Abstract
Mammalian male germ cell development takes place in the testis under the influence of a variety of somatic cells and an incompletely defined paracrine and endocrine influences. Since it is not recapitulated well in vitro, researchers studying spermatogenesis often manipulate the germline by creating transgenic or knockout mice or by administering pharmaceutical agonists/antagonists or inhibitors. The effects of these types of manipulations on germline development can often be determined following microscopic imaging, both of stained and immunostained testis sections. Here, we describe approaches for microscopic analysis of the developing male germline, provide detailed protocols for a variety of immunostaining approaches, and discuss transgenic fluorescent reporter lines for studying the early stages of spermatogenesis.
Keywords: Prospermatogonia, Spermatogonia, Testis, Immunofluorescence, Immunohistochemistry
1. Development of the male mouse germline
The male germline is established in mice as primordial germ cells (PGCs) that colonized the fetal testis at ~embryonic day (E)10.5 become prospermatogonia (also termed gonocytes) following sex determination at ~E11 (DiNapoli and Capel, 2008; McLaren, 2003). Over the next several days, prospermatogonia proliferate and become surrounded by somatic Sertoli cells to form the nascent testis, or seminiferous cords. By ~E15.5, fetal prospermatogonia stop dividing and become mitotically arrested in G0 of the cell cycle until after birth (Vergouwen et al., 1991; Western et al., 2008). At postnatal days (P)1–2, prospermatogonia resume mitosis and transition into type A spermatogonia. This initial spermatogonial population is heterogeneous as early as P3, and can be characterized as either undifferentiated (Aundiff) or differentiating (Adiff) (Kluin et al., 1984; Niedenberger et al., 2015; Yoshida et al., 2006). This heterogeneity is evident in neonatal spermatogonia based on differences in morphology (Drumond et al., 2011; Kluin and de Rooij, 1981), abundance of specific mRNAs (Yoshida et al., 2004, 2006; Hermann et al., 2015), expression of protein fate markers (Niedenberger et al., 2015; Hermann et al., 2015; Busada et al., 2014, 2015), and the ability to seed spermatogenesis in recipient testes following transplantation (McLean et al., 2003). Over the next few days the numbers of spermatogonia continue to increase, resulting in formation of the foundational pool of spermatogonial stem cells (SSCs), undifferentiated progenitors that are poised to differentiate, and STRA8+/KIT+ differentiating spermatogonia (Niedenberger et al., 2015; Busada and Geyer, 2015; Yang and Oatley, 2014).
Timing during the progression of spermatogenesis is remarkably precise, such that specific cell types appear on predictable days during the ‘first round of spermatogenesis’ that begins in the neonatal mouse testis at ~P3, with some strain variation. Type A spermatogonia initiate the differentiation program as A1 spermatogonia form in response to all-trans retinoic acid (ATRA, reviewed in (Busada and Geyer, 2015; Griswold, 2016)). Following several divisions (A2–4, In, B), male germ cells enter meiosis as preleptotene spermatocytes as early as ~P8, and successively become leptotene (~P10), zygotene (~P12), and pachytene spermatocytes (~P14). The first haploid round spermatids are formed by ~P20, which then undergo dramatic morphogenetic changes during spermiogenesis to form condensed spermatids, which are released from the seminiferous epithelium as testicular sperm by ~P35 (Oakberg, 1956; Clermont and Trott, 1969; Bellve et al., 1977). Based on this defined temporal appearance of specific identifiable germ cell subtypes, the developing testis provides an excellent model system to study the progression of spermatogenesis.
Researchers can use mice to study the physiologic roles of various gene products, signaling pathways, and environmental influences on early germ cell development in vivo by generating transgenic and knockout models and by treating mice with hormones, agonists/antagonists, inhibitors, and various toxicants. However, there are small numbers of germ cells in the developing testis, and it is difficult to isolate them with high levels of purity. Therefore, brightfield and fluorescent-based microscopy provide the best means to determine the outcomes of these manipulations on the various phases of germline development. In this article, we will describe current methods and tools for imaging and immunostaining prospermatogonia and spermatogonia, review transgenic models for germ cell imaging, and discuss needs that we feel should be addressed by researchers in the field in the future.
2. Harvesting and preparing fetal and neonatal testes for imaging
The removal of fetal and neonatal mouse testes must be done carefully using fine tip forceps and micro-dissection spring scissors. The attached epididymis serves as a convenient means to grasp the tissue while avoiding nicking or crushing delicate testicular tissue. A stereoscope is not required, but can be useful for dissecting fetal testes and for removing epididymal tissue as well as the overlying connective tunica vaginalis and albuginea. Testes can then be used for a variety of downstream applications such as isolation of total testicular DNA, RNA, or protein, germ or somatic cell isolations, or fixation for various imaging modalities, as described below.
2.1. Fixation for imaging
Light microscopy has been used in numerous studies to carefully characterize the various stages of male germ cell development, and specific morphologic criteria have been assigned to germ cell types at each stage of their development. These include characteristic positioning within the seminiferous epithelium as well as differences in nuclear diameter and chromatin appearance (Kluin et al., 1984; Drumond et al., 2011; Kluin and de Rooij, 1981; Chiarini-Garcia and Russell, 2001). However, these accurate determinations are only feasible in sections from samples that have been properly prepared. Testicular morphology is best maintained for light microscopy following thorough fixation, and there are many different types of fixatives that have been used in the literature. The most commonly used fixative for histological analysis is Bouin’s solution, which contains paraformaldehyde, picric acid, and acetic acid. The acetic acid component causes characteristic condensation of nuclear chromatin, resulting in subtle differences that can be used to define different germ cell types. Bouin’s solution rapidly and thoroughly penetrates tissue, and for mouse testes immersion fixation for ≤24 h is sufficient (P0–4 ≈ 2 h, P5–12 ≈ 4–6 h, P13-adult ≈ 12–24 h). Following fixation, the tissue must be thoroughly washed in 1X PBS to remove as much of the picric acid as possible prior to routine paraffin embedding, sectioning, and staining.
While fixatives such as Bouin’s solution preserve tissue organization and cellular structure exceedingly well, they often do a rather poor job of retaining epitopes for subsequent antibody-based immunohistochemical analyses. In addition, picric acid autofluoresces, which makes Bouin’s-fixed samples generally unsuitable for fluorescence-based immunostaining. If immunostaining is the desired goal, then one testis (or a portion of one testis) should be immersion-fixed in 4% PFA (see supplemental file, protocol 1) using the incubation times outlined above for Bouin’s fixation. Following fixation in 4% PFA, testes washed in 1X PBS and then either dehydrated in ethanol for paraffin embedding or incubated in sucrose prior to cryosectioning.
2.2. Seminiferous cord and tubule whole mount preparation
Testis sections provide a limited two-dimensional view of germ cells within the testis cords, either in longitudinal or cross-sections, and spatial organization within the cords is often lost. Therefore, testis whole (“in toto”) mounts are useful to examine relationships between adjacent germ cells and assess the length of interconnected chains of spermatogonia. These are prepared by first detunicating testes and carefully cutting them into thirds, and testis cords can be gently teased apart using forceps. These pieces are then permeabilized prior to immunostaining; we follow a standard immunostaining protocol (see supplemental file, protocol 2), with the caveat that all incubation times are increased to facilitate their penetration into interior cells within this thicker tissue (see supplemental file, protocol 3).
3. Immunostaining
Specific proteins can be detected within isolated cells or a tissue using a variety of immunostaining approaches. These provide a relative comparison of steady-state protein abundance, determination of subcellular localization, and identification of which cell type(s) express that protein. This technique is dependent upon a specific antibody-antigen interaction, and specificity should be verified by using appropriate negative controls such as no primary antibody control, pre-binding the antibody to the immunizing peptide or recombinant protein (if available), and verifying that antibody recognizes a single band on western blot in a lysate from the tissue of interest. Most applications employ indirect immunostaining, in which a primary antibody is applied to tissue followed by incubation with a secondary antibody that is raised against the host species of the primary antibody and is conjugated to a fluorophore or other molecule enabling detection.
3.1. Immunohistochemistry (IHC) on paraffin sections and indirect immunofluorescence (IIF) on cryosections
Each of the staining techniques (IHC and IIF) has distinct advantages and disadvantages. Paraffin-embedded tissues and labeled slides can be stored at room temperature indefinitely, and enormous numbers of formaldehyde-fixed human normal and diseased samples are available through the pathology departments of most hospitals for retrospective analysis. An advantage of IHC on paraffin-embedded sections is that cellular morphology is well-maintained. However, epitopes are often lost in these samples (usually causing reduced sensitivity), and the use of 3,3′-diaminobenzidine (DAB)-based chromogen labeling makes co-labeling proteins with antibodies from different hosts extremely difficult, and these images cannot be combined to provide a 3-dimensional Z-stack (as with confocal imaging). IIF on fixed cryosections does have the advantage of increased sensitivity, but sections are less amenable to histological analysis and careful characterization of nuclear morphology, which has proven quite useful for discriminating between different spermatogonial subtypes (Drumond et al., 2011; Kluin and de Rooij, 1981; Chiarini-Garcia and Russell, 2001). We have optimized reliable protocols for DAB-based IHC on Bouin’s- and PFA-fixed paraffin-embedded testis sections (see supplemental file, protocol 4) as well as IIF on PFA-fixed cryosections (see supplemental file, protocol 2).
3.2. Co-immunostaining with IIF
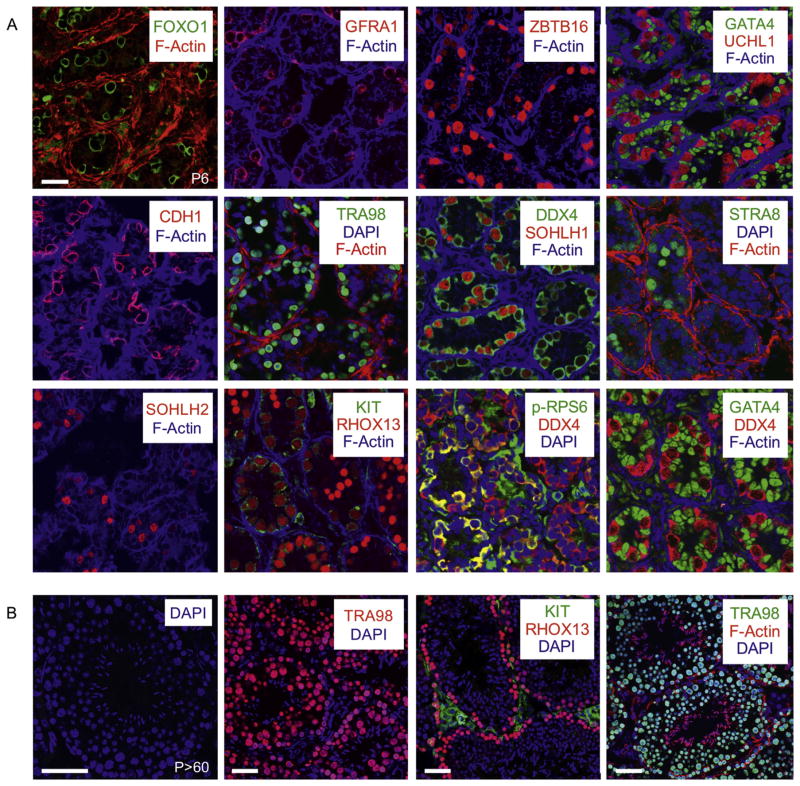
Simultaneous staining for up to three different proteins provides distinct advantages for studying spermatogenesis, as it allows researchers to mark all germ cells (using DDX4 or TRA98, for example), identify specific spermatogonial fate (e.g. undifferentiated spermatogonia with GFRA1 or differentiating spermatogonia with KIT) and then detect a third protein of interest. This allows for assignment of novel protein expression in specific cell types as well as straightforward analysis of changes in the germ cell populations in transgenic, knockout, or chemically- or hormonally-treated animals. A wide variety of excellent specific antibodies are available through commercial vendors or from academic and government research laboratories for the detection of testicular germ and somatic cell types in both PFA-fixed cryosections (Fig. 1) as well as paraffin-embedded sections of the mammalian testis (see Fig. 2). We favor using cryosections in most cases, because in our experience they have better epitope retention.
Fig. 1.
Immunostaining PFA-fixed cryosections from neonatal and adult testes. (A) IIF was performed on cryosections from P6 testes, and specific primary antibodies used for each experiment are identified on each panel, with the colour of the text corresponding to the fluorescent secondary antibody employed. F-actin is labeled in all images using fluorescently-conjugated phalloidin. Antibodies used were: FOXO1 (Cell Signaling Technology, #2880), GFRA1 (R&D Systems, AF560), ZBTB16 (Santa Cruz Biotechnology, sc-22839), GATA4 (Santa Cruz Biotechnology, sc-1237), UCHL1 (Cell Signaling Technology, #D3T2E), CDH1 (Cell Signaling Technology, #3195), TRA98 (Abcam, ab82527), DDX4 (Abcam, ab13840), SOHLH1 (Pangas et al., 2006), STRA8 (Abcam, ab49602), SOHLH2 (Ballow et al., 2006), KIT (Santa Cruz Biotechnology, sc-1494), RHOX13 (Geyer and Eddy, 2008), phospho-RPS6 (Cell Signaling Technology, #5364). (B) IIF was performed on cryosections from P > 60 testes, and specific antibodies are indicated on each panel. Scale bars = 60 μm.
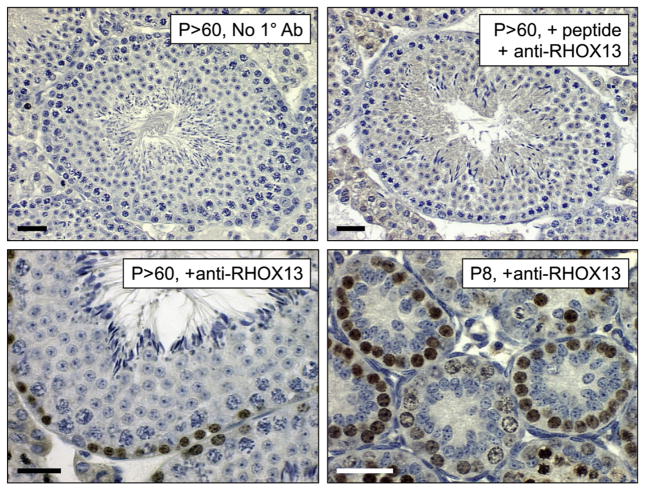
Fig. 2.
Immunostaining Bouin’s-fixed paraffin sections from neonatal and adult testes. IHC was performed on Bouin’s-fixed paraffin sections using anti-RHOX13 (Geyer and Eddy, 2008) without HIER. Ages are indicated on each image. The top row contains control images (no primary antibody in top left, and pre-incubation of primary antibody with the immunizing peptide used for its generation for 30 min at RT on the top right). Scale bars = 60 μm.
To perform co-immunostaining, primary antibodies must be chosen that are generated in distinct host species. These primary antibodies are incubated together either simultaneously or sequentially on testis sections. Following stringency washes, fluorescently-conjugated secondary IgG H + L antibodies are applied to the sections. It is important to choose secondary antibodies that will not interact with one another; goat anti-rabbit and donkey anti-goat secondary antibodies would not work together because the donkey anti-goat would recognize the goat anti-rabbit as a goat protein and bind nonspecifically. This issue can be avoided by first incubating the section with the donkey anti-goat secondary antibody, then washing the sections, and then adding the goat anti-rabbit secondary. Secondary antibodies must be fluorescently-conjugated to fluorophores that are spectrally distinct (fluorescence excitation and emission spectra do not overlap) to be distinguishable from one another. We usually avoid using secondary antibodies conjugated to blue fluorophores because they are less photostable, and generally do not result in bright images. For detailed protocols, see supplemental file, protocols 2–3).
3.3. Counterstaining samples for IIF
Testis sections are mounted with an aqueous mounting medium containing photobleaching inhibitors as well as DAPI (4′,6-diamidino-2-phenylindole), which labels nuclei (Kapuscinski and Szer, 1979). DAPI labeling allows for determination of the size, shape, position, and appearance of the nucleus (such as bright vs. dim). This can be used, for example, to easily distinguish prospermatogonia and spermatogonia (large round pale-staining nuclei) from somatic Sertoli cells (smaller ovoid nuclei with distinct bright spots of heterochromatin) within neonatal testis cords.
We often counterstain testis sections with phalloidin that has been fluorescently-conjugated to a far-red fluorophore (e.g. Alexa Fluor-633, Invitrogen) so that it does not interfere with the green and red confocal channels, allowing for those to be used for co-immunostaining with separate antibodies. Phalloidin binds with high affinity to filamentous (F)-actin, which is particularly abundant in peritubular myoid cells in the testis (Tung and Fritz, 1990; Losinno et al., 2012). This staining essentially outlines the testis cords in the neonate and seminiferous tubules in the adult, allowing for diameter measurement facile determination of the position of cells (inside the cords = Sertoli and germ cells, periphery of the cords = peritubular myoid cells, and outside the cords = interstitial cells such as Leydig cells, endothelial cells, and macrophages).
3.4. Fluorescent conjugation of primary antibodies
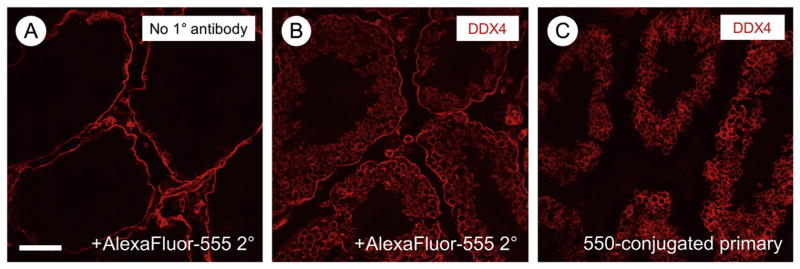
There are many excellent mouse monoclonal antibodies that could be used on mouse tissue sections. However, application of an anti-mouse secondary antibody results in an intense non-specific staining of cells within the testicular interstitium (see Fig. 3A). One way to circumvent this issue is to fluorescently conjugate the primary antibody. We have had the most success labeling 10 μg of primary antibody using the Dylight Fast Conjugation Kits, which allow the addition of a wide variety of fluorescently-colored fluorophores (Abcam, see Fig. 3). One caveat is that signals from directly-conjugated primary antibodies are often fainter, requiring higher concentrations (5–10×) than those used for conventional IIF.
Fig. 3.
Immunostaining results using directly-conjugated mouse monoclonal primary antibody against DDX4. (A) The primary antibody was omitted as a negative control, and the red signal from use of the anti-mouse secondary antibody is visible surrounding the seminiferous tubules and in cells in the interstitium. (B) IIF was performed by separately incubating primary and secondary antibodies. (C) Direct fluorescent labeling of the primary mouse monoclonal antibody results in loss of the nonspecific staining seen in A. Scale bar (in A) = 80 μm.
3.5. Optimizing and troubleshooting immunostaining
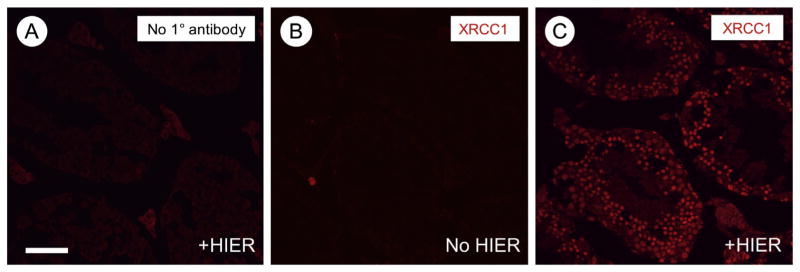
Although similar standard immunostaining protocols are employed in all IHC and IIF techniques, there is often a need for optimization of new antibodies as well as new batches of the same antibody. We have found that the most important variable affecting staining results is the concentration of primary antibody. Therefore, we routinely put 4 tissue sections on each slide, and employ a range of primary antibody dilutions from 1:100 to 1:1000 for incubation at room temperature for 1 h or overnight at 4 °C. If adjusting the primary antibody concentration and incubation time yield no specific signal, heat-induced epitope retrieval (HIER) can be performed in citrate buffer, even on cryosections (see supplemental file, protocol 5). HIER can sometimes restore epitopes that were masked following fixation-induced crosslinking. One example of an antibody that does not work well for IIF without HIER is provided in Fig. 4 (anti-XRCC1, Abcam ab134056).
Fig. 4.
Immunostaining following heat-induced epitope retrieval (HIER). (A) Primary antibody was omitted as a negative control. (B–C) Anti-XRCC1 was detected without (B) and following (C) HIER. Signals are in red. Scale bar (in A) = 80 μm.
3.6. Quantifying germ cells in tissue sections
IIF can be used to assess changes in specific testicular cell populations in genetically-modified mice or following treatments with chemicals or hormones. This can be accomplished in the germ cell population by staining for a pan germ cell marker along with the protein of interest, and the percentage of positive cells can be easily calculated provided enough sections are counted. If the protein of interest is localized to the nucleus, then co-immunostaining should be done for a cytoplasmic fetal and neonatal pan germ cell marker such as DDX4 or DAZL, while if it is cytoplasmic, co-immunostaining should be done for a nuclear pan germ cell marker such as TRA98.
It can be challenging to determine whether a cell should be marked ‘positive’ or ‘negative’ for protein markers in stained sections. To accomplish this, we utilize the threshold tool in Image J software program (U.S. National Institutes of Health, Bethesda, MD, USA). Fluorescent channels must be separated to determine thresholds. We use the threshold tool with the default setting, and then set the lower slider to 255. We determine the threshold for each antibody on a case-by-case basis, and adjust position of the upper slider accordingly. For example, for a protein that is localized to the nucleus, the upper slider is adjusted so that only nuclei are highlighted. It is important to capture all images for each protein using the same microscope settings so that the same threshold settings in ImageJ may be used for each antibody (see reference (Niedenberger et al., 2015) for example thresholds). At least three non-overlapping images are taken, and all germ cells on the images are counted, which for quantitation purposes are defined as cells staining positive for either TRA98 or DDX4. Germ cells that are positive for an additional protein of interest are then counted, and results are presented as the percentage of germ cells that are positive for the protein of interest in the total population.
4. Transgenic mouse models with fluorescent germ cells
Germ cells make up a comparatively small proportion of the overall cell population in the fetal and neonatal testis; the majority are somatic cells such as fibroblasts, Sertoli and Leydig cells, macrophages, peritubular myoid cells, endothelial cells, and leukocytes. A variety of biochemical assays and molecular analyses can be performed on germ cells, but this necessitates their isolation from whole testis. Traditionally, prospermatogonia and spermatogonia have been isolated by taking advantage of their large diameter relative to somatic cells as well as their relative inability to adhere to plastic. Mechanical and enzymatic disruption of testes into single cell suspensions were often followed by short-term incubation in plastic tissue culture dishes. Many somatic cell types would adhere to these dishes, and the non-adherent germ cells would then be separated by the Sta-Put assay, which employs gravity sedimentation through a 2–4% bovine serum albumin (BSA) gradient (Bellve et al., 1977; Bryant et al., 2013). This can be used effectively isolate germ cell populations that are ~80–90% pure, depending on the age of mice used and cell type(s) desired (Geyer et al., 2004; Shima et al., 2004). However, it is a lengthy procedure that requires an apparatus with specialized glassware, a significant amount of optimization, and the requisite expertise to properly identify the resultant isolated cell populations using phase contrast microscopy. Therefore, this approach is not a viable option for most laboratories wishing to study isolated germ cells.
Fluorescence-activated cell sorting provides a much more tractable approach to isolate germ cells, and can be done using antibodies against cell surface markers or using transgenic mice expressing fluorescent reporter proteins. For the latter approach, there are two types of transgen-ic models with fluorescently-labeled germ cells: constitutive and inducible. In the constitutively-expressing models, a promoter segment from a gene with germ cell-restricted expression (at least within the testis) is used to direct the expression of green or red fluorescent reporter proteins such as GFP, mCherry, or tdTomato. Numerous lines of transgenic mice that have been created in this way, and theoretically can be useful for the isolation of male germ cells at various phases of their development. Unfortunately, most are not readily available as certified pathogen-free strains through the Jackson Laboratory, and few have been shown to reliably work well in multiple laboratories over time. Inducible transgenic models generally rely on germ cell-specific expression (again, at least in the testis) of Cre recombinase to delete a DNA sequence preventing fluorescent reporter gene expression. Specific examples of both constitutive and inducible models are described below.
4.1. Transgenic models with constitutive fluorescent reporter expression
Several models have been generated with fluorescently-labeled PGCs, and later, spermatogonia in the neonatal testis. Pou5f1/Oct4-Gfp transgenic lines are in existence ((Ohbo et al., 2003; Porro et al., 2015; Szabo et al., 2002; Yoshimizu et al., 1999), reviewed in (Garcia and Hofmann, 2012)), and at least one may be particularly useful for the isolation of nearly homogeneous populations of PGCs (Szabo et al., 2002). Other transgenic lines that express GFP in PGCs include Prdm1/Blimp1-Gfp (Ohinata et al., 2005), Dppa3/Stella-Gfp (Payer et al., 2006), and Ifitm3/fragilis-Gfp (Tanaka et al., 2004).
Several transgenic lines have been created to fluorescently label spermatogonia, although each line is expressed in only a subset of spermatogonia. In the first example, the Oatley laboratory used the ‘inhibitor of DNA binding gene 4’ (Id4) promoter to direct expression of enhanced GFP (EGFP) in a subset of neonatal prospermatogonia and spermatogonia, and the intensity level is be linked to spermatogonial fate by P6, with ID4-EGFPbright SSCs and ID4-EGFPdim progenitor/differentiating spermatogonia (see whole mount image, Fig. 5) (Hermann et al., 2015; Chan et al., 2014; Helsel et al., 2017). Another laboratory created a transgenic line in which the tdTomato reporter gene was inserted into the 3′ UTR of Id4, and they reported similar results (Sun et al., 2015). Neurog3/Ngn3-eGfp mice exhibit EGFP in a heterogeneous subset of postnatal Aundiff spermatogonia that likely represent undifferentiated progenitors (Yoshida et al., 2004, 2007; Zheng et al., 2009). In the Pou5f1/Oct4-Gfp mice created by the Mann laboratory ((Szabo et al., 2002), JAX strain #004654) and Stra8-eGfp mice (Nayernia et al., 2004), reporter gene expression occurred in a poorly-defined subset of neonatal spermatogonia. In Sohlh1-mCherry-FLAG mice, red fluorescence was detectable in a subset of fetal and neonatal prospermatogonia as well as neonatal spermatogonia in a pattern that resembled but did not fully recapitulate endogenous SOHLH1 expression (Suzuki et al., 2013). Finally, in Dazl-eGfp mice, EGFP expression did not faithfully recapitulate the expression profile of the endogenous DAZL protein in prospermatogonia and spermatogonia, but was instead present in a subset of pachytene spermatocytes and spermatids (Nicholas et al., 2009). In summary, only the Id4-eGfp and Neurog3/Ngn3-eGfp mouse lines are currently in widespread use for studying spermatogonial development.
Fig. 5.
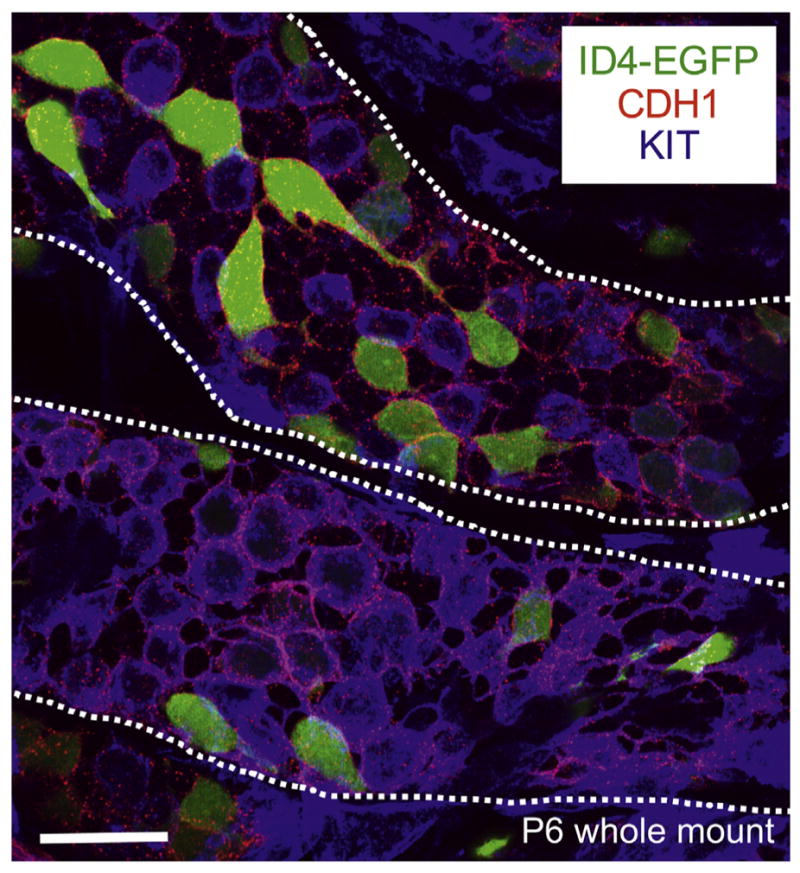
Whole-mount immunostaining of P6 Id4-eGfp testis cords. Maximum intensity Z-stack projection of isolated testis cords from transgenic Id4-eGfp P6 mice (ID4-EGFP epifluorescence in green). Antibody staining was performed for the undifferentiated marker CDH1 (in red) and the differentiating marker KIT (in blue). Cords are outlined with white dashed lines. Scale bar = 25 μm.
4.2. Induced fluorescent reporter expression
A second means of generating fluorescent germ cells is by germ cell Cre recombinase-activated expression of silent fluorescent reporter genes in transgenic mice. The most commonly used models harbor a transgene in the ROSA26 locus in which a lox-STOP-lox cassette lies between a strong promoter and the fluorescent reporter coding sequence. The use of different Cre-recombinase-expressing strains allows researchers to control the cell type(s) that will become fluorescent, and multiple variants are available from the Jackson laboratory. A significant drawback inherent to these models is that when researchers cross 2 lines of hemizygous mice [Gt(ROSA)26Sor and the germ cell-expressing Cre recombinase], only 1/8 of progeny will be male and have both transgenes. This makes these mice rather impractical for many experiments, as there are relatively low numbers of germ cells in the neonatal testis. In addition, this approach requires a robust and reliable Cre-expressing line; unfortunately, few exist that work well in spermatogenesis. Currently, the best Cre-expressing line in prospermatogonia and spermatogonia is Ddx4/Vasa/Mvh-Cre, which is activated in >95% of fetal prospermatogonia as early as E15 ((Gallardo et al., 2007), Jackson Laboratory #006954). This results in deletion of floxed alleles during a quiescent phase of prospermatogonial development, and can be useful for assessing the reproductive phenotype of postnatal germ cell KO mice. One caveat to using this line that has not been communicated well in the literature is that it only works reliably when the Cre transgene is donated by young male breeders <2–3 months of age; as the males age, there is an increase in Cre activation in other tissues, which can lead to lethality (personal experience and personal communications with other spermatogenesis researchers). There is a tamoxifen-inducible version of the Ddx4/Vasa-Cre mice (John et al., 2008), but these have not been cited in many recent publications (Jackson Laboratory, #024760, cryopreserved).
Other Cre-expressing lines active in subsets of postnatal spermatogonia include Stra8-Cre (progenitor and differentiating, Jackson Laboratory, #017490) and Neorog3/Ngn3-Cre (progenitor, (Yoshida et al., 2004)). There is also a tamoxifen-inducible version of the Neorog3/Ngn3-Cre mice (Yoshida et al., 2006), and these have been used with great success by the Yoshida laboratory (Yoshida et al., 2006; Ikami et al., 2015; Nakagawa et al., 2007, 2010).
5. Conclusions
Immunostaining approaches are invaluable tools for those who study spermatogenesis, as they allow for localization of specific proteins and the quantification of different types of germ cells in both WT and genetically- or chemically-treated animal models. These are particularly useful when working with fetal and neonatal testes, which contain small numbers of germ cells that are difficult to isolate, especially in sufficient numbers for many biochemical assays. Our field is in desperate need of transgenic mouse models with fluorescently-labeled germ cells. Specifically, it is critical to have mice in which the entire germline is fluorescently-labeled (e.g. by utilizing the promoter/enhancer elements of genes such as Tra98, Ddx4, Dazl, etc.). It will also be important to generate reliable transgenic lines with specific types of spermatogenic cells labeled (e.g. prospermatogonia and spermatogonia as well as spermatocytes and spermatids). These models will allow for FACS-based isolation of germ cells both at different stages of development and of different types from whole testes. Another pressing need is to develop more antibodies, generated in species other than mice, for the identification of proteins that uniquely mark subsets of each germ cell type (e.g. to distinguish type A1–4 spermatogonia from Intermediate and type B spermatogonia). It is our prediction that the creation of transgenic models for the reliable identification and isolation of specific germ cell types will allow more laboratories to work on mammalian spermatogenesis, which will significantly increase the pace of discovery.
Supplementary Material
Acknowledgments
The authors thank Dr. Brian Hermann (University of Texas at San Antonio) for helpful discussions and Dr. Randy Renegar (East Carolina University) and the members of the Geyer laboratory for critically reading the manuscript. This work was supported by grants from the NIH/NICHD (HD072552 and HD090083) to C.B.G.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2018.01.031.
References
- Ballow D, et al. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294(1):161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Bellve AR, et al. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74(1):68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JM, et al. Separation of spermatogenic cell types using STA-PUT velocity sedimentation. J Vis Exp. 2013;80 doi: 10.3791/50648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busada JT, Geyer CB. The role of retinoic acid (RA) in spermatogonial differentiation. Biol Reprod. 2015;94(1):10. doi: 10.1095/biolreprod.115.135145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busada JT, et al. Retinoic acid induces multiple hallmarks of the prospermatogonia-to-spermatogonia transition in the neonatal mouse. Biol Reprod. 2014;90(3):64. doi: 10.1095/biolreprod.113.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busada JT, et al. Retinoic acid regulates kit translation during spermatogonial differentiation in the mouse. Dev Biol. 2015;397(1):140–149. doi: 10.1016/j.ydbio.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F, et al. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 2014;28(12):1351–1362. doi: 10.1101/gad.240465.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarini-Garcia H, Russell LD. High-resolution light microscopic characterization of mouse spermatogonia. Biol Reprod. 2001;65(4):1170–1178. doi: 10.1095/biolreprod65.4.1170. [DOI] [PubMed] [Google Scholar]
- Clermont Y, Trott M. Duration of the cycle of the seminiferous epithelium in the mouse and hamster determined by means of 3H-thymidine and radioautography. Fertil Steril. 1969;20(5):805–817. doi: 10.1016/s0015-0282(16)37153-9. [DOI] [PubMed] [Google Scholar]
- DiNapoli L, Capel B. SRY and the standoff in sex determination. Mol Endocrinol. 2008;22(1):1–9. doi: 10.1210/me.2007-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drumond AL, Meistrich ML, Chiarini-Garcia H. Spermatogonial morphology and kinetics during testis development in mice: a high-resolution light microscopy approach. Reproduction. 2011;142(1):145–155. doi: 10.1530/REP-10-0431. [DOI] [PubMed] [Google Scholar]
- Gallardo T, et al. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45(6):413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia T, Hofmann MC. Isolation of undifferentiated and early differentiating type A spermatogonia from Pou5f1-GFP reporter mice. Methods Mol Biol. 2012;825:31–44. doi: 10.1007/978-1-61779-436-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CB, Eddy EM. Identification and characterization of Rhox13, a novel X-linked mouse homeobox gene. Gene. 2008;423(2):194–200. doi: 10.1016/j.gene.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CB, et al. Ontogeny of a demethylation domain and its relationship to activation of tissue-specific transcription. Biol Reprod. 2004;71(3):837–844. doi: 10.1095/biolreprod.104.028969. [DOI] [PubMed] [Google Scholar]
- Griswold MD. Spermatogenesis: the commitment to meiosis. Physiol Rev. 2016;96(1):1–17. doi: 10.1152/physrev.00013.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel AR, et al. ID4 levels dictate the stem cell state in mouse spermatogonia. Development. 2017;144(4):624–634. doi: 10.1242/dev.146928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, et al. Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol Reprod. 2015;92(2):54. doi: 10.1095/biolreprod.114.125757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikami K, et al. Hierarchical differentiation competence in response to retinoic acid ensures stem cell maintenance during mouse spermatogenesis. Development. 2015;142(9):1582–1592. doi: 10.1242/dev.118695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GB, et al. Foxo3 is a PI3K-dependent molecular switch controlling the initiation of oocyte growth. Dev Biol. 2008;321(1):197–204. doi: 10.1016/j.ydbio.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuscinski J, Szer W. Interactions of 4′, 6-diamidine-2-phenylindole with synthetic polynucleotides. Nucleic Acids Res. 1979;6(11):3519–3534. doi: 10.1093/nar/6.11.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluin PM, de Rooij DG. A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int J Androl. 1981;4(4):475–493. doi: 10.1111/j.1365-2605.1981.tb00732.x. [DOI] [PubMed] [Google Scholar]
- Kluin PM, Kramer MF, de Rooij DG. Proliferation of spermatogonia and Sertoli cells in maturing mice. Anat Embryol (Berl) 1984;169(1):73–78. doi: 10.1007/BF00300588. [DOI] [PubMed] [Google Scholar]
- Losinno AD, et al. Peritubular myoid cells from rat seminiferous tubules contain actin and myosin filaments distributed in two independent layers. Biol Reprod. 2012;86(5):150, 1–8. doi: 10.1095/biolreprod.111.095158. [DOI] [PubMed] [Google Scholar]
- McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262(1):1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- McLean DJ, et al. Characterization of spermatogonial stem cell maturation and differentiation in neonatal mice. Biol Reprod. 2003;69(6):2085–2091. doi: 10.1095/biolreprod.103.017020. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Nabeshima Y, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell. 2007;12(2):195–206. doi: 10.1016/j.devcel.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, et al. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328(5974):62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayernia K, et al. Stem cell based therapeutical approach of male infertility by teratocarcinoma derived germ cells. Hum Mol Genet. 2004;13(14):1451–1460. doi: 10.1093/hmg/ddh166. [DOI] [PubMed] [Google Scholar]
- Nicholas CR, et al. Characterization of a Dazl-GFP germ cell-specific reporter. Genesis. 2009;47(2):74–84. doi: 10.1002/dvg.20460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenberger BA, Busada JT, Geyer CB. Marker expression reveals heterogeneity of spermatogonia in the neonatal mouse testis. Reproduction. 2015;149(4):329–338. doi: 10.1530/REP-14-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakberg EF. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am J Anat. 1956;99(3):507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- Ohbo K, et al. Identification and characterization of stem cells in prepubertal spermatogenesis in mice. Dev Biol. 2003;258(1):209–225. doi: 10.1016/s0012-1606(03)00111-8. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436(7048):207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Pangas SA, et al. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci U S A. 2006;103(21):8090–8095. doi: 10.1073/pnas.0601083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, et al. Generation of stella-GFP transgenic mice: a novel tool to study germ cell development. Genesis. 2006;44(2):75–83. doi: 10.1002/gene.20187. [DOI] [PubMed] [Google Scholar]
- Porro V, et al. Characterization of Oct4-GFP transgenic mice as a model to study the effect of environmental estrogens on the maturation of male germ cells by using flow cytometry. J Steroid Biochem Mol Biol. 2015;154:53–61. doi: 10.1016/j.jsbmb.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Shima JE, et al. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71(1):319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Sun F, et al. Id4 marks spermatogonial stem cells in the mouse testis. Sci Rep. 2015;5:17594. doi: 10.1038/srep17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Dann CT, Rajkovic A. Generation of a germ cell-specific mouse transgenic CHERRY reporter, Sohlh1-mCherryFlag. Genesis. 2013;51(1):50–58. doi: 10.1002/dvg.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PE, et al. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev. 2002;115(1–2):157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, et al. Regulation of expression of mouse interferon-induced trans-membrane protein like gene-3, Ifitm3 (mil-1, fragilis), in germ cells. Dev Dyn. 2004;230(4):651–659. doi: 10.1002/dvdy.20085. [DOI] [PubMed] [Google Scholar]
- Tung PS, Fritz IB. Characterization of rat testicular peritubular myoid cells in culture: alpha-smooth muscle isoactin is a specific differentiation marker. Biol Reprod. 1990;42(2):351–365. doi: 10.1095/biolreprod42.2.351. [DOI] [PubMed] [Google Scholar]
- Vergouwen RP, et al. Proliferative activity of gonocytes, Sertoli cells and interstitial cells during testicular development in mice. J Reprod Fertil. 1991;93(1):233–243. doi: 10.1530/jrf.0.0930233. [DOI] [PubMed] [Google Scholar]
- Western P, et al. Dynamic regualtion of mitotic arrest in fetal male germ cells. Stem Cells. 2008;26(2):339–347. doi: 10.1634/stemcells.2007-0622. [DOI] [PubMed] [Google Scholar]
- Yang QE, Oatley JM. Spermatogonial stem cell functions in physiological and pathological conditions. Curr Top Dev Biol. 2014;107:235–267. doi: 10.1016/B978-0-12-416022-4.00009-3. [DOI] [PubMed] [Google Scholar]
- Yoshida S, et al. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol. 2004;269(2):447–458. doi: 10.1016/j.ydbio.2004.01.036. [DOI] [PubMed] [Google Scholar]
- Yoshida S, et al. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133(8):1495–1505. doi: 10.1242/dev.02316. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nabeshima Y. A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science. 2007;317(5845):1722–1726. doi: 10.1126/science.1144885. [DOI] [PubMed] [Google Scholar]
- Yoshimizu T, et al. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Develop Growth Differ. 1999;41(6):675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- Zheng K, et al. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev Biol. 2009;9:38. doi: 10.1186/1471-213X-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.