Abstract
The progression of chronic obstructive pulmonary disease (COPD) is associated with marked alterations in circulating immune cell populations, but no studies have characterized alterations in these cell types across the full spectrum of lung function impairment in current and former smokers. In 6,299 subjects from the COPDGene and ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints) studies, we related Coulter blood counts and proportions to cross-sectional forced expiratory volume in 1 second (FEV1), adjusting for current smoking status. We also related cell count measures to 3-year change in FEV1 in ECLIPSE subjects. In a subset of subjects with blood gene expression data, we used cell type deconvolution methods to infer the proportions of immune cell subpopulations, and we related these to COPD clinical status. We observed that FEV1 levels are positively correlated with lymphocytes and negatively correlated with myeloid populations, such as neutrophils and monocytes. In multivariate models, absolute cell counts and proportions were associated with cross-sectional FEV1, and lymphocytes, monocytes, and eosinophil counts were predictive of 3-year change in lung function. Using cell type deconvolution to study immune cell subpopulations, we observed that subjects with COPD had a lower proportion of CD4+ resting memory cells and naive B cells compared with smokers without COPD. Alterations in circulating immune cells in COPD support a mixed pattern of lymphocyte suppression and an enhanced myeloid cell immune response. Cell counts and proportions contribute independent information to models predicting lung function, suggesting a critical role for immune response in long-term COPD outcomes. Cell type deconvolution is a promising method for immunophenotyping in large cohorts.
Keywords: chronic obstructive pulmonary disease, gene expression, immunology, computational biology
Chronic obstructive pulmonary disease (COPD) is associated with profound alterations in immune cells within the lung and in the systemic circulation. The systemic inflammation may reflect “spillover” of inflammatory processes within the lung, primary alterations in the extrapulmonary immune response, or a combination of both processes (1). These alterations affect cell types involved in both the innate (2–6) and adaptive (7–15) immune response. In population studies (16, 17) and a systematic review (18), total counts of peripheral leukocytes were associated with cross-sectional and prospective changes in lung function, but few studies have been performed in large cohorts with detailed cell count data to observe relationships across the full spectrum of lung function. In addition, current smoking (CS) has an independent effect on immune cells (8, 19), and often serves as an important potential confounder of immunologic studies of current and former smokers with COPD.
Cell type quantification by flow cytometry is rarely available from large, population-based studies of COPD. However, novel, cell type “deconvolution” approaches have been shown to infer accurately the relative proportions of immune cell types from genome-wide blood gene expression data (20, 21). Thus, cell type deconvolution is a potentially powerful approach to enable the simultaneous study of many different cell types in large cohorts of subjects with available blood gene expression, but it has not yet been applied to cohorts of subjects with COPD.
We hypothesized that: 1) peripheral immune cell types quantified through Coulter complete blood counts (CBCs) have significant associations to cross-sectional forced expiratory volume in 1 second (FEV1) and prospective FEV1 decline; and 2) cell type deconvolution methods can enable the simultaneous study of multiple immune cell subpopulations in cohorts of smokers with COPD and blood gene expression data. We explored the first hypothesis in two large cohorts of smokers enriched for COPD, the COPDGene and Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) studies, which enabled the characterization of immune cell profiles across the full spectrum of lung function impairment while accounting for CS effects. The large number of study subjects allowed for detailed modeling of the relationship between multiple cell types, CS status, and lung function. To explore the second hypothesis, we used two cell type deconvolution methods to infer immune cell subpopulation proportions in a subset of smokers with blood gene expression data in the ECLIPSE study, and we validated these inferred cell type proportions against measured CBC data. We then compared levels of inferred circulating immune cell subpopulations by COPD status, confirming that inferred estimates of circulating immune cell types, such as monocytes, naive B cells, and resting T memory cells, are altered in the COPD state.
Methods
Study Populations
Recruitment criteria and study protocols for the ECLIPSE and COPDGene studies have been previously reported. COPDGene enrolled 10,192 subjects across the entire GOLD (Global Initiative for Obstructive Lung Disease) spectrum between the ages of 45 and 80 years with at least a 10-pack-year smoking history (22). These subjects completed their phase 1 study visit between 2007 and 2011. As of September 24, 2016, 5,000 subjects had completed their phase 2 5-year follow-up visit, which included all of the data items collected in phase 1, as well as complete blood count data, which was not collected at the phase 1 visit.
The ECLIPSE study was a multicenter study that enrolled subjects aged 40–75 years with COPD and at least a 10-year smoking history (COPD defined by FEV1 <80% of predicted and FEV1/FVC ≤0.7) or who were smokers without COPD (FEV1 >85% and FEV1/FVC >0.7). Details of this study have been previously published (23). Gene expression analyses were performed in a subset of subjects in this study from whom genome-wide gene expression data were generated on the Affymetrix Human U133 Plus2 chip, as previously reported (24). For both COPDGene and ECLIPSE, the institutional review boards of all participating centers approved these studies, and written informed consent was obtained from all subjects.
Phenotype and Covariate Definitions
In COPDGene and ECLIPSE, spirometry was performed before and after administration of 180 μg of albuterol according to international guidelines (25). COPD cases and GOLD stages were defined according to GOLD spirometric criteria (FEV1 % of predicted <80% and FEV1/FVC <0.7) (26). Subjects with preserved ratio impaired spirometry were defined by post-bronchodilator FEV1 % of predicted less than 80% and FEV1/FVC greater than 0.7 (27). COPD blood gene expression subtypes were previously defined by Chang and colleagues (24) using network-based stratification (NBS). Of the four NBS subtypes identified in the original publication, the two most prevalent subtypes were analyzed. These subtypes are referred to as the less-impaired lung function and the more-impaired lung function subtypes. CS status, inhaled corticosteroid use, and oral corticosteroid use were ascertained by questionnaire. In ECLIPSE, only eight subjects reported using oral steroids at baseline, and these were removed from subsequent analyses.
Association of CBC Cell Types with COPD GOLD Stage, Cross-Sectional FEV1, and Prospective Change in FEV1
Using 4,558 subjects with complete CBC and spirometric data from the COPDGene phase 2 visit, we plotted the distribution of neutrophil and lymphocyte counts and proportions against GOLD stage after removing outlying cell count observations greater than ±4 SD from the mean. We tested for univariate association between individual cell counts and proportions with post-bronchodilator FEV1 % of predicted using Wald tests from linear regression (LR) models, and we constructed multivariate regression models relating cell counts and proportions to FEV1 adjusting for CS status and oral and inhaled steroid use reported at baseline.
In 1,741 smokers from the ECLIPSE study with complete covariate, CBC, baseline, and prospective FEV1 measurements, we related absolute cell counts and proportions to post-bronchodilator FEV1 % of predicted levels, as described previously here. For the analysis of 3-year change in FEV1 % of predicted levels, we calculated the difference between the first and last available post-bronchodilator FEV1 % of predicted measurement in all study subjects (calculated as: last measurement − first measurement; i.e., negative values represent decline in lung function). To determine the association between CBC measurements and change in FEV1, we used multivariate LR models with change in FEV1 as the response variable, adjusting for baseline FEV1, days of follow-up, inhaled corticosteroid use at baseline, and smoking status at the first and last study visit. Smoking status was represented in four groups (i.e., current smokers at first and last visit, former smokers at first and last visit, current smoker at first visit and former smoker at last visit, and former smoker at first visit and current smoker at last visit). Models were constructed to analyze cell types individually as well as in the presence of other cell types in the same model. Subjects with less than 1,000 days between their first and last spirometric measurements were excluded from analysis.
Gene Expression
Sample preparation and quality control procedures for genome-wide gene expression data in ECLIPSE have been previously described (28). Standard quality control and quantile normalization were performed. Gene expression data used in this study are accessible via GEO (ECLIPSE GSE76705).
Cell Type Deconvolution and Association of Inferred Cell Types with COPD Status
Cell type deconvolution was performed in 221 ECLIPSE subjects with complete genome-wide gene expression and covariate data. Cell type proportions were inferred using two methods—CIBERSORT (21) and the method of Abbas and colleagues (20) using LR (least squares). Cell type reference expression profiles were used from the LM22 pure-cell dataset obtained on December 21, 2015 from the CIBERSORT website (https://cibersort.stanford.edu). A detailed description is provided in the data supplement.
After obtaining cell type estimates of the 22 cell types from both methods, we organized groups of similar cell types into broader categories to create estimates of additional aggregated groups: CD4+ cells, T cells, B cells, lymphocytes, and monocytes/macrophages. We performed this aggregation by summing individual cell type values for cell types within each category. The 22 inferred cell type proportions and aggregated cell type estimates were tested for association with COPD status and COPD molecular subtypes using the Wilcoxon-Mann-Whitney test. Significant cell type associations were considered to be those with a Wilcoxon-Mann-Whitney test P value less than 0.05 for both CIBERSORT and LR.
Prediction Models for COPD Status and COPD Molecular Subtypes
Classification of subjects according to COPD status or NBS molecular subtype using estimated cell type quantities, CBC quantities, and clinical covariates was performed in 221 subjects from ECLIPSE using the support vector machine (SVM) implementation in the e1071 package (29). Validation within ECLIPSE involved performing one round of partitioning in which half of the subjects was used in the training set and the other half was used in the validation set. Probabilities were returned from the SVM and used with the R package ROCR to generate receiver operating characteristic plots and calculate areas under the curves (AUCs) (30).
Additional details regarding study cohorts and statistical methods are included in the supplemental Materials in the data supplement.
Results
Relating Circulating Immune Cells to Cross-Sectional FEV1 and CS
We examined CBC data from 4,558 smokers from the COPDGene phase 2 visit and an additional 1,741 smokers with more than 1,000 days of spirometric follow-up data in the ECLIPSE study. The clinical characteristics and cell type distributions of analyzed subjects in both studies are shown in Table 1. The CONSORT diagram for the analyses of cross-sectional and longitudinal data in ECLIPSE is shown in Figure E1 in the data supplement, and comparison of characteristics of ECLIPSE analyzed and excluded subjects is shown in Table E1.
Table 1.
Characteristics of Analyzed Subjects in COPDGene and Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints Study
| COPDGene | ECLIPSE | |
|---|---|---|
| N | 4,558 | 1,741 |
| Age, yr | 65.5 (8.7) | 61.9 (7.9) |
| Sex, % female | 50 | 36 |
| Race, % African American | 27 | 0 |
| FEV1 % of predicted | 78.3 (24.9) | 55.0 (26.1) |
| FEV1/FVC | 0.67 (0.15) | 0.66 (0.21) |
| COPD, % GOLD 2–4 | 35 | 84 |
| Pack-years | 44.0 (24.0) | 45.4 (26.5) |
| Current smoking, % | 37 | 39 |
| BMI | 28.9 (6.3) | 26.6 (5.3) |
| Oral steroids, % | 2 | 0 |
| Inhaled steroids, % | 24 | 59 |
| Neutrophil, % | 59.4 (10.0) | 63.9 (8.2) |
| Neutrophil, 1,000 cells/μl | 4.3 (1.6) | 5.0 (1.6) |
| Lymphocyte, % | 29.4 (9.4) | 26.8 (7.6) |
| Lymphocyte, 1,000 cells/μl | 2.0 (0.7) | 2.0 (0.6) |
| Monocyte, % | 8.1 (2.4) | 6.2 (2.1) |
| Monocyte, 1,000 cells/μl | 0.6 (0.2) | 0.5 (0.2) |
| Eosinophil, % | 2.6 (1.7) | 2.8 (1.7) |
| Eosinophil, 1,000 cells/μl | 0.2 (0.1) | 0.2 (0.1) |
| Basophil, % | 0.6 (0.5) | 0.3 (0.2) |
| Basophil, 1,000 cells/μl | 0.03 (0.04) | 0.03 (0.02) |
Definition of abbreviations: BMI = body mass index; COPD = chronic obstructive pulmonary disease; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; FEV1 = forced expiratory volume in 1 second; FVC = forced vital capacity; GOLD = Global Initiative for Obstructive Lung Disease.
Data are mean (SE) unless otherwise indicated.
LR relating the absolute amount and percentage of five cell types to FEV1 % of predicted indicated that neutrophils, lymphocytes, monocytes, and eosinophils are strongly correlated with FEV1, and there are differences in the pattern of association between absolute counts and cell proportions with COPD severity (Table 2 and Table E2). Boxplots showing the amount of each cell type by GOLD stage for COPDGene and ECLIPSE are shown in Figures E2 and E3.
Table 2.
Relationship of Cell Type Counts and Proportions to Forced Expiratory Volume in 1 Second % of Predicted in 4,558 Smokers in COPDGene
| Cell Type Count (1,000 Cells/μl) |
Cell Type Proportion |
|||
|---|---|---|---|---|
| β | P value | β | P value | |
| Neutrophils | −3.53 (0.22) | <0.001 | −0.48 (0.04) | <0.001 |
| Lymphocytes | 2.34 (0.51) | <0.001 | 0.58 (0.04) | <0.001 |
| Monocytes | −21.84 (1.88) | <0.001 | −0.29 (0.16) | 0.06 |
| Eosinophils | −21.25 (3.06) | <0.001 | −0.54 (0.22) | 0.02 |
| Basophils | −33.28 (9.81) | 0.001 | 0.09 (0.7) | 0.89 |
Definition of abbreviations: β = change in forced expiratory volume in 1 second per unit change in cell count or proportion; COPD = chronic obstructive pulmonary disease.
Each row corresponds to a separate univariate model.
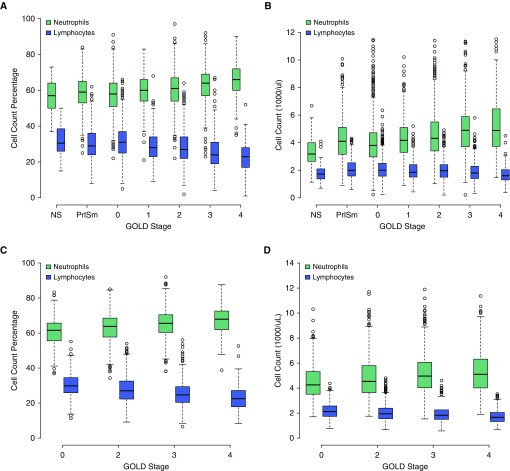
Given the predominance of neutrophils and lymphocytes in blood, we examined the absolute counts and percentages of these cell types across GOLD stages, and we observed two phenomena. First, with increasing COPD severity, the proportion of neutrophils increases and lymphocytes decreases. However, in terms of absolute cell counts, the number of neutrophils increases while the total number of lymphocytes remains relatively stable (Figures 1A and 1C), suggesting that the observed changes in neutrophil and lymphocyte proportions associated with COPD severity are primarily driven by an increase in the number of circulating neutrophils. The same pattern is present in ECLIPSE subjects (Figures 1B and 1D).
Figure 1.
Neutrophil and lymphocyte counts and proportions stratified by GOLD (Global Initiative for Obstructive Lung Disease) spirometric stage. As GOLD stage increases, the relative proportions of peripheral neutrophils and lymphocytes increase and decrease, respectively ([A] COPDGene and [C] ECLIPSE [Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints]). This phenomenon is driven by an increase in absolute neutrophil count, whereas the absolute amount of lymphocytes remains relatively stable across GOLD stages ([B] COPDGene and [D] ECLIPSE). COPD = chronic obstructive pulmonary disease; NS = nonsmoker; PrISm = preserved ratio impaired spirometry (i.e., subjects with forced expiratory volume in 1 second [FEV1]/forced vital capacity >0.7, but FEV1 % predicted <80%).
We evaluated these relationships in a series of models in COPDGene relating cell count, cell proportion, and CS to FEV1 % of predicted while adjusting for inhaled and oral steroid use (Table 3). The models explaining the largest proportion of variance in FEV1, after adjusting for model complexity, included both cell counts and proportions, demonstrating that both measures have independent association to FEV1. Both lymphocyte and neutrophil absolute counts and percentages were significantly associated with FEV1 across most models. The addition of monocyte counts and proportions to the models did not affect the association between neutrophil quantifications and FEV1 (data not shown).
Table 3.
Multivariate Models of the Relationship between Forced Expiratory Volume in 1 Second % of Predicted and Neutrophil and Lymphocyte Quantifications
| Neutrophil Count | Neutrophil % | Lymphocyte Count | Lymphocyte % | Current Smoking | Neutrophil Count × Current Smoking | Neutrophil % × Current Smoking | Lymphocyte Count × Current Smoking | Lymphocyte % × Current Smoking | Adjusted R2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cell proportions | — | 0.39 (0.10)* | — | 0.75 (0.11)* | — | — | — | — | — | 0.274 |
| Cell counts | −2.19 (0.20)* | — | 1.72 (0.44)* | — | — | — | — | — | — | 0.275 |
| Cell counts + current smoking | −2.73 (0.27)* | — | 1.79 (0.58)* | — | −5.74 (2.48)* | 1.21 (0.40)* | — | −0.06 (0.90) | — | 0.276 |
| Cell proportions + counts + current smoking | −2.47 (0.62)* | 0.74 (0.14)* | −0.41 (1.32) | 0.94 (0.16)* | −10.99 (18.91) | 0.24 (1.03) | 0.13 (0.23) | 1.85 (2.07) | −0.1 (0.27) | 0.285 |
Adjusted R2 is the variance explained adjusted for model complexity. Cell counts are in units of 1,000 cells/μl. Each row corresponds to a separate multivariate model. Each model is also adjusted for oral and inhaled steroid use.
P < 0.05.
Relating Circulating Immune Cells to Prospective, 3-Year Change in Lung Function
Because the CBC data in COPDGene were obtained at visit 2, longitudinal FEV1 analysis measures for this cohort were not available. We performed longitudinal analysis for 3-year change in FEV1 % of predicted in 1,741 smokers from the ECLIPSE study who were not taking oral steroids at baseline. In an analysis of single cell type measures, lymphocyte, monocyte, and eosinophil counts and proportions were significantly associated with change in FEV1 (Table 4). Higher monocyte levels at baseline were associated with greater FEV1 decline, and the opposite pattern was observed for eosinophils. Neutrophil proportions, but not counts, were significantly associated with lung function decline. Larger neutrophil proportions were associated with more lung function decline, with the opposite relationship observed for lymphocyte proportion.
Table 4.
Relationship of Cell Type Counts and Proportions at Baseline to 3-Year Change in Forced Expiratory Volume in 1 Second % of Predicted in 1,741 Smokers in the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints Study
| Cell Type Count (1,000 Cells/μl) |
Cell Type Proportion |
|||
|---|---|---|---|---|
| β | P Value | β | P Value | |
| Neutrophils | −0.09 (0.12) | 0.48 | −0.05 (0.02) | 0.031 |
| Lymphocytes | 0.63 (0.32) | 0.05 | 0.07 (0.03) | 0.008 |
| Monocytes | −3.41 (1.08) | 0.002 | −0.32 (0.09) | 0.001 |
| Eosinophils | 4.16 (1.43) | 0.004 | 0.34 (0.11) | 0.003 |
| Basophils | −8.65 (12.8) | 0.50 | −0.50 (1.00) | 0.62 |
Definition of abbreviations: β = change in forced expiratory volume in 1 second (FEV1) over 3 years per unit change in cell count or proportion; COPD = chronic obstructive pulmonary disease.
Change in FEV1, calculated as: last visit value − first visit value (i.e., negative values indicate decline in FEV1). Models analyze one cell type at a time. All models are adjusted for FEV1 % of predicted at baseline, number of days of follow-up, inhaled corticosteroid use at baseline, and smoking status at baseline and at last study visit.
Table 5 demonstrates that, for multivariate models, including counts and proportions of these four cell types, cell counts, but not proportions, showed significant associations to change in FEV1. Absolute counts of monocytes, eosinophils, and lymphocytes were significantly associated with FEV1 decline (P = 0.0003, P = 0.0004, and P = 0.02, respectively). Higher levels of monocytes were associated with larger amounts of FEV1 decline, and the opposite pattern was present for lymphocytes and eosinophils.
Table 5.
Multivariate Models Relating Cell Type Counts and Proportions at Baseline to 3-Year Change in Forced Expiratory Volume in 1 Second % of Predicted in 1,741 Smokers in the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints Study
| Neutrophil Count | Neutrophil % | Lymphocyte Count | Lymphocyte % | Monocyte Count | Monocyte % | Eosinophil Count | Eosinophil % | |
|---|---|---|---|---|---|---|---|---|
| Cell proportions | — | 1.03 (1.03) | — | 1.11 (1.03) | — | 0.69 (1.04) | — | 1.43 (1.05) |
| Cell counts | 0 (0.13) | — | 0.76 (0.32)* | — | −4.64 (1.18)* | — | 4.98 (1.46)* | — |
Definition of abbreviation: COPD = chronic obstructive pulmonary disease.
P < 0.05.
Cell counts are in units of 1,000 cells/μl. Each row corresponds to a separate multivariate regression model. Models adjusted for baseline forced expiratory volume in 1 second % of predicted, number of days of follow-up, inhaled corticosteroid use at baseline, and current smoking status at baseline and last measurement.
Association of Inferred Lymphocyte Subpopulations to COPD and COPD Subtypes
In a subset of 221 subjects from ECLIPSE with complete genome-wide blood gene expression and covariate data (subject characteristics shown in Table E3), we used cell type deconvolution to estimate the proportion of immune cell subpopulations in each study subject, and we related these proportions to COPD case–control status.
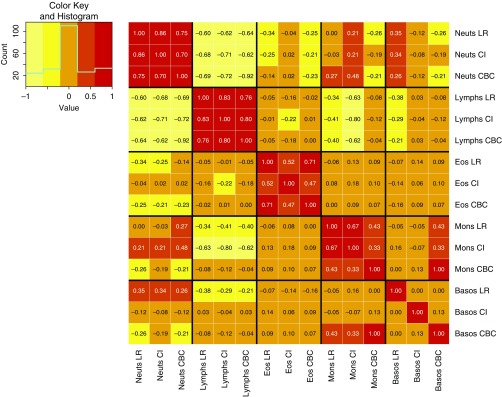
To first assess the performance of cell type deconvolution in blood gene expression from smokers, we examined results from applying two methods that have been previously validated for the detection of immune cell types: CIBERSORT and the LR method of Abbas (20, 21). To benchmark these algorithms against known cell type quantifications, we compared their neutrophil, aggregated lymphocyte, aggregated monocyte, eosinophil, and basophil quantifications against concurrently drawn CBCs (Figure 2). Both methods showed high correlation to neutrophils and lymphocytes (Spearman r ranges from 0.7 to 0.8, P < 0.001), with weaker correlations for eosinophils and monocytes. Correlation with basophils was low for both methods. For the inferred proportions of neutrophils and lymphocytes, the correlation between methods was high (0.86 and 0.83, respectively).
Figure 2.
Correlation between inferred cell subpopulation proportions and complete blood count (CBC) cell type proportions. Spearman correlation between estimated cell subpopulation proportions from two deconvolution methods and CBC proportions. Basos = basophils; CI = CIBERSORT; Eos = eosinophils; LR = linear regression; Mons = monocytes; Neuts = neutrophils; Lymph = lymphocytes.
We compared the inferred cell type proportions by COPD case–control status and observed that, relative to smoker controls, subjects with COPD had significantly lower levels of aggregated lymphocytes, aggregated T cells, CD4+ resting memory cells, and naive B cells, and increased levels of monocytes (Table 6).
Table 6.
Inferred Immune Cell Types Significantly Associated with Chronic Obstructive Pulmonary Disease status
| Linear Regression |
CIBERSORT |
|||||
|---|---|---|---|---|---|---|
| Controls | Cases | P Value | Controls | Cases | P Value | |
| Aggregated monocytes | −0.41 (0.93) | 0.25 (0.96) | 5.4E-07 | −0.35 (0.98) | 0.20 (0.94) | 6.3E-05 |
| Monocytes | −0.33 (0.99) | 0.21 (0.95) | 4.9E-05 | −0.32 (0.97) | 0.19 (0.94) | 2.1E-04 |
| Naive B cells | 0.21 (0.89) | −0.13 (1.06) | 2.4E-03 | 0.15 (0.92) | −0.11 (1.04) | 1.5E-03 |
| Aggregated T cells | 0.24 (0.99) | −0.15 (0.99) | 5.1E-03 | 0.40 (0.98) | −0.22 (0.94) | 3.7E-05 |
| CD4 memory resting T cells | 0.25 (1.05) | −0.16 (0.95) | 6.4E-03 | 0.23 (1.25) | −0.15 (0.79) | 0.01 |
| Aggregated lymphocytes | 0.24 (0.94) | −0.13 (1.01) | 0.02 | 0.25 (0.97) | −0.13 (0.98) | 0.02 |
Cell count values correspond to the estimated cell proportions normalized to a mean of 0 and SE of 1. See Figures E4 and E5 for boxplot distributions. Values are mean (SD) unless otherwise indicated. P values calculated using Wilcoxon rank sum test. The following cell types were not analyzed because they were detected in less than 10% of samples: M1 macrophages; activated dendritic cells; activated mast cells; γ-δ T cells; and T follicular helper cells.
Inferred Cell Type Proportions Predict COPD Blood Gene Expression Subtypes
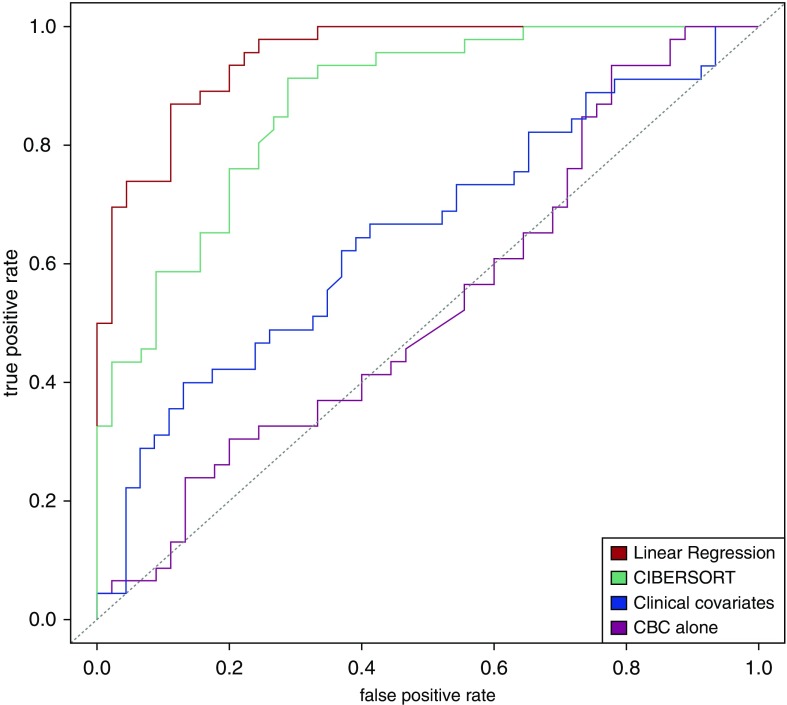
In a previous publication, the ECLIPSE blood gene expression data had been used to define COPD molecular subtypes that differed in clinical characteristics and blood gene expression patterns, and we demonstrated that these subtypes could not be recovered using CBC data alone (24). To determine whether these molecular subgroups can be accurately predicted from inferred cell count proportions, we trained SVM classifiers to predict NBS subtype and COPD case–control status using CBC data, clinical covariates, or inferred cell type proportions. Figure 3 demonstrates that predictive models for NBS subtypes using inferred cell type proportions classified subjects by COPD molecular subtype with high accuracy (AUC = 0.95), and demonstrated better performance than models using only CBC cell type quantities (AUC = 0.53) or clinical covariates (AUC = 0.65). Predictive models for COPD case–control status using inferred cell type proportions also showed statistically significant, but less powerful, predictive performance (AUC = 0.71), with the cell type subpopulation models still outperforming the models using CBC data (Table E4).
Figure 3.
Performance of predictive models for COPD molecular subtypes using CBCs, inferred cell subpopulation proportions, and clinical covariates. Receiver operating characteristic curves demonstrate the predictive performance of support vector machine classifiers using complete blood count data, inferred cell subpopulation data, and clinical covariates for COPD molecular subtypes in 221 Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints subjects. Clinical covariates are age, sex, and pack-years.
We compared levels of the inferred cell type proportions between the more-impaired lung function and less-impaired lung function COPD molecular subtypes, and observed that the list of immune cell types that were significantly different between COPD molecular subtypes was more extensive than between COPD cases and controls. This list included T-regulatory cells, CD4 resting and activated memory T cells, memory B cells, aggregated T cells, aggregated B cells, dendritic cells, and monocytes (Table E5, Figures E4 and E5).
Discussion
Using two large cohorts enriched for subjects with COPD, we characterized alterations in circulating immune cell types associated with cross-sectional FEV1 and longitudinal FEV1 decline. The main findings are: 1) the predominant peripheral immune cell type alteration associated with increasing COPD severity is an increase in the absolute count of neutrophils; 2) monocytes and eosinophils have strong multivariate associations to prospective change in FEV1; and 3) cell type estimates from gene expression deconvolution methods show good accuracy for some cell types.
Prior immunologic studies of COPD have identified important associations with increased innate immune activation and progression of COPD, including neutrophil stimulation (31, 32), alveolar macrophage immune surveillance (33), protease/matrikine activation (34), and activation of the dendritic cell/macrophage axis (35). Most of these studies have been performed in murine models or small-to-moderate-sized study samples with limited ability to control for the effects of CS. Our findings complement and extend previous results by demonstrating that: 1) the decrease in overall lymphocyte proportion in COPD is primarily driven by an increase in absolute neutrophil counts; 2) absolute counts and proportions of immune cell types have independent, statistically significant associations to lung function; and 3) absolute monocyte and eosinophil counts are predictive of COPD progression. This point extends previous observations relating total peripheral leukocyte count to cross-sectional and longitudinal lung function (16–18) by implicating specific myeloid cell types. The fact that monocytes were associated with cross-sectional FEV1 and prospective FEV1 decline, whereas neutrophils only showed multivariate association to cross-section FEV1, is an interesting finding. Previous work has demonstrated an M2 predominant transcriptomic signature in airway macrophages from subjects with COPD, but it is not clear if these disease-related alterations in lung-resident macrophages are supported by replenishment from systemic monocytes or, rather, expansion of lung-resident populations (36). Although our findings do not directly address this question, the association of circulating monocytes with FEV1 decline suggests that systemic monocyte populations do play a role in COPD-related pathobiology. Although circulating neutrophils are clearly associated with COPD severity, they were not an independent predictor of decline after accounting for other cell types and covariates. However, precise mechanistic hypotheses are beyond the scope of this epidemiologic study, and would require detailed assessment of both the lung and systemic compartments.
To study immune cell subpopulations not quantified by CBC, we explored the use of cell type deconvolution to quantify 22 distinct cell subpopulations in a subset of ECLIPSE subjects with blood genome-wide gene expression data. These methods are a promising alternative for estimating cell type proportions in large study samples with available expression data. However, these approaches have not been widely applied in smokers with COPD. Our findings demonstrate that, in smokers enriched for COPD, the deconvolution approaches studied yielded consistent and reasonably reliable estimates of neutrophil and lymphocyte cell proportions with mixed performance for other cell types. Inferred cell type proportions enabled significantly better prediction of externally defined COPD molecular subtypes than CBC and clinical data alone, providing indirect evidence that these inferred proportions capture meaningful information on the cell type composition of bulk blood expression samples. These data provide proof of concept of the feasibility of using cell type deconvolution to study immune cell subpopulations in large cohorts of smokers with available blood gene expression data.
Analysis of the cell type deconvolution data supports previous observations of an overall decrease in lymphocytes and T cells in the COPD state, coupled with an increase in monocytes. When we studied deconvolved cell types in previously defined COPD molecular subtypes, differences in cell type proportions were more pronounced, with the more severely affected subtype characterized by increased monocytes, T-regulatory cells, memory T cells, and memory B cells, as well as decreased total lymphocytes. This pattern is consistent with the expected behavior of T-regulatory cells, which play a critical immunomodulatory role by suppressing other lymphocyte populations in part through IL-10 and transforming growth factor-β signaling (37). Overall, these findings provide additional support for the model that the circulating immune response to CS and COPD is characterized by distinct aspects of suppression of the adaptive immune response and a chronic increase in myeloid cell types, and suggest that, within smokers, there are patterns of coordinated immune response that can be used to identify clinically distinct subgroups of subjects.
The main strength of this study is that peripheral cell type quantifications were available from a large number of smokers with a broad range of lung function from two independent studies. The study design also enabled the study of immune cell alterations adjusting for CS, a major confounder of the relationship between immune cell alterations and COPD severity. The use of cell type deconvolution to study the immune response in COPD is novel, and enabled the simultaneous study of a large number of lymphocyte subpopulations. Because CBC quantifications and blood gene expression were available in the same ECLIPSE subjects from the same time point, we could benchmark our deconvolution approaches against a known standard.
This study also has important limitations. Our analyses were based on a single CBC measurement, and do not explicitly account for measurement error in the CBC. We adjusted for the most common medications in this cohort that could affect cell counts (oral and inhaled corticosteroids), but we did not adjust for other less commonly used medications that can alter CBC measures. We did not have access to immune cells in the lung or specific lung compartments in our study subjects, thus we could not relate the blood observations to the lung compartment. We also were not able to characterize immune cell functional states through cytokine profiling or quantification of response to antigenic stimulation. Although we observed significant associations between circulating immune cell subpopulations and COPD, further study is required to determine the pathophysiological significance of these observations. We did not have flow cytometry values against which we could benchmark our cell type deconvolution estimates, but we did have Coulter counter data available, and our deconvolution results were generated using methods that have been previously validated against flow cytometry for immune cell populations (20, 21).
In conclusion, analysis of CBCs and proportions in over 6,000 subjects from the COPDGene and ECLIPSE studies demonstrated that cross-sectional FEV1 is associated with alterations in multiple circulating immune cell types, including total neutrophil count. COPD disease progression, as quantified by decline in FEV1, is associated with increased absolute monocyte counts and decreased lymphocyte and eosinophil counts at baseline. Cell type deconvolution is a viable approach to simultaneously study multiple immune cell populations in smokers with COPD. Future studies to characterize COPD-related alterations in more fine-grained immune cell types will benefit from quantification of both cell type proportions and absolute counts.
Footnotes
This work was supported by the University of Colorado School of Medicine Medical Student Research Track, National Heart, Lung, and Blood Institute (NHLBI) grants R01HL089897, R01HL089856, R01HL124233, and R01HL126596; the COPDGene study (NCT00608764) is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, GSK, Siemens, and Sunovion; the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints Study (GSK code SCO104960) was funded by GSK.
Author Contributions: Study concept and design—E.H.-S., R.P.B., and P.J.C.; acquisition, analysis, or interpretation of data—E.H.-S., R.P.B., and P.J.C.; drafting of the manuscript—E.H.-S., R.P.B., and P.J.C.; critical revision of the manuscript for important intellectual content—all authors; statistical analysis—E.H.-S., R.P.B., and P.J.C.; obtained funding—R.P.B. and P.J.C.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2017-0373OC on December 5, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Barnes PJ, Celli BR. Systemic manifestations and comorbidities of COPD. Eur Respir J. 2009;33:1165–1185. doi: 10.1183/09031936.00128008. [DOI] [PubMed] [Google Scholar]
- 2.Oudijk E-JD, Nijhuis EHJ, Zwank MD, van de Graaf EA, Mager HJ, Coffer PJ, et al. Systemic inflammation in COPD visualised by gene profiling in peripheral blood neutrophils. Thorax. 2005;60:538–544. doi: 10.1136/thx.2004.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaguliene N, Zemaitis M, Lavinskiene S, Miliauskas S, Sakalauskas R. Local and systemic neutrophilic inflammation in patients with lung cancer and chronic obstructive pulmonary disease. BMC Immunol. 2013;14:36. doi: 10.1186/1471-2172-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sapey E, Stockley JA, Greenwood H, Ahmad A, Bayley D, Lord JM, et al. Behavioral and structural differences in migrating peripheral neutrophils from patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183:1176–1186. doi: 10.1164/rccm.201008-1285OC. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson GC, Seemungal TAR, Patel IS, Bhowmik A, Wilkinson TMA, Hurst JR, et al. Airway and systemic inflammation and decline in lung function in patients with COPD. Chest. 2005;128:1995–2004. doi: 10.1378/chest.128.4.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Eeden SF, Lawrence E, Sato Y, Kitagawa Y, Hogg JC. Neutrophils released from the bone marrow by granulocyte colony-stimulating factor sequester in lung microvessels but are slow to migrate. Eur Respir J. 2000;15:1079–1086. doi: 10.1034/j.1399-3003.2000.01516.x. [DOI] [PubMed] [Google Scholar]
- 7.Arnson Y, Shoenfeld Y, Amital H. Effects of tobacco smoke on immunity, inflammation and autoimmunity. J Autoimmun. 2010;34:J258–J265. doi: 10.1016/j.jaut.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 8.de Jong JW, van der Belt-Gritter B, Koëter GH, Postma DS. Peripheral blood lymphocyte cell subsets in subjects with chronic obstructive pulmonary disease: association with smoking, IgE and lung function. Respir Med. 1997;91:67–76. doi: 10.1016/s0954-6111(97)90070-6. [DOI] [PubMed] [Google Scholar]
- 9.Zhu X, Gadgil AS, Givelber R, George MP, Stoner MW, Sciurba FC, et al. Peripheral T cell functions correlate with the severity of chronic obstructive pulmonary disease. J Immunol. 2009;182:3270–3277. doi: 10.4049/jimmunol.0802622. [DOI] [PubMed] [Google Scholar]
- 10.Silver JS, Kearley J, Copenhaver AM, Sanden C, Mori M, Yu L, et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat Immunol. 2016;17:626–635. doi: 10.1038/ni.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts MEP, Higgs BW, Brohawn P, Pilataxi F, Guo X, Kuziora M, et al. CD4+ T-cell profiles and peripheral blood ex-vivo responses to T-cell directed stimulation delineate COPD phenotypes. Chronic Obstr Pulm Dis. 2015;2:268–280. doi: 10.15326/jcopdf.2.4.2015.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majori M, Corradi M, Caminati A, Cacciani G, Bertacco S, Pesci A. Predominant TH1 cytokine pattern in peripheral blood from subjects with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 1999;103:458–462. doi: 10.1016/s0091-6749(99)70471-9. [DOI] [PubMed] [Google Scholar]
- 13.Hodge G, Nairn J, Holmes M, Reynolds PN, Hodge S. Increased intracellular T helper 1 proinflammatory cytokine production in peripheral blood, bronchoalveolar lavage and intraepithelial T cells of COPD subjects. Clin Exp Immunol. 2007;150:22–29. doi: 10.1111/j.1365-2249.2007.03451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morissette MC, Parent J, Milot J. Perforin, granzyme B, and FasL expression by peripheral blood T lymphocytes in emphysema. Respir Res. 2007;8:62. doi: 10.1186/1465-9921-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Urbanowicz RA, Tighe PJ, Todd I, Corne JM, Fairclough LC. Differential activation of killer cells in the circulation and the lung: a study of current smoking status and chronic obstructive pulmonary disease (COPD) PLoS One. 2013;8:e58556. doi: 10.1371/journal.pone.0058556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sparrow D, Glynn RJ, Cohen M, Weiss ST. The relationship of the peripheral leukocyte count and cigarette smoking to pulmonary function among adult men. Chest. 1984;86:383–386. doi: 10.1378/chest.86.3.383. [DOI] [PubMed] [Google Scholar]
- 17.Chan-Yeung M, Abboud R, Buncio AD, Vedal S. Peripheral leucocyte count and longitudinal decline in lung function. Thorax. 1988;43:462–466. doi: 10.1136/thx.43.6.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan WQ, Man SFP, Senthilselvan A, Sin DD. Association between chronic obstructive pulmonary disease and systemic inflammation: a systematic review and a meta-analysis. Thorax. 2004;59:574–580. doi: 10.1136/thx.2003.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoonhorst SJM, Timens W, Koenderman L, Lo Tam Loi AT, Lammers J-WJ, Boezen HM, et al. Increased activation of blood neutrophils after cigarette smoking in young individuals susceptible to COPD. Respir Res. 2014;15:121. doi: 10.1186/s12931-014-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One. 2009;4:e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, et al. ECLIPSE Investigators. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) Eur Respir J. 2008;31:869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 24.Chang Y, Glass K, Liu Y-Y, Silverman EK, Crapo JD, Tal-Singer R, et al. COPD subtypes identified by network-based clustering of blood gene expression. Genomics. 2016;107:51–58. doi: 10.1016/j.ygeno.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Standardization of spirometry, 1994 update: American Thoracic Society. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 26.Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report: GOLD Executive Summary. Am J Respir Crit Care Med. 2017;195:557–582. doi: 10.1164/rccm.201701-0218PP. [DOI] [PubMed] [Google Scholar]
- 27.Wan ES, Hokanson JE, Murphy JR, Regan EA, Make BJ, Lynch DA, et al. COPDGene Investigators. Clinical and radiographic predictors of GOLD-unclassified smokers in the COPDGene study. Am J Respir Crit Care Med. 2011;184:57–63. doi: 10.1164/rccm.201101-0021OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh D, Fox SM, Tal-Singer R, Bates S, Riley JH, Celli B. Altered gene expression in blood and sputum in COPD frequent exacerbators in the ECLIPSE cohort. PLoS One. 2014;9:e107381. doi: 10.1371/journal.pone.0107381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:27. [Google Scholar]
- 30.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 31.Hoenderdos K, Condliffe A. The neutrophil in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2013;48:531–539. doi: 10.1165/rcmb.2012-0492TR. [DOI] [PubMed] [Google Scholar]
- 32.Quint JK, Wedzicha JA. The neutrophil in chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2007;119:1065–1071. doi: 10.1016/j.jaci.2006.12.640. [DOI] [PubMed] [Google Scholar]
- 33.Barnes PJ. Alveolar macrophages as orchestrators of COPD. COPD. 2004;1:59–70. doi: 10.1081/COPD-120028701. [DOI] [PubMed] [Google Scholar]
- 34.Gaggar A, Weathington N. Bioactive extracellular matrix fragments in lung health and disease. J Clin Invest. 2016;126:3176–3184. doi: 10.1172/JCI83147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim EY, Battaile JT, Patel AC, You Y, Agapov E, Grayson MH, et al. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nat Med. 2008;14:633–640. doi: 10.1038/nm1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlahos R, Bozinovski S. Role of alveolar macrophages in chronic obstructive pulmonary disease. Front Immunol. 2014;5:435. doi: 10.3389/fimmu.2014.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vignali DAA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]