Abstract
The egg yolk precursor protein vitellogenin is widely used as a biomarker of estrogen exposure in male fish. However, standardized methodology is lacking and little is known regarding the reproducibility of results among laboratories using different equipment, reagents, protocols, and data analysis programs. To address this data gap we tested the reproducibility across laboratories to evaluate vitellogenin gene (vtg) expression and assessed the value of using a freely available software data analysis program. Samples collected from studies of male fathead minnows (Pimephales promelas) exposed to 17a-ethinylestradiol (EE2) and minnows exposed to processed wastewater effluent were evaluated for vtg expression in 4 laboratories. Our results indicate reasonable consistency among laboratories if the free software for expression analysis LinRegPCR is used, with 3 of 4 laboratories detecting vtg in fish exposed to 5 ng/L EE2 (n ¼ 5). All 4 laboratories detected significantly increased vtg levels in 15 male fish exposed to wastewater effluent compared with 15 male fish held in a control stream. Finally, we were able to determine that the source of high interlaboratory variability from complementary deoxyribonucleic acid (cDNA) to quantitative polymerase chain reaction (qPCR) analyses was the expression analysis software unique to each real-time qPCR machine. We successfully eliminated the interlaboratory variability by reanalyzing raw fluorescence data with independent freeware, which yielded cycle thresholds and polymerase chain reaction (PCR) efficiencies that calculated results independently of proprietary software. Our results suggest that laboratories engaged in monitoring programs should validate their PCR protocols and analyze their gene expression data following the guidelines established in the present study for all gene expression biomarkers.
Keywords: Real-time polymerase chain reaction variability, Fathea Minnow, Bioassay, Estrogen, Wastewater, Interlaboratory
INTRODUCTION
Wastewater treatments plants [1], biosolids runoff [2], and concentrated animal feeding operations [3] release complex mixtures of endocrine-disrupting chemicals (EDCs) such as estrogens into the environment. Evidence exists that some EDCs feminize male fish and impact the reproductive health of aquatic life [4,5]. Furthermore, exposure to estrogenic pollutants has resulted in population-level effects [4,6] and has even induced transgenerational effects in aquatic organisms [4,7,8]. Thus, characterizing estrogenic effects is of national interest to regulatory agencies and the general public. However, challenges remain because standardized methods of quantifying estrogenicity have not been developed. The lack of standardization complicates interpretations among laboratories participating in screening programs. In our view, the first step in this process is developing a method of quantifying estrogenicity that can be used consistently among many laboratories.
Numerous bioassays exist for evaluating estrogenicity. Identification of testis–ova in histological preparations of male testis has been associated with estrogenic contaminant exposures [5,9–11]. However, the utility of testis–ova as a biomarker of estrogenicity has recently been questioned because of observations of testis–ova in historic samples [5] and in fish from reference sites [12]. Thus, identification of markers that can be specifically linked to recent estrogenic exposures is of interest. Estrogen-responsive gene or protein expression in body tissues results directly from estrogen receptor activation and thus may be more definitive evidence of estrogen exposure. Estrogen-responsive proteins can be quantified by enzyme immunoassay [13,14], western blots [13,15], and 2-dimensional protein arrays [16], and can be localized immunohistochemically [17,18] in a variety of tissues. Estrogen-responsive genes can be monitored individually using real-time quantitative polymerase chain reaction (qPCR) [19–21], reverse transcription- (RT) polymerase chain reaction (PCR) [19,20], and deoxyribonucleic acid (DNA) microarrays [22]. Estrogenic effects are also increasingly being evaluated by comparing metabolite profiles of exposed and unexposed fish [23,24].
Of these methods to measure estrogenic exposure, qPCR may be the most readily adaptable for use in monitoring within a regulatory framework. Advances in PCR technology now permit quantification of specific transcripts using fluorescent markers bound to the PCR products [19]. The relative fluorescence in the target gene is normalized to a reference gene known not to be affected by the exposure of interest. The resulting metric is then compared across experimental groups or fish collected from contaminated and reference sites [25]. It has been found that qPCR assays improve with RT-PCR, with increased sensitivity, throughput, and ease of use [19]. In our investigations, we employed qPCR technology to evaluate estrogen-responsive gene expression and compared the results among 4 laboratories analyzing the same samples.
Biological responses to estrogen exposures include the induction of vitellogenin (vtg) in male fish, a gene that codes for egg yolk precursor proteins normally found in sexually maturing female fish [5,13,26]. Vitellogenin is widely applied as a biomarker of estrogen exposures [26,27]. However, standardized methods and estimates of variability in vtg levels among laboratories analyzing the same samples are lacking. Without an understanding of the sources and magnitude of interlaboratory variability and how these affect conclusions, only tentative predictions can be made regarding the accuracy and precision of vtg as a monitoring endpoint.
Evaluation of interlaboratory variability is a common practice in the biomedical sciences where production of consistent results is of utmost importance in human disease diagnoses [28,29]. This practice has also been applied in the environmental sciences, especially when public health is of concern, such as in the tracking of fecal bacteria [30,31]. As articulated by Hultman et al. [32], this practice should also be applied to quantify and identify sources of interlaboratory variability in vtg expression in fish [32]. To our knowledge, evaluation of interlaboratory variability in vtg expression has not been accomplished to date.
The objectives of the present study were to: 1) contribute to the development of a standard operating procedure for quantifying estrogenic gene expression to be used in a whole effluent toxicity context [33], and 2) evaluate the combined effects of different analysts, laboratory equipment, reagents/kits, and gene expression analysis software on variability in estrogenic gene expression. Thus, we aimed to establish a standardized approach to analyzing vtg expression data in EDC-exposed male fish and provide recommendations to minimize interlaboratory variability. The results suggest that implementation of a large-scale vtg monitoring program may be possible following our guidelines.
MATERIALS AND METHODS
Experimental designs and sample handling
Both ribonucleic acid (RNA; batch 1) and complementary (c)DNA (batch 2 and batch 3) from the samples were used to estimate interlaboratory variability resulting from various points along a typical qPCR workflow. The total RNA or cDNA from male fathead minnows (Pimephales promelas) was sourced from 3 studies designed to satisfy other research objectives.
For batch 1, the RNA samples were from the study by Reddy et al. in 2015 conducted at the US Environmental Protection Agency (USEPA) in Cincinnati Ohio [34] and consisted of 5 treatment replicates (1 fish/replicate, n ¼ 5) of male fathead minnow exposed for 48 h in a static renewal to 0, 1, or 2 ng/L ethinylestradiol (EE2) in reconstituted moderately hard water [34]. These samples represent typical laboratory exposure studies in which there is maximal control over the environmental conditions.
For batch 2, the cDNA samples were from 5 treatment replicates (8 fish/replicate, n ¼ 5) of male fathead minnow exposed in cages for 14 d to 0, 2.5, 5, 10, or 20 ng/L EE2 in outdoor aquatic mesocosms. The mesocosm setup and experimental design follows Schwindt et al. [4] except that the fish were exposed every other day in a semistatic renewal. The mesocosms used in this study were artificial ecosystems with fluctuating photoperiod, water temperature, productivity, and nutrients [35]. Thus the mesocosms represent an exposure venue with minimal control over environmental conditions while still allowing treatment replication.
For batch 3, cDNA samples were from 15 male fathead minnows exposed in cages for 5 d to stream water at a reference site or to wastewater effluent in the Denver, Colorado (USA) metropolitan area. A reference site was chosen on Clear Creek, a tributary of the South Platte River, near the mouth of Clear Creek Canyon in Golden, Colorado that was not directly subjected to wastewater effluent. The wastewater treatment plant (WWTP) receives approximately 454 106 L/d and has primary and secondary treatment (activated sludge) with ammonification and discharges into the South Platte River.
Fifteen adult male fathead minnows obtained from the USEPA Aquatic Research Facility in Cincinnati, Ohio, were caged in the river within 50 m downstream of the WWTP discharge in 15-cm diameter 150-cm polyvinylchloride (PVC) pipes that were capped on either end. Sections of PVC were removed and replaced with stainless steel mesh to allow water flow. Cages were anchored in a slow-moving section of the stream using metal fence posts and metal wire. Limitations in the number of fish available prevented the use of multiple cages per site. We deployed the fish on a Thursday and retrieved them the following Tuesday (5-d exposure). All hardware was removed at the end of the experiment. Treatment of all animals followed USEPA and Colorado State University Institutional Animal Care and Use Committee (IACUC) protocols.
RNA and cDNA preparation
The RNA for batch 1 and cDNA batch 2 was extracted, prepared, and assessed as previously described [4,34]. For batch 3, RNA was extracted with TRI Reagent1 (Molecular Research Center) following the manufacturer’s protocol. The integrity of batch 3 samples was assessed using an Agilent 2100 BioAnalyzer and Agilent RNA 6000 Nano Kits. All batch 3 RNA samples used in the present study were of high quality, with an RNA Integrity Number (RIN) 8. The RNA was reverse transcribed as previously described [19].
Samples were shipped overnight on dry ice to laboratories participating in the study. The laboratories were located at the University of Massachusetts (Amherst, MA, USA); USEPA, Office of Research and Development (Cincinnati, OH), the USEPA Region 5 laboratory (Chicago, IL), and the USEPA Region 8 laboratory (Denver, CO). The RNA concentrations were assessed on all samples at each laboratory using a NanoDrop1 spectrophotomer with A260:A280 ratios > 1.8.
Real-time qPCR and vtg mRNA expression analysis
All laboratories used QuantumRNATM 18S universal primers (18S ribosomal [r]RNA; Ambion/Life Technologies) to normalize vtg expression. The vtg primer sequences used by all laboratories are CGAAGGCACGCTGATAGA and TGA-CAAGCCAACAGCAAGAG, as reported in Biales et al. [19].
The USEPA Region 8 and Office of Research and Development laboratories followed Biales et al. [19] for cDNA synthesis and qPCR analysis using a DNA Engine Opticon1 2 system (BioRad) and HS DyNAmoTM SYBR1 Green qPCR kit (Thermo Fisher Scientific). After each run, the USEPA Office of Research and Development calculated the relative expression using the 2−ΔΔCt method [36] from fluorescence data processed by the qPCR machine. The USEPA Region 8 and Office of Research and Development laboratories used RT-PCR methods previously published (USEPA Region 5 laboratory used Applied Biosystems Power SYBR Green PCR Master Mix, Thermo Fisher Scientific, when it received cDNA). The USEPA Region 8 laboratory imported the non-baseline-subtracted raw fluorescence data into LinRegPCR freeware 37, calulated new Ct values, and then calculated relative expression using the 2−ΔΔCt method.
Both the University of Massachusetts and USEPA Region 5 laboratories used the StepOnePlus™ real time PCR System and the Power SYBR Green RNA-to-Ct™ 1-Step Kit (Applied Biosystems/Life Technologies). For calculation of relative expression, the USEPA Region 5 laboratory followed the protocol from the manufacturer included with the StepOnePlus™ expression analysis software. The University of Massachusetts laboratory used the 2−ΔΔCt method for relative expression calculations from Ct values exported from the StepOnePlus™ expression analysis software. For batches 2 and 3 (when cDNA was received), the University of Massachusetts conducted the qPCR using the FastStart Universal SYBR Green Master (with Rox Reference Dye; Roche). See the Supplemental Data, Table S1 for a listing of equipment, volumes, and primers used by all 4 laboratories.
Interlaboratory variability
Experimental samples were analyzed in duplicate or triplicate depending on available sample concentrations. We hypothesized that most interlaboratory variability was because of the proprietary data processing software, specifically how PCR efficiencies are calculated by each manufacturer, based on the discussion in Ramakers et al. [37]. To evaluate interlaboratory variability, we tested the influence of each PCR machine and how it processed the fluorescence data. Using batch 2 data from the mesocosm study, each laboratory analyzed the fluorescence data according to their in-house protocols. Each laboratory then exported the non-baseline–corrected raw fluorescence data, as recommended in Ramakers et al. [37], for analysis with standalone freeware (LinRegPCR). New Ct and PCR efficiencies were estimated using LinRegPCR for each laboratory, and relative expression was calculated using the 2−ΔΔCt method.
Data analysis
The vtg expression data were loge transformed to meet normality and equal variance assumptions. Analysis of covariance (Proc MIXED, SAS software Ver 9.3, ©2012, SAS Institute) was used to assess the effect of experimental treatment (EE2 or field site) on vtg expression levels with laboratory included as a covariate. Interlaboratory and treatment effects were subjected to a Tukey adjustment because of the large number of pairwise comparisons. Type I error was set at a the 4 laboratories are presented (e.g., 5.3 < t < 8.4, 0.001 < p < 0.005). The evaluation was restricted to the batch that had the greatest number of exposure concentrations and therefore provided the opportunity to examine the response curve.
RESULTS
The vtg expression levels for each laboratory and measured EE2 concentrations are presented in Table 1. When each laboratory employed its own methods of data processing and calculation of relative expression, significant interlaboratory variability was observed. In the laboratory exposure (batch 1), the laboratory (f3,43 ¼ 222, p < 0.001) and treatment (f2,43 ¼ 12.2, p < 0.0001) affected vtg expression. All laboratories differed significantly at all EE2 levels (6.3 < t43 < 22.8, p < 0.0001), and only the USEPA Region 5 laboratory significantly detected vtg at 2 ng/L, relative to controls (t43 ¼ 5.7, p ¼ 0.04; Figure 1A). For all laboratories, inter- and intra-assay coefficient of variation was <7% estimated from a standard pool of high-expressing vtg cDNA or RNA.
Table 1.
Vitellogenin (vtg) Expression Levels (vtg/18S rRNA) in Male Fathead Minnows (Pimephales promelas) (mean ± SEM)
| Treatment (Nominal EE2) | EE2* | aORD vtg | bR5 vtg | cR8 vtg | dUM vtg | |
|---|---|---|---|---|---|---|
| Batch 1 | 48 h; 0 ng/L | 0 | 7×10−9 ± 3 × 10−9 | 68.5 ± 49 | 2 × 10−6 ± 6 × 10−7 | eN/D |
| 48 h; 1 ng/L | 1.33 | 4×10−8 ±2 × 10−8 | 1.7 × 103 ± 1.3 × 103 | 6 × 10−6 ± 2 × 10−6 | N/D | |
| 48 h; 2 ng/L | 2.23 | 6× 10−7 ± 3 × 10−7 | 6.6 × 104 ± 4.7 × 104** | 5 × 10−3 ± 2 × 10−3 | 0.2 ± 0.01 | |
| Batch 2 | 14 d; 0 ng/L | 0 | N/D | 1.5 × 104 ± 1.6 × 104 | 0.06 ± 0.03 | N/D |
| 14 d; 2.5 ng/L | 1.5 | N/D | 3 × 103 ± 8 × 102 | 0.01 ± 0.002 | 0.008 ± 5 × 10−4 | |
| 14 d; 5 ng/L | 3.7 | 0.01 ± 7.7 × 10−3** | 1 × 105 ± 4 × 104** | 0.2 ± 0.07 | 0.16 ± 0.03** | |
| 14 d; 10ng/L | 7.9 | 0.06 ± 0.04 | 6 × 105 ± 1.2 × 105 | 0.7 ± 0.1** | 1.14 ± 0.34 | |
| 14 d; 20ng/L | 16.6 | 0.06 ± 0.05 | 1 × 106±1 × 105 | 1.8 ± 0.7 | 2.15 ± 0.48 | |
| Batch 3 | 5 d; Reference Site | fN/A | 3 × 10−3 ± 3 × 10−3 | 30.6 ± 12.0 | 8 × 10−3 ± 4 × 10−4 | 10−3 ± 10−5 |
| 5 d; WWTP Site | N/A | 64.4 ± 27** | 7×105 ± 1 × 105** | 0.9 ± 0.2** | 2.5 ± 0.9** | |
Measured levels of 17α-Ethinylestradiol (EE2) Concentrations (ng/L)
Office of Research and Development, US Environmental Protection Agency, Cincinnati, OH
US Environmental Protection Agency, Region 5 Laboratory, Chicago, IL
US Environmental Protection Agency, Region 8 Laboratory, Golden, CO
University of Massachusetts, Amherst, MA
Not Detected
Not Available
Indicates significantly different (p < 0.05) from controls (0 ng/L) or from the Reference Site.
Figure 1.
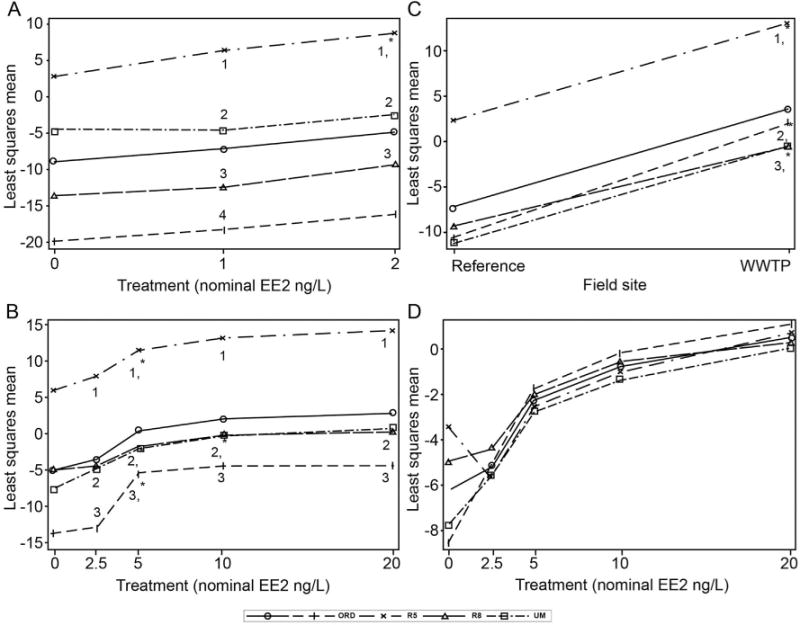
Interactive effects of different laboratories analyzing the same male fathead minnow (Pimephales promelas) liver tissue for vitellogenin (vtg) gene expression. Fathead minnows were exposed to either 17α-ethinylestradiol (EE2) in reconstituted moderately hard water (A), EE2 in outdoor aquatic mesocosms filled with lake water (B), reference site or wastewater treatment plant (WWTP) effluent water (C), or, in the case of (D), the effect of independent analysis software on the same vtg expression data shown in (B). Different numbers indicate statistical differences among laboratories at a given level of EE2 (A and B) or at the WWTP (C) (p < 0.05). Asterisks indicate where vtg expression first differed significantly from 0 ng/L EE2 (A and B) or if it was significantly different from the reference site (C) (p < 0.05). No interlaboratory differences in vtg gene expression at any level of EE2 were found in (D). Data are least squares means, which are adjusted means for all terms in the statistical model and are different from arithmetic means. The open circles with solid lines are the mean expression data for all laboratories. Participating laboratories are: ORD = US Environmental Protection Agency (USEPA) Office of Research and Development, Cincinnati, OH; R5 = USEPA Region 5 Laboratory, Chicago, IL, R8 = USEPA Region 8 Laboratory, Golden, CO; UM = University of Massachusetts, Amherst, MA, USA.
In the mesocosm samples (batch 2), a significant treatment by laboratory interaction was also found (f12,75 ¼ 2.4, p ¼ 0.01; Figure 1B; note that the asterisks indicate the first concentration at which vtg differed significantly from the control for each laboratory). The lowest exposure detected was a concentration of 5 ng/L. The USEPA Office of Research and Development, University of Massachusetts, and USEPA Region 5 laboratories all significantly detected vtg at 5 ng/L EE2 (5.3 < t75 < 8.4, 0.0001 < p < 0.0002), and the USEPA Region 8 laboratory detected it at 10 ng/L (t75 ¼ 4.4, p ¼ 0.005). A significant difference in vtg expression among laboratories was found at 2.5, 10, and 20 ng/L (3.8 < t75 < 17, 0.0001 < p < 0.04); however, University of Massachusetts and USEPA Region 8 vtg gene expression did not differ from each other at any of the concentrations (p > 0.05). The USEPA Region 5 laboratory was significantly different from all laboratories at 5 ng/L (12 < t75 < 17, p < 0.0001) but no difference was found among USEPA Office of Research and Development, University of Massachusetts, and USEPA Region 8 (p > 0.05).
A significant laboratory by field site interaction was found in fish exposed to stream water or wastewater-dominated stream water (batch 3; f3,104 = 4.7, p = 0.004). All laboratories significantly detected vtg at the WWTP (12 < t104 < 17, p < 0.0001), but the levels of vtg also varied significantly among laboratories. The vtg expression produced by the USEPA Region 5 laboratory was significantly higher than that of the other laboratories (15 < t104 < 18, p < 0.0001). The USEPA Office of Research and Development laboratory was higher than the USEPA Region 8 and University of Massachusetts laboratories (3.3 < t104 < 3.6, 0.0004 < p < 0.001; Figure 1C).
In the batch 2 samples, when the Ct and PCR efficiencies for all laboratories were estimated with the same independent freeware and relative expression was subsequently calculated, the effect of EE2 remained (f4,70 ¼ 45.3, p < 0.0001), but interlaboratory variability was reduced below significance (f3,70 ¼ 1.85, p ¼ 0.15; Figure 1D).
DISCUSSION
We successfully developed a procedure for processing data for vtg expression, to be used in evaluating estrogenic effluent. Samples were analyzed at 4 laboratories across the United States, and we found the results to be reproducible and sensitive, as discussed in the following paragraphs. Our findings provide an initial step for the potential development of a nationwide monitoring plan, given the concerns over exposures of fish to estrogenic contaminants.
In the present study, we have demonstrated reasonable similarity among laboratories in terms of the minimum transcription levels of vtg induction. Within the conditions tested in the present study, multiple laboratories can produce the same result using different analysts, reagents, equipment, and even different methods of relative expression quantification. We have also demonstrated that significantly high interlaboratory variability can be eliminated through the standardization of data analysis techniques and that the minimum inductions levels that can be detected can be improved. These guidelines are to: 1) export non-baseline–subtracted raw fluorescence data to a spreadsheet; 2) import the raw fluorescence data into software that is independent of the PCR machine manufacturer, such as LinRegPCR [37]; 3) following the software protocol, estimate new Ct and PCR efficiencies; and 4) calculate relative expression using the 2-DDCt method [36]. Given these guide-lines, our results suggest that implementation of large-scale vtg monitoring programs may be possible because interlaboratory variability can be eliminated from a statistical standpoint. Even further, our guidelines do not require that laboratories alter anything about their PCR protocols, reagents, or methods and only require excellent technical competence among the analysts performing the assays.
A critical assumption in qPCR methodologies where quantification of relative expression is a goal is that PCR efficiency be constant over time and among different samples. However, measured PCR efficiencies range from 1.8 to 2.0 [38], and that difference in efficiency can lead to a 4 × error in the relative gene expression calculation [37]. This error can clearly result in false-positive or false-negative results leading to erroneous conclusions. In this present example of relative estrogenicity and vtg expression in fish, researchers, resource managers, or regulatory officials could make economically significant decisions or recommendations based on bad data. Fortunately, this problem can be alleviated by applying our recommendations that are derived from the work by Ramakers et al. [37].
Although our methods were effective at identifying and minimizing interlaboratory variability, limits in statistical power are apparent given that only one laboratory (USEPA Region 5) detected vtg at 2 ng/L in batch 1. We expect that with a larger sample the other laboratories would have detected an increase in vtg at 2 ng/L. Likewise, in the mesocosm experiment, one laboratory did not detect a significant increase in vtg at 5 ng/L with a sample size of 5, yet prior work using the mesocosms and employing the same analyst found significantly increased vtg expression at 5 ng/L (3.2 ng/L measured) with n ¼ 7 replicates per treatment [4]. Although we are encouraged by the ability to reduce interlaboratory variability, studies designed without sufficient statistical power could still result in false-negative results. An excellent discussion of statistical power and experimental design within the context of vtg expression analysis is presented by Flick et al. [39] who state that, if possible, a power analysis should be conducted before samples are collected. Thus, as always, experimental design is of utmost importance and should be optimized prior to conducting interlaboratory investigations.
Of significant interest among ecotoxicologists is the identification of biomarkers of contaminant exposure that are: 1) specific, 2) reproducible within and among laboratories, 3) predictive of higher level effects, such as reproductive failure, and 4) sensitive to environmentally relevant concentrations. Few biomarkers meet those expectations, especially (3). However, vtg comes close to satisfying these criteria. It is relatively specific because it is only expressed following estrogen receptor activation. The reproducibility of vitellogenin within laboratories has been evaluated for both gene expression [19] and plasma protein concentrations [13] and is easily estimated by running technical replicates of the same pool of tissue in each assay. The resulting measures of variability among those replicates give inter- and intra-assay variation within the laboratory. In the present study, the inter- and intra-assay variation was minimal (<7% coefficient of variation), indicating a high level of precision and technical competence within each laboratory. Of increasing interest, however, is estimating and minimizing interlaboratory variability. The utility of a given biomarker such as vtg is clearly increased if multiple laboratories can produce similar results on the same samples. The present study provides an initial indication that interlaboratory variability can easily be minimized through the use of a few simple and freely available tools. Therefore, our results strongly support the use of vtg gene induction in fish as a reliable biomarker of estrogenic exposure.
As far as satisfying the third criteria of vtg being predictive of higher level effects, more work is needed. For example, Schwindt et al. [4] found elevated vtg at environmentally relevant EE2 concentrations in fish exposed as adults but no reproductive effects. Conversely, fish exposed early in life and then removed from the EE2 exposure showed reproductive effects but no vtg induction. In this case, one could conclude that vtg offers little predictive capacity for higher level effects. However, the work by Miller et al. [40] demonstrated that androgen-induced vtg suppression in females led to reduced egg production. The investigators then parameterized a population model with the experimental data, and reductions in vtg in females were observed. In this case, the vtg suppression was predictive of higher level effects on the population. Identification of biomarkers that can predict higher level effects is the prime motivation behind the concept of adverse outcome pathways [41]. This area of investigation aims to tie an initiating event, such a chemical binding to a receptor and inducing gene expression, to ecological changes that are meaningful to resource managers and risk assessors.
To our knowledge, this is the first study to evaluate and identify sources of interlaboratory variability in vtg gene expression. By using the same software and the same methods of relative expression calculation, investigators can minimize or even eliminate interlaboratory variability in vtg expression. These results indicate that vtg expression analysis can be reproducible within and among laboratories and suggest that large-scale monitoring programs may be possible using the methods described.
Supplementary Material
Acknowledgments
The authors thank A. Schwindt for his contributions to the present study. Research at the University of Massachusetts was supported by a HATCH award to K. Arcaro.
Disclaimer—Although the present study was subjected to review by the US Environmental Protection Agency and approved for publication, it may not necessarily reflect official Agency policy nor should mention of commercial products be considered endorsement. Any use of trade firm or product names is for descriptive purposes only and does not imply endorsement by the US Government. The present study was performed under the auspices of Colorado State University protocol numbers 10-1685A and 12-3501A.
Footnotes
Data availability—Nonsensitive datasets are made publicly accessible via the Environmental Dataset Gateway, at https://edg.epa.gov/metadata/catalog/main/home.page
References
- 1.Vajda AM, Barber LB, Gray JL, Lopez EM, Woodling JD, Norris DO. Reproductive disruption in fish downstream from an estrogenic wastewater effluent. Environ Sci Technol. 2008;42:3407–3414. doi: 10.1021/es0720661. [DOI] [PubMed] [Google Scholar]
- 2.Smith SR. Organic contaminants in sewage sludge (biosolids) and their significance for agricultural recycling. Philos Trans R Soc A Math Phys Eng Sci. 2009;367:4005–4041. doi: 10.1098/rsta.2009.0154. [DOI] [PubMed] [Google Scholar]
- 3.Yost EE, Meyer MT, Dietze JE, Williams CM, Worley-Davis L, Lee B, Kullman SW. Transport of steroid hormones, phytoestrogens, and estrogenic activity across a swine lagoon/sprayfield system. Environ Sci Technol. 2014;48:11600–11609. doi: 10.1021/es5025806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwindt AR, Winkelman DL, Keteles K, Murphy M, Vajda AM. An environmental oestrogen disrupts fish population dynamics through direct and transgenerational effects on survival and fecundity. J Appl Ecol. 2014;51:582–591. [Google Scholar]
- 5.Schwindt AR, Kent ML, Ackerman LK, Simonich SLM, Landers DH, Blett T, Schreck CB. Reproductive abnormalities in trout from western U.S. national parks. Trans Am Fish Soc. 2009;138:522–531. [Google Scholar]
- 6.Kidd KA, Blanchfield PJ, Mills KH, Palace VP, Evans RE, Lazorchak JM, Flick RW. Collapse of a fish population after exposure to a synthetic estrogen. Proc Natl Acad Sci U S A. 2007;104:8897–8901. doi: 10.1073/pnas.0609568104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwindt AR. Parental effects of endocrine disrupting compounds in aquatic wildlife: Is there evidence of transgenerational inheritance? Gen Comp Endocrinol. 2015;219:152–164. doi: 10.1016/j.ygcen.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Bhandari RK, vom Saal FS, Tillitt DE. Transgenerational effects from early developmental exposures to bisphenol A or 17α-ethinylestradiol in medaka, Oryzias latipes. Sci Rep. 2015;5:9303. doi: 10.1038/srep09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jobling S, Coey S, Whitmore JG, Kime DE, Van Look KJW, McAllister BG, Beresford N, Henshaw AC, Brighty G, Tyler CR, Sumpter JP. Wild intersex roach (Rutilus rutilus) have reduced fertility. Biol Reprod. 2002;67:515–524. doi: 10.1095/biolreprod67.2.515. [DOI] [PubMed] [Google Scholar]
- 10.Woodling JD, Lopez EM, Maldonado TA, Norris DO, Vajda AM. Intersex and other reproductive disruption of fish in wastewater effluent dominated Colorado streams. Comp Biochem Physiol C Toxicol Pharmacol. 2006;144:10–15. doi: 10.1016/j.cbpc.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 11.Kipfer S, Segner H, Wenger M, Wahli T, Bernet D. Long-term estrogen exposure of whitefish Coregonus lavaretus induces intersex but not Lake Thun-typical gonad malformations. Dis Aquat Organ. 2009;84:43–56. doi: 10.3354/dao02031. [DOI] [PubMed] [Google Scholar]
- 12.Bahamonde PA, Munkittrick KR, Martyniuk CJ. Intersex in teleost fish: Are we distinguishing endocrine disruption from natural phenomena? Gen Comp Endocrinol. 2013;192:25–35. doi: 10.1016/j.ygcen.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Schwindt AR, Feist GW, Schreck CB. Stress does not inhibit induced vitellogenesis in juvenile rainbow trout. Environ Biol Fishes. 2007;80:453–463. [Google Scholar]
- 14.Heppell S, Sullivan C. Gag (Mycteroperca microlepis) vitellogenin: Purification, characterization and use for enzyme-linked immunosorbent assay (ELISA) of female maturity in three species of grouper. Fish Physiol Biochem. 1999;20:361–374. [Google Scholar]
- 15.Tyler CR, van Aerle R, Hutchinson TH, Maddix S, Trip H. An in vivo testing system for endocrine disruptors in fish early life stages using induction of vitellogenin. Environ Toxicol Chem. 1999;18:337–347. [Google Scholar]
- 16.Shrader EA, Henry TR, Greeley MS, Bradley BP. Proteomics in zebrafish exposed to endocrine disrupting chemicals. Ecotoxicology. 2003;12:485–488. doi: 10.1023/b:ectx.0000003034.69538.eb. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi K, Tamotsu S, Yasuda K, Oishi T. Vitellogenin-immunohistochemistry in the liver and the testis of the medaka, Oryzias latipes, exposed to 17β-estradiol and p-nonylphenol. Zoolog Sci. 2005;22:453–461. doi: 10.2108/zsj.22.453. [DOI] [PubMed] [Google Scholar]
- 18.Arukwe A, Nilsen BM. Immunohistochemical analysis of the vitellogenin response in the liver of Atlantic salmon exposed to environmental oestrogens. Biomarkers. 1999;4:373–380. doi: 10.1080/135475099230750. [DOI] [PubMed] [Google Scholar]
- 19.Biales AD, Bencic DC, Flick RW, Lazorchak J, Lattier DL. Quantification and associated variability of induced vitellogenin gene transcripts in fathead minnow (Pimphales promelas) by quantitative real-time polymerase chain reaction assay. Environ Toxicol Chem. 2007;26:287–296. doi: 10.1897/06-213r.1. [DOI] [PubMed] [Google Scholar]
- 20.Biales AD, Bencic DC, Lazorchak JL, Lattier DL. A quantitative real-time polymerase chain reaction method for the analysis of vitellogenin transcripts in model and nonmodel fish species. Environ Toxicol Chem. 2007;26:2679–2686. doi: 10.1897/07-101.1. [DOI] [PubMed] [Google Scholar]
- 21.Miracle A, Ankley G, Lattier D. Expression of two vitellogenin genes (vg1 and vg3) in fathead minnow (Pimephales promelas) liver in response to exposure to steroidal estrogens and androgens. Ecotoxicol Environ Saf. 2006;63:337–342. doi: 10.1016/j.ecoenv.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Filby AL, Thorpe KL, Maack G, Tyler CR. Gene expression profiles revealing the mechanisms of anti-androgen- and estrogen-induced feminization in fish. Aquat Toxicol. 2007;81:219–231. doi: 10.1016/j.aquatox.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Ekman D, Teng Q, Villeneuve D, Kahl M, Jensen K, Durhan E, Ankley G, Collette T. Profiling lipid metabolites yields unique information on sex- and time-dependent responses of fathead minnows (Pimephales promelas) exposed to 17α-ethynylestradiol. Metabolomics. 2009;5:22–32. [Google Scholar]
- 24.Ekman DR, Teng Q, Villeneuve DL, Kahl MD, Jensen KM, Durhan EJ, Ankley GT, Collette TW. Investigating compensation and recovery of fathead minnow (Pimephales promelas) exposed to 17alpha-ethynylestradiol with metabolite profiling. Environ Sci Technol. 2008;42:4188–4194. doi: 10.1021/es8000618. [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl MW, Georgieva TM, Georgiev IP, Ontsouka E, Hageleit M, Blum JW. Real-time RT-PCR quantification of insulin-like growth factor (IGF)-1, IGF-1 receptor, IGF-2, IGF-2 receptor, insulin receptor, growth hormone receptor, IGF-binding proteins 1, 2 and 3 in the bovine species. Domest Anim Endocrinol. 2002;22:91–102. doi: 10.1016/s0739-7240(01)00128-x. [DOI] [PubMed] [Google Scholar]
- 26.Arukwe A, Goksøyr A. Eggshell and egg yolk proteins in fish: Hepatic proteins for the next generation: Oogenetic, population, and evolutionary implications of endocrine disruption. Comp Hepatol. 2003;2:4. doi: 10.1186/1476-5926-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kime DE, Nash JP, Scott AP. Vitellogenesis as a biomarker of reproductive disruption by xenobiotics. Aquaculture. 1999;177:345–352. [Google Scholar]
- 28.Damond F, Benard A, Ruelle J, Alabi A, Kupfer B, Gomes P, Rodes B, Albert J, Böni J, Garson J, Ferns B, Matheron S, Chene G, Brun-Vezinet F. Quality control assessment of Human Immunodeficiency Virus type 2 (HIV-2) viral load quantification assays: Results from an international collaboration on HIV-2 infection in 2006. J Clin Microbiol. 2008;46:2088–2091. doi: 10.1128/JCM.00126-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsden SC, Daly S, Geilenkeuser W-J, Duncan G, Hermitte F, Marubini E, Neumaier M, Orlando C, Palicka V, Paradiso A, Pazzagli M, Pizzamiglio S, Verderio P. EQUAL-quant: An international external quality assessment scheme for real- time PCR. Clin Chem. 2006;52:1584–1591. doi: 10.1373/clinchem.2005.066019. [DOI] [PubMed] [Google Scholar]
- 30.Ebentier DL, Hanley KT, Cao Y, Badgley BD, Boehm AB, Ervin JS, Goodwin KD, Gourmelon M, Griffith JF, Holden PA, Kelty CA, Lozach S, McGee C, Peed LA, Raith M, Ryu H, Sadowsky MJ, Scott EA, Domingo JS, Schriewer A, Sinigalliano CD, Shanks OC, Van De Werfhorst LC, Wang D, Wuertz S, Jay JA. Evaluation of the repeatability and reproducibility of a suite of qPCR-based microbial source tracking methods. Water Res. 2013;47:6839–6848. doi: 10.1016/j.watres.2013.01.060. [DOI] [PubMed] [Google Scholar]
- 31.Shanks OC, Sivaganesan M, Peed L, Kelty CA, Blackwood AD, Greene MR, Noble RT, Bushon RN, Stelzer EA, Kinzelman J, Anan’Eva T, Sinigalliano C, Wanless D, Griffith J, Cao Y, Weisberg S, Harwood VJ, Staley C, Oshima KH, Varma M, Haugland RA. Interlaboratory comparison of real-time PCR protocols for quantification of general fecal indicator bacteria. Environ Sci Technol. 2012;46:945–953. doi: 10.1021/es2031455. [DOI] [PubMed] [Google Scholar]
- 32.Hultman MT, Rundberget JT, Tollefsen KE. Evaluation of the sensitivity, responsiveness and reproducibility of primary rainbow trout hepatocyte vitellogenin expression as a screening assay for estrogen mimics. Aquat Toxicol. 2015;159:233–244. doi: 10.1016/j.aquatox.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Chapman PM. Whole effluent toxicity testing—Usefulness, level of protection, and risk assessment. Environ Toxicol Chem. 2000;19:3–13. [Google Scholar]
- 34.Reddy TV, Flick R, Lazorchak JM, Smith ME, Wiechman B, Lattier DL. Experimental paradigm for in-lab proxy aquatic studies under conditions of static, non flow-through chemical exposures. Environ Toxicol Chem. 2015;34:2796–2802. doi: 10.1002/etc.3121. [DOI] [PubMed] [Google Scholar]
- 35.Schwindt AR. PhD thesis. Colorado State University; Fort Collins, CO, USA: 2013. The population ecology of fathead minnows (Pimephales promelas) in estrogen contaminated environments. [Google Scholar]
- 36.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 37.Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 38.Kainz P. The PCR plateau phase—Towards an understanding of its limitations. Biochim Biophys Acta Gene Struct Expr. 2000;1494:23–27. doi: 10.1016/s0167-4781(00)00200-1. [DOI] [PubMed] [Google Scholar]
- 39.Flick RW, Bencic DC, See MJ, Biales AD. Sensitivity of the vitellogenin assay to diagnose exposure of fathead minnows to 17α-ethynylestradiol. Aquat Toxicol. 2014;152:353–360. doi: 10.1016/j.aquatox.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 40.Miller DH, Jensen KM, Villeneuve DL, Kahl MD, Makynen EA, Durhan EJ, Ankley GT. Linkage of biochemical responses to population-level effects: A case study with vitellogenin in the fathead minnow (Pimephales promelas) Environ Toxicol Chem. 2007;26:521–527. doi: 10.1897/06-318r.1. [DOI] [PubMed] [Google Scholar]
- 41.Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrrano JA, Tietge JE, Villeneuve DL. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem. 2010;29:730–741. doi: 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


