Abstract
Selective repression of the antifibrotic gene CXCL10 contributes to tissue remodeling in idiopathic pulmonary fibrosis (IPF). We have previously reported that histone deacetylation and histone H3 lysine 9 (H3K9) methylation are involved in CXCL10 repression. In this study, we explored the role of H3K27 methylation and the interplay between the two histone lysine methyltransferases enhancer of zest homolog 2 (EZH2) and G9a in CXCL10 repression in IPF. By applying chromatin immunoprecipitation, Re-ChIP, and proximity ligation assays, we demonstrated that, like G9a-mediated H3K9 methylation, EZH2-mediated histone H3 lysine 27 trimethylation (H3K27me3) was significantly enriched at the CXCL10 promoter in fibroblasts from IPF lungs (F-IPF) compared with fibroblasts from nonfibrotic lungs, and we also found that EZH2 and G9a physically interacted with each other. EZH2 knockdown reduced not only EZH2 and H3K27me3 but also G9a and H3K9me3, and G9a knockdown reduced not only G9 and H3K9me3 but also EZH2 and H3K27me3. Depletion and inhibition of EZH2 and G9a also reversed histone deacetylation and restored CXCL10 expression in F-IPF. Furthermore, treatment of fibroblasts from nonfibrotic lungs with the profibrotic cytokine transforming growth factor-β1 increased EZH2, G9a, H3K27me3, H3K9me3, and histone deacetylation at the CXCL10 promoter, similar to that observed in F-IPF, which was correlated with CXCL10 repression and was prevented by EZH2 and G9a knockdown. These findings suggest that a novel and functionally interdependent interplay between EZH2 and G9a regulates histone methylation-mediated epigenetic repression of the antifibrotic CXCL10 gene in IPF. This interdependent interplay may prove to be a target for epigenetic intervention to restore the expression of CXCL10 and other antifibrotic genes in IPF.
Keywords: histone methylation, CXCL10, gene expression, lung fibroblast, pulmonary fibrosis
Clinical Relevance
Idiopathic pulmonary fibrosis (IPF) is a fatal respiratory disease characterized by the accumulation of activated lung fibroblasts and excessive deposition of collagen, leading to distortion of the alveolar architecture and loss of lung function. Selective repression of the antifibrotic gene CXCL10 contributes to this tissue remodeling in IPF. We report that a novel interdependent interplay between the two histone lysine methyltransferases enhancer of zest homolog 2 (EZH2) and G9a regulates histone methylation–mediated epigenetic repression of the CXCL10 gene in IPF, but the machinery for CXCL10 expression remains intact. Thus, the interplay between EZH2 and G9a may represent a viable target for epigenetic intervention to reactivate CXCL10 and other silenced antifibrotic genes in IPF.
Idiopathic pulmonary fibrosis (IPF) is a deadly respiratory disease of unknown etiology with a median survival of 3–4 years and a lack of effective therapy (1). IPF is characterized by the accumulation of activated lung fibroblasts (myofibroblasts) that are ultimately responsible for the excessive deposition of collagen, leading to distortion of the alveolar architecture, loss of lung function, and ultimately death (2). Fibroblasts isolated from idiopathic pulmonary fibrosis lungs (F-IPF; activated lung fibroblasts) are phenotypically different from fibroblasts isolated from nonfibrotic lungs (F-NL) in that they express higher levels of α-smooth muscle actin (α-SMA) (3) and lower levels of antifibrotic genes such as cyclooxygenase-2 (COX-2) (4–6), Thy-1 (7, 8), caveolin-1 (9), and C-X-C motif chemokine 10 (CXCL10) (or interferon-γ–inducible protein of 10 kD) (10, 11). Recent studies strongly suggest that epigenetic dysregulation plays a key role in the targeted repression of these antifibrotic genes (4, 5, 7, 10, 12).
CXCL10 is a strong inhibitor of angiogenesis (13) and can be induced by IL-1β and IFN-γ (10), but not by the profibrotic cytokine transforming growth factor-β1 (TGF-β1) (14). It has been shown that lung fibroblasts and tissues from patients with IPF and the murine model of bleomycin-induced pulmonary fibrosis express significantly less CXCL10 than normal cells and tissues and that CXCL10 inhibits angiogenesis, lung fibroblast migration and proliferation, and subsequent lung fibrosis (11, 15–17). In contrast, blockade of CXCL10 increases pulmonary fibrosis (15, 16), and genetically targeting the CXCL10 receptor CXCR3 results in increased mortality with progressive interstitial fibrosis in bleomycin-induced pulmonary fibrosis (18). These observations strongly suggest that CXCL10 deficiency, as a result of gene repression, plays a key role in aberrant tissue remodeling and the development of pulmonary fibrosis.
TGF-β1 is a major activator of lung fibroblasts both in vitro (19) and in vivo (20). Although TGF-β1 treatment reduces the expression of antifibrotic genes (e.g., caveolin-1) in human lung fibroblasts (9), whether it leads to alterations of epigenetic modifications of antifibrotic genes similar to those observed in F-IPF has not been explored.
Methylation of histone proteins at specific lysine residues plays a major role in the regulation of gene expression and repression. The most well-characterized histone methylations are histone H3 lysine 27 trimethylation (H3K27me3) and histone H3 lysine 9 di-/trimethylation (H3K9me2/3), which are generally correlated with gene repression (21). H3K27me3 is catalyzed by the histone lysine methyltransferase (KMT) enhancer of zest homolog 2 (EZH2), the catalytic subunit of polycomb repressive complex 2 (PRC2) (22). In contrast, histone lysine demethylases (KDMs), such as KDM6A and KDM6B (also known as jumonji domain-containing protein 3 [JMJD3]), are enzymes capable of specifically demethylating H3K27me3 at gene promoters to derepress gene transcription (23, 24). H3K9me2/3 is catalyzed by H3K9-specific KMTs such as G9a and G9a-like protein (25). It has been shown that G9a can methylate H3K27 in vitro and in vivo (26, 27), suggesting a cross-talk between H3K9 and H3K27 methylations. A recent study has also demonstrated that G9a enzymatic activity can mediate EZH2 recruitment to regulate the repression of a subset of genes that are common targets of both enzymes (28), thus providing direct proof of a functional interplay between G9a and EZH2. We have previously demonstrated that histone deacetylation and H3K9 methylation, but not DNA methylation, are involved in CXCL10 repression in IPF (10). However, whether H3K27 methylation by EZH2 impacts CXCL10 repression in IPF and whether EZH2 enzymatic activity is required for G9a-mediated gene repression remain unknown. In this study, we explored the role of EZH2-mediated H3K27 methylation and the interplay between EZH2 and G9a in CXCL10 epigenetic repression in F-IPF. We found that EZH2 and G9a physically interact with each other at the CXCL10 promoter in F-IPF and that the interplay between EZH2 and G9a plays a key role in the epigenetic repression of CXCL10 in IPF. Some of the results of these studies were previously reported in the form of an abstract (29).
Methods
Cell Culture
F-IPF and F-NL were supplied by Dr. Feghali-Bostwick (University of Pittsburgh) and were described before (10, 30). IL-1β (R&D Systems) was used to induce CXCL10 expression (10). For TGF-β1 treatment, F-NL were incubated with TGF-β1 (R&D Systems) for 3 days before analyses or treatment with IL-1β and siRNAs.
Chromatin Immunoprecipitation and Re-ChIP
Chromatin immunoprecipitation (ChIP) was performed using the ChIP-IT Express Kit and the Re-ChIP-IT Express Kit (Active Motif), as described elsewhere (5, 10), with antibodies against H3K27me3, H3K9me3, EZH2, acetylated and total H3 and H4 (Merck Millipore), and G9a (Santa Cruz Biotechnology). The Re-ChIP was conducted with chromatin from the first ChIP using antibodies against the secondary targets.
JMJD3 Overexpression
F-IPF (80% confluent) were incubated with pMSCV vectors encoding human JMJD3 (pMSCV-JMJD3) or a mutant variant lacking catalytic activity (pMSCV-mJMJD3) (Addgene) (31) or with empty vectors (1 μg), premixed 2:1 with TransFast (Promega) for 72 hours before being treated with IL-1β for 4 and 24 hours. mJMJD3 and JMJD3 overexpression was assessed by Western blotting with an antibody recognizing both JMJD3 and mJMJD3 (Santa Cruz Biotechnology).
Inhibitor Study and siRNA Transfection
F-IPF were pretreated with 10 nM EZH2 inhibitor 3-Deazaneplanocin A (DZNep; Cayman Chemical) (32) for 3 days before being treated with IL-1β for 4 and 24 hours for epigenetic analyses and for CXCL10 mRNA and protein expression by real-time RT-PCR and ELISA, respectively (10).
F-IPF and F-NL were transfected with predesigned siRNAs against human EZH2 (Hs_EZH2_3 FlexiTube siRNA, target sequence CAGACGAGCTGATGAAGTAAA) and G9a (Hs_BAT8_3 FlexiTube siRNA, target sequence CACCATGAACATCGATCGCAA), as well as ALLStars Negative Control siRNA (Qiagen) using HiPerFect Transfection Reagent (Qiagen) (5). Cells were then serum starved for 24 hours before treatment with TGF-β1 and IL-1β for further analyses.
Proximity Ligation Assay
Serum-starved F-IPF on glass coverslips were fixed in 4% paraformaldehyde and treated with 0.3% Triton X-100. EZH2 and G9a interaction was detected with Duolink In Situ Red Starter Kit Mouse/Rabbit (Olink Bioscience) following the manufacturer’s instructions. Briefly, after blocking, slides were incubated with primary mouse anti-EZH2 antibody (1:200, clone AC22; Merck Millipore) and rabbit anti-G9a antibody (1:200, ab133482; Abcam) overnight at 4°C. Slides were washed and then incubated with proximity ligation assay (PLA) probes Anti-Mouse MINUS and Anti-Rabbit PLUS (Olink Bioscience), respectively, for 1 hour at 37°C. After unbound probes were removed, slides were incubated with Duolink Ligase for 30 minutes at 37°C followed by rolling circle amplification. Amplified DNA was detected by Duolink In Situ Detection Reagent Red. Nuclei were stained with DAPI. Images were taken with a Nikon Eclipse 90i fluorescence imaging microscope (Nikon Instruments).
Statistical Analysis
Data were presented as mean ± SEM of experiments with three to six separate cell lines. Data were examined for normality using Shapiro-Wilk and Pearson’s chi-square tests. ANOVA was used to evaluate statistical significance of the mean values between F-NL and F-IPF, which was followed by Student’s t test for comparisons at specific time points. P < 0.05 was regarded as statistically significant.
Results
H3K27me3 Is Increased at the CXCL10 Promoter in F-IPF and Is Reduced by EZH2 Inhibition and JMJD3 Overexpression
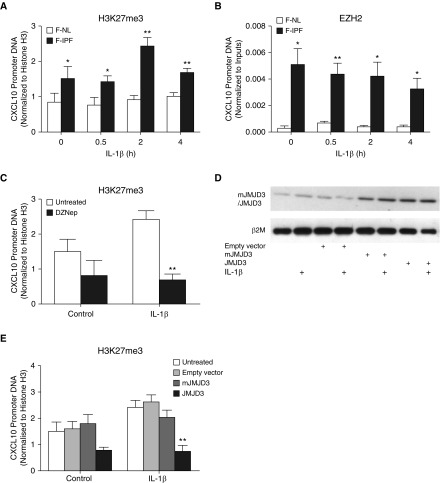
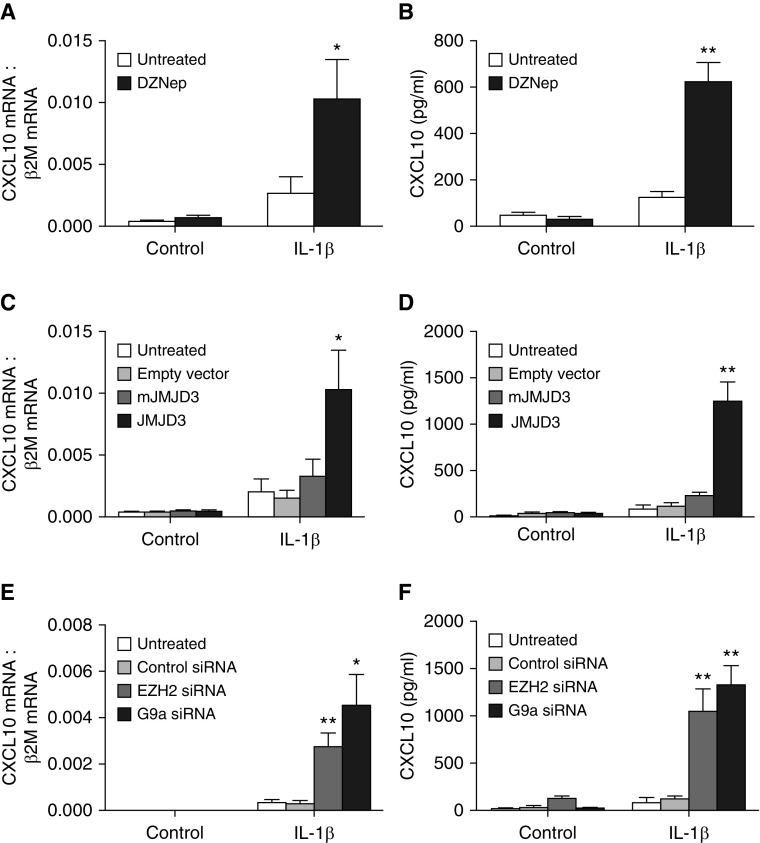
We have previously shown that H3K9me3 is increased at the CXCL10 promoter in F-IPF compared with F-NL (10). Because H3K27me3 is another typical histone methylation mark linked to repressive chromatin, we next analyzed its presence at the CXCL10 promoter by ChIP assay. We observed that under both unstimulated and IL-1β–stimulated conditions, H3K27me3 at the CXCL10 promoter was markedly elevated in F-IPF compared with F-NL (Figure 1A). In addition, the association of EZH2, the KMT responsible for H3K27me3, with the CXCL10 promoter was also consistently increased in F-IPF compared with F-NL with or without IL-1β treatment (Figure 1B). The data suggest that H3K27, like H3K9, is also hypermethylated at the CXCL10 promoter in F-IPF as a result of increased recruitment of EZH2. To determine whether EZH2 inhibitors and the specific H3K27me3 demethylase JMJD3 could modulate H3K27me3 at the CXCL10 promoter, we examined the effect of the EZH2 inhibitor DZNep and overexpression of JMJD3 in F-IPF cells. Treatment of the cells with DZNep, alone or with IL-1β, reduced H3K27me3 at the CXCL10 promoter by 45% and 70%, respectively (Figure 1C), compared with untreated cells. Transfection of F-IPF with pMSCV-mJMJD3 and pMSCV-JMJD3 showed increased expression of mutated and wild-type JMJD3, respectively, whereas transfection with empty vector and treatment with IL-1β, either alone or with the vectors, had no effect on JMJD3 expression (Figure 1D). Furthermore, overexpression of wild-type JMJD3, but not the mutated mJMJD3 lacking JMJD3 enzymatic activity, in both control and IL-1β–treated cells markedly reduced H3K27me3 association with the CXCL10 promoter (Figure 1E). The results thus suggest that EZH2 is critically involved in H3K27me3 at the CXCL10 promoter in F-IPF and that inhibition of EZH2 and overexpression of JMJD3 can reduce H3K27me3 and impact CXCL10 repression.
Figure 1.
Histone H3 lysine 27 trimethylation (H3K27me3) is increased and modulated at the CXCL10 promoter in fibroblasts from idiopathic pulmonary fibrosis lungs (F-IPF). (A and B) Confluent and serum-starved fibroblasts from nonfibrotic lungs (F-NL) and F-IPF cells were treated with IL-1β (1 ng/ml) for the times indicated. F-IPF cells were incubated with (C) 3-deazaneplanocin A (DZNep) (10 nM) or (D and E) transfected with empty vector, pMSCV-mJMJD3, or pMSCV-JMJD3 before being treated with IL-1β (1 ng/ml) for a further (C and E) 4 hours or (D) 24 hours. Chromatin immunoprecipitation was conducted with specific antibodies against (A, C, and E) H3K27me3, (B) enhancer of zest homolog 2 (EZH2), and (A, B, C, and E) total histone H3. The associated CXCL10 promoter DNA was amplified by real-time RT-PCR, and the amount was calculated and normalized to (A, C, and E) total histone H3 or (B) input control. Data are expressed as mean ± SEM of experiments with six separate F-NL and/or F-IPF cell lines performed in duplicate. *P < 0.05, **P < 0.01 compared with corresponding F-NL or untreated F-IPF cells. (D) Total cell lysates were collected for Western blot analysis of mutant variant of jumonji domain-containing protein 3 (mJMJD3) and jumonji domain-containing protein 3 (JMJD3) overexpression with β2-microglobulin (β2M) as the loading control. This is representative of three separate experiments with different F-IPF cell lines.
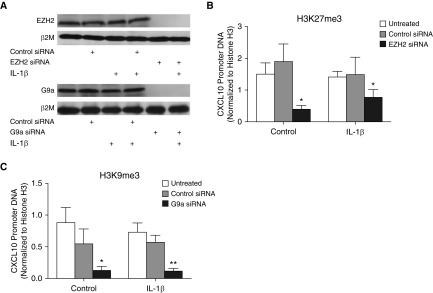
H3K27me3 and H3K9me3 at the CXCL10 Promoter Are Reduced by EZH2 and G9a siRNAs
Figure 1C and our previous study (10) show that inhibitors of EZH2 and G9a reduce H3K27me3 and H3K9me3, respectively, at the CXCL10 promoter. To further validate the role of EZH2 and G9a in mediating CXCL10 repression in F-IPF, EZH2 and G9a siRNAs were applied. As shown in Figure 2A, EZH2 and G9a siRNAs, but not the control siRNA, completely abolished their respective target gene expression. Like the EZH2 inhibitor DZNep, EZH2 siRNA, but not the control siRNA, either alone or with IL-1β, reduced H3K27me3 at the CXCL10 promoter by 73% and 44%, respectively, compared with untreated cells (Figure 2B). Similarly, like the G9a inhibitor BIX-01294 (10), G9a siRNA, but not the control siRNA, either alone or with IL-1β, reduced H3K9me3 at the CXCL10 promoter by 85% and 83%, respectively, compared with untreated cells (Figure 2D). The data confirm the effect of the EZH2 and G9a inhibitors in Figure 1C and in our previous study (10) and suggest a key role for EZH2 and G9a in mediating H3K27me3 and H3K9me3, respectively, at the CXCL10 promoter in F-IPF.
Figure 2.
EZH2 and G9a knockdown alters repressive histone methylation at the CXCL10 promoter in F-IPF. F-IPF cells were transfected with control siRNA, EZH2 siRNA, or G9a siRNA before being treated with IL-1β (1 ng/ml) for a further (A) 24 hours or (B and C) 4 hours. (A) Total cell lysates were collected for Western blot analysis of EZH2 and G9a with β2M as the loading control. This is representative of three separate experiments with different F-IPF cell lines. Chromatin immunoprecipitation was conducted with specific antibodies against (B) H3K27me3, (C) H3K9me3, and (B and C) total histone H3. (B and C) The associated CXCL10 promoter DNA was amplified by real-time RT-PCR, and the amount was calculated and normalized to total histone H3. Data are expressed as mean ± SEM of experiments with six separate F-IPF cell lines performed in duplicate. *P < 0.05, **P < 0.01 compared with corresponding untreated cells.
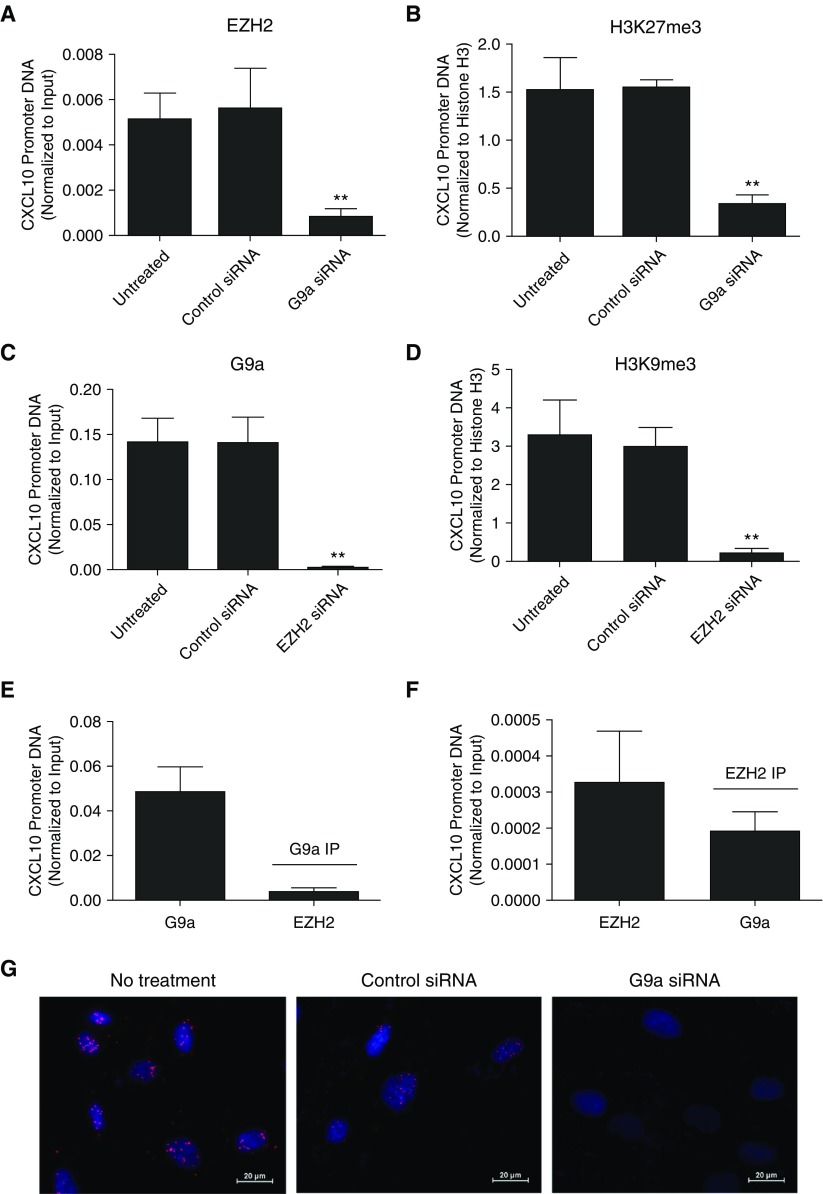
Cross-Talk between EZH2 and G9a at the CXCL10 Promoter in F-IPF
To explore the cross-talk between EZH2 and G9a, we first applied ChIP to assess whether G9a knockdown with siRNA could affect EZH2-induced H3K27me3 and vice versa. We found that G9a knockdown significantly reduced EZH2 and H3K27me3 at the CXCL10 promoter (Figures 3A and 3B); similarly, EZH2 knockdown with siRNA also led to a significant reduction of G9a and H3K9me3 at the CXCL10 promoter (Figures 3C and 3D). In addition, the Re-ChIP assay revealed the presence of EZH2 in the immunoprecipitate (IP) of G9a (Figure 3E) and the presence of G9a in the IP of EZH2 (Figure 3F) at the CXCL10 promoter in unstimulated F-IPF, suggesting physical interactions between the two KMTs. To further confirm physical interaction between EZH2 and G9a proteins, PLA was conducted in unstimulated F-IPF. As shown in Figure 3G, strong red PLA signals were detected in untreated and control siRNA-transfected F-IPF; however, the signals were markedly reduced when G9a was knocked down. Because PLA signals were produced when the two PLA probes bound, which took place only if both EZH2 and G9a were closer than 40 nm, these observations strongly suggest that EZH2 and G9a physically interact with each other and regulate repressive histone methylation at the CXCL10 promoter in F-IPF in an interdependent manner.
Figure 3.
EZH2 and G9a interact with each other to regulate histone methylation at the CXCL10 promoter in F-IPF. F-IPF cells were (A–D) transfected with control siRNA, G9a siRNA, or EZH2 siRNA or (E and F) untreated. Chromatin pellets were (A–F) extracted and immunoprecipitated with specific antibodies against (A and F) EZH2, (C and E) G9a, (B) H3K27me3, (D) H3K9me3, and (B and D) total histone H3. The immunoprecipitates (IPs) of (E) G9a and (F) EZH2 were immunoprecipitated again with antibodies against (E) EZH2 and (F) G9a. The associated CXCL10 promoter DNA was amplified by real-time RT-PCR, and the amount was calculated and normalized to (B and D) total histone H3 or (A, C, E, and F) input control. Data are expressed as mean ± SEM of experiments with six separate F-IPF cell lines performed in duplicate. **P < 0.01 compared with untreated cells. Serum-starved F-IPF cells grown on sterile glass coverslips were fixed in 4% paraformaldehyde. In situ interaction between EZH2 and G9a was detected by proximity ligation assay with primary mouse anti-EZH2 antibody and rabbit anti-G9a antibody as well as by proximity ligation assay probes Anti-Mouse MINUS and Anti-Rabbit PLUS. (G) Amplified DNA was detected by Duolink In Situ Detection Reagents Red, and DAPI (blue) was used to identify the nuclei. Images are representative of three separate experiments with different F-IPF cell lines. Scale bars: 20 μm.
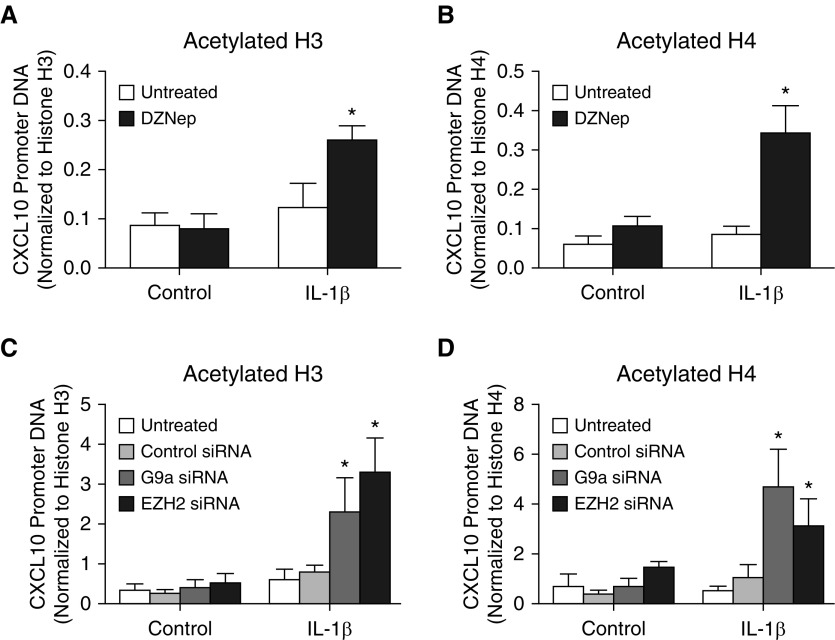
Histone Acetylation at the CXCL10 Promoter in F-IPF Is Increased by EZH2 and G9a Inhibition
To further validate the role of EZH2 and G9a in mediating CXCL10 epigenetic repression in F-IPF, the effect of EZH2 and G9a inhibition on histone acetylation was assessed. As shown in Figure 4, treatment of F-IPF cells with either DZNep or IL-1β alone did not significantly alter histone H3 and H4 acetylation at the CXCL10 promoter. However, like G9a inhibition by BIX-01294 (10), EZH2 inhibition markedly increased histone H3 and H4 acetylation in cells treated with both DZNep and IL-1β compared with cells treated with IL-1β alone (Figures 4A and 4B). Similarly, treatment of F-IPF with G9a siRNA or EZH2 siRNA alone had no significant effect on histone H3 and H4 acetylation without cytokine stimulation; however, marked increases were observed with IL-1β stimulation compared with untreated cells (Figures 4C and 4D). These data strongly suggest that G9a and EZH2 play a key role in mediating not only repressive histone methylation but also histone deacetylation at the CXCL10 promoter in F-IPF.
Figure 4.
EZH2 and G9a inhibition and knockdown promote histone acetylation at the CXCL10 promoter in F-IPF. F-IPF cells were (A and B) incubated with DZNep (10 nM) or (C and D) transfected with control siRNA, G9a siRNA, or EZH2 siRNA in culture medium for 48 hours and in serum-free medium for 24 hours before being treated with IL-1β (1 ng/ml) for a further 4 hours. The protein–DNA complexes were then cross-linked by formaldehyde treatment, and chromatin pellets were extracted. Acetylated and total histone (A and C) H3 and (B and D) H4 were immunoprecipitated with specific antibodies. The associated CXCL10 promoter DNA was amplified by real-time RT-PCR, and the amount was calculated and normalized to total histone H3 and H4. Data are expressed as mean ± SEM of experiments with six separate F-IPF cell lines performed in duplicate. *P < 0.05 compared with corresponding untreated cells.
CXCL10 Expression in F-IPF Is Restored by EZH2 and G9a Inhibition
To investigate whether EZH2 and G9a inhibition could lead to CXCL10 gene derepression, F-IPF cells were treated with DZNep or JMJD3 overexpression, either alone or with the potent CXCL10 inducer IL-1β. DZNep and JMJD3 overexpression alone had no effect on CXCL10 mRNA expression, as analyzed by real-time RT-PCR, whereas marked increases in CXCL10 mRNA were observed when cells were treated with either DZNep or JMJD3 overexpression together with IL-1β compared with IL-1β alone (Figures 5A and 5C). ELISA analysis revealed no increase in CXCL10 protein expression when cells were treated with DZNep and slight increases when cells were treated with empty vector or overexpression of mJMJD3 and JMJD3 compared with untreated control cells. However, when cells were treated with DZNep or JMJD3 overexpression together with IL-1β, marked increases in CXCL10 protein expression were observed compared with cells treated with IL-1β alone (Figures 5B and 5D). Furthermore, knocking down EZH2 and G9a with siRNAs alone did not markedly increase CXCL10 mRNA and protein expression. However, when cells were then stimulated with IL-1β, significant increases in CXCL10 mRNA and protein were observed compared with cells treated with IL-1β alone (Figures 5E and 5F). The results thus show that EZH2 and G9a inhibition not only removes repressive histone modifications at the CXCL10 promoter but also restores the capacity of F-IPF cells to express CXCL10 and therefore strongly suggest a central role for EZH2 and G9a interplay in mediating CXCL10 epigenetic repression in IPF.
Figure 5.
EZH2 and G9a inhibition and knockdown restore CXCL10 expression in F-IPF. F-IPF cells were (A and B) incubated with DZNep (10 nM) or (C and D) transfected with empty vector, pMSCV-mJMJD3, or pMSCV-JMJD3 or (E and F) transfected with control siRNA, EZH2 siRNA, or G9a siRNA in culture medium for 48 hours and in serum-free medium for 24 hours before being treated with IL-1β (1 ng/ml) for a further (A, C, and E) 4 hours or (B, D, and F) 24 hours. Total RNA was isolated, and mRNA levels of CXCL10 and the internal control β2M were determined by real-time RT-PCR. (A, C, and E) Results are calculated as the ratio of CXCL10 mRNA and β2M mRNA. (B, D, and F) Medium was collected, and CXCL10 concentration was analyzed by ELISA. Data are expressed as mean ± SEM of six separate experiments performed in duplicate. *P < 0.05, **P < 0.01 compared with corresponding untreated cells.
TGF-β1–induced Epigenetic Repression of CXCL10 in F-NL Is Mediated by an Interdependent Cross-talk between EZH2 and G9a
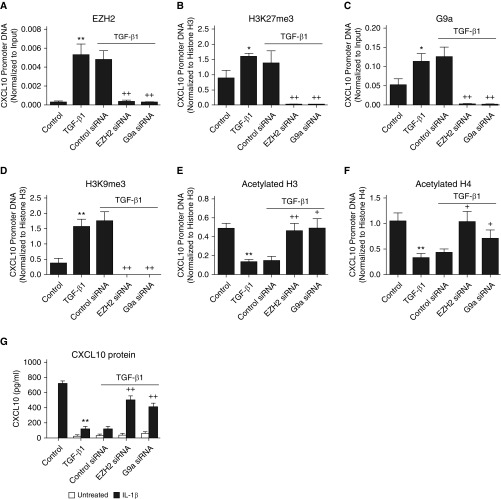
Because TGF-β1 is a potent profibrotic cytokine that may play a key role in the phenotypic changes observed in F-IPF compared with F-NL, we next explored whether TGF-β1 treatment of F-NL could lead to repressive histone modifications at the CXCL10 promoter and CXCL10 repression, similar to those observed in F-IPF, and to assess the role of the cross-talk between EZH2 and G9a in this process. As shown in Figure 6, TGF-β1 treatment significantly increased the association of EZH2 and G9a to the CXCL10 promoter in F-NL (Figures 6A and 6C). This was mirrored by corresponding increases of H3K27me3 and H3K9me3 (Figures 6B and 6D). Knocking down EZH2 with siRNA markedly reduced TGF-β1–induced association of not only its target protein EZH2 (Figure 6A) but also G9a to the CXCL10 promoter (Figure 6C). Likewise, knocking down G9a with siRNA markedly reduced TGF-β1–induced association of not only its target protein G9a (Figure 6C) but also EZH2 to the CXCL10 promoter (Figure 6A). Similar cross-inhibition was also observed for both H3K27me3 and H3K9me3 with either EZH2 siRNA or G9a siRNA (Figures 6B and 6D). In addition, TGF-β1 treatment also led to significant reduction of active histone H3 acetylation (Figure 6E) and H4 acetylation (Figure 6F) at the CXCL10 promoter in F-NL; however, this was largely prevented by siRNA knockdown of either EZH2 or G9a (Figures 6E and F). Furthermore, TGF-β1 treatment also induced a significant reduction of IL-1β–induced CXCL10 production in F-NL compared with cells treated with IL-1β alone, and this downregulation was also prevented by EZH2 or G9a siRNA knockdown (Figure 6G). These observations suggest that the interdependent cross-talk between EZH2 and G9a also plays a key role in TGF-β1–induced epigenetic repression of CXCL10 in F-NL and thereby further support the importance of EZH2 and G9a in mediating CXCL10 epigenetic repression in IPF.
Figure 6.
Transforming growth factor-β1 (TGF-β1)–induced epigenetic repression of CXCL10 in fibroblasts from F-NL is mediated by an interdependent cross-talk between EZH2 and G9a. (A–F) F-NL cells were transfected with control siRNA, EZH2 siRNA, or G9a siRNA in culture medium for 72 hours before incubation with TGF-β1 (2 ng/ml) in culture medium for 48 hours and in serum-free medium for 24 h before (G) being treated with IL-1β (1 ng/ml) in the presence of TGF-β1 for a further 24 h. The protein–DNA complexes were then cross-linked by formaldehyde treatment, and chromatin pellets were extracted. (A) EZH2, (B) H3K27me3, (C) G9a, (D) H3K9me3, acetylated histone (E) H3 and (F) H4, and total histone (B, D, and E) H3 and (F) H4 were immunoprecipitated with specific antibodies. The associated CXCL10 promoter DNA was amplified by real-time RT-PCR, and the amount was calculated and normalized to (A and C) input control or (B, D, E, and F) total histone H3 and H4. (G) CXCL10 concentration in the medium was analyzed by ELISA. Data are expressed as mean ± SEM of six separate experiments performed in duplicate. *P < 0.05, **P < 0.01 compared with control cells; +P < 0.05, ++P < 0.01 compared with TGF-β1 alone.
Discussion
We have previously demonstrated that H3K9 methylation is involved in CXCL10 repression in IPF (10). The main novel findings of the present study are that EZH2-mediated H3K27me3 was also significantly increased at the CXCL10 promoter in F-IPF and that EZH2 and G9a interacted with each other at the CXCL10 promoter, as demonstrated by Re-ChIP assay. More importantly, we found that EZH2 siRNA markedly reduced not only EZH2 and H3K27me3 but also G9a and H3K9me3 and that G9a siRNA markedly reduced not only G9a and H3K9me3 but also EZH2 and H3K27me3 at the CXCL10 promoter. EZH2 and G9a siRNAs also led to increased histone acetylation and restored CXCL10 expression in F-IPF. Furthermore, treatment of F-NL with TGF-β1 largely recapitulated EZH2- and G9a-mediated repressive histone modifications and CXCL10 repression observed in F-IPF. To the best of our knowledge, this is the first report to suggest that a functionally interdependent cross-talk between EZH2 and G9a regulates the epigenetic repression of antifibrotic genes in any fibrotic condition.
It is well established that EZH2- and G9a-mediated histone hypermethylation contributes to the epigenetic silencing of tumor suppressor genes. We have previously shown that the G9a specific inhibitor BIX-01294 reduces H3K9 methylation and the recruitment of additional chromatin modifiers but increases histone H3 and H4 acetylation at the CXCL10 promoter in F-IPF (10). In this study, we report that the EZH2 inhibitor DZNep not only reduced H3K27me3 as expected but also increased histone H3 and H4 acetylation at the CXCL10 promoter. Similarly, H3K27me3 was also reduced by overexpression of the H3K27me3-specific demethylase JMJD3 (Figures 1 and 4). The data suggest that EZH2, like G9a, also plays a role in CXCL10 epigenetic repression in IPF. However, epigenetic inhibitors may display off-target effects, as demonstrated by a recent study showing inhibition of both EZH2 and the H3K9 KMT SETDB1 by DZNep (32). It is therefore necessary to apply siRNA knockdown to validate the findings with epigenetic inhibitors. We also revealed that successful siRNA knockdown of EZH2 and G9a recapitulated the effects of EZH2 and G9a inhibitors in this (Figures 2 and 4) and previous (10) studies. This is consistent with recent findings that EZH2 and G9a knockdown reduces H3K27me3 and H3K9me3 at the COX-2 promoter and restores COX-2 expression in F-IPF cells (5), as well as that G9a knockdown reduces H3K9 methylation at the promoters of the epithelial cell adhesion molecule and E-cadherin and restores their expression in lung cancer cells (33) and human breast cancer cells (34). As a result of the removal of epigenetic repression by inhibition and knockdown of EZH2 and G9a, active epigenetic modifications, such as histone acetylation, and the capability of F-IPF to express CXCL10 in response to IL-1β were restored. These observations thus suggest that EZH2- and G9a-mediated histone methylation is closely associated with the epigenetic repression of a group of antifibrotic genes including COX-2 and CXCL10 in IPF and that inhibition and knockdown of these two KMTs switch local epigenetic status from repressive to active in response to IL-1β. However, although G9a and EZH2 are important in the epigenetic repression of antifibrotic genes in IPF and G9a and EZH2 are upregulated in a number of different cancers to maintain the malignant phenotype (35, 36), our observations suggest that increased expression is not necessarily a prerequisite for their repressive effect. In fact, we have previously reported that there is no difference in the global expression of EZH2 and G9a between F-IPF and F-NL (5, 10). The increase of H3K27me3 and H3K9me3 at the CXCL10 promoters in F-IPF is therefore likely due to gene promoter–specific recruitment of EZH2 and G9a.
Although H3K27 and H3K9 methylation is catalyzed by different lysine-specific KMTs, they share common functions in gene silencing and have similar roles in the control of many cellular processes. Cross-talk between the two methylation marks has also been suggested. For instance, G9a has been shown to methylate H3K27 in vitro and in vivo (26, 27). However, the underlying mechanisms were unclear until a recent study demonstrated that G9a interacts physically and functionally with PRC2 and that G9a enzymatic activity mediates the recruitment of EZH2 to specific target genes to enable H3K27me3, thus providing direct proof of an interplay between the two main histone lysine methylation mechanisms to ensure epigenetic gene silencing (28). To find out whether this interplay is involved in the epigenetic repression of antifibrotic genes in IPF and whether EZH2 can regulate G9a-mediated gene repression through H3K9me3, we applied a Re-ChIP assay to explore the physical interaction between EZH2 and G9a at the CXCL10 promoter in F-IPF. The findings that EZH2 was detected in the IP of G9a and G9a was detected in the IP of EZH2 suggest that the two KMTs colocalize at the CXCL10 promoter (Figure 3). This was further confirmed by the PLA demonstrating that EZH2 and G9a physically interact with each other in the nuclei of F-IPF (Figure 3). We also demonstrated that EZH2 siRNA markedly reduced not only EZH2 and H3K27me3 but also G9a and H3K9me3 at the CXCL10 promoter and vice versa with G9a siRNA. Because the EZH2 and G9a siRNAs we used knocked down the expression of only their respective specific genes, but not the other (5), and because the expression of PRC2 core members, including EZH2, was not affected by the absence of G9a (28), our data suggest that the local presence of EZH2 and G9a is required for the recruitment, not the expression, of each other to the CXCL10 promoter and that the two KMTs act cooperatively and interdependently to regulate CXCL10 epigenetic repression in IPF. This is further supported by the observations that histone acetylation was increased at the CXCL10 promoter and that CXCL10 expression was restored in response to IL-1β by the knockdown of either EZH2 or G9a with siRNA.
We noticed that EZH2 siRNA- and G9a siRNA-induced reduction in H3K27 and H3K9 methylation was not compensated by other H3K27 and H3K9 KMTs. This is unlikely due to the off-target effects of the siRNAs on other KMTs. Several lines of evidence indicate that the depletion of one subunit of the H3K9 KMT complex or the PRC2 complex can lead to the destabilization of other subunits at the protein but not the mRNA level. For instance, depletion of G9a leads to the destabilization of other H3K9 KMTs, such as G9a-like protein, SETDB1, and SUV39H1, and results in reduced H3K9me2/me3 on G9a target genes, indicating that the integrity of these KMTs is interdependent (37). Similarly, depletion of EZH2 leads to the destabilization of other core components of the PRC2 complex, such as SUZ12 and EED, and results in reduced H3K27me3 on PRC2 target genes (38). It is therefore possible that the destabilization of the other KMTs, as a result of EZH2 and G9a depletion, may also contribute to the reduction in H3K27 and H3K9 methylation in siRNA-transfected cells. Further investigation is required to confirm this.
It is increasingly recognized that F-IPF represent a persistently activated phenotype of lung fibroblasts and play a key role in the development and progression of IPF and that these cells are epigenetically reprogrammed so that a group of antifibrotic genes, including CXCL10, COX-2, and Thy-1, are epigenetically repressed in a way similar to the epigenetic silencing of tumor suppressor genes in cancers. We provide further evidence that dysregulated histone methylation contributes to CXCL10 epigenetic repression in F-IPF by comparing the cells with F-NL. However, one clear limitation of this comparative study is that we explored the epigenetic events at the end of the disease process. Although the results are novel and valuable in the understanding of the epigenetic mechanisms involved in the epigenetic repression of antifibrotic genes in IPF, little is known about whether similar repressive histone modifications and mechanisms can be established in vitro with factors that promote persistent fibroblast activation and pulmonary fibrogenesis. We report that treatment of F-NL with the profibrotic cytokine TGF-β1 increased EZH2, G9a, H3K27me3, H3K9me3, and histone deacetylation at the CXCL10 promoter, similar to that observed in F-IPF. These epigenetic modifications were correlated with the repression of IL-1β–induced CXCL10 and were prevented by EZH2 and G9a siRNAs. These findings strongly suggest that TGF-β1 treatment of lung fibroblasts can recapitulate the epigenetic events associated with CXCL10 repression in F-IPF and further support an important role of the interdependent cross-talk between EZH2 and G9a in mediating CXCL10 epigenetic repression in IPF. It is worth noting that although IL-1β is regarded as a profibrotic cytokine (39), it exhibits antifibrotic effects such as induction of antifibrotic genes COX-2 (4, 5) and CXCL10 (10) in normal lung fibroblasts and inhibition of TGF-β1–induced myofibroblast formation and collagen synthesis (40). However, in TGF-β1–treated F-NL, the capability to express antifibrotic genes COX-2 (41) and CXCL10 (present study) in response to IL-1β is markedly reduced, similar to that observed in F-IPF (4, 5, 10), resulting in unopposed profibrotic effects of both TGF-β1 and IL-1β. Our observations provide early evidence that repressive epigenetic modifications can be introduced, at least to some extent, by TGF-β1 treatment, leading to the repression of IL-1β–induced antifibrotic genes in IPF. However, further studies are needed to understand the order of epigenetic events and the mechanisms responsible for the maintenance of these epigenetic modifications and the profibrotic phenotype of the cells. Because antifibrotic genes are important to maintaining the nonfibrotic phenotype of lung fibroblasts, it is plausible that epigenetic inhibitors, by preventing antifibrotic gene repression, could contribute to the maintenance of the phenotype of F-NL. Preliminary data derived from our ongoing studies indeed show that the HDAC inhibitors suberoylanilide hydroxamic acid (vorinostat) and LBH589 as well as the G9a inhibitor BIX-01294 reduce TGF-β1–induced expression of type I collagen and α-SMA in F-NL and that depletion of G9a and EZH2 reduces basal-level expression of type I collagen and α-SMA in F-IPF (unpublished data), but the role that antifibrotic genes, such as CXCL10 and COX-2, play in this process remains to be investigated.
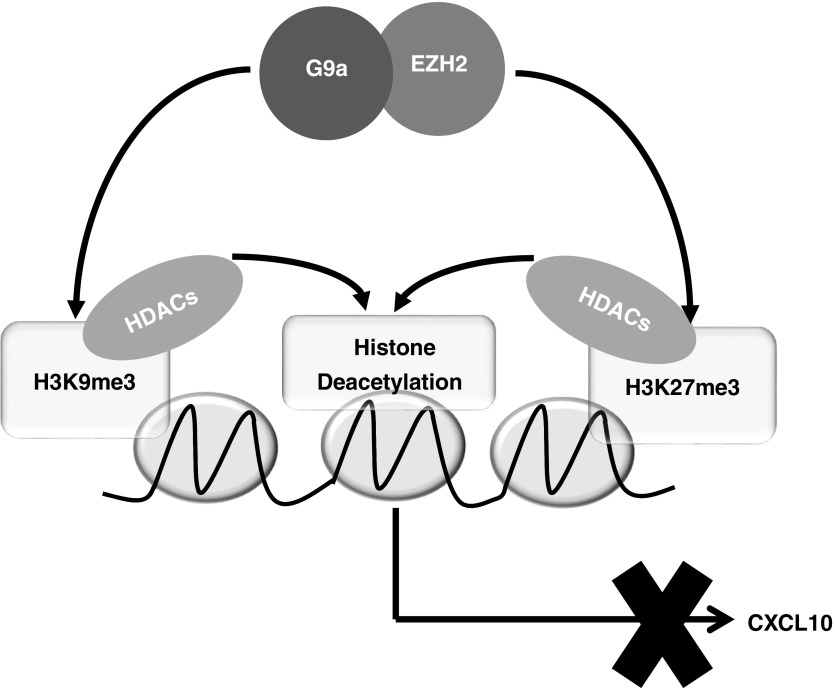
In conclusion, our data strongly suggest that a novel, interdependent interplay between EZH2 and G9a plays a key role in the regulation of the epigenetic repression of CXCL10 and potentially other antifibrotic genes in IPF (Figure 7). Our data also suggest that the machinery for the expression of these genes in IPF remains intact and functional and can mediate derepression once the repressive epigenetic modifications are removed. Thus, the interplay between EZH2 and G9a may represent a viable target to reactivate silenced antifibrotic genes for the therapeutic development for this fatal disease.
Figure 7.
A hypothetical model depicting the role of the interplay between EZH2 and G9a in the regulation of CXCL10 gene repression in idiopathic pulmonary fibrosis. EZH2 and G9a physically and interdependently interact with each other in F-IPF. EZH2- and G9a-mediated H3K27me3 and H3K9me3 result in the recruitment of HDACs to the CXCL10 promoter. This then leads to histone deacetylation, causing reinforced epigenetic silencing of the CXCL10 gene in F-IPF. Inhibition or depletion of either EZH2 or G9a leads to the removal of repressive H3K27me3, H3K9me3, and histone deacetylation, resulting in chromatin derepression and CXCL10 gene reexpression in F-IPF.
Footnotes
Supported by two grants (G0600890 and MR/K003259/1) from the Medical Research Council.
Author Contributions: Conception and design: W.R.C., O.J.B., A.P., and L.P.; analysis and interpretation: W.R.C., O.J.B., A.P., and L.P.; drafting the work or revising it critically for important intellectual content: W.R.C., O.J.B., A.P., G.J., A.J.K., and L.P.; and final approval of the version to be published: W.R.C., O.J.B., A.P., G.J., A.J.K., and L.P.
Originally Published in Press as DOI: 10.1165/rcmb.2017-0286OC on October 20, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ratner M. Landmark approvals in idiopathic pulmonary fibrosis. Nat Biotechnol. 2014;32:1069–1070. doi: 10.1038/nbt1114-1069. [DOI] [PubMed] [Google Scholar]
- 2.Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roach KM, Wulff H, Feghali-Bostwick C, Amrani Y, Bradding P. Increased constitutive αSMA and Smad2/3 expression in idiopathic pulmonary fibrosis myofibroblasts is KCa3.1-dependent. Respir Res. 2014;15:155. doi: 10.1186/s12931-014-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coward WR, Watts K, Feghali-Bostwick CA, Knox A, Pang L. Defective histone acetylation is responsible for the diminished expression of cyclooxygenase 2 in idiopathic pulmonary fibrosis. Mol Cell Biol. 2009;29:4325–4339. doi: 10.1128/MCB.01776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coward WR, Feghali-Bostwick CA, Jenkins G, Knox AJ, Pang L. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J. 2014;28:3183–3196. doi: 10.1096/fj.13-241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilborn J, Crofford LJ, Burdick MD, Kunkel SL, Strieter RM, Peters-Golden M. Cultured lung fibroblasts isolated from patients with idiopathic pulmonary fibrosis have a diminished capacity to synthesize prostaglandin E2 and to express cyclooxygenase-2. J Clin Invest. 1995;95:1861–1868. doi: 10.1172/JCI117866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, et al. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008;39:610–618. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders YY, Kumbla P, Hagood JS. Enhanced myofibroblastic differentiation and survival in Thy-1(−) lung fibroblasts. Am J Respir Cell Mol Biol. 2007;36:226–235. doi: 10.1165/rcmb.2006-0178OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang XM, Zhang Y, Kim HP, Zhou Z, Feghali-Bostwick CA, Liu F, et al. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J Exp Med. 2006;203:2895–2906. doi: 10.1084/jem.20061536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coward WR, Watts K, Feghali-Bostwick CA, Jenkins G, Pang L. Repression of IP-10 by interactions between histone deacetylation and hypermethylation in idiopathic pulmonary fibrosis. Mol Cell Biol. 2010;30:2874–2886. doi: 10.1128/MCB.01527-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keane MP, Arenberg DA, Lynch JPR, III, Whyte RI, Iannettoni MD, Burdick MD, et al. The CXC chemokines, IL-8 and IP-10, regulate angiogenic activity in idiopathic pulmonary fibrosis. J Immunol. 1997;159:1437–1443. [PubMed] [Google Scholar]
- 12.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Epigenetic regulation of myofibroblast differentiation by DNA methylation. Am J Pathol. 2010;177:21–28. doi: 10.2353/ajpath.2010.090999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strieter RM, Kunkel SL, Arenberg DA, Burdick MD, Polverini PJ. Interferon γ-inducible protein 10 (IP-10), a member of the C-X-C chemokine family, is an inhibitor of angiogenesis. Biochem Biophys Res Commun. 1995;210:51–57. doi: 10.1006/bbrc.1995.1626. [DOI] [PubMed] [Google Scholar]
- 14.Happel C, Steele AD, Finley MJ, Kutzler MA, Rogers TJ. DAMGO-induced expression of chemokines and chemokine receptors: the role of TGF-β1. J Leukoc Biol. 2008;83:956–963. doi: 10.1189/jlb.1007685. [DOI] [PubMed] [Google Scholar]
- 15.Tager AM, Kradin RL, LaCamera P, Bercury SD, Campanella GS, Leary CP, et al. Inhibition of pulmonary fibrosis by the chemokine IP-10/CXCL10. Am J Respir Cell Mol Biol. 2004;31:395–404. doi: 10.1165/rcmb.2004-0175OC. [DOI] [PubMed] [Google Scholar]
- 16.Keane MP, Belperio JA, Arenberg DA, Burdick MD, Xu ZJ, Xue YY, et al. IFN-γ-inducible protein-10 attenuates bleomycin-induced pulmonary fibrosis via inhibition of angiogenesis. J Immunol. 1999;163:5686–5692. [PubMed] [Google Scholar]
- 17.Jiang D, Liang J, Campanella GS, Guo R, Yu S, Xie T, et al. Inhibition of pulmonary fibrosis in mice by CXCL10 requires glycosaminoglycan binding and syndecan-4. J Clin Invest. 2010;120:2049–2057. doi: 10.1172/JCI38644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang D, Liang J, Hodge J, Lu B, Zhu Z, Yu S, et al. Regulation of pulmonary fibrosis by chemokine receptor CXCR3. J Clin Invest. 2004;114:291–299. doi: 10.1172/JCI16861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolodsick JE, Peters-Golden M, Larios J, Toews GB, Thannickal VJ, Moore BB. Prostaglandin E2 inhibits fibroblast to myofibroblast transition via E prostanoid receptor 2 signaling and cyclic adenosine monophosphate elevation. Am J Respir Cell Mol Biol. 2003;29:537–544. doi: 10.1165/rcmb.2002-0243OC. [DOI] [PubMed] [Google Scholar]
- 20.Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J. Adenovector-mediated gene transfer of active transforming growth factor-β1 induces prolonged severe fibrosis in rat lung. J Clin Invest. 1997;100:768–776. doi: 10.1172/JCI119590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 22.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 24.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 27.Wu H, Chen X, Xiong J, Li Y, Li H, Ding X, et al. Histone methyltransferase G9a contributes to H3K27 methylation in vivo. Cell Res. 2011;21:365–367. doi: 10.1038/cr.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mozzetta C, Pontis J, Fritsch L, Robin P, Portoso M, Proux C, et al. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell. 2014;53:277–289. doi: 10.1016/j.molcel.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Coward WR, Feghali-Bostwick CA, Jenkins RG, Knox AJ, Pang L. A central role for G9a and EZH2 In the epigenetic repression of IP-10 in idiopathic pulmonary fibrosis [abstract] Am J Respir Crit Care Med. 2014;189:A5580. doi: 10.1096/fj.13-241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilewski JM, Liu L, Henry AC, Knauer AV, Feghali-Bostwick CA. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am J Pathol. 2005;166:399–407. doi: 10.1016/S0002-9440(10)62263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JK, Kim KC. DZNep, inhibitor of S-adenosylhomocysteine hydrolase, down-regulates expression of SETDB1 H3K9me3 HMTase in human lung cancer cells. Biochem Biophys Res Commun. 2013;438:647–652. doi: 10.1016/j.bbrc.2013.07.128. [DOI] [PubMed] [Google Scholar]
- 33.Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 34.Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo Y, Shen L, Suzuki S, Kurokawa T, Masuko K, Tanaka Y, et al. Alterations of DNA methylation and histone modifications contribute to gene silencing in hepatocellular carcinomas. Hepatol Res. 2007;37:974–983. doi: 10.1111/j.1872-034X.2007.00141.x. [DOI] [PubMed] [Google Scholar]
- 36.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 38.Fiskus W, Pranpat M, Balasis M, Herger B, Rao R, Chinnaiyan A, et al. Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells. Mol Cancer Ther. 2006;5:3096–3104. doi: 10.1158/1535-7163.MCT-06-0418. [DOI] [PubMed] [Google Scholar]
- 39.Gasse P, Mary C, Guenon I, Noulin N, Charron S, Schnyder-Candrian S, et al. IL-1R1/MyD88 signaling and the inflammasome are essential in pulmonary inflammation and fibrosis in mice. J Clin Invest. 2007;117:3786–3799. doi: 10.1172/JCI32285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mia MM, Boersema M, Bank RA. Interleukin-1β attenuates myofibroblast formation and extracellular matrix production in dermal and lung fibroblasts exposed to transforming growth factor-β1. PLoS One. 2014;9:e91559. doi: 10.1371/journal.pone.0091559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabasa M, Royo D, Molina-Molina M, Roca-Ferrer J, Pujols L, Picado C, et al. Lung myofibroblasts are characterized by down-regulated cyclooxygenase-2 and its main metabolite, prostaglandin E2. PLoS One. 2013;8:e65445. doi: 10.1371/journal.pone.0065445. [DOI] [PMC free article] [PubMed] [Google Scholar]