Abstract
The last step in the synthesis of lignin and suberin has been proposed to be catalyzed by peroxidases, although other proteins may also be involved. To determine which peroxidases are involved in the synthesis of lignin and suberin, five peroxidases from tomato (Lycopersicon esculentum) roots, representing the majority of the peroxidase activity in this organ, have been partially purified and characterized kinetically. The purified peroxidases with isoelectric point (pI) values of 3.6 and 9.6 showed the highest catalytic efficiency when the substrate used was syringaldazine, an analog of lignin monomer. Using a combination of transgenic expression and antibody recognition, we now show that the peroxidase pI 9.6 is probably encoded by TPX1, a tomato peroxidase gene we have previously isolated. In situ RNA hybridization revealed that TPX1 expression is restricted to cells undergoing synthesis of lignin and suberin. Salt stress has been reported to induce the synthesis of lignin and/or suberin. This stress applied to tomato caused changes in the expression pattern of TPX1 and induced the TPX1 protein. We propose that the TPX1 product is involved in the synthesis of lignin and suberin.
Plants have a large number of peroxidase isoenzymes that may differ by more than 50% in amino acid sequence (Welinder, 1992). Peroxidases (EC 1.11.1.7) are oxidoreductases that catalyze the oxidation of a diverse group of organic compounds using hydrogen peroxide as the ultimate electron acceptor (Dawson, 1988). Peroxidases have been suggested to be involved in various metabolic steps such as auxin catabolism (Normanly et al., 1995), the formation of isodi-Tyr bridges in the cross-linking of cell wall proteins (Schnabelrauch et al., 1996), the cross-linking of pectins by diferulic bridges (Amaya et al., 1999), and the oxidation of cinnamyl alcohols prior to their polymerization during lignin and suberin formation (Roberts et al., 1988; Whetten et al., 1998). In addition to the common aromatic matrix, suberin contains an aliphatic domain that makes this polymer highly hydrophobic (Kolattukudy, 1980). This difference between lignin and suberin accounts for the distinct functions proposed and, consequently, their different distribution throughout plant tissues (Wallace and Fry, 1994).
Lignin is present in vascular plants and is mainly synthesized in cells to become part of the transport system. Suberin is synthesized in cells from the endodermis and exodermis of the root, where it strengthens the cell wall and contributes to control the water movement. In aerial parts, suberin is also considered to be a component of the wound- and pathogen-induced plant defense response (Mohan et al., 1993). The last catalytic step in the synthesis of the lignin and the aromatic domain of suberin is the oxidation of cinnamyl alcohols, and this is catalyzed by a peroxidase and/or laccase enzyme (Whetten et al., 1998). The involvement of a specific peroxidase in this catalytic step has been largely examined due to the interest in the control of the metabolic steps involved in the synthesis and composition of these polymers (Bernards and Lewis, 1998).
From early studies, peroxidases were classified as acidic and basic isoenzymes according to their pI values. Some reports suggested that peroxidases with acidic pI values were responsible for the oxidation of cinnamyl alcohols during the ligno-suberization (Mohan et al., 1993). However, the involvement of basic peroxidases in lignin biosynthesis has been reported (Liu and Ger, 1997). The oxidation of syringaldazine, a lignin monomer analog, by a particular peroxidase was suggested to be indicative of its involvement in the synthesis of lignin and suberin (Pang et al., 1989; Catesson, 1992; Christensen et al., 1998). Other studies have used the expression of specific peroxidase genes in lignifying or suberizing plant tissues as the argument for their role in these biosynthetic pathways (Mohan et al., 1993; Christensen et al., 1998). Transgenic approaches have also been used to provide information on the role of several peroxidases in lignin synthesis, although the information is not always consistent (Whetten et al., 1998). As indicated by Lewis and Yamamoto (1990), there must be a confluent information from kinetics, structural, and gene expression studies for involving a peroxidase in a specific metabolic step.
A tomato (Lycopersicon esculentum) peroxidase gene, TPX1, encoding a basic isoenzyme, is specifically expressed in roots, and its expression was up-regulated after treatment with 100 mm NaCl (Botella et al., 1994a). In addition, TPX1 transcripts were induced in vascular tissue of aerial parts after wounding (Botella et al., 1994b). Recently, we reported an increase in lignin content of tomato leaves of transgenic plants overexpressing TPX1 (Mansouri et al., 1999). These results suggested that the cell wall-targeted peroxidase TPX1 might be involved in the ligno-suberization of the root and the aerial parts.
Metabolic responses to salt stress are complex, since many processes such as carbon metabolism, accumulation of compatible osmolytes, ion partitioning, energy metabolism, and growth are modified (Bohnert and Sheveleva, 1998). Specifically, at the cellular level it has been shown that salt stress affects the cell wall by both a decrease in cell expansion (Iraki et al., 1989) and an increase in polymerization of monolignols of the root (Cruz et al., 1992).
To determine which peroxidase was involved in the process of synthesis of lignin and/or suberin in tomato roots, five peroxidase isoenzymes representing the majority of the total activity in this organ were purified. Two of the purified isoenzymes showed the highest catalytic efficiency with syringaldazine as the substrate. In the present study, we show that one of these peroxidases is encoded by TPX1. Immunological studies and in situ RNA localization in control and salinized plants support a role for TPX1 in ligno-suberization in tomato roots.
MATERIALS AND METHODS
Plant Material
Tomato (Lycopersicon esculentum L. Mill. cv Pera) seedlings were grown for 2 weeks in vermiculite and then transferred to hydroponic culture, where growth and NaCl treatment were performed as previously described (Botella et al., 1994a). Root tissue was sampled from 2-month-old plants with and without treatments with 100 mm NaCl in the culture medium. Tobacco (Nicotiana tabacum) transgenic plants overexpressing TPX1 were obtained by the following procedure: The TPX1 cDNA was transferred from Bluescript pBSII (Stratagene, La Jolla, CA) cloning vector to the binary plasmid pKYLX71 (Schardl et al., 1987) using PCR. Specific primers with targeted restriction sites were used to include sequence encoding the signal peptide in the insert of the expression cassette. The PKYLX71-TPX1 vector was transferred to the LBA4404 strain of Agrobacterium tumefaciens by electroporation. Transgenic tobacco plants were generated following standard procedures (Horsch et al., 1985).
Peroxidase Isoenzyme Isolation from Tomato Roots and Partial Purification
Tomato roots were homogenized in a mortar at 0°C to 4°C with 10 mm sodium phosphate, pH 6.0, containing 1 m KCl (1:4, w/v) and 25% (w/v) polyvinylpyrrolidone. Homogenates were shaken at 25°C for 2 h and then centrifuged at 1,500g for 10 min. Supernatants were used as crude extracts for further purification. This crude extract was gradually brought to 80% (w/v) ammonium sulfate at 0°C to 4°C and centrifuged at 10,000g for 30 min. The precipitate was resuspended in 1.5 mL of 0.1 m sodium phosphate, pH 7.2, containing NaCl 0.1 m and dialyzed overnight against the same buffer at 0°C to 4°C.
This extract was loaded on a Sephacryl S-200 chromatography column previously equilibrated with 0.1 m sodium phosphate, pH 7.2, and eluted with 100 mL of the same buffer. As a result, one peak of peroxidase activity was eluted. The fractions with this activity were pooled and dialyzed overnight against 10 mm Tris-Cl, pH 8.0, at 0°C to 4°C.
The dialyzed pool was loaded on a DEAE Sephacel chromatography column equilibrated with 10 mm Tris-Cl, pH 8.0. Three peaks of peroxidase activity were detected in the elution of the column with the washing buffer. When each of them was monitored on an isoelectric focusing (IEF) gel, showed pI values of 9.6, 8.2, and 7.5. Further elution of the DEAE Sephacel column with a linear gradient (0.01–0.2 m NaCl in the equilibrium buffer) gave three peaks in the profile. The first and the second peak corresponded to a mixture of peroxidases in the IEF gel. The third activity peak included two peroxidases with pI values of 6.5 and 3.6. This last peak was dialyzed against 25 mm phosphate, pH 6.0, and aliquots were loaded in two different columns: one with DEAE Sephacel and the other with SP-Sephadex C50. Elution from the DEAE Sephacel with 25 mm phosphate, pH 6.0, gave a single peak corresponding to the pI 6.5 peroxidase, and the elution from the SP-Sephadex C50 column with 25 mm phosphate, pH 6.0, gave a peak corresponding to the pI 3.6 peroxidase.
Peroxidase Extraction from Tobacco Seedlings
Tobacco seedlings were homogenized in a mortar at 0°C to 4°C with 50 mm potassium phosphate, pH 6.0, (1:1.5, w/v) with 25% (w/v) polyvinylpyrrolidone. The extract was centrifuged at 15,000 rpm for 30 min at 4°C. Supernatants were used as soluble extracts of peroxidases, then lyophilized and resuspended in 50 mm potassium phosphate buffer, pH 6.0, to concentrate the protein. The pellet was subjected to reextraction until the supernatants contained no detectable peroxidase activity. Peroxidases ionically bound to the cell wall were obtained by extracting the pellet with the same buffer containing 1 m KCl after shaking for 2 h at 4°C. Supernatants containing the peroxidases ionically bound to the cell wall were dialyzed overnight against 25 mm phosphate, pH 6.0, and then lyophilized and resuspended in 50 mm phosphate, pH 6.0, to obtain a detectable peroxidase activity.
Peroxidase Activity, PAGE, and IEF
Peroxidase activity was determined with o-dianisidine as a substrate in a 1-mL reaction mixture containing 0.63 mm o-dianisidine, 2.8 mm H2O2, and 50 mm sodium phosphate, pH 5.7; the activity was measured following the continuous increase in A460 at 37°C (ε460 nm: 11.3 mm−1 cm−1) (Quesada et al., 1990). When syringaldazine was the substrate, the activity was determined at 30°C in a 1-mL reaction mixture containing syringaldazine 0.156 mm (or various concentration for substrate curves), 0.05 mm H2O2, in all cases, and 100 mm sodium phosphate, pH 7.4, following the absorbance increase at 530 nm (ε530 nm of 27 mm−1 cm−1) as described by Goldberg et al. (1983). One unit of enzyme activity (U) is defined as the amount of enzyme that oxidizes 1 μmol of phenolic substrate at the temperature and pH specified for each reaction (Goldberg et al., 1983). Protein was monitored in the chromatographic procedures by their A280 and A260 (Stoscheck, 1990), and in the extracts by the method of Bradford (1976) using bovine serum albumin as the standard. Cationic PAGE was performed on 7.5% (w/v) polyacrylamide gels at 3 mA gel−1 (Reisfeld et al., 1962). IEF of soluble extracts and those ionically bound to cell walls were performed on a pH range of 8.0 to 10 on vertical polyacrylamide gel slabs (Robertson et al., 1987). Gels were stained for peroxidase activity using the substrate o-dianisidine (Quesada et al., 1990).
Immunological Methods
The purified peroxidase of pI 9.6 was used as immunogen to produce antisera. A fraction of the purified isoenzyme containing 20 μg of protein was mixed with equal volume of a Freund's complete adjuvant as previously described (Quesada et al., 1990). The homogeneous emulsion was injected subcutaneously to the rabbit. Prior to the experiment, samples of preimmune serum were obtained. The antiserum was obtained and the antibodies purified as described by Quesada et al. (1990). ELISA assays were performed according to the method of Tigier et al. (1991). Western blot from cationic PAGE and IEF polyacrylamide slabs on a Immobilon polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA) were performed using 0.7% (w/v) acetic acid at 100 V and 250 mA for 45 min. Membranes were processed as described for tissue printing.
Tissue Printing of Root Sections and in Situ Hybridization
Tomato roots were hand-sectioned 15 to 30 mm from the tip and impressions were made on Immobilon PVDF membranes (Millipore) (Peyrano et al., 1997). Antisera anti-pI 9.6 peroxidase (dilution 1:250) was used as primary antibody, and the reaction was detected as described in the amplified alkaline phosphatase goat-anti-rabbit immunoblot assay kit from Bio-Rad (Hercules, CA).
Root sections (5 mm long) from the apical, middle, and basal part of the root were collected and immediately fixed in formaldehyde. Tissue sections (10 μm thick) and in situ hybridizations were carried out as previously described by Coen et al. (1990). Probes for in situ hybridization were labeled with digoxigenin-11-UTP. TPX1 insert was cloned in Bluescript II SK (−) plasmid. Plasmid pTPX1 was linearized with a restriction enzyme that cuts the flanking polylinker region farther from the T3 promoter (XhoI), and 0.6 mg was used as a template to synthesize digoxigenin-labeled antisense RNA using T3 polymerase (no unlabeled UTP was used in the reaction). Sense RNA probe was synthesized as described above but using T7 RNA polymerase and pTPX1 previously cut with BamHI as a template, and used as a negative control. The RNA probes were subjected to mild alkaline hydrolysis by heating at 60°C for 45 min in 100 mm carbonate buffer (pH 10.2), and about 4% of each labeling reaction in 40 μL of hybridization buffer (Ingham et al., 1985) was used as a probe for each slide and incubated at 50°C overnight.
RESULTS
Kinetic Studies of Tomato Root Peroxidases
A large number of isoenzymes of tomato roots were visualized in an IEF gel using the substrate o-dianisidine. Five peroxidases with pI values of 9.6, 8.2, 7.5, 6.5, and 3.6 represented the majority of the peroxidase activity. Each of these peroxidases were purified using ammonium sulfate fractionation and several chromatographic steps (see “Materials and Methods”). The kinetic parameters of the purified isoenzymes are shown in Table I. Peroxidases with pI values of 9.6 and 3.6 showed the highest values of the catalytic efficiency for syringaldazine (Table I). kcat/Km measures the enzyme catalytic efficiency with a particular substrate, which provides valuable information when comparing several enzyme forms for the same substrate.
Table I.
Kinetic properties of the purified peroxidase isoenzymes from tomato roots using syringaldazine as a substrate
| Isoenzyme | Vmax | Km | [Et] | kcat | kcat/Km |
|---|---|---|---|---|---|
| pI | μm s−1 | μm | s−1 | μm−1 s−1 | |
| 9.6 | 6.83 | 11.4 | 0.41 | 17.0 | 1.50 |
| 8.2 | 2.52 | 14.7 | 0.21 | 12.0 | 0.82 |
| 7.5 | 0.41 | 8.0 | 0.25 | 1.6 | 0.20 |
| 6.5 | 1.40 | 46.3 | 0.11 | 12.7 | 0.27 |
| 3.6 | 12.30 | 26.0 | 0.32 | 38.4 | 1.47 |
Vmax, ΔA530·s−1/ɛ (for syrindaldazine); ɛ, molar extinction coefficient; [Et], enzyme concentration; A430/ɛ (for peroxidase); kcat, Vmax/[Et]; kcat/Km, catalytic efficiency.
Antibodies against pI 9.6 Isoenzyme Recognize Other Tomato Root Peroxidases with Different Affinity
Polyclonal antibodies were obtained against the purified pI 9.6 isoenzyme, and the cross-reactivity with all of the purified peroxidases was evaluated using ELISA (Table II). Peroxidases of pI 9.6 and 3.6 had the highest cross-reactivity with the antibodies. This high level of recognition of pI 9.6 and 3.6 isoenzymes is an indication of the similarity of the two proteins at the immunological level despite their different pI values. The pattern of recognition of the different peroxidases by the antibodies (Table II) correlated somewhat with the catalytic efficiency for syringaldazine (Table I).
Table II.
Percentage of cross-reaction of the different peroxidase isoenzymes from tomato roots with the rabbit polyclonal antibodies raised against the pI 9.6 isoenzyme using a quantitative ELISA method
| Isoenzyme | A405 | Protein | Cross-Reaction |
|---|---|---|---|
| pI | mg mL−1 | % | |
| 9.6 | 0.152 | 1.80 | 100 |
| 8.2 | 0.089 | 1.40 | 77 |
| 7.5 | 0.057 | 0.86 | 48 |
| 6.5 | 0.039 | 0.73 | 40 |
| 3.6 | 0.156 | 1.84 | 100 |
The pI 9.6 Peroxidase Is Encoded by TPX1
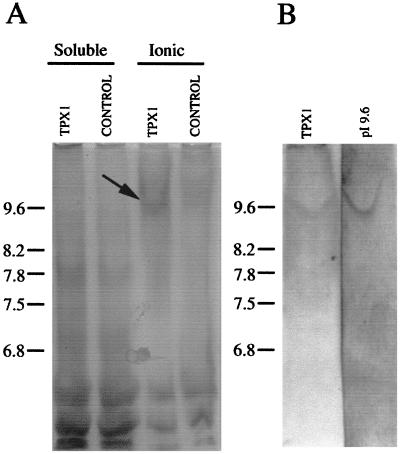
To determine whether TPX1 corresponds to any of the basic peroxidases purified, we used transgenic tobacco plants that overexpress TPX1. Higher total peroxidase activity was detected in the cell wall fraction of the transgenic plants compared with untransformed plants, which suggests that TPX1 is located in the cell wall. A new peroxidase with a pI value of 9.6 appeared in leaves of tobacco plants transformed with TPX1 under the control of the cauliflower mosaic virus 35S promoter (Fig. 1A). This peroxidase was not detected in leaves of tobacco transformed with the empty vector. Moreover, the polyclonal antibodies raised against the purified peroxidase with a pI of 9.6 recognized the new protein in tobacco plants overexpressing TPX1. This new protein recognized in transgenic tobacco plants has the same pI value (9.6) of the purified protein from tomato root (Fig. 1B). Although the theoretical pI value deduced from the TPX1 sequence is 7.5 (Botella et al., 1994b), we propose that TPX1 encodes the native tomato root peroxidase of pI 9.6. Putative glycosylation of TPX1 and the conformation of the native protein could account for this difference.
Figure 1.
IEF in the pH 8.0 to 10 range of extracts prepared from the aerial part of tobacco seedlings developed for peroxidase activity and cross-reacted with the antibodies against the peroxidase of pI 9.6. A, Peroxidase activity of the soluble and ionic extracts from transgenic (TPX1) and control tobacco lines. B, Western blot of the IEF gel of the ionic extract. One unit of enzyme is defined as the amount of enzyme forming 1 μmol product min−1. The same activity (3 units) was loaded in each lane.
TPX1 Expression Is Restricted to Cells Undergoing Synthesis of Lignin and Suberin
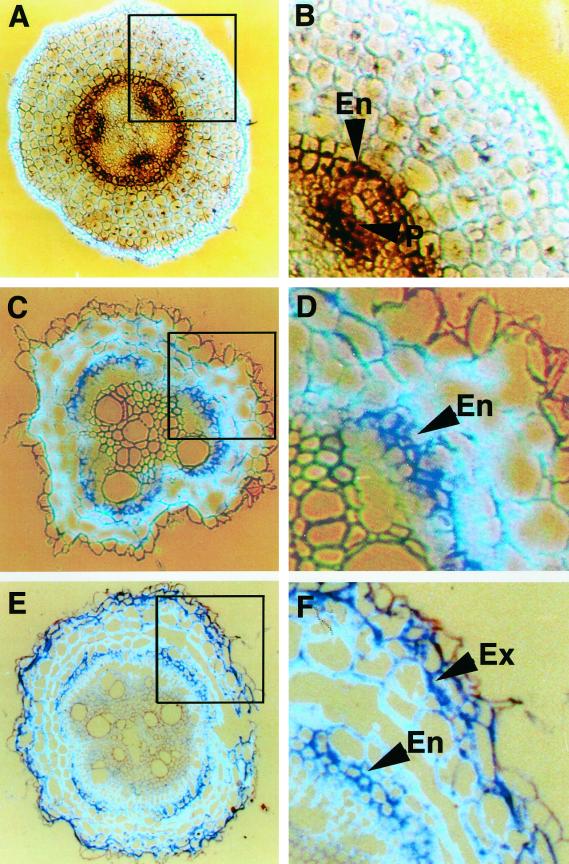
The expression of TPX1 was studied in tomato roots using in situ mRNA hybridization. In the apical zone of the root (last 0.5 cm from the root tip), TPX1 transcripts were detected in the endodermis and the protoxylem (Fig. 2, A and B). In the medium zone (8–10 cm from the tip), TPX1 transcripts were detected in the endodermis and the hypodermis (Fig. 2, C and D). In most species, these cells deposit suberin and are called exodermis (Peterson, 1988). In the basal zone (13–15 cm from the tip), TPX1 expression was also detected in the endodermis and the exodermis (Fig. 2, E and F).
Figure 2.
TPX1 expression in tomato roots by in situ hybridization. Transversal sections of hydroponically grown tomato roots were probed with digoxigenin-labeled antisense TPX1 RNA. A, Expression of TPX1 (RNA signal was detected as a dark brown or purple color) in the apical zone of the root (last 0.5 cm from the root tip). B, Detail of A showing that the expression is restricted to the endodermis (En) and protoxylem (P). C, Expression of TPX1 (purple) in the medium zone of the root (8–10 cm from the root tip). D, Detail of C showing the expression in the endodermis (En). E, Expression of TPX1 in the basal zone of the root (13–15 cm from the root tip). F, Detail of E revealing expression in the endodermis (En) and the exodermis (Ex).
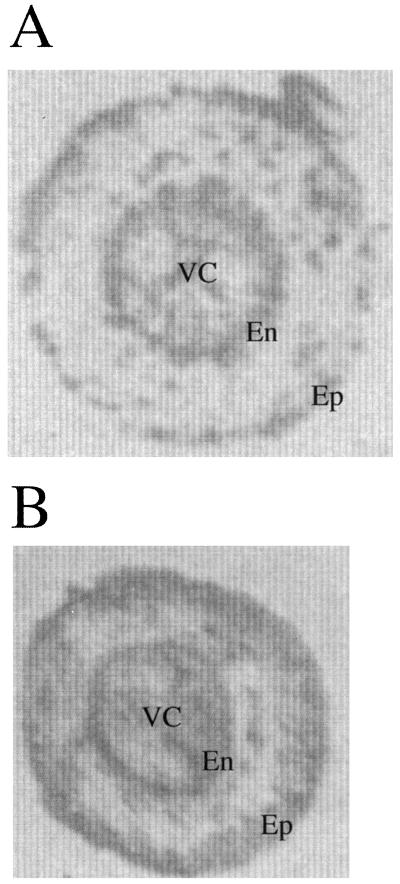
Root tissue printing studies were performed using TPX1 antibodies as shown in Figure 3A. Root sections corresponded to the medium zone of plants grown under identical conditions as those used in the in situ mRNA studies. Antibodies were associated to the endodermis, exodermis, and vascular cylinder. Because TPX1 antibodies recognize other peroxidases from tomato root, the signals may represent the mixture of several peroxidases.
Figure 3.
Tissue printing of hand-sectioned tomato roots made to react with the antibodies raised against the pI 9.6 isoenzyme. A, Control. B, Salinized. En, Endodermis; Ep, epidermis; VC, vascular cylinder.
Changes of TPX1 Expression under Salt Stress
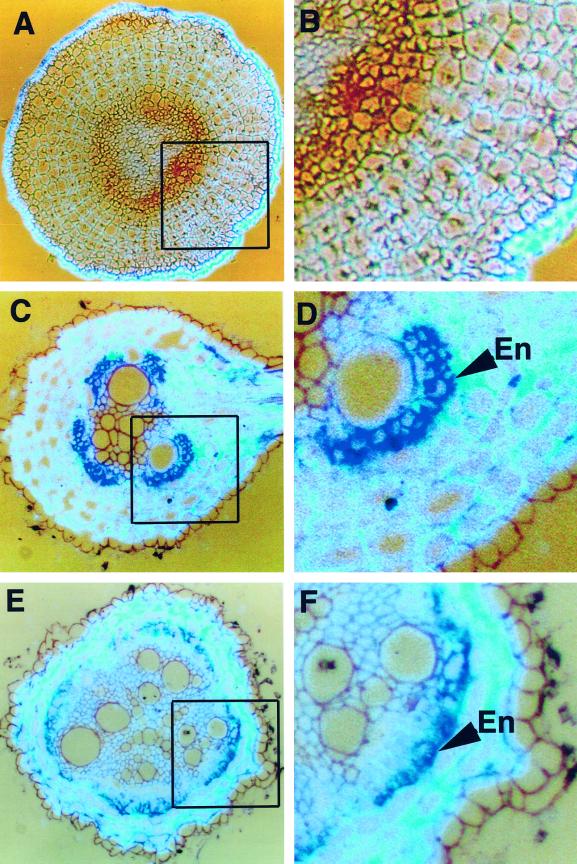
Salt stress produces an alteration of water transport as a consequence of an increase in the amount of lignin and suberin in the root (Cruz et al., 1992). In tomato, it has been reported that NaCl treatment produces both an increase in TPX1 transcripts in tomato roots (Botella et al., 1994a) and a decrease in the hydraulic conductance of roots (Peyrano et al., 1997). To investigate whether TPX1 expression is affected by salt stress at the cellular level, we also analyzed the expression of TPX1 using in situ hybridization in tomato roots after treatment with 100 mm NaCl. The root sections analyzed were equivalent to those shown in Figure 2. In the apical zone of the root, NaCl treatment abolished TPX1 expression in both the endodermis and the protoxylem (Fig. 4, A and B). In the medium zone of the root, an increase of TPX1 transcripts in the endodermis was observed after NaCl treatment (Fig. 4, C and D). This increasein TPX1 expression is consistent with previous results obtained by northern analysis (Botella et al., 1994a). In the basal part of the root, TPX1 expression was repressed in the exodermis; however, expression in the endodermis appeared unaffected (Fig. 4, E and F).
Figure 4.
Analysis of TPX1 expression in root sections of tomato plants treated with 100 mm NaCl for 24 h. A, Expression of TPX1 (dark brown) in the apical zone of the root (last 0.5 cm from the root tip). B, Detail of A showing that the expression is completely abolished in the endodermis and protoxylem after NaCl treatment. C, Expression of TPX1 (purple) in the medium zone of the root (8–10 cm from the root tip). D, Detail of C showing TPX1 expression in the endodermis (En). E, Expression of TPX1 in the basal zone of the root (13–15 cm from the root tip). F, Detail of E revealing that TPX1 expression is limited to the endodermis (En).
pI 9.6 Isoenzyme Is Increased after Salt Treatment
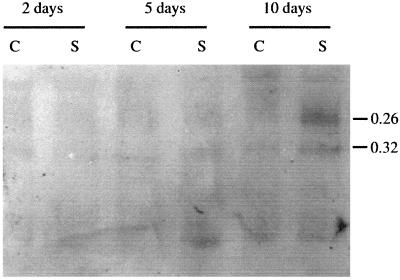
Antibodies against pI 9.6 peroxidase isoenzyme were used to determine if the changes detected in TPX1 transcripts after salt stress were followed by changes in the amount of this protein. However, the lack of specificity of the antibodies obtained required the separation of TPX1 prior to immunodetection (Table II). Purified peroxidases separated in a PAGE system at basic pH were identified by their mobility. Peroxidases with RF values of 0.32 and 0.26 in this gel corresponded to the peroxidases TPX1 (pI 9.6) and pI 8.2. Western analysis of tomato root proteins in control and salinized plants showed an increase in pI 9.6 and pI 8.2 peroxidases 10 d after salt treatment (Fig. 5). Increase of total peroxidase protein recognized by pI 9.6 antibodies of salt-treated roots was observed (Fig. 3B), however, tissue printing is not as quantitative as western-blot analysis.
Figure 5.
Western blot of cationic PAGE of tomato root extracts at different days after NaCl treatment. C, Control; S, salinized. Peroxidases of RF values of 0.26 and 0.32 correspond to the 8.2 and 9.6 pI isoforms, respectively.
DISCUSSION
Multigene families of peroxidases have been found in all species studied thus far; however, it remains difficult to assign a specific peroxidase to an in vivo function. Recently, the reactions catalyzed by peroxidases have been analyzed in depth at both the structural and the mechanistic level, with new insights obtained from site-directed mutagenesis (Smith and Veitch, 1998). However, the information available does not explain the occurrence and physiological significance of the multiple peroxidase isoenzymes. An approach used to uncover the function of peroxidases has been the development of transgenic plants either overexpressing or underexpressing a specific peroxidase gene (Sherf et al., 1993; Kajita et al., 1994; McIntyre et al., 1996; Lagrimini et al., 1997). However, this approach has failed to provide definitive information and the in vivo role of any peroxidase remains elusive.
Several reports have shown that peroxidases present in tissues undergoing lignification display in vitro activity toward syringaldazine (Catesson, 1992; Christensen et al., 1998). Among tomato root peroxidases, the peroxidase of pI 9.6 displays a high affinity for syringaldazine (Km of 11.4 μm) and the highest value of catalytic efficiency jointly with the pI 3.6 peroxidase, 1.5 μm−1 s−1 and 1.47 μm−1 s−1, respectively (Table I). The values for catalytic efficiency are used to evaluate the preference of an enzyme for different substrates and represents the rate constant of the reaction to form the enzyme-substrate complex (Fersht, 1985).
We have previously characterized the tomato gene TPX1, which encodes a basic peroxidase (Botella et al., 1993). TPX1 is expressed in tomato roots and in vascular tissue after wounding (Botella et al., 1994a, 1994b). Overexpression of TPX1 in tobacco and the use of antibodies against the pI 9.6 isoenzyme suggest that this peroxidase is the product of TPX1. In addition, it unequivocally confirms TPX1 cell wall targeting, since the antibodies only recognized a protein extracted at high ionic strength when cell wall proteins are being extracted. This was expected because the 5′ sequence corresponding to the predicted signal peptide was included in the expression cassette of the binary vector (see “Materials and Methods”).
In situ RNA studies of TPX1 expression established a correlation between the expression of this gene and the synthesis of lignin and suberin in specific root cells. Root formation is a highly controlled developmental process determined by the metabolic activities of the different cell types. In some of these cells the synthesis of lignin and suberin occurs at specific times or in different cells (Peterson, 1988; Lewis and Yamamoto, 1990). Lignin synthesis occurs in the protoxylem, whose final destination is to become part of the root vascular system. Once the xylem vessels are formed, the synthesis of lignin stops. Synthesis of suberin mainly occurs in the endodermis. It has also been reported that synthesis of suberin also occurs in the exodermis, a layer of root cells that extends from the epidermis and to the cortical cells (Peterson, 1988). TPX1 is expressed in cells undergoing active lignin and suberin synthesis.
The expression pattern of TPX1 was modified by NaCl treatment. An increased TPX1 expression in the endodermis and the exodermis are in agreement with previous physiological changes observed in tomato root under salt stress, in which a decreased water conductance was reported as result of the salt stress (Peyrano et al., 1997). In other plant species, diminished water conductance in the roots after osmotic stress has been explained by the increased ligno-suberization of this organ (Cruz et al., 1992).
Several reports on transgenic plants with diminished expression of specific peroxidases did not reduce the amount of lignin (McIntyre et al., 1996; Lagrimini et al., 1997). The possibility of several peroxidases involved in the synthesis of lignin and/or suberin in a plant species has been argued to explain the lack of effect in lignin content in transgenic plants underexpressing a specific peroxidase (Sherf et al., 1993). A different transgenic approach is to overexpress a peroxidase gene. We have reported that TPX1 overexpression in tomato significantly increased the lignin content in transgenic tomato plants (Mansouri et al., 1999). This information, together with the results from the present study, further supports the involvement of TPX1 in ligno-suberization in tomato root.
ACKNOWLEDGMENT
We thank Dr. D. Bradley for advice on in situ hybridization experiments.
Footnotes
This research was supported by Comisión Interministerial de Ciencia y Tecnología (grant no. BIO94–0622–CO2–01) from the Ministerio de Educación y Ciencia, Spain, and by Consejo Investigaciones Científicas y Técnicas Provincia de Córdoba, Secretaría Ciencia y Técnica-Universidad Nacional Río Cuarto, and Consejo Nacional de Investigaciones Científicas y Técnicas from Argentina. M.Q. was the recipient of a fellowship from Consejo Investigaciones Científicas y Técnicas Provincia de Córdoba, Córdoba, Argentina, and she acknowledges the Agencia Esparíola de Cooperación Internacional, Spain, for short-term financial support to stay in Málaga, Spain.
LITERATURE CITED
- Amaya I, Botella MA, Calle M, Medina MI, Heredia A, Bressan RA, Hasegawa PM, Quesada MA, Valpuesta V. Improved germination under osmotic stress or tobacco plants overexpressing a cell wall peroxidase. FEBS Lett. 1999;457:80–84. doi: 10.1016/s0014-5793(99)01011-x. [DOI] [PubMed] [Google Scholar]
- Bernards MA, Lewis NG. The macromolecular aromatic domain in suberized tissue: a changing paradigm. Phytochemistry. 1998;47:915–933. doi: 10.1016/s0031-9422(98)80052-6. [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Sheveleva E. Plant stress adaptations: making metabolism move. Curr Opin Plant Biol. 1998;1:267–274. doi: 10.1016/s1369-5266(98)80115-5. [DOI] [PubMed] [Google Scholar]
- Botella MA, Quesada MA, Hasegawa PM, Valpuesta V. Nucleotide sequences of two peroxidase genes from tomato (Lycopersicon esculentum) Plant Physiol. 1993;103:665–666. doi: 10.1104/pp.103.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella MA, Quesada MA, Kononowicz AK, Bressan RA, Pliego F, Hasegawa PM, Valpuesta V. Characterization and in situ localization of a salt-induced tomato peroxidase mRNA. Plant Mol Biol. 1994a;25:105–114. doi: 10.1007/BF00024202. [DOI] [PubMed] [Google Scholar]
- Botella MA, Quesada MA, Medina MI, Pliego F, Valpuesta V. Induction of a tomato peroxidase gene in vascular tissue. FEBS Lett. 1994b;347:195–198. doi: 10.1016/0014-5793(94)00542-7. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Catesson AM. Plant peroxidases and cell differentiation: cyto- and histological aspects. In: Penel C, Gaspar T, Greppin H, editors. Plant Peroxidases 1980–1990, Topics and Detailed Literature on Molecular, Biochemical and Physiological Aspects. Switzerland: University of Geneva; 1992. pp. 117–124. [Google Scholar]
- Christensen JH, Bauw G, Welinder KG, Van Montagu M, Boerjan W. Purification and characterization of peroxidases correlated with lignification in poplar xylem. Plant Physiol. 1998;118:125–135. doi: 10.1104/pp.118.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Carpenter R. Floricula: a homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Cruz RT, Jordan WR, Drew MC. Structural changes and associated reduction of hydraulic conductance in roots of Sorghum bicolor L. following exposure to water deficit. Plant Physiol. 1992;99:203–212. doi: 10.1104/pp.99.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson JH. Probing structure-function relations in heme-containing oxygenases and peroxidases. Science. 1988;240:433–439. doi: 10.1126/science.3358128. [DOI] [PubMed] [Google Scholar]
- Fersht A. Enzyme Structure and Mechanism. W.H. New York: Freeman; 1985. pp. 105–106. [Google Scholar]
- Goldberg R, Catesson AM, Czaninski Y. Some properties of syringaldazine oxidase, a peroxidase specifically involved in the lignification processes. Z Pflanzenphysiol. 1983;110:267–279. [Google Scholar]
- Horsch RB, Fry FE, Hoffman NL, Eicholtz D, Rojers SJ, Fraley RT. A simple and general method of transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Howard KR, Ish-Horowicz D. Transcription pattern of the Drosophila segmentation gene hairy. Nature. 1985;318:439–445. [Google Scholar]
- Iraki NM, Bressan RA, Hasegawa PM, Carpita NC. Alteration of the physical and chemical structure of the primary cell wall of growth-limited plant cells adapted to osmotic stress. Plant Physiol. 1989;91:39–47. doi: 10.1104/pp.91.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita S, Osakabe K, Katayama Y, Kawai S, Matsumoto Y, Hata K, Morohoshi N. Agrobacterium-mediated transformation of poplar using disarmed binary vector and the overexpression of a specific member of a family of poplar peroxidase genes in transgenic poplar cell. Plant Sci. 1994;103:231–239. [Google Scholar]
- Kolattukudy PE. Biopolyester membranes of plants: cutin and suberin. Science. 1980;208:990–1000. doi: 10.1126/science.208.4447.990. [DOI] [PubMed] [Google Scholar]
- Lagrimini LM, Gingas V, Finger F, Rothstein S, Liu TTY. Characterization of antisense transformed plants deficient in the tobacco anionic peroxidase. Plant Physiol. 1997;114:1187–1196. doi: 10.1104/pp.114.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NG, Yamamoto E. Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- Liu ZH, Ger MJ. Changes of enzyme activity during pollen germination in maize, and possible evidence of lignin synthesis. Aust J Plant Physiol. 1997;24:329–335. [Google Scholar]
- Mansouri IE, Mercado JA, Santiago-Doménech N, Pliego-Alfaro F, Valpuesta V, Quesada MA. Biochemical and phenotypical characterization of transgenic tomato plants overexpressing a basic peroxidase. Physiol Plant. 1999;106:355–362. [Google Scholar]
- McIntyre CL, Bettenay HM, Manners JM. Strategies for the suppression of peroxidase gene expression in tobacco. II. In vivo suppression of peroxidase activity in transgenic tobacco using ribozyme and antisense constructs. Transgenic Res. 1996;5:263–270. doi: 10.1007/BF01972880. [DOI] [PubMed] [Google Scholar]
- Mohan R, Bajar AM, Kolattukudy PE. Induction of a tomato anionic peroxidase gene (tap1) by wounding in transgenic tobacco and activation of tap1/GUS and tap2/GUS chimeric gene fusions in transgenic tobacco by wounding and pathogen attack. Plant Mol Biol. 1993;21:341–354. doi: 10.1007/BF00019949. [DOI] [PubMed] [Google Scholar]
- Normanly J, Slovin JP, Cohen JD. Rethinking auxin biosynthesis and metabolism. Plant Physiol. 1995;107:323–329. doi: 10.1104/pp.107.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang A, Catesson AM, Francesch C, Rolando C, Goldberg R. On substrate specificity of peroxidases involved in the lignification process. J Plant Physiol. 1989;135:325–329. [Google Scholar]
- Peterson CA. Exodermal Casparian bands: their significance for ion uptake by roots. Physiol Plant. 1988;72:204–208. [Google Scholar]
- Peyrano G, Taleisnik E, Quiroga M, de Forchetti SM, Tigier H. Salinity effects on hydraulic conductance, lignin content and peroxidase activity in tomato roots. Plant Physiol Biochem. 1997;35:387–393. [Google Scholar]
- Quesada MA, Tigier HA, Bukovac MJ, Valpuesta V. Purification of an anionic isoperoxidase from peach seeds and its immunological comparison with other anionic isoperoxidases. Physiol Plant. 1990;79:623–628. doi: 10.1111/j.1399-3054.1990.tb00035.x. [DOI] [PubMed] [Google Scholar]
- Reisfeld RA, Lewis VJ, Williams DE. Disc electrophoresis of basic proteins and peptides on polyacrylamide gels. Nature. 1962;195:281–283. doi: 10.1038/195281a0. [DOI] [PubMed] [Google Scholar]
- Roberts E, Kutchan T, Kolattukudy PE. Cloning and sequencing of cDNA for a highly anionic peroxidase from potato and the induction of its mRNA in suberizing potato tubers and tomato fruits. Plant Mol Biol. 1988;11:15–26. doi: 10.1007/BF00016010. [DOI] [PubMed] [Google Scholar]
- Robertson EF, Dannelly K, Malloy PJ, Reeves HC. Rapid isoelectric focusing in vertical polyacrylamide minigel system. Anal Biochem. 1987;167:290–294. doi: 10.1016/0003-2697(87)90166-7. [DOI] [PubMed] [Google Scholar]
- Schardl CL, Byrd AD, Benzion G, Altshuler MA, Hildebrand DF, Hunt AG. Design and construction of a versatile system for the expression of foreign genes in plants. Gene. 1987;61:1–11. doi: 10.1016/0378-1119(87)90359-3. [DOI] [PubMed] [Google Scholar]
- Schnabelrauch LS, Kieliszewski M, Upham BL, Alizedeh H, Lamport DTA. Isolation of pI 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. Plant J. 1996;9:477–489. doi: 10.1046/j.1365-313x.1996.09040477.x. [DOI] [PubMed] [Google Scholar]
- Sherf BA, Bajar AM, Kolattukudy PE. Abolition of an inducible highly anionic peroxidase activity in transgenic tomato. Plant Physiol. 1993;101:201–208. doi: 10.1104/pp.101.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AT, Veitch NC. Substrate binding and catalysis in heme peroxidases. Curr Opin Chem Biol. 1998;2:269–278. doi: 10.1016/s1367-5931(98)80069-0. [DOI] [PubMed] [Google Scholar]
- Stoscheck CM. Quantitation of proteins. In: Deutscher MP, editor. Methods in Enzymology. 182. Guide to Protein Purification. San Diego: Academic Press; 1990. pp. 50–68. [DOI] [PubMed] [Google Scholar]
- Tigier HA, Quesada MA, Heredia A, Valpuesta V. Partial deglycosylation of an anionic isoperoxidase from peach seeds-effect on enzyme activity, stability and antigenicity. Physiol Plant. 1991;83:144–148. [Google Scholar]
- Wallace G, Fry SC. Phenolic components of the plant cell wall. Int Rev Cytol. 1994;151:229–267. doi: 10.1016/s0074-7696(08)62634-0. [DOI] [PubMed] [Google Scholar]
- Welinder KG. Superfamily of plant, fungal and bacterial peroxidases. Curr Opin Struct Biol. 1992;2:388–392. [Google Scholar]
- Whetten RW, MacKay JJ, Sederoff RR. Recent advances in understanding lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:585–609. doi: 10.1146/annurev.arplant.49.1.585. [DOI] [PubMed] [Google Scholar]