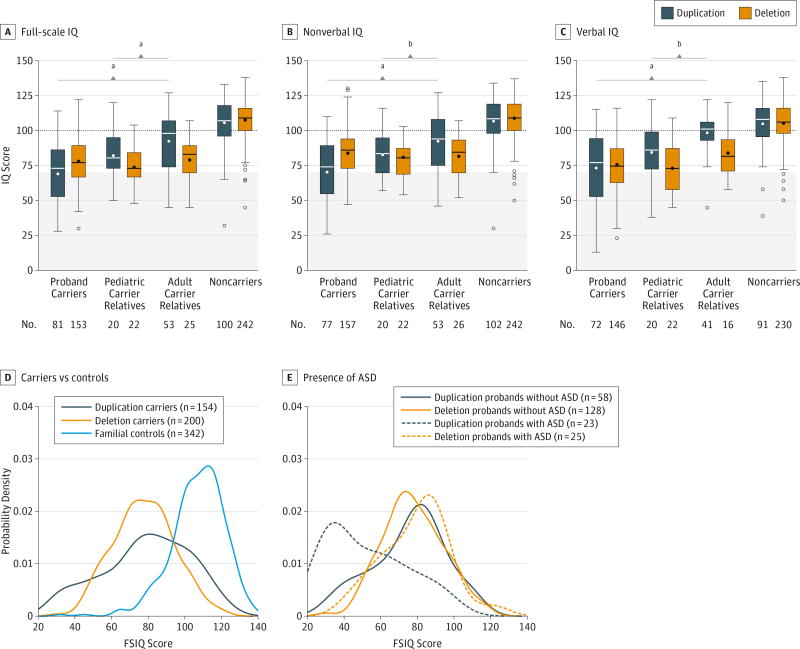
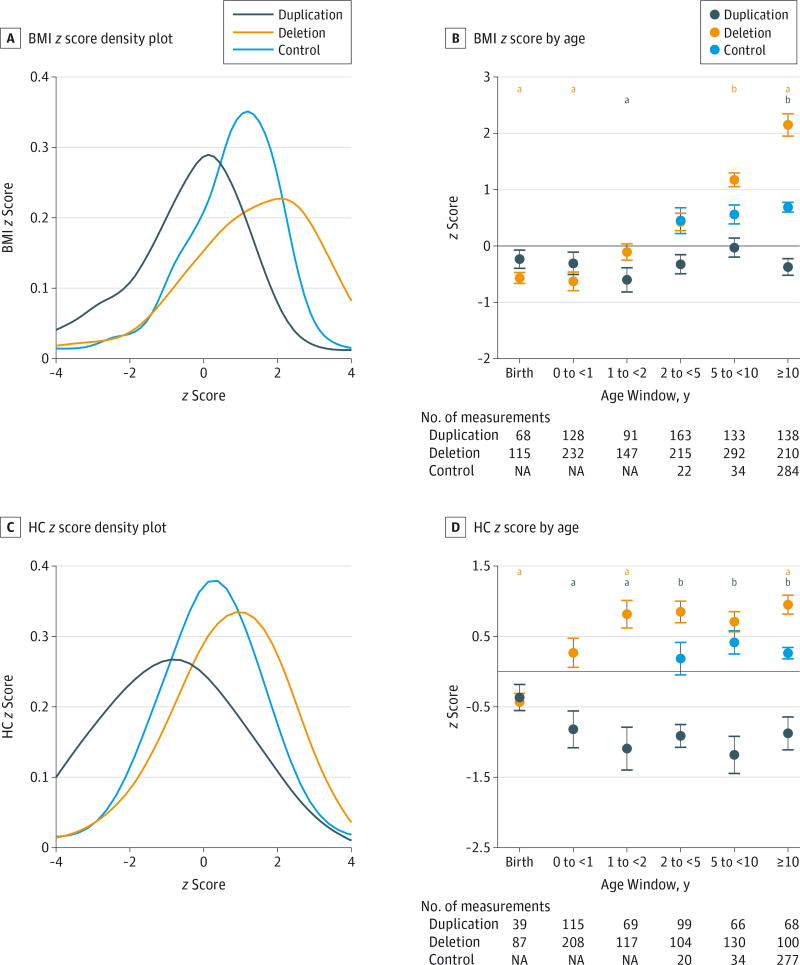
Debra D’Angelo
Debra D’Angelo, MS
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Sébastien Lebon
Sébastien Lebon, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Qixuan Chen
Qixuan Chen, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Sandra Martin-Brevet
Sandra Martin-Brevet, MS
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
LeeAnne Green Snyder
LeeAnne Green Snyder, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Loyse Hippolyte
Loyse Hippolyte, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Ellen Hanson
Ellen Hanson, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Anne M Maillard
Anne M Maillard, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
W Andrew Faucett
W Andrew Faucett, MS
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Aurélien Macé
Aurélien Macé, MS
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Aurélie Pain
Aurélie Pain, MS
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Raphael Bernier
Raphael Bernier, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Samuel J R A Chawner
Samuel J R A Chawner, MA
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Albert David
Albert David, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Joris Andrieux
Joris Andrieux, MD, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Elizabeth Aylward
Elizabeth Aylward, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Genevieve Baujat
Genevieve Baujat, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Ines Caldeira
Ines Caldeira, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Philippe Conus
Philippe Conus, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Carrina Ferrari
Carrina Ferrari, MS
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Francesca Forzano
Francesca Forzano, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Marion Gérard
Marion Gérard, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Robin P Goin-Kochel
Robin P Goin-Kochel, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Ellen Grant
Ellen Grant, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Jill V Hunter
Jill V Hunter, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Bertrand Isidor
Bertrand Isidor, MD, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Aurélia Jacquette
Aurélia Jacquette, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Aia E Jønch
Aia E Jønch, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Boris Keren
Boris Keren, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Didier Lacombe
Didier Lacombe, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Cédric Le Caignec
Cédric Le Caignec, MD, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Christa Lese Martin
Christa Lese Martin, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Katrin Männik
Katrin Männik, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Andres Metspalu
Andres Metspalu, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Cyril Mignot
Cyril Mignot, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Pratik Mukherjee
Pratik Mukherjee, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Michael J Owen
Michael J Owen, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Marzia Passeggeri
Marzia Passeggeri, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Caroline Rooryck-Thambo
Caroline Rooryck-Thambo, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Jill A Rosenfeld
Jill A Rosenfeld, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Sarah J Spence
Sarah J Spence, MD, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Kyle J Steinman
Kyle J Steinman, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Jennifer Tjernagel
Jennifer Tjernagel, MS
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Mieke Van Haelst
Mieke Van Haelst, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Yiping Shen
Yiping Shen, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Bogdan Draganski
Bogdan Draganski, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Elliott H Sherr
Elliott H Sherr, MD, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
David H Ledbetter
David H Ledbetter, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Marianne B M van den Bree
Marianne B M van den Bree, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Jacques S Beckmann
Jacques S Beckmann, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
John E Spiro
John E Spiro, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Alexandre Reymond
Alexandre Reymond, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Sébastien Jacquemont
Sébastien Jacquemont, MD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1,
Wendy K Chung
Wendy K Chung, MD, PhD
1Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York (D’Angelo, Chen); Pediatric Neurology Unit, Department of Pediatrics, Lausanne University Hospital, Lausanne, Switzerland (Lebon); Department of Medical Genetics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland (Martin-Brevet, Hippolyte, Maillard, Macé, Pain, Caldeira, Jønch, Passeggeri, Beckmann, Jacquemont); Clinical Research Associates, New York, New York (Snyder, Tjernagel); Department of Psychiatry, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Hanson); Genomic Medicine Institute, Geisinger Clinic, Danville, Pennsylvania (Faucett); Swiss Institute of Bioinformatics, University of Lausanne, Lausanne, Switzerland (Macé, Beckmann); Department of Psychiatry and Behavioral Science, University of Washington, Seattle (Bernier); Medical Research Council Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, Wales (Chawner, Owen, van den Bree); Service de Génétique Médicale, Faculté de Médecine, Centre Hospitalier Universitaire (CHU) Nantes, Institut National de la Santé et de la Recherche Medicale (INSERM) Unités Mixtes de Recherche 957, Nantes, France (David, Isidor, Lacombe, Le Caignec); Institut de Génétique Médicale, Hospital Jeanne de Flandre, Centre Hospitalier Régional Universitaire (CHRU) de Lille, Lille, France (Andrieux); Center for Integrative Brain Research, Children’s Research Institute, Seattle, Washington (Aylward); Département de Génétique, Hôpital Necker– Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), Paris, France (Baujat); INSERM U1163, Hôpital Necker–Enfants Malades, Paris, France (Baujat); Institut Imagine, Université Paris Descartes-Sorbonne Paris Cité, Paris, France (Baujat); Department of Psychiatry, Cery Hospital, CHU Vaudois and University of Lausanne, Lausanne, Switzerland (Conus, Ferrari); Division of Medical Genetics, Galliera Hospital, Genova, Italy (Forzano); Departement de Génétique, AP-HP, Hôpital Robert Debré, Université Paris VII-Paris Diderot, Paris, France (Gérard); Section of Psychology, Department of Pediatrics, Baylor College of Medicine, Houston, Texas (Goin-Kochel); Department of Radiology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Grant, Mignot); Department of Radiology, Baylor College of Medicine, Houston, Texas (Hunter); Département de Génétique et de Cytogénétique, Unité fonctionnelle de Génétique Clinique, AP-HP, Hôpital Pitié-Salpêtrière, Paris, France (Jacquette); Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France (Jacquette); Groupe de Recherche Clinique, Déficience Intellectuelle et Autisme, Université Pierre-et-Marie-Curie, Paris, France (Jacquette); Department of Genetics and Cytogenetics, Groupe Hospitalier Pitié Salpêtrière, AP-HP, Paris, France (Keren); Service de Génétique Médicale, CHU de Bordeaux, Bordeaux, France (Lacombe, Rooryck-Thambo); Autism and Developmental Medicine Institute, Geisinger Health System, Danville, Pennsylvania (Martin, Ledbetter); Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland (Männik, Reymond); Estonian Genome Center, University of Tartu, Tartu, Estonia (Männik, Metspalu); Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Metspalu); Department of Radiology and Biomedical Imaging, University of California, San Francisco (Mukherjee); Laboratoire Maladies Rares: Génétique et Métabolisme, Université de Bordeaux, Bordeaux, France (Rooryck-Thambo); Signature Genomic Laboratories, LLC, PerkinElmer, Inc, Spokane, (Rosenfeld); Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts (Spence); Department of Neurology, Seattle Children’s Research Institute and University of Washington, Seattle (Steinman); Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, the Netherlands (Van Haelst); Genetic Diagnostic Laboratory, Department of Laboratory Medicine, Children’s Hospital, Boston, Massachusetts (Shen); Laboratoire de Recherche en Neuroimagerie, Department for Clinical Neurosciences, Centre Hospitalo-Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (Draganski); Department of Neurology, University of California, San Francisco (Sherr); Simons Foundation, New York, New York (Spiro); CHU Sainte-Justine Research Center, Montreal, Canada (Jacquemont); Department of Pediatrics, Université de Montréal, Montreal, Quebec, Canada (Jacquemont); Division of Molecular Genetics, Department of Pediatrics, Columbia University, New York, New York (Chung); Department of Medicine, Columbia University, New York, New York (Chung)
1