409, SDWH31409, HLJJY1409, HLJLYG1410, SDWH(F)1410, SDLS(C)1410, SDSG(A)1410 SDSG(B)1410, SDJN(D)1410, SDJN(E)1412 HLJSH1412, JSZL, SDJNa-14, SDLY-14, NMG2015, HB150309, SDLK150320, SDQZ150319, JL150418 NK150425, SDQZ150414, SDZC150416, HB150528, SDZC150514, SDZC150601, HLJSH150609, HLJSH150702, HLJHEB150710, SDJNi2-15, SDJNi3-15, SDLW2-15, SDZB2-15, SDJNi1-15, SDJNi4-15, SDJNi5-15, SDLW1-15, SDLY-15, SDMZ1-15 SDTA2-15, SDTA3-15, SDWK-15, HeN1505) belong to subgenotype 2.1d. The other nine isolates (SDQU1408, SDLW1410, SDZC1411, JL2015, SDZC150526, SDHS9150129, SDHS10150129 SD19-15, SDJNi6-15) belong to subgenotype 2.1b.
The subgenotype 2.1d also include 16 previously reported isolates: SX-04, HuZ2-05, SH2-05, ZJ7.2005, ZS1-08, Zj0801, SDQS11, GD176/2011, GD45/2011, 1-19/HeB-2011, 1-5/HeN-2011, 2-41/HeB-2011 2-55/HeB-2011, 2-31/HeN-2011, 1-21/HeB-2012 and SDTA1-13. The detailed information on these isolates is shown in Table 2.
Table 2.
Characteristics of the subgenotype 2.1d CSFV isolates collected between 2004 and 2013
| No | Virus strain | Collection date | Collection area | Accession no |
|---|---|---|---|---|
| 1 | SX-04 | 2004 | Zhejiang | EF683623 |
| 2 | HuZ2-05 | 2005 | Zhejiang | EF683606 |
| 3 | SH2-05 | 2005 | Shanghai | EF683621 |
| 4 | ZJ7.2005 | 2005 | Zhejiang | DQ907714 |
| 5 | ZS1-08 | 2008 | Zhejiang | FJ607779 |
| 6 | Zj0801 | 2008.03 | Zhejiang | FJ529205 |
| 7 | SDQS11 | 2011 | Hunan | JQ001834 |
| 8 | GD176/2011a | 2011 | Guangdong | KT853107 |
| 9 | GD45/2011a | 2011 | Guangdong | KT853111 |
| 10 | 1-19/HeB-2011 | 2011.07 | Hebei | KC809983 |
| 11 | 1-5/HeN-2011 | 2011.10 | Henan | KC809981 |
| 12 | 2-41/HeB-2011 | 2011.10 | Hebei | KC809984 |
| 13 | 2-55/HeB-2011 | 2011.10 | Hebei | KC809979 |
| 14 | 2-31/HeN-2011 | 2011.11 | Henan | KC809982 |
| 15 | 1-21/HeB-2012 | 2012.03 | Hebei | KC809980 |
| 16 | SDTA1-13 | 2013.12 | Shangdong | KT953592 |
CSFV strains were isolated in Guangdong province by other labs
Nt and aa analysis of E2 of 2014–2015 isolates
The E2 gene is 1119 nt long and encodes 373 aas. Compared with genotype 2 isolates, including Paderborn (2.1a), HEBZ (2.1b), GDPY.20-08 (2.1c), Zj0801 (2.1d), LAL-290 (2.2) and Alfort (2.3), the nine new 2.1b isolates had 85.8%-94.9% nt and 90.1%-97.3% aa identity, and the 52 new 2.1d isolates showed 86.9%-97.3% nt and 90.6%-98.4 aa identity, which were higher than with genotype 1 isolates of Shimen (1.1), Brescia90 (1.2), CSF0306 (1.3), CSF0705 (1.4), or subgenotype 3.4 isolate of P97. Furthermore, the nine new 2.1b isolates had greater similarity to subgenotype 2.1b isolate of HEBZ than to other genotype 2 isolates of Paderborn (2.1a), GDPY.20-08 (2.1c), or Zj0801 (2.1d). Similarly, the 52 new 2.1d isolates had greater similarity to subgenotype 2.1d isolate of Zj0801 than to other genotype 2 isolates of Paderborn (2.1a), HEBZ (2.1b), or GDPY.20-08 (2.1c). The detailed results are shown in (Table 3).
Table 3.
Analysis of nucleotide (nt) and amino acid (aa) identity of E2 gene between the 61 new isolates (including 9 2.1b isolates and 52 2.1d isolates) collected in 2014–2015 and other 11 representative CSFV isolates (%)
| Identities | Shimen (1.1) | Brescia 90 (1.2) | CSF0306 (1.3) | CSF0705 (1.4) | Paderborn (2.1a) | HEBZ (2.1b) | |
|---|---|---|---|---|---|---|---|
| 2.1b | nt | 82.8-84.1 | 81.8-83.4 | 83.5-84.4 | 83.8-85.2 | 93.2-94.8 | 93.5-94.9 |
| aa | 87.8-89.9 | 88.5-91.2 | 89.0-91.2 | 88.5-90.6 | 94.6-97.3 | 95.2-97.3 | |
| 2.1d | nt | 83.1-84.4 | 82.4-83.6 | 82.5-83.5 | 82.9-84.1 | 93.1-94.3 | 95.1-96.5 |
| aa | 89.1-91.0 | 89.0-90.9 | 88.5-90.3 | 88.7-90.3 | 94.6-97.3 | 95.4-97.6 | |
| Identities | GDPY.20-08 (2.1c) | Zj0801 (2.1d) | LAL-290 (2.2) | Alfort (2.3) | P97 (3.4) | ||
| 2.1b | nt | 90.3-92.1 | 93.2-94.6 | 85.8-86.7 | 87.7-88.2 | 80.9-82.3 | |
| aa | 94.4-96.8 | 94.4-96.5 | 90.1-92.0 | 92.2-93.3 | 87.9-90.1 | ||
| 2.1d | nt | 90.3-91.4 | 95.8-97.3 | 85.3-86.4 | 86.9-88.0 | 81.7-82.4 | |
| aa | 94.1-96.8 | 96.2-98.4 | 89.5-91.7 | 90.6-92.8 | 88.7-90.3 | ||
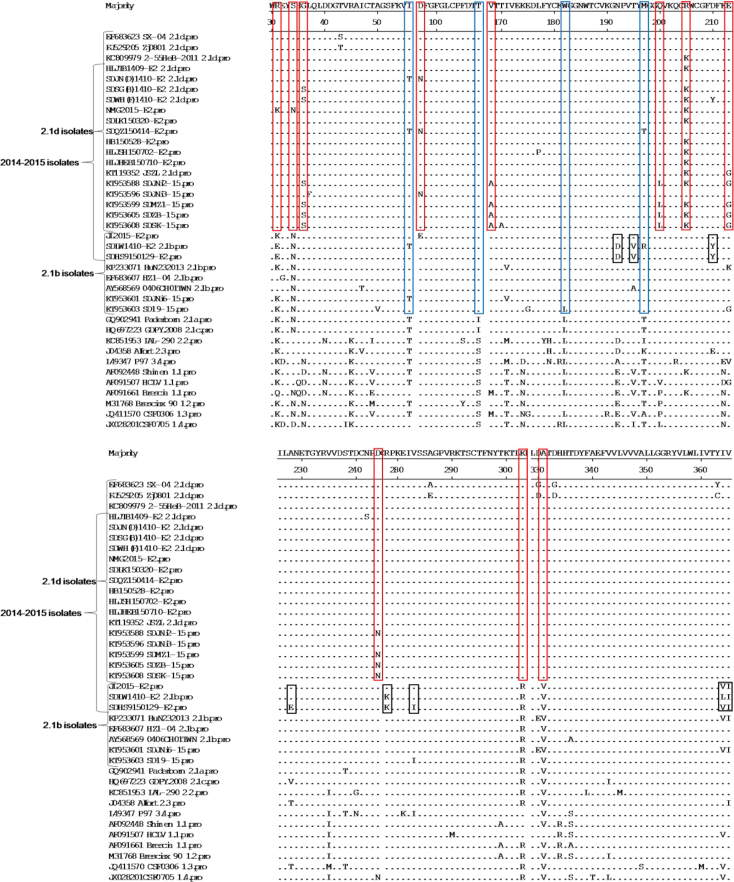
According to the results of aa analysis of E2, the subgenotype 2.1d isolates, including 52 isolates collected in 2014–2015 and 16 previous isolates, showed several unique molecular characteristics, including aa R at position 31 (R31), S34, K205, K303 and A331, compared with all other isolates. Some subgenotype 2.1d isolates also showed unique aa substitutions, including G/D/N36S, D97N, V/M168A, Q/V/P200L, E/N213G, and D244N. In addition, most subgenotype 2.1d and 2.1b isolates shared four consistent aa substitutions at positions T56I, I/S108T, L182W, and T/K/A197M compared with other subgenotype isolates. We also found that most new subgenotype 2.1b isolates collected in 2014–2015 showed unique molecular characteristics, including aa D at positions D192, V195, Y210, E228, K278, I283, VLA364, and I365 (Fig. 2).
Fig. 2.
Amino acid sequence alignments of E2 genes of 38 isolates collected in 2014–2015 and 27 reference CSFV strains. A-C - the unique molecular characteristics (R31, S34, S36, N97, A168, L200, K205, G213, N244, K303, and A331) of 2.1d isolates are indicated by red boxes (□). The consistent molecular characteristics (T56, T108, W182, and M197) of 2.1d and 2.1b isolates are indicated by blue boxes (□).The unique molecular characteristics (D192, V195, Y210, E228, K278, I283, V/L/A364, and I365) of the new 2.1b isolates collected in 2014–2015 are indicated by black boxes (□)
Some aa sites of E2 protein were further analysed. In position 205 of E2 protein, most new 2.1d isolates collected in 2014–2015 showed aa K, and other 2.1d isolates were aa R, which was consistent with the 2.1b and other subgenotype isolates. In addition, most 1.1 isolates showed the same aa S, K, and K with 2.1d isolates in positions 34, 205, and 303 of E2 protein, respectively. In position 331 of E2 protein, all the 2.1d isolates showed aa A, and the other subgenotype isolates showed aa V, except for subgenotype 1.1 isolate HCLV, a Chinese lapinised vaccine strain (C-stain), which showed the same aaA with 2.1d isolates (Fig. 2).
Geographical distribution of subgenotype 2.1d isolates
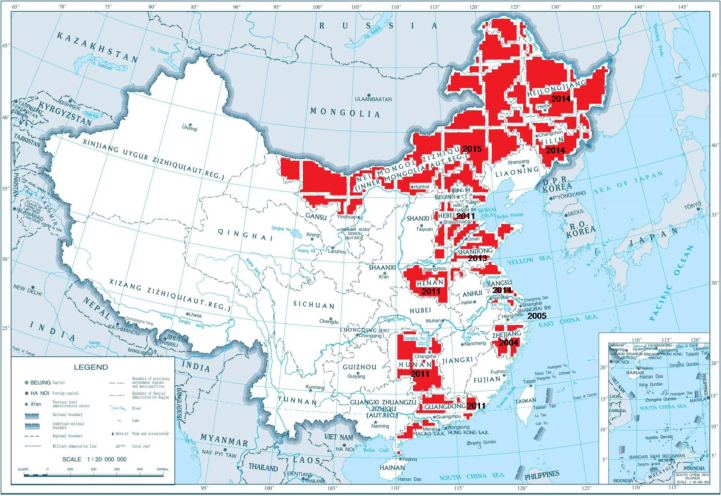
The 52 new 2014–2015 subgenotype 2.1d isolates were distributed over seven provinces (Shandong, Jilin, Heilongjiang, Jiangsu, Hebei, Henan, and Inner Mongolia) of China. However, most strains were isolated in Shandong province. The other 16 subgenotype 2.1d strains were isolated in six provinces (Hebei, Henan, Hunan, Zhejiang, Shanghai, and Guangdong) between 2004 and 2013. These provinces in which the emergence of the 2.1d isolates were confirmed formed a large, relatively connected area on the map of China (Fig. 3).
Fig. 3.
Distribution and emergence time of the CSFV subgenotype 2.1d isolates in China. The 2.1d isolates emerged in the provinces marked red
Discussion
CSF is a devastating, highly contagious, often fatal disease of swine, causing huge economic losses to pig farms in numerous areas of the world (5, 18). In 1954, the C-strain vaccine was successfully developed. The vaccine was used in many endemic countries, and large CSF outbreaks had been rare (14). However, although the intensive control programmes have been implemented for over 20 years, CSF has not been completely controlled since it is sporadic or endemic in many countries of the world (1, 3, 13, 19). Recently, a new cluster of 2.1c virus subgenotype was identified in South China (10). Our laboratory also reported a new subgenotype 2.1d in China last year (30). The 2.1d isolates were epidemic in Shandong province, which has caused heavy economic losses since 2014 (8). In the present study, we reported 18 new CSFV isolates collected in 2015. The phylogenetic relationship, genetic diversity, and epidemic status of all the new CSFV isolates collected in 2014–2015 were fully analysed.
The full-length E2 sequence can be used for classification of CSFV isolates (22). Therefore, the phylogenetic tree was constructed based on the 224 full-length E2 gene sequences, including 61 isolates collected in 2014–2015 and 163 reference CSFV strains (Fig. 1). The result showed that CSFV isolates could be divided into three genotypes (1, 2, and 3) and several subgenotypes (1.1-1.4, 2.1a-2.1d, 2.2, 2.3 and 3.4). Among the 61 new CSFV isolates, 52 isolates belong to subgenotype 2.1d and the other 9 isolates belong to 2.1b. The 2.1d isolates were clearly in an independent branch, close to 2.1b isolates, and were located at the end of the evolutionary tree. Interestingly, the C-strain, HCLV, belong to the 1.1 subgenotype, located at the other end of the evolutionary tree. This result was consistent with a previous report, which showed that the prevalent CSFV isolates shifted away from the vaccine strain (2). This might have resulted from their adaptive abilities to the selection forces within the host (2). Recently, Gong et al. (6) reported that subgenotype 2.1 isolates of CSFV could be divided into 10 sub-subgenotypes (2.1a-2.1j) and the 2.1d-2.1j were newly identified (6). However, in this report, many 2.1 isolates, especially the newly emerged 2014–2015 isolates, were not included. So we do not agree with this classification completely.
The E2 nt and aa sequences of the 2014–2015 isolates were compared with 11 representative CSFV isolates, including Shimen (1.1), Brescia90 (1.2), CSF0306 (1.3), CSF0705 (1.4), Paderborn (2.1a), HEBZ (2.1b), GDPY.20-08 (2.1c), Zj0801 (2.1d), LAL-290 (2.2), Alfort (2.3), and P97 (3.4) (Table 3). The results indicated that the nine new 2.1b isolates and 52 2.1d isolates showed the highest identity with 2.1b isolate HEBZ and 2.1d isolate Zj0801 respectively, which suggests the accuracy of the classification of these new isolates. In the present study, the 2.1d isolates showed many unique molecular characteristics in E2 protein (R31, S34, S 36, N97,A168, L200G213, N244, K205, K303, and A331), and most of these characteristics were reported previously (8, 30) (Fig. 2). Furthermore, the 2.1d isolates also shared the same aa with the 2.1b isolates in some positions of E2 (T56, T108, W182, and M197) (Fig. 2). Previous reports have found that natural recombination occurrs in CSFV isolates (7, 9, 29). In this study, we did not find evidence that the 2.1d isolates are derived from recombination between 2.1b and 1.1 or other subgenotype isolates after recombination analysis by the SimPlot programme (12) (data not shown). Although we speculated that the newly emerged 2.1d isolates may have diverged from 2.1b isolates, the internal evidence needs further exploration. In addition, we also found some unique molecular characteristics (D192, V195, Y210, E228, K278, I283, and V/L/A364 and I365) of the new 2.1b isolates (Fig. 2). Whether these isolates will diverge into a new subgenotype is worthy of attention and vigilance.
As we all know, E2 is the most antigenic protein of CSFV and is involved in virus neutralisation. Four antigenic domains, A (86-176aas), B (1-83aas), C (1-110aas), and D (86-110aas), have been mapped on E2 (28). Domain A was subdivided into A1, A2, and A3.The aa substitutions reported in the present study were located in all these four domains. Whether these substitutions could affect the structure and function of E2 needs to be further studied. However, the six cysteines at positions 4, 48, 103, 129, 139, and 167, which were essential for binding by monoclonal antibodies of the four domains, had no variation in E2 protein of the 2014–2015 isolates (28). In addition, the potential N-glycosylation sites in E2 protein of these isolates were consistent with previous isolates.
Previous reports showed that several CSFV subgenotypes (1.1, 2.1, 2.2, and 2.3) existed in mainland China, and subgenotype 2.1b had become predominant within the last 10 years (2, 27). In 2014–2015, the new 2.1d isolates were epidemic in some districts of China (8, 30). All the 2.1d isolates were distributed over 11 provinces (Shandong, Jilin, Heilongjiang, Jiangsu, Hebei, Inner Mongolia, Henan, Hunan, Zhejiang, Shanghai, and Guangdong), and these areas together formed a defined region on the map of China (Fig. 3). In addition, the epidemic regions of 2.1d isolates seem to be grossly undetermined because most cases were not notified by farmers, and some 2.1d strains isolated by other laboratories were not reported in a timely manner. If all these new strains were isolated and reported, the epidemic areas would be larger and the trend of geographical cluster could be more obvious. Furthermore, most 2.1d isolates, together with some 2.1b isolates, were isolated simultaneously. The earliest subgenotype 2.1d strain, SX-04, was isolated in 2004. This indicated that the 2.1d isolates emerged more than 10 years ago, evolved continuously with subgenotype 2.1b isolates, and were epidemic until recently. Taken together, the presented results indicate that the 2.1d isolates may have diverged from 2.1b isolates.
It is well known that the C-strain, which belonged to subgenotype 1.1, was widely used in China. The vaccination may influence the divergence of CSFV through recombination or point mutation (9). In addition, positive selection pressure may act on the divergence of CSFV under C-strain vaccination, and several positively selected sites are found in E2 protein (8, 9, 21). Previous reports indicated that the surface structural proteins of CSFV of the vaccine-related groups contain more positive sites than other proteins of the vaccine-related groups and all proteins of the non-vaccine-related groups, suggesting that the difference was from immune selection (9, 11). In the present study, we did not find the evidence that the aa substitution of the new isolates was related to C-strain immune pressure. However, considering the fact that all clinical samples were collected from C-strain immunised pig farms, we speculated that the C-strain may promote the divergence and appearance of these new isolates. Of course, the internal evolution mechanism needs further research.
In summary, we analysed the phylogenetic relationship, genetic diversity, and epidemic status of the new 2014–2015 CSFV isolates. Most of these isolates belong to 2.1d and others belong to 2.1b. The 2.1d isolates and new 2.1b isolates showed unique molecular characteristics. The 2.1d isolates appeared more than 10 years ago, evolved continuously with subgenotype 2.1b isolates, and were epidemic until recently. We speculate that the 2.1d isolates might have diverged from 2.1b isolates. This comprehensive analysis may provide new insights into the prevention and control of CSF.
Footnotes
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: The research and the article were financed with the funds of the State Key Laboratory of Veterinary Biotechnology (No. SKLVBF201612), the National Natural Science Foundation of China (No. 31502097), the Key Programme Foundation of Higher Education of Educational Commission of Henan Province (No. 15A230026) and the Foundation of Nanyang Normal University (No. 15082).
Animal Rights Statement: The authors declare that the experiments on animals were conducted in accordance with local Ethical Committee laws and regulations as regards care and use of laboratory animals.
References
- 1.Bhaskar N., Ravishankar C., Rajasekhar R., Sumod K., Sumithra T.G., John K., Mini M., Ravindran R., Shaji S., Aishwarya J.. Molecular typing and phylogenetic analysis of classical swine fever virus isolates from Kerala, India. Virus Dis. 2015;26:260–266. doi: 10.1007/s13337-015-0271-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N., Hu H., Zhang Z., Shuai J., Jiang L., Fang W.. Genetic diversity of the envelope glycoprotein E2 of classical swine fever virus: recent isolates branched away from historical and vaccine strains. Vet Microbiol. 2008;127:286–299. doi: 10.1016/j.vetmic.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Chen N., Li D., Yuan X., Li X., Hu H., Zhu B., Wan X., Fang W.. Genetic characterization of E2 gene of classical swine fever virus by restriction fragment length polymorphism and phylogenetic analysis. Virus Genes. 2010;40:389–396. doi: 10.1007/s11262-010-0465-8. [DOI] [PubMed] [Google Scholar]
- 4.Deng M.C., Huang C.C., Huang T.S., Chang C.Y., Lin Y.J., Chien M.S., Jong M.H.. Phylogenetic analysis of classical swine fever virus isolated from Taiwan. Vet Microbiol. 2005;106:187–193. doi: 10.1016/j.vetmic.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Edwards S., Fukusho A., Lefevre P.C., Lipowski A., Pejsak Z., Roehe P., Westergaard J.. Classical swine fever: the global situation. Vet Microbiol. 2000;73:103–119. doi: 10.1016/s0378-1135(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 6.Gong W., Wu J., Lu Z., Zhang L., Qin S., Chen F., Peng Z., Wang Q., Ma L., Bai A., Guo H., Shi J., Tu C.. Genetic diversity of subgenotype 2.1 isolates of classical swine fever virus. Infect Genet Evol. 2016;41:218–226. doi: 10.1016/j.meegid.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 7.He C.Q., Ding N.Z., Chen J.G., Li Y.L.. Evidence of natural recombination in classical swine fever virus. Virus Res. 2007;126:179–185. doi: 10.1016/j.virusres.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Hu D., Lv L., Gu J., Chen T., Xiao Y., Liu S.. Genetic Diversity and Positive Selection Analysis of Classical Swine Fever Virus Envelope Protein Gene E2 in East China under C-Strain Vaccination. Front Microbiol. 2016;7:85. doi: 10.3389/fmicb.2016.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji W., Niu D.D., Si H.L., Ding N.Z., He C.Q.. Vaccination influences the evolution of classical swine fever virus. Infect Genet Evol. 2014;25:69–77. doi: 10.1016/j.meegid.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Jiang D.L., Gong W.J., Li R.C., Liu G.H., Hu Y.F., Ge M., Wang S.Q., Yu X.L., Tu C.. Phylogenetic analysis using E2 gene of classical swine fever virus reveals a new subgenotype in China. Infect Genet Evol. 2013;17:231–238. doi: 10.1016/j.meegid.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Leifer I., Ruggli N., Blome S.. Approaches to define the viral genetic basis of classical swine fever virus virulence. Virology. 2013;438:51–55. doi: 10.1016/j.virol.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C.. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo T.R., Liao S.H., Wu X.S., Feng L., Yuan Z.X., Li H., Liang J.J., Meng X.M., Zhang H.Y.. Phylogenetic analysis of the E2 gene of classical swine fever virus from the Guangxi Province of southern China. Virus genes. 2011;42:347–354. doi: 10.1007/s11262-011-0578-8. [DOI] [PubMed] [Google Scholar]
- 14.Luo Y., Li S., Sun Y., Qiu H.J.. Classical swine fever in China: a minireview. Vet Microbiol. 2014;172:1–6. doi: 10.1016/j.vetmic.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Lv C., Yang Q., Gao X., Yao Y., Li X., Xiao Y., Tian K.. Complete genome sequence of a classical swine fever virus isolate belonging to new subgenotype 2.1d from Henan Province, Central China. Genome Announc. 2016;4 doi: 10.1128/genomeA.00093-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyers G., Thiel H.J.. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 17.Moennig V., Plagemann P.G.. The pestiviruses. Adv Virus Res. 1992;41:53–98. doi: 10.1016/s0065-3527(08)60035-4. [DOI] [PubMed] [Google Scholar]
- 18.Moennig V., Floegel-Niesmann G., Greiser-Wilke I.. Clinical signs and epidemiology of classical swine fever: a review of new knowledge. Vet J. 2003;165:11–20. doi: 10.1016/s1090-0233(02)00112-0. [DOI] [PubMed] [Google Scholar]
- 19.Munoz-Gonzalez S., Perez-Simo M., Munoz M., Bohorquez J.A., Rosell R., Summerfield A., Domingo M., Ruggli N., Ganges L.. Efficacy of a live attenuated vaccine in classical swine fever virus postnatally persistently infected pigs. Vet Res. 2015;46:78. doi: 10.1186/s13567-015-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paton D.J., McGoldrick A., Greiser-Wilke I., Parchariyanon S., Song J.Y., Liou P.P., Stadejek T., Lowings J.P., Bjorklund H., Belak S.. Genetic typing of classical swine fever virus. Vet Microbiol. 2000;73:137–157. doi: 10.1016/s0378-1135(00)00141-3. [DOI] [PubMed] [Google Scholar]
- 21.Perez L.J., Diaz de Arce H., Perera C.L., Rosell R., Frias M.T., Percedo M.I., Tarradas J., Dominguez P., Nunez JI., Ganges L.. Positive selection pressure on the B/C domains of the E2-gene of classical swine fever virus in endemic areas under C-strain vaccination. Infect Genet Evol. 2012;12:1405–1412. doi: 10.1016/j.meegid.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 22.Postel A., Schmeiser S., Bernau J., Meindl-Boehmer A., Pridotkas G., Dirbakova Z., Mojzis M., Becher P.. Improved strategy for phylogenetic analysis of classical swine fever virus based on full-length E2 encoding sequences. Vet Res. 2012;43:50. doi: 10.1186/1297-9716-43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postel A., Schmeiser S., Perera C.L., Rodriguez L.J., Frias-Lepoureau M.T., Becher P.. Classical swine fever virus isolates from Cuba form a new subgenotype 1.4. Vet Microbiol. 2013;161:334–338. doi: 10.1016/j.vetmic.2012.07.045. [DOI] [PubMed] [Google Scholar]
- 24.Sun S.Q., Yin S.H., Guo H.C., Jin Y., Shang Y.J., Liu X.T.. Genetic typing of classical swine fever virus isolates from China. Transbound Emerg Dis. 2013;60:370–375. doi: 10.1111/j.1865-1682.2012.01346.x. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu C., Lu Z., Li H., Yu X., Liu X., Li Y., Zhang H., Yin Z.. Phylogenetic comparison of classical swine fever virus in China. Virus Res. 2001;81:29–37. doi: 10.1016/s0168-1702(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 28.van Rijn P.A., Miedema G.K., Wensvoort G., van Gennip H.G., Moormann R.J.. Antigenic structure of envelope glycoprotein E1 of hog cholera virus. J Virol. 1994;68:3934–3942. doi: 10.1128/jvi.68.6.3934-3942.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber M.N., Streck A.F., Silveira S., Mosena A.C., Silva M.S., Canal C.W.. Homologous recombination in pestiviruses: identification of three putative novel events between different subtypes/genogroups. Infect Genet Evol. 2015;30:219–224. doi: 10.1016/j.meegid.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Leng C., Feng L., Zhai H., Chen J., Liu C., Bai Y., Ye C., Peng J., An T., Kan Y., Cai X., Tian Z., Tong G.. A new subgenotype 2.1d isolates of classical swine fever virus in China. Infect Genet Evol. 2014;2015;34:94–105. doi: 10.1016/j.meegid.2015.05.031. [DOI] [PubMed] [Google Scholar]