Abstract
Importance
The single-nucleotide polymorphism rs1344706 in the zinc finger protein 804A gene (ZNF804A) shows genome-wide association with schizophrenia and bipolar disorder. Little is known regarding the expression of ZNF804A and the functionality of rs1344706.
Objectives
To characterize ZNF804A expression in human brain and to investigate how it changes across the life span and how it is affected by rs1344706, schizophrenia, bipolar disorder, and major depressive disorder.
Design, Setting, and Participants
Molecular and immunochemical methods were used to study ZNF804A messenger RNA (mRNA) and ZNF804A protein, respectively. ZNF804A transcripts were investigated using next-generation sequencing and polymerase chain reaction–based methods, and ZNF804A protein was investigated using Western blots and immunohistochemistry. Samples of dorsolateral prefrontal cortex and inferior parietal lobe tissue were interrogated from 697 participants between 14 weeks’ gestational age and age 85 years, including patients with schizophrenia, bipolar disorder, or major depressive disorder.
Main Outcomes and Measures
Quantitative measurements of ZNF804A mRNA and immunoreactivity, and the effect of diagnosis and rs1344706 genotype.
Results
ZNF804A was expressed across the life span, with highest expression prenatally. An abundant and developmentally regulated truncated ZNF804A transcript was identified, missing exons 1 and 2 (ZNF804AE3E4) and predicted to encode a protein lacking the zinc finger domain. rs1344706 influenced expression of ZNF804AE3E4 mRNA in fetal brain (P = .02). In contrast, full-length ZNF804A showed no association with genotype (P > .05). ZNF804AE3E4 mRNA expression was decreased in patients with schizophrenia (P = .006) and increased in those with major depressive disorder (P < .001), and there was a genotype-by-diagnosis interaction in bipolar disorder (P = .002). ZNF804A immunoreactivity was detected in fetal and adult human cerebral cortex. It was localized primarily to pyramidal neurons, with cytoplasmic as well as dendritic and nuclear staining. No differences in ZNF804A-immunoreactive neurons were seen in schizophrenia or related to rs1344706 (P > .05).
Conclusions and Relevance
rs1344706 influences the expression of ZNF804AE3E4, a novel splice variant. The effect is limited to fetal brain and to this isoform. It may be part of the mechanism by which allelic variation in ZNF804A affects risk of psychosis. ZNF804A is translated in human brain, where its functions may extend beyond its predicted role as a transcription factor.
The single-nucleotide polymorphism (SNP) rs1344706 within the zinc finger protein 804A gene (ZNF804A; GenBank NM_194250.1) (2q32.1) was the first to show unequivocal genome-wide association with schizophrenia and with a broader psychosis phenotype including bipolar disorder.1 These findings have been replicated2–4 and confirmed in meta-analyses,5,6 and they beg the question of what the roles of ZNF804A and rs1344706 are in health and disease.
The gene’s nomenclature reflects an amino-terminal sequence domain with homology to other members of the Cys2His2 family of zinc finger-binding transcription factors.7,8 However, empirical evidence for this role is lacking, other than preliminary data suggesting that ZNF804A affects the expression of some genes.9–11 Regarding the influence of rs1344706, the SNP (T → G, where T is both the major allele and the risk allele) resides in the second intron. As such, it is a noncoding polymorphism of unknown functionality, although some studies have reported imaging or behavioral correlates of the allele12–23 including differences in neural activity,12,14,20,22 white matter,18,19 and cognition.16,17 In molecular terms, the underlying mechanism likely involves a differential effect of the 2 alleles on gene regulation and transcript expression. The available data show no clear influence of rs1344706 on ZNF804A messenger RNA (mRNA) in adult brain,2,5,24 but an effect in the second trimester in utero has been reported.25
Despite these findings, many questions remain. First, for several psychosis risk genes, there is evidence that the risk SNPs affect specific (and previously unidentified) transcript isoforms via alternative splicing or promoter use.26 To our knowledge, this possibility has not been investigated for ZNF804A. Second, it is not clear whether ZNF804A expression is altered in schizophrenia or other psychiatric disorders.2,27 Third, whether ZNF804A is translated in human brain has yet to be shown. These issues are critical for understanding the pathophysiological significance of the genetic association of ZNF804A with psychosis. Herein, we report identification of a novel ZNF804A isoform in human dorsolateral prefrontal cortex (DLPFC) and map its expression across the life span. Notably, this isoform is selectively affected by rs1344706 in fetal brain, and it is also affected by diagnosis and diagnosis-bygenotype interactions. In contrast, the canonical ZNF804A transcript exhibits no genotype effects and only minor diagnostic differences. We also show that ZNF804A immunoreactivity is present in the human cerebral cortex, localized primarily to pyramidal neurons, and with no changes observed in schizophrenia.
Methods
Human Tissue
We studied ZNF804A transcripts in DLPFC from the National Institute of Mental Health (NIMH)/Lieber Institute for Brain Development series, including a lifetime cohort of nonpsychiatric controls from second trimester to 85 years old and patients with schizophrenia, bipolar disorder, and major depressive disorder (Table; see the article by Lipska et al28 for additional methods regarding demographic, clinical, and pathological descriptions). A subset of these postmortem fetal, infant, child, and adolescent brain tissue samples was provided by the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders. Another subset of the adult samples was donated to this study from the Stanley Brain Collection of the Stanley Medical Research Institute (see the article by Torrey et al29 for additional details on this collection). These samples were curated, processed, and dissected with methods identical to those used for the samples collected by the NIMH/Lieber Institute for Brain Development.
Table. Participants Used for Studies of ZNF804A Transcripts in Dorsolateral Prefrontal Cortex.
| Characteristic | Controls |
Patients With Diagnosis |
||||
|---|---|---|---|---|---|---|
| Fetal (n = 43) |
Aged 0-18 y (n = 73) |
Aged >18 y (n = 210) |
Bipolar Disorder (n = 61) |
Major Depressive Disorder (n = 135) |
Schizophrenia (n = 175) |
|
| Female, % | 48.8a | 32.8 | 29.0 | 41.0 | 42.2a | 37.1 |
| Age, mean, y | 14-20b | 8.6 | 44.2 | 44.8 | 45.7 | 50.2a |
| White, %c | 11.6a | 61.6a | 41.4 | 83.6a | 85.9a | 54.3 |
| pH | 6.10a | 6.43a | 6.55 | 6.36a | 6.35a | 6.41a |
| RNA integrity number | 8.81a | 8.22 | 8.25 | 7.99a | 8.03a | 7.84a |
| Postmortem interval, h | 2.6a | 23.9a | 30.7 | 32.9 | 37.9a | 38.5a |
| Age at onset, mean, y | NA | NA | NA | 23.0 | 31.1 | 22.4 |
| Duration of illness, mean, y | NA | NA | NA | 21.7 | 14.4 | 27.5 |
Abbreviation: NA, not applicable.
P < .05 compared with adult controls.
Expressed as range in gestational weeks.
Assessment of race made by medical examiner and next of kin.
Gray matter was dissected out, snap frozen, and stored at −80°C prior to RNA extraction, which was carried out using standard methods. The DLPFC tissue from this series was also used for protein extraction and Western blotting. The second brain series, used for immunohistochemistry, came from the Stanley Medical Research Institute, who provided free-floating sections of inferior parietal lobe from men with schizophrenia and male control participants (eTable 1 in the Supplement).30 For this series, after brief formalin fixation of the cerebral hemisphere, 3- to 5-cm coronal slabs were cut, formalin fixed for 3 to 15 weeks, and stored in buffered 30% sucrose. All blocks containing inferior parietal lobe were serially cryostat sectioned at 60 μm. Participants from both series were genotyped for rs1344706 using TaqMan assay C_2834835_10 (Life Technologies). Immunohistochemistry was also carried out on slide-mounted sections cut from formalin-fixed, wax-embedded blocks of frontal, temporal, or visual cortex from 5 additional adults and of frontal cortex from 2 neonates. The NIMH/Lieber Institute for Brain Development brains were collected with audiotaped oral informed consent from legal next of kin under National Institutes of Health institutional review board protocol 90-M-1042. Studies in Oxford, United Kingdom, were approved by Oxfordshire National Health Service Research Ethics Committee B (#O02.040).
RNA Sequencing and 5′ Rapid Amplification of Complementary DNA Ends
We used RNA sequencing (RNAseq) to identify potentially new expressed transcripts of ZNF804A and to guide the construction of relevant primers for 5′ rapid amplification of complementary DNA (cDNA) ends (5′ RACE). We performed RNAseq on Clontech pooled poly(A)+ mRNA from fetal and adult human brain to identify potential 5′ cDNA ends. After fragmentation, reverse transcriptase and random primers copied the cleaved fragments into first-strand cDNA. Second-strand cDNA was synthesized using T4 DNA polymerase, T4 polynucleotide kinase, and Klenow DNA polymerase, adding a single adenine base using a 3′ to 5′ exo− Klenow fragment, and ligating the paired-end adapters using T4 DNA ligase. An index (up to 12 nucleotides) was inserted into Illumina adapters so multiple samples could be sequenced in 1 lane of an 8-lane flow cell. Products were purified and enriched with polymerase chain reaction (PCR) to create the final cDNA library for highthroughput sequencing using the HiSeq 2000 system (Illumina). Results were mapped to GRCh37/hg19 using the TopHat2 splice-aware alignment software.31,32
We used the RNAseq read alignment data to design primer pairs to amplify ZNF804A transcripts. We first performed 5′ RACE in the commercial fetal and adult brain poly(A)+ mRNA samples to validate the RNAseq results, with ZNF804A-specific antisense primers binding at exon 4, using SMART RACE cDNA Amplification and Advantage 2 PCR kits (Clontech). The poly(A)+ mRNAs were reverse transcribed according to the manufacturer’s protocol. The PCR amplification profile was as follows: 94°C for 2 minutes; 30 cycles of 94°C for 30 seconds, 70°C for 30 seconds, and 72°C for 2 minutes; and 72°C for 10 minutes after the last cycle. When necessary, nested PCR was done using gene-specific primers and a nested universal primer A. Having confirmed the 5′ cDNA ends, we used end-to-end PCR to validate the transcripts in the 2 samples by Advantage 2 PCR kits (Clontech) following the manufacturer’s recommendations. All products were cloned into pCR4-TOPO (Invitrogen) and sequenced.
After confirming the presence of a truncated transcript of ZNF804A in the commercial fetal and adult brain poly(A)+ mRNA samples, we performed end-to-end PCR in the DLPFC series (Table) to establish the presence of the truncated transcript in an independent sample and in a specific brain region.
Quantitative Reverse Transcription–PCR
Based on the results of the RNAseq, 5′ RACE, and end-to-end PCR, we designed primers to selectively detect the canonical ZNF804A transcript (Applied Biosystems; forward: TCTCAGCAAGAACGGGAACAA; reverse: CCAGAGCTTTTGCTATGGTATTTTC; probe: ACTCTGGACTATGCTGAGAA) and the newly identified transcript (Applied Biosystems; forward: CAAGCCAAAATGCGAGAAAATATT; reverse: CCTTGTCGAGAGGTAAACACAACA; probe: TTGTTAGAAGTGGATTGTCATGA), using Primer Express software (Applied Biosystems), for quantitative analyses by quantitative reverse transcription–PCR carried out using the TaqMan Gene Expression Assay on an ABI Prism 7900 system (Applied Biosystems) by the standard curve method. Samples were quantified in triplicate, with an initial denaturing step of 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C and 60 seconds at 60°C. Expression was normalized to the geometric mean of 3 housekeeping genes (β-actin: assay Hs99999903_m1; β2-microglobulin: assay Hs99999907_m1; and β-glucuronidase: assay Hs99999908_m1). These analyses were conducted in the DLPFC samples (Table).
Western Blotting
Methods, antibodies, and experimental controls are detailed in the eAppendix in the Supplement. Briefly, protein extraction and Western blotting were carried out using standard techniques. We included not only protein extracts from adult and fetal human brain but also protein extracts from transfected and nontransfected HEK293 cells9 and competent bacteria expressing recombinant ZNF804A. The main anti-ZNF804A antibody used was a polyclonal antibody mapping to an internal epitope of human ZNF804A (D-14; Santa Cruz Biotechnology).
Immunohistochemistry
Immunohistochemistry was carried out as described in the eAppendix in the Supplement, using the avidin-biotin complex–diaminobenzidine method. Given the findings (see Results), we investigated whether ZNF804A immunoreactivity was affected by schizophrenia or rs1344706 using the sections of inferior parietal lobe (eTable 1 in the Supplement). We focused our analyses on lamina III pyramidal neurons, measuring their density, size, and staining intensity (eAppendix in the Supplement).
Statistical Analysis
Quantitative reverse transcription–PCR analyses were conducted in R version 3.0.1 software (R Foundation) using linear modeling, treating log2-transformed expression levels of each transcript as the outcome. Statistical models investigating age-related expression patterns in control participants covaried for RNA integrity number (RIN), sex, postmortem interval (PMI), and race. Subsequent models exploring the influence of these biological variables on the expression of ZNF804A were adjusted for age as a B-spline with knots at 1, 10, 20, and 50 with an offset at birth to capture nonlinear patterns in expression. Statistical models investigating diagnosis-related expression patterns in adults covaried for sex, race, pH, PMI, and RIN; diagnosis-related expression fold changes were relative to the control group. Statistical models investigating genotype-by-diagnosis interactions included main-effect regression terms for genotype and diagnosis as well as an interaction term between the 2 variables to capture genotype effects within each diagnosis group. The statistical models for the genotype effect in fetal samples adjusted only for RIN and race, as the PMI of these samples was short (mean, 2.6 hours) and many samples were missing information on pH. All reported P values were calculated from t statistics computed from the log fold change and its standard error from each multiple regression model; therefore, they represent covariateadjusted P values.
For the immunohistochemical study, group comparisons were by unpaired t tests, and the influence of confounding variables was examined using Pearson correlations. Because of the small sample size (eTable 1 in the Supplement), G carriers were compared with TT homozygotes for statistical analysis.
Results
Identification of a Novel Truncated Transcript of ZNF804A
Using RNAseq, 5′ RACE, end-to-end PCR, and quantitative reverse transcription–PCR, we identified a novel human transcript of ZNF804A in fetal and adult whole-brain RNA and in DLPFC. It skips the first 2 exons and has a new 5′ untranslated region in intron 2 (approximately 20 kilobases downstream of rs1344706) (eFigure 1 in the Supplement). This isoform, denoted ZNF804AE3E4, remains in frame and is predicted to encode an approximately 123-kDa protein that lacks the N-terminal 135 amino acids of ZNF804A, including the zinc finger domain located at residues 57 to 81. Given these findings, our quantitative analyses comprised separate assays for the 2 transcripts.
Effects of Age, Sex, and Other Factors on ZNF804A Transcripts Across the Life Span
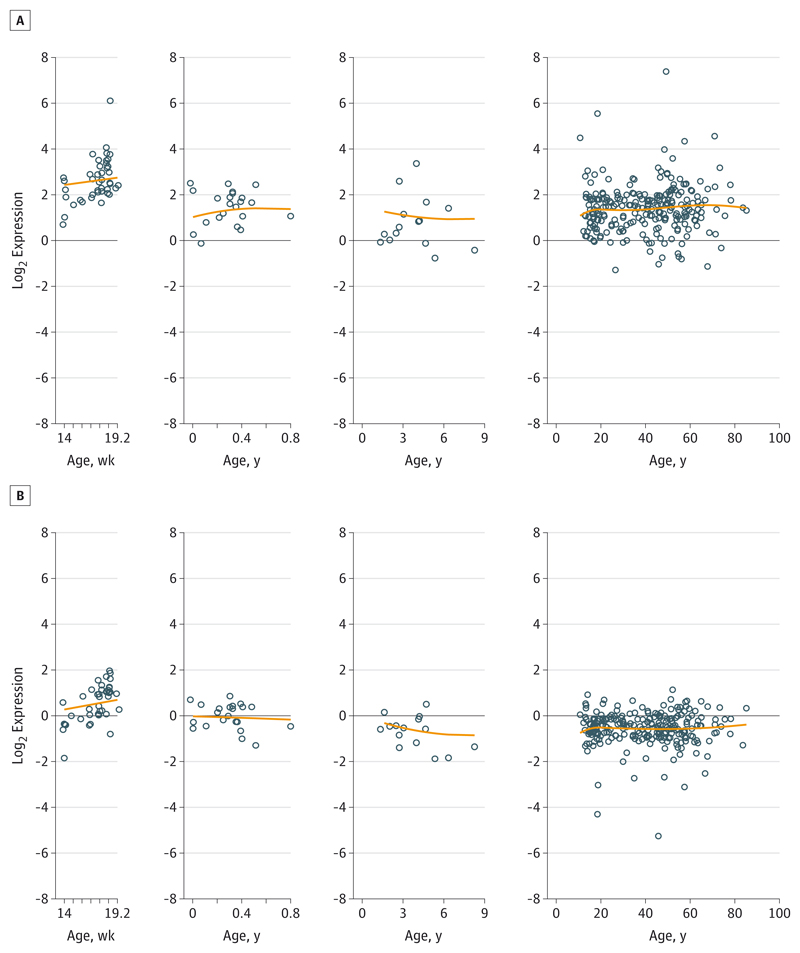
Both ZNF804A transcripts were detected across the entire age range (Figure 1), being preferentially expressed fetally relative to postnatal life (ZNF804A: 1.98-fold higher expression, P < .001; ZNF804AE3E4: 2.63-fold higher expression, P < .001). Overall, ZNF804AE3E4 is more abundant than full-length ZNF804A mRNA (3.79-fold higher expression, P < .001). Their lifetime expression trajectories were similar—both transcripts decreased by similar amounts during infancy and childhood (11% decrease per year on average; full length: P < .001; truncated: P = .01) but remained constant across adulthood.
Figure 1. Expression Trajectories for ZNF804AE3E4 and ZNF804A Messenger RNA Across the Life Span.
Life-span expression trajectories for the novel truncated transcript isoform ZNF804AE3E4 (A) and full-length ZNF804A (B) messenger RNA in human dorsolateral prefrontal cortex. Circles indicate observed data; lines, smoothing spline within each age stage.
There was no main effect of sex on either transcript across the life span, but ZNF804A mRNA was more highly expressed in men compared with women (1.20-fold higher expression, P = .02). There were significant effects of both RIN and PMI on the full-length transcript (RIN: P < .001; PMI: P = .03), but only RIN was associated with levels of the truncated transcript (P < .001). There were differences in ZNF804AE3E4 expression levels between the small set of Hispanic participants (n = 7; 1.90-fold lower expression, P = .02) and Asian participants (n = 6; 1.85-fold higher expression, P = .04) compared with white participants; there were no differences between African American and white participants or any racial differences in the expression of the full-length transcript.
Both ZNF804A Transcripts Are Associated With Diagnosis
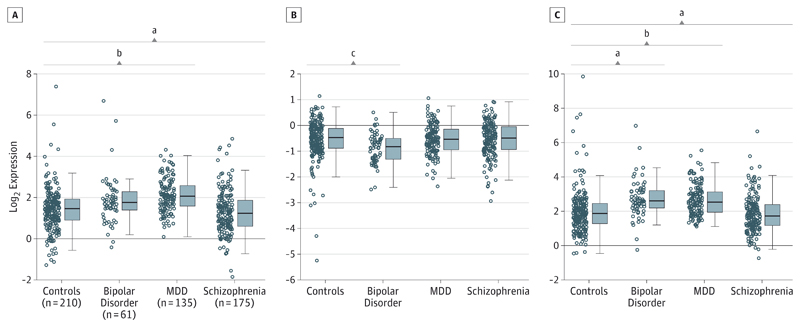
We examined whether the 2 transcripts are differentially expressed in patients with schizophrenia, bipolar disorder, and major depressive disorder in DLPFC. As shown in Figure 2, ZNF804AE3E4 mRNA was increased in major depressive disorder (1.45-fold higher expression, P < .001) and decreased in schizophrenia (1.22-fold lower expression, P = .006). In contrast, full-length ZNF804A mRNA was unaltered in schizophrenia and major depressive disorder and reduced in bipolar disorder (1.17-fold lower expression, P = .01). The ZNF804AE3E4 to ZNF804A mRNA ratio was associated with all 3 disorders: it was elevated in bipolar disorder (P = .002) and major depressive disorder (P < .001) and decreased in schizophrenia (P = .001). There were no significant diagnosis-by-sex interactions affecting either transcript.
Figure 2. Diagnostic Effects on Expression of ZNF804A Transcripts in Dorsolateral Prefrontal Cortex.
A, ZNF804AE3E4 messenger RNA (mRNA) shows elevated expression in major depressive disorder (MDD) but decreased expression in schizophrenia and no change in bipolar disorder compared with controls. B, Full-length ZNF804A mRNA shows decreased expression in bipolar disorder and no change in schizophrenia and MDD. C, Ratio of ZNF804AE3E4 to ZNF804A mRNA. Compared with controls, the ratio is greater (ie, relative increase in ZNF804AE3E4 mRNA) in bipolar disorder and MDD but lower in schizophrenia. Boxes indicate interquartile range; horizontal lines in boxes, median; and whiskers, 1.5 × interquartile range. The postmortem brain samples involved are from the National Institute of Mental Health/Lieber Institute for Brain Development series (Table).
a P < .01 for each diagnostic group vs controls in 1 linear model (see Methods).
b P < .001 for each diagnostic group vs controls in 1 linear model (see Methods).
c P < .05 for each diagnostic group vs controls in 1 linear model (see Methods).
Expression of ZNF804A mRNA did not correlate with antipsychotic or antidepressant exposure overall or within each diagnostic group (all P > .10), nor were these medications significant in multivariate regression models. ZNF804AE3E4 mRNA was associated with antidepressant use across all participants (1.32-fold increase, P = .001) but not within any diagnostic group. Medication history did not bias the observed fold changes of expression between diagnostic groups, but the fact that only 413 participants (279 of 371 with a psychiatric diagnosis [75.2%]) had a recorded medication history reduced statistical significance (not shown).
Psychosis Risk SNP rs1344706 Affects Fetal ZNF804AE3E4 mRNA Expression
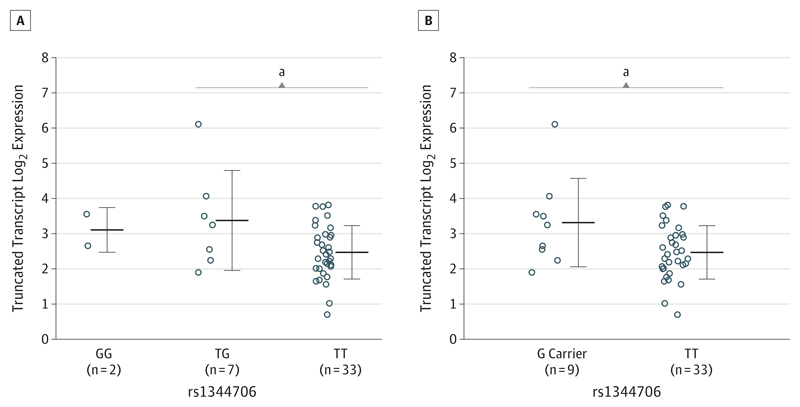
The SNP rs1344706 did not affect expression of either transcript when all participants were analyzed together. However, in a planned analysis restricted to the fetal brains given the finding by Hill and Bray,25 we found a significant effect of rs1344706 on ZNF804AE3E4 mRNA (F = 6.07; P = .02) (Figure 3), with participants homozygous for the risk T allele (n = 33) showing lower expression than G carriers (n = 9) (P = .03). In contrast, full-length ZNF804A mRNA showed no allelic effects in fetal or adult samples.
Figure 3. Genotype Effect of rs1344706 on ZNF804AE3E4 Messenger RNA in Human Fetal Brain.
Homozygotes for the risk T allele are compared with GG homozygotes and TG heterozygotes (A) and with G carriers (B). Homozygotes for the risk T allele have lower expression than G carriers or TG heterozygotes. Center horizontal lines indicate mean; error bars, standard deviation.
a P < .05.
In post hoc comparisons, we also identified 1 genotype-by-diagnosis interaction: in the bipolar disorder group, TT homozygotes had lower expression of ZNF804AE3E4 mRNA (P = .002) (eFigure 2 in the Supplement).
Detection and Distribution of ZNF804A Immunoreactivity
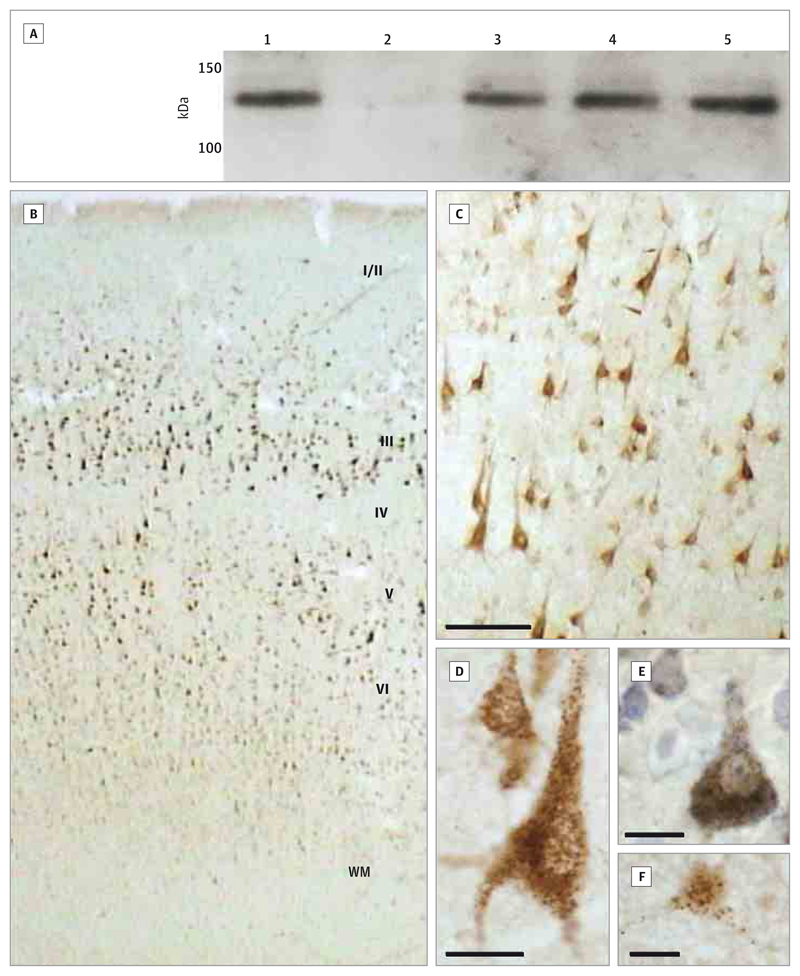
Western blots showed that ZNF804A immunoreactivity is present in adult and fetal brain (Figure 4A and eFigure 3 in the Supplement). A single band was identified at approximately 140 kDa, similar to that observed in ZNF804A-transfected cells and bacteria9 and corresponding to the predicted molecular weight of 137 kDa. Signal was abolished by the blocking peptide, and the P-13 mouse antibody produced the same banding pattern (eFigure 4 in the Supplement).
Figure 4. ZNF804A Immunoreactivity in Human Brain.
A, Western blot. Lane 1 is HEK293 cells transfected with ZNF804A; lane 2, nontransfected HEK293 cells; lane 3, fetal cortex; and lanes 4 and 5, adult frontal cortex. eFigure 3 in the Supplement shows a full-length blot and the β-actin loading control, and eFigure 4 in the Supplement shows the D-14 blocking peptide control and a blot using a different anti-ZNF804A antibody. B, ZNF804A immunoreactivity through the inferior parietal cortex. Roman numerals indicate the cortical layers; WM, white matter. C, Layer III of the inferior parietal cortex (scale bar = 100 μm). D, Pyramidal neurons in layer III of the temporal cortex (scale bar = 10 μm). E, Layer III of the inferior parietal cortex with Nissl counterstain (scale bar = 10 μm). F, Frontal cortex of a neonate (scale bar = 10 μm). eFigure 5 in the Supplement shows the immunohistochemistry peptide block control.
Immunohistochemistry showed staining for ZNF804A in human cerebral cortex sections (Figure 4B-F), especially in lamina III and concentrated over pyramidal neurons. Immunoreactivity was strong in the cytoplasm and proximal dendrites, often with a prominent granular appearance (Figure 4D-F). Some staining of nuclei was also observed. Other cell populations did not show consistent immunolabeling. Findings were similar in all cortical regions examined and were comparable between free-floating and slide-mounted sections. The pyramidal cell immunoreactivity was abolished by preincubation with the blocking peptide (eFigure 5 in the Supplement).
There were no differences in the density, size, or staining intensity of ZNF804A-immunoreactive neurons between patients with schizophrenia and controls or between rs1344706 genotypes (eTable 2 in the Supplement). Although staining intensity correlated inversely with storage time, the latter did not differ between diagnostic or genotype groups, and the group comparisons remained negative if staining intensity was included as a covariate.
Discussion
The association between rs1344706, located in an intron of ZNF804A, and psychosis is statistically robust but biologically unexplained, reflecting the lack of understanding of the roles of ZNF804A and the unknown functionality of the SNP. Herein, we report findings that advance both these questions. We identify a novel mRNA isoform, ZNF804AE3E4, in human brain that appears selectively modulated by rs1344706 during fetal life. In contrast, full-length ZNF804A mRNA shows no genotype effects. In consequence, we propose that the ZNF804AE3E4 isoform mediates, at least partly, the association of rs1344706 with psychosis. The relative expression of ZNF804AE3E4 mRNA compared with the full-length transcript was decreased in schizophrenia and increased in major depressive disorder and bipolar disorder. We also provide the first evidence, to our knowledge, that ZNF804A is translated in human brain and is localized primarily to pyramidal neurons.
Prior to this study, only 1 ZNF804A transcript had been reported in human brain, although a rare isoform with a putative extra exon was identified in lymphoblasts of some individuals.33 Using RNAseq, a powerful technology for identifying novel splice variants,34,35 and confirming by 5′ RACE and end-to end PCR, we discovered the truncated isoform ZNF804AE3E4, which lacks exons 1 and 2 and has a 5′ extension of exon 3. This transcript was abundant and remains in frame, suggesting that it represents an alternative proteincoding transcript of potential functional significance. Both mRNAs were more highly expressed in fetal brain but continue to be expressed throughout life, relative to endogenous control gene levels.
As risk genes for psychiatric disorders are discovered, revealing the mechanism for association becomes important. ZNF804A is typical of many such associations because the risk SNP is noncoding, and without a coding variant in linkage disequilibrium any functionality is likely mediated via an effect on gene expression. A recent study provided initial evidence for this in fetal but not adult brain.25 Our data confirm this observation and, critically, show that the effect of rs1344706 is on ZNF804AE3E4 mRNA rather than on full-length ZNF804A. The primers used in the earlier study amplify both variants, so the present data are both a replication and a clarification. Our finding that a psychosis risk SNP selectively affects expression of 1 transcript variant of a gene adds to an increasing list of such findings36–42 and supports the proposal that this may be a common feature, particularly for fetally enriched isoforms.26 It is also relevant that splice variants are being recognized as showing functional and therapeutically relevant differences from the canonical isoform,43–45 including examples from other schizophrenia-associated genes,39,46 and being mediated by various molecular mechanisms.44,47,48 As well as having an effect in fetal brain, rs1344706 also influenced expression within a diagnostic group, namely ZNF804AE3E4 mRNA in bipolar disorder. The latter was not a planned analysis and hence the finding is preliminary, but it is notable that, as in fetal brains, the risk allele is associated with lower expression. The mechanism is unclear; presumably, rs1344706 interacts with other bipolar disorder–specific factors (whether genetic, epigenetic, or epiphenomenal) to modulate ZNF804A expression.
In addition to the influences of genotype, main effects of diagnosis were found. These again primarily affected ZNF804AE3E4 mRNA rather than the full-length transcript. The diagnostic alterations do not appear to be linked directly to the genetic predisposition: the transcripts change in opposite directions in the 2 disorders with which ZNF804A is associated (decreased in schizophrenia, increased in bipolar disorder), and expression is also affected in major depressive disorder, for which there is no significant evidence of association with ZNF804A. The altered expression in the 3 disorders may in part result from antidepressant use (see Results), but this does not substantially explain the findings. We also did not see relationships with other disease-related variables (eg, duration, age at onset). Thus, the explanations for differential expression of ZNF804AE3E4 mRNA in these disorders and the reduction of ZNF804A mRNA in bipolar disorder remain unclear and warrant additional research.
The presence of ZNF804A immunoreactivity indicates that the gene is not only transcribed but also translated in human brain. ZNF804A immunoreactivity was not concentrated in the nucleus as expected for a classic transcription factor and as reported in rodent neural progenitors.9 Its abundance in the cytoplasm suggests additional roles for ZNF804A in human brain; it also raises the possibility of cross-reactivity of the antibody to another protein, although this is unlikely given the experimental controls used. Neither rs1344706 nor schizophrenia affected our measures of ZNF804A immunoreactivity, so the penetrance of genotype and diagnostic effects through to ZNF804A protein remains to be determined. In any event, understanding the pathophysiological significance of ZNF804A is hindered by uncertainty about the translation and function of ZNF804AE3E4. The variant is predicted to encode a 123-kDa protein; although no separate band of this size was seen on Western blots, it could be subsumed within the approximately 137-kDa band corresponding to full-length ZNF804A if ZNF804AE3E4 undergoes posttranslational modifications that increase its apparent molecular weight. If it is translated, ZNF804AE3E4 may well contribute to the non–transcription factor–related functions postulated here (because it lacks a zinc finger domain), but it does not contain strong sequence motifs or homologies that might give positive clues in this regard.
Conclusions
We describe a novel, truncated transcript of ZNF804A, ZNF804AE3E4. Unlike the full-length transcript, ZNF804AE3E4 is regulated in human fetal brain by the genome-wide psychosis risk allele rs1344706. This finding suggests that ZNF804AE3E4 may be a critical isoform mediating the association and draws attention to the importance of alternative transcripts, and the fetal period, in the processes by which genetic variation affects neurodevelopmental psychiatric disorders. The fact that ZNF804A is translated into protein is also significant, increasing the likelihood that the gene has functional and pathophysiological roles in the brain. Equally, our data highlight the complexities involved when trying to identify the biological basis of a genetic association with a psychiatric disorder.
Supplementary Material
Funding/Support
This work was supported by the Lieber Institute for Brain Development and by a strategic award from the Wellcome Trust (Drs Weinberger and Harrison). Dr Cousijn was supported by a studentship from the Wellcome Trust supervised by Dr Harrison and Anna C. Nobre, PhD, University of Oxford, Oxford, United Kingdom.
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Tao and Cousijn contributed equally, and Drs Hyde and Kleinman contributed equally. Drs Harrison and Hyde had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Tao, Cousijn, Maher, Weinberger, Harrison, Hyde, Kleinman.
Acquisition, analysis, or interpretation of data: Tao, Cousijn, Jaffe, Burnet, Edwards, Eastwood, Shin, Lane, Walker, Weinberger, Harrison, Hyde, Kleinman.
Drafting of the manuscript: Tao, Jaffe, Weinberger, Harrison, Hyde.
Critical revision of the manuscript for important intellectual content: Cousijn, Jaffe, Burnet, Edwards, Eastwood, Shin, Lane, Walker, Maher, Weinberger, Harrison, Hyde, Kleinman.
Statistical analysis: Tao, Cousijn, Jaffe, Shin, Weinberger, Harrison, Hyde.
Obtained funding: Cousijn, Harrison, Hyde, Kleinman.
Administrative, technical, or material support: Tao, Edwards, Eastwood, Lane, Walker, Hyde, Kleinman.
Study supervision: Burnet, Eastwood, Weinberger, Harrison, Hyde, Kleinman.
Conflict of Interest Disclosures: None reported.
Additional Contributions: Amy Deep-Soboslay, MEd, and Llewellyn B. Bigelow, MD, Lieber Institute for Brain Development, Johns Hopkins University, Baltimore, Maryland, assisted in clinical diagnosis and demographic characterization, and Chao Li, PhD, Mary M. Herman, MD, Juan C. Troncoso, MD, Michelle Mighdoll, and Li Chen, Department of Psychiatry, University of Oxford, Oxford, United Kingdom, provided excellent technical assistance. H. Ronald Zielke, PhD, Robert D. Vigorito, MS, PA, and Robert M. Johnson, BS, National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders, University of Maryland, Baltimore, provided fetal, child, and adolescent brain specimens for this study, and Maree Webster, PhD, Stanley Medical Research Institute, Chevy Chase, Maryland, provided adult brain specimens from the Stanley Medical Research Institute Brain Collection. We thank the families of all of the decedents for the generous donation of tissue dedicated to this research. No compensation was received from the funders for these contributions.
Contributor Information
Ran Tao, Lieber Institute for Brain Development, Johns Hopkins University, Baltimore, Maryland.
Helena Cousijn, Department of Psychiatry, University of Oxford, Oxford, United Kingdom.
Andrew E. Jaffe, Lieber Institute for Brain Development, Johns Hopkins University, Baltimore, Maryland.
Joo Heon Shin, Lieber Institute for Brain Development, Johns Hopkins University, Baltimore, Maryland.
Paul J. Harrison, Department of Psychiatry, University of Oxford, Oxford, United Kingdom.
References
- 1.O’Donovan MC, Craddock N, Norton N, et al. Molecular Genetics of Schizophrenia Collaboration Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 2.Riley B, Thiselton D, Maher BS, et al. Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of Schizophrenia sample. Mol Psychiatry. 2010;15(1):29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwab SG, Kusumawardhani AA, Dai N, et al. Indonesian Schizophrenia Genetics Consortium Association of rs1344706 in the ZNF804A gene with schizophrenia in a case/control sample from Indonesia. Schizophr Res. 2013;147(1):46–52. doi: 10.1016/j.schres.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R, Lu SM, Qiu C, et al. Population-based and family-based association studies of ZNF804A locus and schizophrenia. Mol Psychiatry. 2011;16(4):360–361. doi: 10.1038/mp.2010.55. [DOI] [PubMed] [Google Scholar]
- 5.Williams HJ, Norton N, Dwyer S, et al. Molecular Genetics of Schizophrenia Collaboration (MGS); International Schizophrenia Consortium (ISC); SGENE-plus; GROUP Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry. 2011;16(4):429–441. doi: 10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R, Yan JD, Valenzuela RK, et al. Further evidence for the association of genetic variants of ZNF804A with schizophrenia and a meta-analysis for genome-wide significance variant rs1344706. Schizophr Res. 2012;141(1):40–47. doi: 10.1016/j.schres.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Tadepally HD, Burger G, Aubry M. Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes and effector domains. BMC Evol Biol. 2008;8:176. doi: 10.1186/1471-2148-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Razin SV, Borunova VV, Maksimenko OG, Kantidze OL. Cys2His2 zinc finger protein family: classification, functions, and major members. Biochemistry (Mosc) 2012;77(3):217–226. doi: 10.1134/S0006297912030017. [DOI] [PubMed] [Google Scholar]
- 9.Girgenti MJ, LoTurco JJ, Maher BJ. ZNF804a regulates expression of the schizophrenia-associated genes PRSS16, COMT, PDE4B, and DRD2. PLoS One. 2012;7(2):e32404. doi: 10.1371/journal.pone.0032404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill MJ, Jeffries AR, Dobson RJ, Price J, Bray NJ. Knockdown of the psychosis susceptibility gene ZNF804A alters expression of genes involved in cell adhesion. Hum Mol Genet. 2012;21(5):1018–1024. doi: 10.1093/hmg/ddr532. [DOI] [PubMed] [Google Scholar]
- 11.Umeda-Yano S, Hashimoto R, Yamamori H, et al. The regulation of gene expression involved in TGF-β signaling by ZNF804A, a risk gene for schizophrenia. Schizophr Res. 2013;146(1–3):273–278. doi: 10.1016/j.schres.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 12.Esslinger C, Walter H, Kirsch P, et al. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324(5927):605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- 13.Donohoe G, Morris DW, Corvin A. The psychosis susceptibility gene ZNF804A: associations, functions, and phenotypes. Schizophr Bull. 2010;36(5):904–909. doi: 10.1093/schbul/sbq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasetti R, Sambataro F, Chen Q, Callicott JH, Mattay VS, Weinberger DR. Altered cortical network dynamics: a potential intermediate phenotype for schizophrenia and association with ZNF804A. Arch Gen Psychiatry. 2011;68(12):1207–1217. doi: 10.1001/archgenpsychiatry.2011.103. [DOI] [PubMed] [Google Scholar]
- 15.Voineskos AN, Lerch JP, Felsky D, et al. The ZNF804A gene: characterization of a novel neural risk mechanism for the major psychoses. Neuropsychopharmacology. 2011;36(9):1871–1878. doi: 10.1038/npp.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M, Xu Z, Zhai J, et al. Evidence of IQ-modulated association between ZNF804A gene polymorphism and cognitive function in schizophrenia patients. Neuropsychopharmacology. 2012;37(7):1572–1578. doi: 10.1038/npp.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hargreaves A, Morris DW, Rose E, et al. ZNF804A and social cognition in patients with schizophrenia and healthy controls. Mol Psychiatry. 2012;17(2):118–119. doi: 10.1038/mp.2011.102. [DOI] [PubMed] [Google Scholar]
- 18.Kuswanto CN, Woon PS, Zheng XB, et al. Genome-wide supported psychosis risk variant in ZNF804A gene and impact on cortico-limbic WM integrity in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(3):255–262. doi: 10.1002/ajmg.b.32032. [DOI] [PubMed] [Google Scholar]
- 19.Wassink TH, Epping EA, Rudd D, et al. Influence of ZNF804a on brain structure volumes and symptom severity in individuals with schizophrenia. Arch Gen Psychiatry. 2012;69(9):885–892. doi: 10.1001/archgenpsychiatry.2011.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linden DE, Lancaster TM, Wolf C, et al. ZNF804A genotype modulates neural activity during working memory for faces. Neuropsychobiology. 2013;67(2):84–92. doi: 10.1159/000344001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefanis NC, Hatzimanolis A, Avramopoulos D, et al. Variation in psychosis gene ZNF804A is associated with a refined schizotypy phenotype but not neurocognitive performance in a large young male population. Schizophr Bull. 2013;39(6):1252–1260. doi: 10.1093/schbul/sbs110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurin K, Rasetti R, Sambataro F, et al. Effects of ZNF804A on neurophysiologic measures of cognitive control. Mol Psychiatry. 2013;18(8):852–854. doi: 10.1038/mp.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cousijn H, Rijpkema M, Harteveld A, et al. Schizophrenia risk gene ZNF804A does not influence macroscopic brain structure: an MRI study in 892 volunteers. Mol Psychiatry. 2012;17(12):1155–1157. doi: 10.1038/mp.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guella I, Sequeira A, Rollins B, et al. Evidence of allelic imbalance in the schizophrenia susceptibility gene ZNF804A in human dorsolateral prefrontal cortex. Schizophr Res. 2014;152(1):111–116. doi: 10.1016/j.schres.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill MJ, Bray NJ. Evidence that schizophrenia risk variation in the ZNF804A gene exerts its effects during fetal brain development. Am J Psychiatry. 2012;169(12):1301–1308. doi: 10.1176/appi.ajp.2012.11121845. [DOI] [PubMed] [Google Scholar]
- 26.Kleinman JE, Law AJ, Lipska BK, et al. Genetic neuropathology of schizophrenia: new approaches to an old question and new uses for postmortem human brains. Biol Psychiatry. 2011;69(2):140–145. doi: 10.1016/j.biopsych.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, Chen Q, Ye T, et al. Evidence of sex-modulated association of ZNF804A with schizophrenia. Biol Psychiatry. 2011;69(10):914–917. doi: 10.1016/j.biopsych.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Lipska BK, Deep-Soboslay A, Weickert CS, et al. Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol Psychiatry. 2006;60(6):650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 29.Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The Stanley Foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44(2):151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- 30.Stanley Medical Research Institute. Inferior parietal collection. [Accessed July 4];2014 http://www.stanleyresearch.org/dnn/Default.aspx?tabid=199.
- 31.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada T, Hashimoto R, Yamamori H, et al. Expression analysis of a novel mRNA variant of the schizophrenia risk gene ZNF804A. Schizophr Res. 2012;141(2–3):277–278. doi: 10.1016/j.schres.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Griffith M, Griffith OL, Mwenifumbo J, et al. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7(10):843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- 35.Ameur A, Zaghlool A, Halvardson J, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18(12):1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 36.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16(2):129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 37.Law AJ, Lipska BK, Weickert CS, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci U S A. 2006;103(17):6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sartorius LJ, Weinberger DR, Hyde TM, Harrison PJ, Kleinman JE, Lipska BK. Expression of a GRM3 splice variant is increased in the dorsolateral prefrontal cortex of individuals carrying a schizophrenia risk SNP. Neuropsychopharmacology. 2008;33(11):2626–2634. doi: 10.1038/sj.npp.1301669. [DOI] [PubMed] [Google Scholar]
- 39.Huffaker SJ, Chen J, Nicodemus KK, et al. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med. 2009;15(5):509–518. doi: 10.1038/nm.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakata K, Lipska BK, Hyde TM, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci U S A. 2009;106(37):15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Straub RE, Lipska BK, Egan MF, et al. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol Psychiatry. 2007;12(9):854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- 42.Hyde TM, Lipska BK, Ali T, et al. Expression of GABA signaling molecules KCC2, NKCC1, and GAD1 in cortical development and schizophrenia. J Neurosci. 2011;31(30):11088–11095. doi: 10.1523/JNEUROSCI.1234-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12(10):715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majumdar S, Grinnell S, Le Rouzic V, et al. Truncated G protein-coupled mu opioid receptor MOR-1 splice variants are targets for highly potent opioid analgesics lacking side effects. Proc Natl Acad Sci U S A. 2011;108(49):19778–19783. doi: 10.1073/pnas.1115231108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrie ES, Smith RM, Sanford JC, Sadee W. mRNA transcript diversity creates new opportunities for pharmacological intervention. Mol Pharmacol. 2012;81(5):620–630. doi: 10.1124/mol.111.076604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Law AJ, Wang Y, Sei Y, et al. Neuregulin 1-ErbB4-PI3K signaling in schizophrenia and phosphoinositide 3-kinase-p110δ inhibition as a potential therapeutic strategy. Proc Natl Acad Sci U S A. 2012;109(30):12165–12170. doi: 10.1073/pnas.1206118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nag K, Sultana N, Kato A, Hirose S. Headless splice variant acting as dominant negative calcitonin receptor. Biochem Biophys Res Commun. 2007;362(4):1037–1043. doi: 10.1016/j.bbrc.2007.08.107. [DOI] [PubMed] [Google Scholar]
- 48.Córdoba-Chacón J, Gahete MD, Duran-Prado M, et al. Identification and characterization of new functional truncated variants of somatostatin receptor subtype 5 in rodents. Cell Mol Life Sci. 2010;67(7):1147–1163. doi: 10.1007/s00018-009-0240-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.