Abstract
Three D-cyclin genes are expressed in the apical meristems of snapdragon (Antirrhinum majus). The cyclin D1 and D3b genes are expressed throughout meristems, whereas cyclin D3a is restricted to the peripheral region of the meristem, especially the organ primordia. During floral development, cyclin D3b expression is: (a) locally modulated in the cells immediately surrounding the base of organ primordia, defining a zone between lateral organs that may act as a developmental boundary; (b) locally modulated in the ventral petals during petal folding; and (c) is specifically repressed in the dorsal stamen by the cycloidea gene. Expression of both cyclin D3 genes is reduced prior to the cessation of cell cycle activity, as judged by histone H4 expression. Expression of all three D-cyclin genes is modulated by factors that regulate plant growth, particularly sucrose and cytokinin. These observations may provide a molecular basis for understanding the local regulation of cell proliferation during plant growth and development.
The shoot apical meristem, which gives rise to the aerial part of the plant, has classically been divided into two or more zones (Clowes, 1961). Cells in the central zone are considered to be undifferentiated and can divide indefinitely. Cell proliferation in the central zone causes the displacement of cells into the peripheral zone, where they divide more quickly (Lyndon and Robertson, 1976) and contribute to the formation of the stem and of lateral organs such as leaves, bracts, and floral organs. While most cells in the stem and mature organs eventually cease division, some tissues (e.g. vascular cambium) retain the capacity for continued proliferation throughout the lifetime of the plant. Other tissues, such as secondary meristems, may remain dormant for some time before re-activation.
The molecular basis for localized differences in cell proliferation within the meristem is unclear, but could, as in other organisms, involve changes in the length of the cell division cycle. For example, the duration of the cell cycle in Drosophila is increased by the successive addition of G2 and then G1 phases (Edgar and O'Farrell, 1989). An alternative, but not exclusive, control mechanism operates later in larval development, when entry into and exit from the cycle is modulated, leading to localized differences in cell proliferation rates (Edgar and Lehner, 1996).
Entry into the cell cycle in both animals and yeast is under the control of external signals which act through regulating the level of G1 cyclins (Matsushime et al., 1991; for review, see Pines, 1995). These specific G1 cyclins in turn activate cyclin-dependent protein kinases (CDKs) and thus drive cell cycle progression. Examples include the Cln genes in budding yeast and the D-cyclin genes in animals (for review, see Sherr, 1995). Plant D-cyclins were recently identified by functional complementation in a yeast strain deficient for G1 cyclins (Soni et al., 1995). On restimulation of growth-arrested plant suspension-cultured cells, transcription from the Arabidopsis cyclin D3 gene is induced by the plant growth substance cytokinin, while the Arabidopsis cyclin D2 gene responds to carbon source availability (Soni et al., 1995). The observation that overexpression of cyclin D3 allows leaf explants to proliferate in the absence of cytokinin (Riou-Khamlichi et al., 1999) supports the idea that plant D-cyclins can also modulate cell cycle activity in response to extracellular signals.
To investigate further the role of D-cyclins in plant development, we isolated three D-cyclin genes from snapdragon (Antirrhinum majus) whose expression is modulated by plant growth substances. Cyclin D1 and cyclin D3b are expressed in both the peripheral and the central regions of meristems, whereas cyclin D3a is expressed only in the peripheral region, suggesting that differences in the regulation of cell proliferation between these two regions may involve the differential expression of D-cyclin genes. Furthermore, we show that cyclin D3b expression is modulated at several key stages during floral development, including: (a) in boundary zones between primordia; (b) during petal morphogenesis; and (c) in the dorsal anther as a result of cycloidea activity. D-Cyclin expression, therefore, discriminates between cell populations with different fates and could provide a link between the regulation of the cell division cycle and the pattern of plant development.
MATERIALS AND METHODS
Gene Isolation
A cDNA library prepared from poly(A+) RNA isolated from young wild-type snapdragon (Antirrhinum majus) inflorescences was screened as described in Simon et al. (1994) using random-primed cDNA probes containing the cyclin box of Arabidopsis D-type cyclins (Soni et al., 1995). Inserts from representatives of each hybridization class were subcloned into pGEM4Z (Promega, Madison, WI) and sequenced on an automated sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA). Sequences were compiled and conceptually translated and aligned using the GCG programs PILEUP and PRETTYBOX (Genetics Computer Group, Madison, WI).
Southern Analysis
DNA was extracted from plants using a modified CTAB preparation, essentially as described by Coen et al. (1986), separated, transferred to Hybond NX membranes, and hybridized with each radiolabeled cyclin gene.
Northern Analysis
Seedlings were germinated in Murashige and Skoog liquid medium (with or without Suc) for 8 to 9 d at 25°C. Plant growth substances were added to the liquid medium, and seedlings were harvested 6 to 24 h later. RNA extractions were performed as described by Vervoerd et al. (1989) with the extraction buffer from Soni and Murray (1994). RNA gel-blot analysis was described previously (Murray et al., 1987). An estimate of loading was obtained by methylene blue staining: membranes were soaked in 1× sodium chloride/sodium phosphate/EDTA (buffer) (SSPE), in 0.02% (w/v) methylene blue prepared in 0.3 m sodium acetate, pH 5.2, and rinsed in 20% (v/v) ethanol. After hybridization, washes were performed in 0.1% SSC, 0.1% (w/v) SDS at 42°C and 55°C for 10 min (except for histone H4, the wash was performed at 42°C for 10 min). Relative amounts of specific mRNAs were estimated using the public domain NIH Image program (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image), and normalized to the loading controls.
RNA in Situ Hybridization
Probes for the cyclin D1 gene were made by amplifying a 400- or a 1,000-bp fragment from the plasmid pVAL8 with the specific primers pVAL8T3 (5′-AATTAACCCTCACTAAAGGGATCACTGCTCAAACATGCTGAG-3′) and pVAL8T7 (5′-CAAGCTCTAATACGACTCACTATAGGGAGAAGAGCTCTCTTTGTC-3′), or with pVAL8T7 and the forward pBC sequencing primer. The resulting products, which lack the poly(A+) tail and contain terminal T3 and T7 RNA polymerase priming sites, were purified on WIZARD columns and eluted in 50 μL of water, 5 μL of which was used to make sense and antisense probes. The 3′ portion of cyclin D3b gene, lacking a poly(A+) tail, was cloned as an EcoRI fragment in both orientations into the pBS vector to produce pVAL54 and pVAL55. After linearization with HindIII, these plasmids generated sense and antisense templates for T7 polymerase. A 1.2-kb PCR product spanning the entire cyclin D3a cDNA (which was already truncated at the 3′ end) was cloned in the EcoRV site of pBCKS(−) to produce pVAL34. Linearization with BamHI or HindIII provided templates for T3 or T7 polymerases to generate antisense or sense RNA probes, respectively. Digoxigenin labeling of RNA probes, tissue preparation, and in situ hybridization were as described in Bradley et al. (1993) and Fobert et al. (1994). The specificity of the cyclin D3a and cyclin D3b probes was assessed by dot-blot hybridization of each fluorescein-labeled sense RNA with digoxigenin-labeled antisense RNAs. Hybridization, processing, and development of the dot blots were essentially as described by Boehinger Mannheim (Basel).
RESULTS
Isolation and Characterization of D-Cyclins from Snapdragon
A cDNA library was screened at low stringency with three D-cyclin probes of Arabidopsis, Arath;cycD3, Arath;cycD2, and Arath;cycD1 (Soni et al., 1995). Eight clones were isolated with the Arath;cycD3 probe that, based on restriction analysis and Southern hybridization, were considered to represent the same gene, cyclin D3a. The longest clone was 1,130 bp and contained an open reading frame of 1,026 bp. Another cDNA clone, isolated with the Arath;cycD2 probe, contained a novel cyclin-like gene more closely related to the plant D3 cyclin family than to the D2 and was called cyclin D3b. The missing 5′ end was rescued from the cDNA library by PCR amplification, and three independent clones were sequenced. The reconstructed sequence of the cyclin D3b cDNA contained a 1,092-bp open reading frame that potentially encodes a 364-amino acid protein. A single clone, isolated with the Arath;cycD1 probe, was named cyclin D1 and contained a 993-bp open reading frame that potentially encodes a 331-amino acid protein.
To assess the level of similarity between the genes, the nucleic acid sequences were aligned using the LaserGene program and the GCS program BestFit. Pairwise comparison of the whole gene sequences revealed that cyclin D1 and cyclin D3a were 31% identical and cyclin D1 and cyclin D3b were 27% identical. The cyclin D3a and D3b genes were most similar, sharing 43% identity overall, but a region of about 750 bp within the coding regions displayed 66% identity. However, even within this region, the maximum length of continuous identity was less than 15 bp (with one exception of 17 bp).
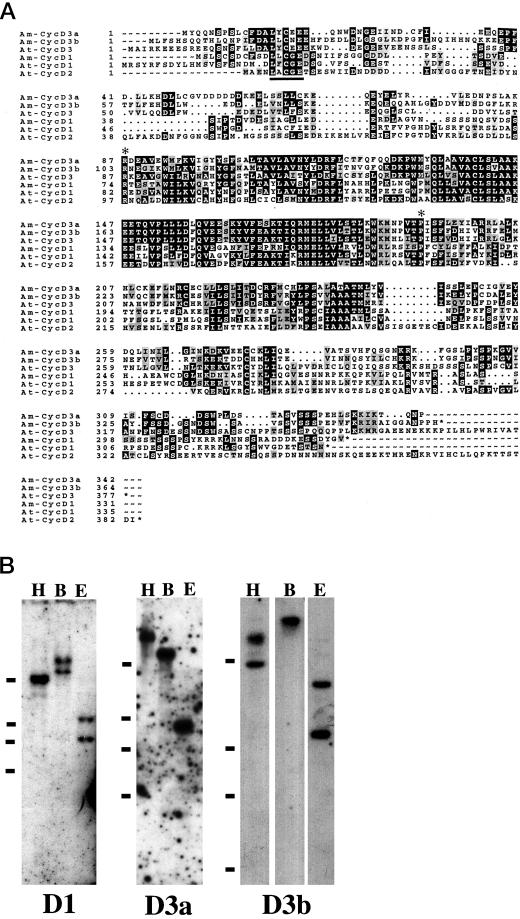
The deduced amino acid sequences of the snapdragon D-cyclin genes show similarities with mammalian D-type cyclins and Arabidopsis D-cyclins (Fig. 1A). The cyclin D1 gene shares about 49% identity at the amino acid level with Arath;cycD1 and about 29% and 26% identities with cyclin D3a and D3b, respectively. Cyclin D3a and cyclin D3b are 72% similar to each other at the amino acid level. The cyclin D3a and D3b genes show 66% and 70% identity, respectively, with Arath;cycD3 (Soni et al., 1995; Renaudin et al., 1996). The snapdragon cyclins D3 and D1 share several structural characteristics typical of D-cyclins. They lack the mitotic “destruction motif” RxxL(x)2–4xxN, which is involved in the degradation of mitotic cyclins by the ubiquitin-mediated pathway during anaphase (Glotzer et al., 1991). Instead, they contain PEST motifs rich in Pro, Glu, Ser, and Thr, which are typical of proteins with rapid turnover (Rogers et al., 1986). A putative retinoblastoma (Rb)-binding motif (LxCxE, where x is any amino acid) is located near the N terminus of all three snapdragon D-cyclins; similar motifs are also found in mammalian D-cyclins (Dowdy et al., 1993; Kato et al., 1993). An additional Rb-binding core motif is located downstream at residue 64 in cyclin D3a, but the significance of this remains to be determined.
Figure 1.
A, Predicted amino acid sequences of snapdragon cyclin D1, cyclin D3a (Am-CycD3a), and cyclin D3b (Am-CycD3b), illustrating conserved (black) and similar (gray) residues with D-cyclins from Arabidopsis (At-CycD1, 2, and 3; Soni et al., 1995). The extent of the cyclin box is indicated by two asterisks. Dots indicate gaps introduced to maximize sequence alignment. The retinoblastoma interaction motif is underlined. EMBL accession numbers are cyclin D1, AJ250396; cyclin D3a, AJ250397; and cyclin D3b, AJ250398. B, Southern analysis of snapdragon genomic DNA cut with HindIII (H), BamHI (B), and EcoRI (E) and probed with cyclin D1 (D1), cyclin D3a (D3a), and cyclin D3b (D3b). The positions of the 2-, 3-, 4-, and 10-kb molecular mass markers are indicated by bars.
Southern blotting of genomic DNA using each D-cyclin gene as a probe revealed one or two bands, depending on the restriction enzyme used to cut the DNA (Fig. 1B). Cyclin D1 probes recognized a single band of about 10 kb when DNA was restricted with HindIII, but two bands (at about one-half the intensity) when EcoRI and BamHI were used. An EcoRI site is present within the cDNA, which is consistent with these results. A very faint second band was sometimes observed, suggesting the presence of a structurally related gene in the genome. The cyclin D3a probe recognized a single band in DNA restricted with all three enzymes. The cyclin D3b probe recognized a single high-Mr band in DNA restricted with BamHI, but two bands in both HindIII and EcoRI cut DNA. These results are consistent with the cyclin D3b probe recognizing a single gene.
D-Cyclin Expression Is Modulated by Substances Affecting Plant Cell Proliferation
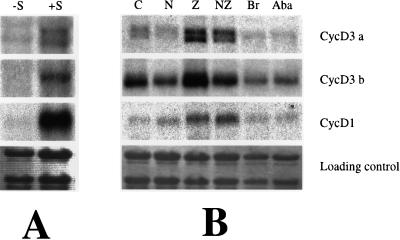
D-cyclins are proposed to transduce extracellular growth signals by altering the potential for cell proliferation (Sherr, 1995). If this model is correct, then substances such as Suc, which affect plant growth, might alter cyclin gene expression. To test this hypothesis, we germinated seedlings in liquid culture in light conditions with or without additional Suc. Post-germination growth of snapdragon seedlings is absolutely dependent on added Suc even in the presence of light, presumably because carbon dioxide uptake is restricted under these conditions. Cyclin mRNA levels increased 24 h after the addition of Suc (Fig. 2A, lanes −S and +S). Estimates of the relative induction varied from approximately 2-fold for cyclin D3a, to 3-fold for cyclin D3b, to 9-fold for cyclin D1. Light is not required for this stimulation, since plants grown in either light or dark conditions responded in a similar way (data not shown).
Figure 2.
Expression of D-cyclin genes in response to plant growth substances. A, Seedlings grown in Murashige and Skoog liquid medium in the dark for 8 d and then for a further 24 h without Suc (−S) or with Suc (+S). B, Seedlings grown in Murashige and Skoog medium with Suc in the light for 9 d and then for a further 24 h with added growth regulators as follows: none (C); 1 μm naphthylacetic acid (N); 1 μm zeatin (Z); 1 μm naphthylacetic acid and 1 μm zeatin (NZ); 1 μm 22-23 homo-brassinolide (Br); and abscisic acid (Aba). Northern blots were sequentially probed with cyclin D3a, cyclin D3b, and cyclin D1.
To examine the effects of exogenous plant growth substances, seedlings were germinated in the light with Suc for 9 d and then for another day with either no added hormone (Fig. 2B, lane C) or with various growth substances (Fig. 2B, lanes N, Z, NZ, Br, and Aba). Two of the three D-cyclin genes were stimulated by cytokinin: a 2.5-fold induction was observed for cyclin D3a and a 3-fold induction for cyclin D1. The apparent increase shown by cyclin D3b was found to be insignificant after being normalized against the loading controls. The effect of auxin is more complex—it stimulates cyclin D1 expression (2-fold), but for cyclin D3b, auxin alone is very weakly inhibitory (about 90% of the control level) and slightly antagonizes the stimulatory effects of cytokinin. cyclin D3a levels are not significantly affected by auxin. Both abscisic acid and brassinolide reduced the relative amounts of cyclin D3a mRNAs to 45% and 25%, respectively, and cyclin D3b mRNA to 60% and 85%, but, surprisingly, they both appeared to slightly stimulate cyclin D1 mRNA accumulation (1.3- and 1.7-fold increases). Plants grown under conditions expected to support active growth contain two different-sized transcripts that are recognized by the cyclin D3a gene, but only one when growth is inhibited by lack of Suc or by inhibitors such as abscisic acid. The two transcripts could represent two products of the cyclin D3a gene or may indicate the expression of a closely related gene.
D-Cyclin Expression Reveals the Presence of Two Distinct Populations of Cells in Apical Meristems
The spatial and temporal expression patterns of cyclin D3 and D1 genes were studied during shoot development by RNA in situ hybridization using digoxigenin-labeled antisense RNA probes (Figs. 3–6). Sense probes were used as controls and produced no significant signal (Figs. 3 and 6; data not shown).
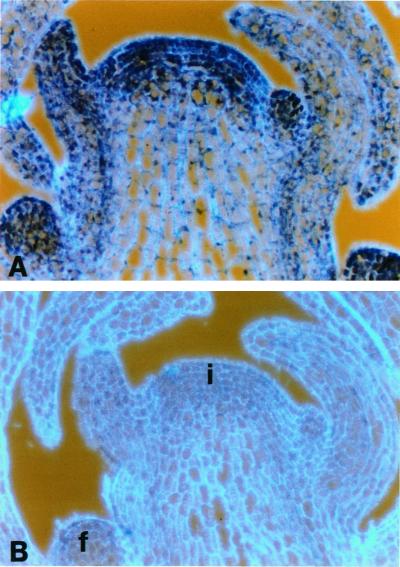
Figure 3.
RNA in situ hybridization of consecutive longitudinal sections of an inflorescence apex probed with cyclin D1. Sections were probed with digoxigenin-labeled antisense RNA (A) and sense (B) probes and viewed under bright-field optics, which gives a purple-blue color. The cell walls, revealed by epifluorescence, appear light blue. i, Inflorescence meristem; b, bracts; f, floral meristems.
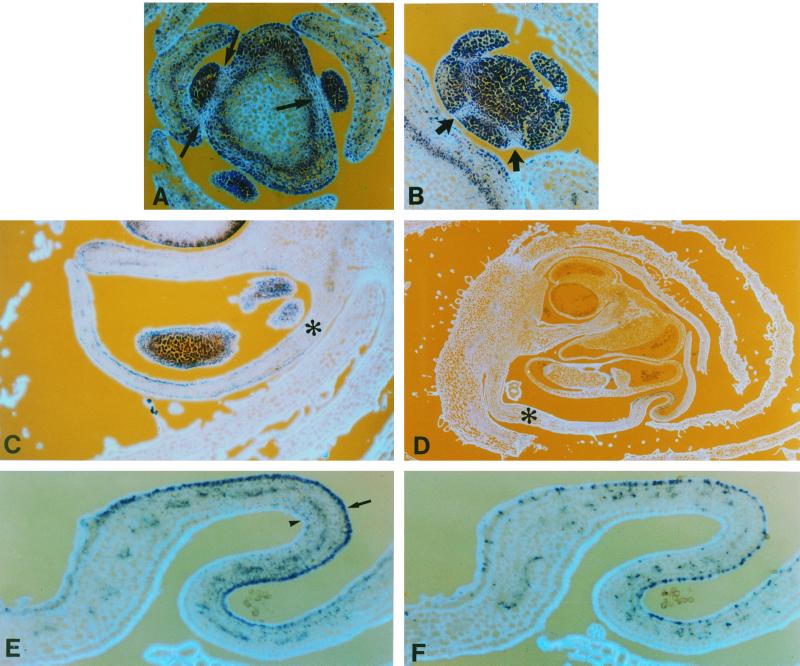
Figure 6.
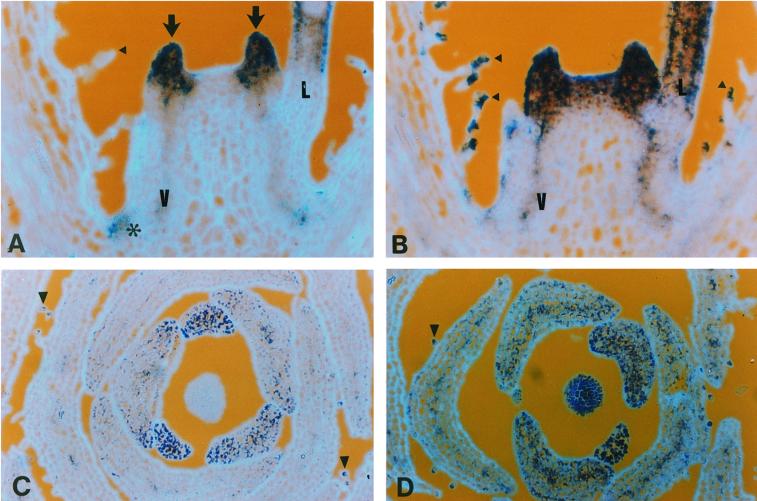
Patterns of cyclin D3b expression during floral development. Sections through young floral meristems at about node 7 (A) and 14 (B) showing regions of reduced cyclin D3b expression (arrows) surrounding the young meristems (A) and sepal primordia (B). Sections through floral buds before (C) and during (D) the initiation of petal folding, probed with cyclin D3b gene. Ventral petals are indicated with an asterisk. E and F, Near consecutive sections of folded regions of the ventral petal shown in D probed with cyclin D3b (E) or histone H4 (F); the inner surface of the petal is indicated by an arrow and the outer surface by an arrowhead.
Cyclin D1 transcripts are present throughout the plant, with somewhat higher levels in meristems and in cells associated with the vascular procambial tissues (Fig. 3A). The gene is also expressed in the root at a low level but in the developing anther sacs and carpels expression levels are elevated (data not shown). Probes derived from a small 400-bp fragment or from the whole gene produce identical patterns (data not shown). Control sections probed with a sense probe produced no signal under similar conditions (Fig. 3B).
The transcripts of the two cyclin D3 genes were also found in all shoot meristems but were restricted to specific regions. Consecutive longitudinal sections of young vegetative meristems probed alternately with cyclin D3a (Fig. 4A) or cyclin D3b (Fig. 4B) revealed that the two cyclin D3 genes were expressed in different but overlapping domains in the shoot apex. Cyclin D3a was expressed in the extreme periphery of the meristem and in the leaf primordia. A lower level of transcript was also detected in the young leaves and in secondary meristems formed in the axils of more mature leaves. Cyclin D3b was expressed throughout the vegetative meristem but also in young primordia, in young leaves, and, at lower levels, in vascular tissues and in secondary meristems. Serial transverse sections of inflorescence meristems probed with the cyclin D3 genes (Fig. 4, C and D) indicated that cyclin D3a transcripts were not detected in the central region of the inflorescence meristem (Fig. 4C), whereas cyclin D3b transcripts were abundant within all cells in the top six to seven layers of the apical dome. Expression of both cyclin D3a and cyclin D3b genes was initially uniform within young bract primordia and very young floral primordia, but as the bracts increased in size, cyclin D3a expression declined and was restricted to the edges of the bracts and cells associated with vascular tissues. Cyclin D3b expression was maintained for longer and was more evenly dispersed throughout the bract. Organ identity affects how the cyclin D3 genes are expressed during the development of the multicellular trichomes: only cyclin D3b was expressed in the developing trichomes found on leaves, but developing bract trichomes expressed both cyclins D3 (Fig. 4D). The mature trichomes do not contain either cyclin D3 transcript, suggesting that cyclin D3 transcripts are present only when trichomes cells are proliferating.
Figure 4.
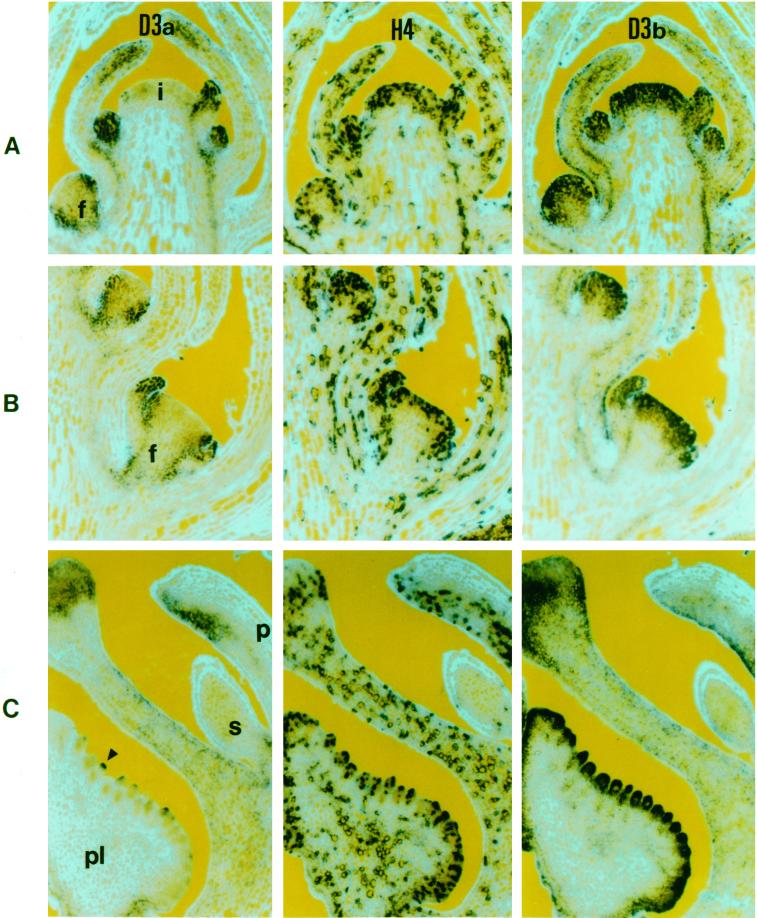
RNA in situ hybridization of apices probed with cyclin D3a and cyclin D3b. Sections were probed with digoxigenin-labeled antisense RNA and viewed under bright-field optics which gives a purple-blue color. The cell walls, revealed by epifluorescence, appear light blue. A and B, Consecutive longitudinal sections of a vegetative meristem. A, Cyclin D3a transcripts are located mainly in the primordia (arrows) and the periphery of the meristem, with lower amounts in the young leaves (L), secondary meristems (*), and vascular tissues (v). B, Cyclin D3b is expressed throughout the central and peripheral regions of the meristem, in young leaves, secondary meristems, vascular tissues, and in trichomes (arrowheads). C and D, Transverse sections through the tip of an inflorescence apex and surrounding bracts probed with the cyclin D3a gene (C) and cyclin D3b (D). Arrowheads indicate trichomes.
Since the two cyclin D3 genes display a significant degree of similarity within the central portion of the gene (66% identity over a region of about 600 bp), the specificity of the RNA probes was tested by reciprocally hybridizing one to the other on dot blots under conditions that closely mimicked in situ hybridization conditions. Whereas both antisense probes strongly recognize the cognate sense probe, the reaction with the non-cognate probe was judged to be less than 1% (data not shown). The lack of cross-reaction under these conditions is not surprising, since the regions of continuous identical sequence between cyclin D3a and D3b are very small (less than 10 bp with three exceptions; one each of 17, 15, and 12 bp). Mismatches in the RNA-RNA duplex are sensitive to RNase A digestion, and the short duplexes thus produced are likely to be unstable. The cyclin D1 gene sequence has a more limited region, about 300 bp long, with 61% and 63% identity to cyclin D3a and D3b, respectively. The fragments of continuous sequence identity, in this case, are all less than 10 bp and are unlikely to permit significant cross-hybridization. The distinct hybridization patterns of the three genes on Southern blots also indicate that the three genes do not readily cross-hybridize.
Cyclin D3 Genes Are Expressed during All Phases of the Cell Cycle
We have shown previously that cells within meristems are asynchronous as to their progression through the cell cycle so that genes expressed during particular phases of the cell cycle give rise to a spotty pattern (Fobert et al., 1994, 1996). Both cyclin D3 probes produced relatively uniform staining within a given region, suggesting that cyclin D3 gene expression remains relatively constant during the cell cycle. To define the extent to which cyclin D3 expression was restricted to a particular phase of the cell cycle, we used histone H4 expression as a marker for cells in S-phase. The histone genes are useful because their expression is not restricted to actively dividing cells, but is also in any cell where DNA replication occurs. Histone H4 expression is primarily DNA replication dependent in plant and non-plant species (for review, see Stein et al., 1992; Chaubet et al., 1996), and thus, under normal growth conditions, high-level expression of histone H4 is considered to be indicative of the S-phase.
To ascertain if cyclin D3 genes were expressed in a manner dependent on the cell cycle phase, we probed consecutive longitudinal sections with cyclin D3a, cyclin D3b, or histone H4 (Fig. 5, A–C). Within the top eight to 12 cell layers of the meristem and young primordia, only a portion of cells (about 30%–40%) expressed histone H4, but all cells in these layers expressed cyclin D3b and, within the young primordia, all cells expressed cyclin D3a as well. Since S-phase cells account for less than one-half of the population in these areas, as indicated by histone expression, the relatively uniform staining between neighboring cells indicates that D-cyclin transcripts are present in both S-phase and non-S-phase cells. This could be due either to continued transcription throughout the cell cycle or to prolonged stability of the transcripts.
Figure 5.
RNA in situ hybridization of consecutive longitudinal sections of inflorescence apices with RNA probes from the cyclin D3a (D3a), histone H4 (H4), and cyclin D3b (D3b) genes. In A through C, RNA is indicated by the dark-blue color, while epifluorescence was used to reveal calcofluor-stained (light blue) cell walls. A, Inflorescence apex containing the inflorescence meristem (i) and floral meristems at three different stages (f). B, Sections from a florotypic floral meristem (f). C, Sections through an unopened floral bud showing petal (p), staminode (s), carpel wall (c), and placenta (pl).
The presence of cells containing histone transcripts also provides an indication of the level of cell cycle activity within a given region: within actively growing inflorescence and young floral meristems, approximately 38% of the cells contained histone transcript, but this decreased to less than 5% in the central regions of the stem and in older bracts. The highest levels of cyclin D3 transcripts were found in regions also expressing high levels of histone H4. As bracts became more distant from the meristem, both histone H4 and cyclin D3 transcripts became less abundant, but cyclin D3 levels declined more sharply than histone. Occasional bract cells expressed histone H4 until at least node 14, but cyclin D3b transcript was only just detectable within bracts at node 10.
Expression of cyclin D3a was limited to a subdomain of histone expressing cells in the periphery of floral meristems, in young organ primordia (Fig. 5, A and B), and at the extremities of older organs (Fig. 5C). Cyclin D3a transcript levels reduced more rapidly than cyclin D3b, becoming localized to the periphery of bracts, and were barely detectable in bracts at node 8.
Cells within the cortex and pith of the stem had comparatively low levels of both D3-cyclins (Fig. 5). Both transcripts were undetectable in the pith below node 3 (about 12 cells below the tip of the apical meristem), but cyclin D3b transcript remained detectable in the epidermis, underlying cells in the cortex and in vascular traces until at least below node 15. Cyclin D3a expression within the stem was restricted to the vascular traces.
A general trend, that the region marked by cyclin D3 expression was less extensive than that marked by histone, was observed throughout development. In older floral buds (Figs. 5C and 6), regions expressing cyclin D3b tended to be contained within regions of histone-positive cells. Moreover, not all histone-expressing regions also expressed detectable levels of cyclin D3a or D3b; for example, the central region of the placenta was notable in having a relatively high level of histone transcripts while lacking substantial levels of cyclin D3 mRNA.
Cyclin D3b Expression Is Modulated during Floral Development
Floral development in snapdragon is well-defined both morphologically and genetically. To relate the expression of the cyclin D3 genes to floral development, we have used the nodal numbering system as defined by Carpenter et al. (1996). During early floral development, both cyclin D3a and cyclin D3b transcripts were present in all cells of the floral meristem from the time they were first visible (about node 2–3) until about node 6 to 7 (loaf stage meristem). Thereafter, cyclin D3a transcript was progressively restricted to a ring of cells around the periphery of the meristem. In the longitudinal sections shown, this ring appears as staining on the flanks of the floral meristem (Fig. 5A, f). By the time of sepal emergence (about node 10–12; floritypic stage), cyclin D3a was clearly absent from the central region of the floral meristem (Fig. 5B, f), being restricted to the sepal primordia and to a ring of cells about five to seven wide internal to the sepals, where petal primordia were presumed to be initiating (Fig. 5B).
Significant local declines in the level of cyclin D3b transcript were also observed in the cells of the stem immediately adjacent to the base of floral meristems (Figs. 5, A and B, and 6A, arrows). Transverse sections reveal that meristems at this stage have localized decreases in cyclin D3b at the base of sepals (Fig. 6B).
In young petals, cyclin D3b was expressed in all cells but at somewhat lower levels in cells near the base. As petals mature, local variation in cyclin D3 expression becomes more marked. First, expression of cyclin D3b was higher and more prolonged in the cells of the inner petal epidermis of all petals (Fig. 6C). Secondly, expression in the ventral petals is prolonged with respect to the dorsal petals (Fig. 6D; data not shown). Finally, as the ventral petals of the snapdragon flower mature, they develop complex shapes due to the production of folds in the lobes (Fig. 6D). As the petals grow, but before the lobes had started to fold, cells with high cyclin D3b expression became progressively restricted to the inner or adaxial epidermal cells (Fig. 6C). In petals at a more advanced stage, where the folds were already evident, the transcript was restricted to folds in the petal (Fig. 6D). To demonstrate that the cells in these folds were active in the cell cycle, we probed adjacent sections with histone H4 and cyclin D3b. The signals obtained using these two genes showed close correspondence, but, as in meristems, the region containing histone-expressing cells extended somewhat beyond the region with cyclin D3b transcript (Fig. 6, E and F). A consequence of the folding process is that cell proliferation continues in the ventral petal when the dorsal petals have ceased, and this is correlated with prolonged expression of cell-cycle-related genes including histone and D3 cyclins.
Whereas cells in the central regions of meristems tend to contain only cyclin D3b transcripts, cells that express cyclin D3a generally also contain cyclin D3b transcripts. However, a number of regions appear to contain cells that express predominantly cyclin D3a. Such cells tend to be found around the boundaries between floral organs or between auxiliary meristems and the stem (Fig. 5B, compare the floral meristem [f] probed with cyclin D3a and floral meristem probed with cyclin D3b). The relative abundance of the two transcripts also varied and although cyclin D3b tends to be the most abundant, cyclin D3a becomes predominant in regions of the maturing petal (Fig. 5C), indicating that distinct regulatory mechanisms may control the two D3-type cyclin genes.
The Cycloidea Gene Is Required to Repress Cyclin D3b Expression in the Dorsal Stamen
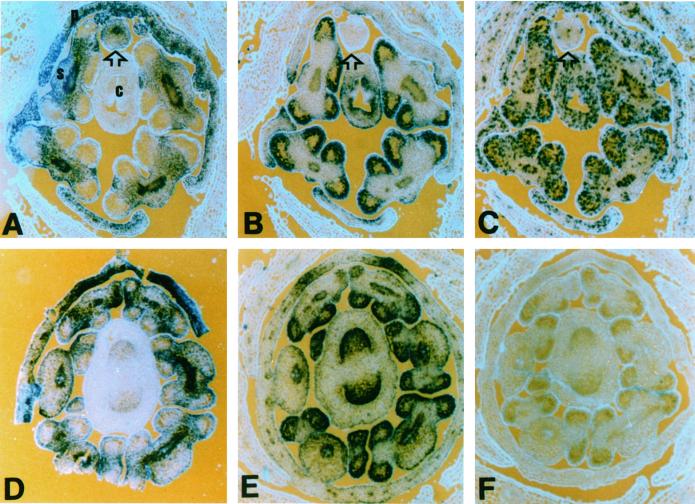
Snapdragon floral meristems form five stamen primordia, but only four give rise to functional stamens. Full development of the dorsal stamen primordium (or staminode) is suppressed by the action of the cycloidea gene (Luo et al., 1996). The four developing stamens expressed high levels of cyclin D3b and histone H4, but the staminode expressed low levels of histone H4 (Figs. 7C and 5C) and low or no detectable cyclin D3b transcripts (Fig. 7B). Similar expression patterns have also been observed with other cell cycle genes including cyclin B1, cyclin B2, cdc2c, and cdc2d (data not shown). The def gene, which is expressed in all petals and stamens (Schwarz-Sommer et al., 1992), continues to be expressed in the staminode even after cell cycle gene expression is repressed (Fig. 7A). This indicates that at least some classes of mRNAs are still present and the arrested development of this organ does not involve the general degradation of all mRNAs. In a cyc-608 mutant strain, all stamen primordia (of which there can be up to six) develop into mature functional stamens. These stamens all express both def (Fig. 7D) and cyclin D3b (Fig. 7E), as well as histone H4 (data not shown). We conclude from these observations that cycloidea function is required, either directly or indirectly, to suppress cyclin D3b and other cell cycle gene expression in the staminode.
Figure 7.
RNA in situ hybridization of transverse sections of wild-type and cycloidea mutant floral buds. A through C, Wild-type flower probed with deficiens (A), cyclin D3b (B), and histone H4 (C). c, Carpels; s, functional stamens; arrowhead, arrested stamen; p, petals; s, sepals. D through F, cycloidea mutant flower probed with deficiens (D), cyclin D3b (E) probes, and a cyclin D3b sense control (F).
DISCUSSION
We have isolated three snapdragon cyclin genes, cyclin D1, cyclin D3a, and cyclin D3b. They clearly group within the D-type cyclins based on sequence similarities and characteristics, including PEST and retinoblastoma-binding motifs. The D-type cyclins display overlapping but distinct expression patterns in meristems, revealing developmentally distinct populations of dividing cells. Furthermore, we show that cyclin D3 expression is spatially and temporally restricted to subsets of proliferating cells and is modulated by exogenous plant growth regulators. Finally, we have demonstrated that the cycloidea gene, which locally regulates cell proliferation, is required to repress transcription of cyclin D3 in the dorsal stamen.
D-Cyclin Transcript Levels Are Independent of Cell Cycle Phase
Probing alternative sections with histone H4, a marker of S-phase (Fobert et al., 1994), and D3 cyclins led us to conclude that the D-cyclin transcripts reported here are not strongly modulated during the cell division cycle. In contrast, phase-dependent expression, as exhibited by the B-type mitotic cyclins (Fobert et al., 1994) cdc2c and cdc2d (Fobert et al., 1996) and by histone H4, produce a stochastic pattern of labeling, because neighboring cells tend to be at different stages of the cell cycle. The differences in transcript levels could be the result of differences in promoter activity, transcript stability, or both, and remain to be determined. Regions active in the cell cycle (as judged by histone expression) are more extensive than regions expressing cyclin D3 genes. Although this may indicate that cyclin D3 genes are not directly concerned with cell cycle progression, it is consistent with the idea that D-cyclins operate in a regulatory pathway upstream of the cell cycle (Murray, 1997) and that locally increased levels of D-cyclin may enhance local potential for proliferation (Riou-Khamlichi et al., 1999). In this scenario, the low basal levels of cell cycle activity observed in the cyclin D3-free regions could be cyclin D3 independent or may utilize other cyclins; for example, the constitutive cyclin D1 gene or some other, as yet unidentified, D-cyclin gene. It seems highly likely that snapdragon contains a more extensive gene family of D-cyclins than we report here. We have been unable to isolate a D2-type cyclin from floral shoot cDNA libraries, but these are known in both Arabidopsis (Soni et al., 1995) and tobacco (Sorrell et al., 1999). In addition, there may be up to three distinct classes of D3 cyclins in many species (J.A.H. Murray, unpublished data).
Cyclin D Transcript Levels Are Modulated during Plant Growth
The in situ analysis demonstrates that D3 cyclin transcript levels vary during development in a manner that is consistent with a role in regulating cell proliferation. The cyclin D3b gene is expressed when cells are first formed in the central region of the apical meristem, and continues to be expressed as they are displaced out into the periphery. As the development of the primordia and stem progresses, cyclin D3b expression is down-regulated in regions with reduced cell cycle activity, but is maintained in those regions that continue to proliferate (such as localized areas of petals and the epidermal layer of the internode). Finally, expression is greatly reduced as cell cycle activity ceases and differentiation occurs. The cyclin D3a gene is not expressed in the central region, but is switched on when cells are incorporated into organ primordia. Expression of these two genes, therefore, discriminates between two distinct populations of cells within all three types of apical meristem: vegetative, inflorescence, and floral. An alternative view, that the tissues expressing cyclin D3b alone are indeterminate while tissues expressing both cyclin D3 genes are determinate structures, does not hold for floral meristems, which are determinate and show the same general pattern as indeterminate meristems. Expression of cyclin D3a is associated with the developmental pathway leading to formation of the lateral organs rather than determinacy per se.
Establishment of developmental boundaries has been associated with locally reduced cell proliferation (Vincent et al., 1995) and may be coincident with decreased cyclin D3 expression around the bases of floral organs. Local modulation of cyclin D3 expression is also evident in the dorsal anther and during petal folding, supporting the notion that localized D-cyclin expression might mediate differential cell proliferation within plant tissues. D-Cyclin expression patterns in Drosophila also anticipate the pattern of cell proliferation (Finley et al., 1996), so that spatial control of their expression may be a widespread phenomenon.
The cycloidea (cyc) gene restricts cell proliferation in the dorsal domain of the floral meristem without changing floral organ identity, and controls floral symmetry by reducing the growth rate, primordia initiation, and subsequent primordial development in the dorsal domain of the flower (Luo et al., 1996). Loss-of-function mutants in cyc have increased numbers of both petal and stamen primordia, suggesting that cyc also restricts cell proliferation during early floral meristem development. Although others have proposed that developmental regulators affect localized cell proliferation, the molecular details of any linkage between the cell cycle genes and these regulators is missing. In this study, we have shown that the cyc gene is required to modulate transcription of cell cycle genes in the domain of the flower, where it is known to have its morphogenetic effects. In the wild-type flower, a reduction in both histone and D-cyclin transcripts in the dorsal stamen primordia is correlated with the early cessation of cell proliferation, whereas in a cyc mutant that does not express any detectable cyc transcript (Luo et al., 1996), all stamen primordia express cyclin D3b and continue to develop, producing functional stamens. The cyc gene is the founding member of a family of putative transcription factors involved in regulating plant growth: the CYC family, which includes the teosinte branched 1 gene required to suppress lateral growth in maize (Doebley et al., 1997). Two other related genes from rice, PCF1 and PCF2, bind to a cis element of the PCNA gene and may be involved in regulating its expression (Kosugi and Ohashi, 1997). Further definition of how the cyc gene product modulates cyclin D3 gene expression may provide insight into the molecular mechanisms underlying the local control of cell proliferation in plant meristems.
Significance of D-Cyclin Expression for Plant Development
We have shown that mitogenic signals stimulate transcription of snapdragon D-cyclins in a fashion similar to that previously reported for Arabidopsis D-cyclins (Soni et al., 1995), and that a gene, cyc, controlling meristem development represses their transcription in a localized domain of the meristem. Perturbation of D-cyclin expression in plants causes abnormal development, uncoupling the link between cytokinin and cell proliferation in culture and disrupting the normal morphology of the meristem (Riou-Khamlichi et al., 1999). Analogous signal transduction pathways that regulate cell proliferation in response to extracellular signals also exist in mammalian cells and lead to the activation of D-cyclin-dependent kinases. These kinases regulate cell proliferation by inactivation of Rb, an inhibitor of S-phase-specific gene transcription. Phosphorylation inactivates this inhibitory function of Rb and allows cells to enter the S-phase (for review, see Elledge et al., 1996). A divergent Rb-like gene that binds plant D-cyclins on the basis of an N-terminal LxCxE motif (Huntley et al., 1998) has recently been isolated from maize (Xie et al., 1996). Therefore, cell proliferation in plants may be mediated by a D-cyclin/Rb pathway similar to that described for mammalian cells (Murray, 1997).
Localized regulation of cell division is important for normal plant development, and we propose that this control could operate, at least in part, via D-cyclin expression. Mutations in cyc (and many other genes affecting development) alter the position at which cells divide, supporting the notion that local regulation of cell division is important for plant morphogenesis. Models of meristem structure (Ledin, 1954; Wardlaw, 1957; Sussex, 1989) have proposed the existence of zones that differ in terms of cell behavior and fate, and our observations suggest that these cells also differ at the molecular level. As cells progress through the peripheral zone, they become divided into two groups: those that contribute to the primordia and those that do not. Cells in the primordia continue to divide and to express both cyclin D3 genes, whereas cells between the primordia form the boundary zone between developing organs and have reduced cyclin D3 expression. Subsequently, groups of cells in or close to the boundary zone may contribute to the formation of auxiliary vegetative and floral meristems, at which stage expression of both cyclin D3 genes is again increased.
The group of D3-type cyclins may play an important role in spatially regulating cell proliferation. In this paper we have shown: (a) that the expression of two cyclin D3 genes is highly dynamic during plant morphogenesis (marked changes occurring at developmental boundaries around organs and meristems and localized expression is associated with petal morphogenesis); (b) that expression is locally modulated by a developmental regulator, the cycloidea gene; and (c) that the combinatorial expression patterns of the cyclin D3a and D3b genes reflect the zonation model of the shoot apical meristem. Together with the observation that ectopic expression of the Arabidopsis cyclin D3 gene perturbs the structure of the shoot apical meristem, these results suggest that D-cyclins may provide a link between developmental regulators and the spatial control of cell proliferation.
ACKNOWLEDGMENTS
We thank Graham Scofield for help during the sequencing of the genes, Margit Menges and Susan Bunneywell for help with preparation of the manuscript, Coral Vincent and Emma Keck for technical advice, and Clive Lloyd, Peter Shaw, Desmond Bradley, and Jan Traas for valuable discussions and comments.
Footnotes
This work was supported by the European Union Science Programme and by the Biotechnology and Biological Sciences Research Council. V.G. was supported by an European Molecular Biology Organsiation Fellowship and subsequently by a joint Biotechnology and Biological Sciences Research Council/Institut National de la Recherche Agronomique Fellowship. P.F. was supported in part by a Natural Sciences and Engineering Research Council of Canada postdoctoral fellowship.
LITERATURE CITED
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes results from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Carpenter R, Copsey L, Vincent C, Doyle S, Magrath R, Coen E. Control of flower development and phyllotaxy by meristem identity genes in Antirrhinum. Plant Cell. 1996;7:2001–2011. doi: 10.1105/tpc.7.12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubet N, Flenet M, Clement B, Brignon P, Gigot C. Identification of cis-elements regulating the expression of an Arabidopsis histone H4 gene. Plant J. 1996;10:425–435. doi: 10.1046/j.1365-313x.1996.10030425.x. [DOI] [PubMed] [Google Scholar]
- Clowes F, editor. Apical Meristems. Oxford: Blackwell Scientific Publications; 1961. [Google Scholar]
- Coen E, Carpender R, Martin C. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell. 1986;47:285–296. doi: 10.1016/0092-8674(86)90451-4. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Dowdy SF, Hinds PW, Louie K, Reed SI, Arnold A, Weinberg RA. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- Edgar B, O'Farrell P. Genetic control of cell division patterns in the Drosophila embryo. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Lehner CF. Developmental control of cell cycle regulators: a fly's perspective. Science. 1996;274:1646–1652. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Winston J, Harper JW. A question of balance: the role of cyclin-kinase inhibitors in development and tumorigenesis. Trends Cell Biol. 1996;6:388–393. doi: 10.1016/0962-8924(96)10030-1. [DOI] [PubMed] [Google Scholar]
- Finley R, Thomas B, Zipursky S, Brent R. Isolation of Drosophila cyclin D, a protein expressed in the morphogenetic furrow before entry into S phase. Proc Natl Acad Sci USA. 1996;93:3011–3015. doi: 10.1073/pnas.93.7.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert PR, Coen ES, Murphy GJP, Doonan JH. Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J. 1994;13:616–624. doi: 10.1002/j.1460-2075.1994.tb06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert PR, Gaudin V, Lunness P, Coen ES, Doonan JH. Distinct classes of the cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell. 1996;8:1465–1476. doi: 10.1105/tpc.8.9.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray A, Kirschner M. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Huntley R, Healy S, Freeman D, Lavender P, de Jager S, Greenwood J, Makker J, Walker E, Jackman M, Xie Q, Bannister AJ, Kouzarides T, Gutierrez G, Doonan JH, Murray JAH. The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with the G1/S regulators and plant cyclin D proteins. Plant Mol Biol. 1998;37:155–169. doi: 10.1023/a:1005902226256. [DOI] [PubMed] [Google Scholar]
- Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. PCF1 and PCF2 specially bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledin R. The vegetative shoot apex of Zea mays. Am J Bot. 1954;41:11–17. [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- Lyndon RF, Robertson ES. The quantitative ultrastructure of the pea shoot apex in relation to leaf initiation. Protoplasma. 1976;87:387–402. [Google Scholar]
- Matsushime H, Roussel MF, Ashmun RA, Sherr CJ. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- Murray JAH. The retinoblastoma protein is in plants. Trends Plant Sci. 1997;2:82–84. [Google Scholar]
- Murray JAH, Scarpa R, Rossi N, Ceasarini G. Antagonistic controls regulate copy number of the yeast 2 microns plasmid. EMBO J. 1987;6:4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. Cyclins, CDKs and cancer. Semin Cancer Biol. 1995;6:63–72. doi: 10.1006/scbi.1995.0009. [DOI] [PubMed] [Google Scholar]
- Renaudin JP, Doonan J, Freeman D, Hashimoto J, Hirt H, Inze D, Jacobs T, Kouchi H, Rouz P, Sauter M, Savour A, Sorrell D, Sundaresan V, Murray J. Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol. 1996;32:1003–1018. doi: 10.1007/BF00041384. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor P, Hansen R, Tetens F, Lonning W, Saedler H, Sommer H. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 1992;11:251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- Simon R, Carpenter R, Doyle S, Coen E. Fimbriata controls flower development by mediating between meristem and organ identity genes. Cell. 1994;78:99–107. doi: 10.1016/0092-8674(94)90576-2. [DOI] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Shah ZH, Murray JAH. A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell. 1995;7:85–103. doi: 10.1105/tpc.7.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni R, Murray JAH. Isolation of intact DNA and RNA from plant tissues. Anal Biochem. 1994;218:474–476. doi: 10.1006/abio.1994.1214. [DOI] [PubMed] [Google Scholar]
- Sorrell DA, Combettes B, Chaubet-Gigot N, Gigot C, Murray JAH. Distinct cyclin D genes show mitotic accumulation or constant levels of transcripts in tobacco Bright Yellow-2 cells. Plant Physiol. 1999;119:343–351. doi: 10.1104/pp.119.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, Stein JL, van Wijnen AJ, Lian JB. Regulation of histone gene expression. Curr Opin Cell Biol. 1992;4:166–173. doi: 10.1016/0955-0674(92)90028-b. [DOI] [PubMed] [Google Scholar]
- Sussex I. Developmental programming of the shoot meristem. Cell. 1989;56:225–229. doi: 10.1016/0092-8674(89)90895-7. [DOI] [PubMed] [Google Scholar]
- Vervoerd TC, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent C, Carpenter R, Coen E. Cell lineage patterns and homeotic gene activity during Antirrhinum flower development. Curr Biol. 1995;5:1449–1458. doi: 10.1016/s0960-9822(95)00282-x. [DOI] [PubMed] [Google Scholar]
- Wardlaw C. On the organization and reactivity of the shoot apex in vascular plants. Am J Bot. 1957;44:176–185. [Google Scholar]
- Xie Q, Sanz-Burgos AP, Hannon GJ, Gutirrez C. Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 1996;15:4900–4908. [PMC free article] [PubMed] [Google Scholar]