This study demonstrated N-myristoylation of 72 proteins and identified hundreds of myristoylated proteins in different membrane networks using a targeted MS approach.
Abstract
N-terminal myristoylation, a major eukaryotic protein lipid modification, is difficult to detect in vivo and challenging to predict in silico. We developed a proteomics strategy involving subfractionation of cellular membranes, combined with separation of hydrophobic peptides by mass spectrometry-coupled liquid chromatography to identify the Arabidopsis thaliana myristoylated proteome. This approach identified a starting pool of 8837 proteins in all analyzed cellular fractions, comprising 32% of the Arabidopsis proteome. Of these, 906 proteins contain an N-terminal Gly at position 2, a prerequisite for myristoylation, and 214 belong to the predicted myristoylome (comprising 51% of the predicted myristoylome of 421 proteins). We further show direct evidence of myristoylation in 72 proteins; 18 of these myristoylated proteins were not previously predicted. We found one myristoylation site downstream of a predicted initiation codon, indicating that posttranslational myristoylation occurs in plants. Over half of the identified proteins could be quantified and assigned to a subcellular compartment. Hierarchical clustering of protein accumulation combined with myristoylation and S-acylation data revealed that N-terminal double acylation influences redirection to the plasma membrane. In a few cases, MYR function extended beyond simple membrane association. This study identified hundreds of N-acylated proteins for which lipid modifications could control protein localization and expand protein function.
INTRODUCTION
N-terminal myristoylation (MYR) is a major eukaryotic protein lipid modification that usually occurs cotranslationally, when the nascent chain emerges from the ribosomal tunnel (Wilcox et al., 1987; Glover et al., 1997; Giglione et al., 2015). The myristoyl moiety is added to the glycine in position 2, after removal of the N-terminal methionine. MYR mainly functions to target proteins to membranes, where myristoylated (MYRed) proteins interact with partners to trigger signal transduction, although the lipid moiety can also have other functions (Turnbull and Hemsley, 2017). MYR is necessary but insufficient for stable protein membrane anchoring, and stronger membrane associations are induced by a proximal second reversible signal (Peitzsch and McLaughlin, 1993; Silvius and l’Heureux, 1994; Bhatnagar and Gordon, 1997) often provided by attachment of another lipid, a palmitate, a polybasic motif region (PBR), or a protein-protein interaction domain (Running, 2014; Turnbull and Hemsley, 2017). However, only MYR is needed to localize proteins to the endomembrane compartment, while an S-acylation (palmitoylation [PAL]) site, if embedded close to a MYR site, contributes to plasma membrane (PM) relocalization (Traverso et al., 2013b). MYR is considered an irreversible modification, but MYRed proteins may undergo dynamic relocalization in response to the second signal (Lanyon-Hogg et al., 2017).
MYR is catalyzed by N-myristoyltransferase (NMT), an enzyme family that adds a C:14:0 lipid to the N termini of a subset of proteins displaying an N-terminal glycine (Gly) unmasked after the action of methionine aminopeptidase (Breiman et al., 2016). There are two NMTs in higher eukaryotes (NMT1 and NMT2) including plants. NMT1 is the most abundant and active catalyst (Giglione et al., 2015), including in Arabidopsis thaliana, in which NMT1 deletion is embryo-lethal and NMT2 deletion produces only a weak phenotype (Pierre et al., 2007). Moreover, Arabidopsis NMT1 regulates the functional and morphological integrity of plant endomembranes (Renna et al., 2013).
MYR of the N-terminal Gly of a protein target only occurs when the context of the residues following the first Gly (Gly2) is favorable (Traverso et al., 2013a). Although superficially a simple constraint, characterization of the MYRed proteome remains complicated because (1) MYR is a difficult modification to predict using available bioinformatics tools, which currently lack sufficient accuracy, sensitivity, and specificity due to the vast prediction space and very limited number of available experimentally validated sequences (Martinez et al., 2008; Traverso et al., 2013a); and (2) commonly used MYR prediction algorithms such as NMT Predictor (Maurer-Stroh et al., 2002a, 2002b) and Myristoylator (Bologna et al., 2004) were primarily developed using data from fungi and animal NMTs, which do not always translate well to predicting plant MYR (Lu and Hrabak, 2013). Nevertheless, score and pattern analysis based on in vitro data of several dozen of peptides assayed with Arabidopsis NMT1 predicted that ∼2% of the entire plant proteome undergoes this MYR (Boisson et al., 2003; Martinez et al., 2008; Traverso et al., 2013a, 2013b). The Arabidopsis genome contains 2587 predicted open reading frames (MGxx set) that fulfilled the first prerequisite for MYR, namely, the presence of an N-terminal Gly residue at position 2 (Gly2). Additional criteria, including the presence of a positive charge on residue 7, narrow the predicted myristoylome to 421 proteins (http://www.i2bc.paris-saclay.fr/maturation/Myristoylome.html) (Boisson et al., 2003; Boisson and Meinnel, 2003; Martinez et al., 2008).
The number of definitively characterized MYRed proteins in plants remains limited, with only 43 identified to date in Arabidopsis. Most of the data that are available originate from in vitro enzymatic tests and/or in vitro transcription-translation experiments performed with a radiolabeled myristate precursor, with direct in vivo demonstration of MYR in only a handful of cases by means of heterologous, transient overexpression of tagged proteins (de Vries et al., 2006; Li et al., 2010; Witte et al., 2010). However, MYRed peptides were not directly detected in these studies, with direct MYRed peptide detection limited to in vitro transcription and translation in wheat germ extracts of AtPOL and AtArf1A1C proteins (Gagne and Clark, 2010; Yamauchi et al., 2010). MYR is very difficult to detect and quantify experimentally in vivo because (1) MYRed N-terminal peptides are hydrophobic and irreversible, and (2) MYRed proteins localize to various cell membranes (Hannoush, 2015). Although radiolabeling techniques allow MYR validation in vivo, they preclude proteome-wide identification of native MYRed proteins. Fatty acid analogs featuring reactive tags and click chemistry have recently been applied to identify dozens of new MYRed proteins in several metazoa (Hannoush, 2015). Unfortunately, this method has not yet been successfully deployed in plant extracts, having been applied only to a single reporter protein overexpressed in Arabidopsis protoplasts and transgenic plants (Boyle et al., 2016). As a result, no proteome-wide in vivo plant myristoylome has been yet reported. The favored approach to detect MYR in plants relies on site-directed mutagenesis of the putative myristoylable N-terminal Gly2 to Ala, which prevents MYR. Differential localization of the two protein constructs is then used as indirect evidence of MYR (Turnbull and Hemsley, 2017). There has yet to be comprehensive, high-throughput identification of the subcellular MYRed proteome (Chen et al., 2004).
Here, we implemented a targeted proteomics approach in various Arabidopsis membrane subproteomes from cultures grown under dark and light conditions to characterize the subcellular localization and MYR status of proteins in vivo. Extensive subfractionation of cell membrane systems vastly increased the dynamic resolution of mass spectrometry (MS). Moreover, biochemical subfractionation was combined with high-resolution separation of hydrophobic peptides by MS-coupled liquid chromatography (LC), with analysis complemented with a newly developed MS data-dependent acquisition method that allowed us to specifically identify MYRed peptides. We compare our empirically derived MS results to predictions based on the genome sequence (e.g., ∼27,500 proteins in the Arabidopsis proteome [most recent estimates from TAIR10 and ARAPORT11], 2587 proteins in the MGxx set, and the subset of 421 in predicted myristoylome). The quantitative MS analysis detected ∼32% of the Arabidopsis proteome, including 51% of the predicted myristoylome, mainly in PM and detergent-resistant membrane (DRM) fractions. The accumulation of identified myristoylome (set of MS-detected proteins with MYR predicted or detected directly) in the PM and DRM fractions did not change significantly under light and dark conditions. We provide direct evidence of MYR in vivo for 72 nonredundant proteins. Of note, 18 of these did not belong to the predicted myristoylome, and in one case an internal MYRed peptide was identified, strongly suggesting that posttranslational MYR occurs in plants.
RESULTS
Arabidopsis Membrane Proteome Subfractionation with Protein Identification and Quantification Using a Dedicated MS-Based Pipeline
To search for MYRed proteins, we focused on subcellular fractions such as the PM and DRM, since they are predicted to be enriched for MYRed proteins (Morel et al., 2006) (see workflow in Supplemental Figure 1). Liquid cultures Arabidopsis cells were grown in dark or light conditions, and microsomal fractions (µZ) were prepared by cell disruption and differential centrifugation for subsequent use for PM, DRM, and Golgi vesicle preparation as previously described (Graham, 2001; Jurgens, 2004; Marmagne et al., 2006, 2007; Morel et al., 2006). To obtain an endomembrane-enriched fraction, the dextran (Dex) phase from the PEG/Dex partition was analyzed (Supplemental Figure 1). Total protein (Tot), Dex, µZ, PM, Golgi vesicle, and DRM fractions under different growth conditions (e.g., TotL, total fraction grown in light) were subjected to quantitative MS (Supplemental Figure 2). After removing ambiguous gene models and assembling ambiguously identified protein families, the entire set of detected proteins consisted of 8837 proteins, providing substantial coverage of the Arabidopsis proteome (32%) and featuring enrichment of membrane-associated proteins.
Specific antibodies against sucrose synthase, RbcL, and ATP synthase (H+-ATPase) were used as markers of the cytosol, plastid, and PM, respectively, and were dissimilarly distributed in the different fractions (Supplemental Figure 3A). We also assessed the accumulation of 20 proteins previously identified in PM and DRM proteomics data sets or annotated based on individual characterization (curated protein locations obtained from The Plant Proteome Database [PPDB; http://ppdb.tc.cornell.edu] and the literature) by label-free quantification, which showed the appropriate predicted distributions (Supplemental Figure 3B). For example, soluble chloroplast markers such as RbcL were identified in the Tot fraction, whereas mitochondrial ATPase subunits and voltage-dependent anion channels were found mainly in the µZD and Dex fractions. As expected, PM and DRM markers were strongly enriched in PM and DRM fractions but less in µZD fractions. Furthermore, since label-free quantification of individual proteins may be biased by peptide ionization and/or abundance related to the complexity of the analyzed fraction, we retrieved the curated localization data of the 2526 proteins from the PPDB and plotted the protein mass of each of the main subcellular fractions against PPDB subcellular protein localization (Supplemental Figure 3C). Approximately eighty percent of proteins mass annotated as PM and DRM were identified in PM and DRM fractions, with ∼10% also retrieved in µZ fractions. Overall, these observations fully agreed with the enrichment observed for individual PM and DRM markers (Supplemental Figure 3B).
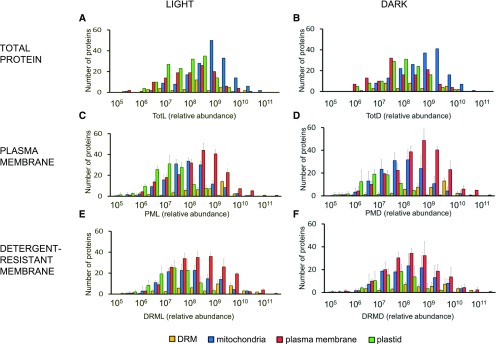
Finally, to determine protein enrichment in different cellular compartments in terms of numbers of identified proteins, the frequency distributions of the relative protein abundance of 5376 identified proteins were analyzed in PM, DRM, and Tot fractions (Figure 1). Protein category abundance within the analyzed samples spanned five orders of magnitude (106–1011). In TotL and TotD (total fraction grown in dark; Figures 1A and 1B), PM- and DRM-annotated proteins peaked at 5 × 108 and 4 × 108, respectively, whereas in PML and PMD (Figures 1C and 1D), PM-annotated proteins spanned the highest abundance categories at 5 × 109, showing approximately one to two orders of magnitude enrichment compared with other cellular fractions. Within the DRML and DRMD fractions (Figures 1E and 1F), DRM-predicted proteins peaked within 8 × 109 to 1 × 1010 frequency bins. The above workflow substantially enriches each subcellular compartment and provides the necessary gain in dynamic resolution for further analysis.
Figure 1.
Protein Category Abundance in Different Subcellular Proteomes.
Frequency distribution of relative protein abundance in Tot ([A] and [B]), PM ([C] and [D]), and DRM ([E] and [F]) purified from light ([A], [C], and [E]) or dark ([B], [D], and [F]) grown Arabidopsis cultures. Relative protein abundance was calculated based on the Σ(Narea), with the total protein population spread across six orders of abundance. Displayed protein populations are based on curated localizations retrieved from previously published PM and DRM proteomes (Marmagne et al., 2007; Kierszniowska et al., 2009; Minami et al., 2009) and extracted from the PPDB. Mitochondria (blue), plastid (green), PM (red), and DRM (yellow). Errors bars indicate sd between 3 biological replicates.
Protein Functional Distribution in the Different Fractions
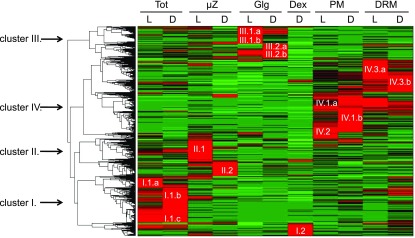
To obtain an integrated view of protein distribution in the various fractions, we performed unsupervised hierarchical clustering to group proteins sharing similar expression profiles using quantitative relative protein abundance (Σ(Narea)) per fraction data. A total of 5376 proteins produced four major clusters (I–IV; Figure 2), corresponding mainly to proteins with strong enrichment in Tot (cluster I, 1328 proteins), µZ (cluster II, 1031 proteins), Golgi (cluster III, 883 proteins), and PM/DRM (cluster IV, 2134 proteins). PM and DRM clusters represented a particular case, as a substantial fraction of clustered proteins were common to both fractions. There was a correlation between cluster functional specialization and subcellular localization, highlighting functional specialization for the PM/DRM fractions. In the PM, the majority of proteins were involved in vesicular and PM transport (38%), signaling (8%), and cell wall biosynthesis and degradation (6%). DRM fractions were enriched for cell organization (46%), modification and homeostasis (21%), and signaling (4%) proteins. Cell wall-related functions and thylakoid contaminants were depleted in the DRM from both light- and dark-grown cells. These data show that the obtained fractions are fully representative of the expected cell compartment.
Figure 2.
Cluster Analysis of Cellular Fractions Reveals Four Distinct Clusters.
For hierarchical clustering, quantitative information based on Σ(Narea) per fraction was used. To ensure meaningful clustering, proteins with a Σ(Narea) <108 in all analyzed fractions were not included in the clustering analysis as they could constitute a source of noise due to the weakly associated quantitative data. Four main protein abundance clusters were identified corresponding to the purified subcellular fractions: Tot (cluster I), µZ (cluster II), Golgi vesicle (Glg; cluster III), PM (clusters IV.1 and IV.2), and DRM (clusters IV.3). In all fractions, each corresponding cluster was subdivided to take in account proteins that showed preferential accumulation in light- or dark-grown cells: TotL (I.1.a, I.1.c), TotD (I.1.b, I.1.c), µzL (II.1), µzD (II.2), GlgL (III.1, III.2.b), GlgD (III.2), Dex (I.2), PML (IV.1.a, IV.2), PMD (IV.1.b), DRML (IV.3.a), and DRMD (IV.3.b). Red indicates increased protein accumulation.
Targeted MS to Identify in Vivo Myristoylated Proteins in Arabidopsis
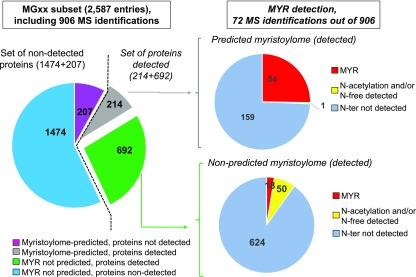
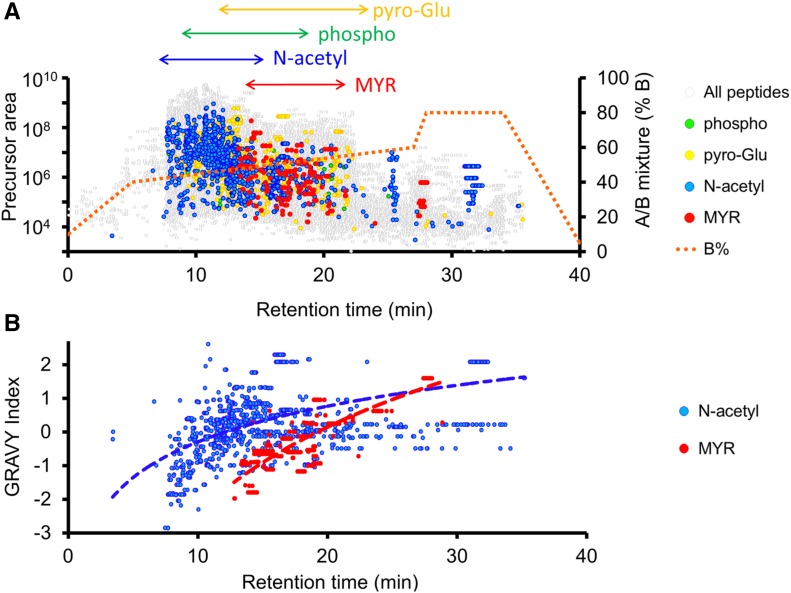
In our study, 906 of the 8837 starting pool of MS-detected proteins contain the MGxx motif (35% of the 2587 in the entire MGxx set), and 214 of these belonged to the predicted myristoylome (Figure 3). The increased dynamic resolution of MS analysis afforded by sample fractionation revealed an important part of the predicted myristoylome (51%) and allowed us to provide direct in vivo proof of the presence of a myristoyl moiety associated with Gly2. To access this rare MYRed peptidome, we developed a targeted MS-based approach (Figure 4). To maximize the chances of detecting highly hydrophobic peptides, the nano-LC peptide separation gradient was elongated (Figure 4A), shifting the retention time of the MYRed peptides toward higher mobile phase concentrations compared with nonmodified peptides or phosphorylated, acetylated, and pyro-Glu peptides (Figure 4A). This shift in MYRed peptides toward less “crowded” regions of the elution profile significantly increased the dynamic resolution of the analysis. The MYRed or non-MYRed peptide elution profiles were closely correlated with their calculated grand average of hydropathy index (GRAVY; Figure 4B), suggesting that the shifted elution of some MYRed peptides toward the end of the gradient could be due to their high hydrophobicity.
Figure 3.
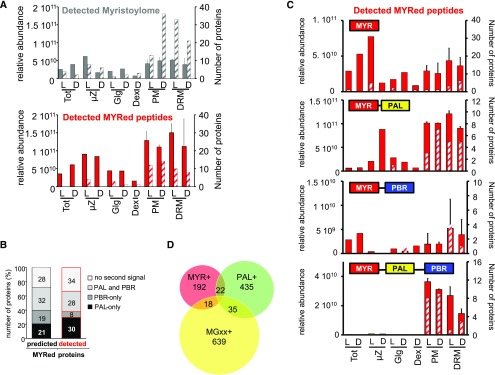
Proteomic Analysis of the MS-Identified Arabidopsis Myristoylome.
The Arabidopsis genome contains 2587 predicted open reading frames with a Gly at position 2 (MGxx set). Within those, 906 proteins were detected by MS (10% of the starting candidate pool of 8837 MS-detected proteins); 214 of these were part of the predicted myristoylome and 692 were not. Of the 214 in the predicted myristoylome, the myristoyl moiety was detected for 54 via targeted MS, 159 lacked identification of the N-terminal peptide, and only one had an N-acetylated or free N terminus. The 207 remaining predicted MYRed proteins were not detected in the MS-identified set. Within the set of 692 MS-identified proteins not predicted to be MYRed (but carrying a Gly2, i.e., in the MGxx set), 18 had a MYRed peptide detected via targeted MS and 50 had an acetyl or free N terminus.
Figure 4.
Identified Protein Modifications and Their Distribution on Nano-LC Elution Gradients.
LC elution profiles used in LTQ-Orbitrap (Velos) for the identification of MYRed peptides were “stretched” within 40 to 60% buffer B (0.1 FA and 5% ACN) to maximize high-hydrophobicity peptide resolution (red dotted line). The maximum MYRed peptides (red dots) were observed within the 12 to 22 min retention time subsequent to the main elution bulk of non-modified peptides (gray). Other frequently observed posttranslational modifications such as phosphorylation (green), pyroglutamylation (yellow), and N-terminal acetylation (blue) are indicated (A). The correlation between GRAVY hydrophobicity index and retention time for MYRed (red) or acetylated (blue) peptides is presented in (B).
Despite this technical optimization, the high complexity of the peptide mixtures precluded identification of low-abundance MYRed peptides. To overcome this limitation, we implemented data-dependent acquisition, which relies on mass inclusion lists generated from the predicted masses of MYRed peptides extracted from the MGxx subset (see Methods). To obtain a complementary identification of MYRed peptides, PM and DRM samples were analyzed in parallel on a time-of-flight (TOF) instrument. As expected, MS/MS spectra of N-MYRed peptides obtained by T-TOF showed better coverage of the b1-MYRed-Gly diagnostic ion (92%) than those observed by LTQ-Orbitrap (Velos) (26%), the latter identifying MYRed peptides based on the presence of b2 and subsequent b-ions. Collision-induced dissociation, which triggers fragmentation on both instruments, led in very rare cases to the fragmentation of the amide bond between the myristate and the N-terminal α-amine group, yielding a 210 D loss. The 268 D MYRed-Gly b1-ions or subsequent MYRed-b(n)-ions were observed in the majority of peptides. This was in contrast to the myristoyl-neutral loss observed on synthetic MYRed peptides by MALDI-TOF, where neutral loss of 210 D was observed within 30 to 40% collision energy (Chen et al., 2004).
Overall, direct site identification revealed 72 MYRed proteins, the most comprehensive data set on the in vivo myristoylome reported to date. The use of LTQ-Orbitrap and T-TOF instruments together yielded complementary coverage of the in vivo N-terminal myristoylome with an overlap of 25 (33%) MYRed peptides identified by both instruments and 23 and 24 (30% and 35%) peptides identified by LTQ-Orbitrap or T-TOF, respectively. Of the 72 experimentally detected MYRed peptides, 54 were part of the predicted Arabidopsis myristoylome, representing 13% of the predicted myristoylome (Martinez et al., 2008; Traverso et al., 2013a). Surprisingly, within the 692 MG-containing proteins (of 906 total identified) that were not part of the predicted Arabidopsis myristoylome (Figure 3), 18 were found to contain a MYRed peptide (Table 1, Figure 3; Supplemental Data Set 1.1), suggesting that a number of proteins still escape MYR prediction.
Table 1. Overview of Identified MYRed Proteins Recovered with the N-Terminal MYRed Peptide.
| Accessiona | Protein Nameb | Clusterc | Pred. Myristoylomed | Observed MYRed Peptidee |
N-Terminal Protein Sequencef | Myristoyl Moiety Identified on Peptideg | MYR-Gly Positionh | |
|---|---|---|---|---|---|---|---|---|
| LTQ-Orbitrap | Triple-Quad | |||||||
| AT3G09540.1 | Pectin lyase-like | IV.3.a | X | X | X | MGNLHGIHRS | C14-gNLHGIHR | 2 |
| AT4G38690.1 | 1-Phosphatidylinositol phosphodiesterase-like | IV.1.b | X | X | MGSHLSKQLE | C14-gSHLSK | 2 | |
| AT4G25970.1 | Phosphatidylserine decarboxylase (PSD3) | IV.1.b | X | X | MGNGNSTETK | C14-gNGNSTETK | 2 | |
| AT5G51570.1 | Hypersensitive induced reaction (HIR3) | III.1.a | X | X | MGNTYCILGG | C14-gNTYcILGGcIEQASVGVVER | 2 | |
| AT5G62740.1 | Hypersensitive induced reaction (HIR4) | IV.1.a | X | X | MGNLFCCVQV | C14-gNLFccVQVDQSTVAIK | 2 | |
| AT1G69840.1 | Hypersensitive induced reaction (HIR1) | IV.1.b | X | MGQALGCIQV | C14-gQALGcIQVDQSNVAIK | 2 | ||
| AT3G01290.1 | Hypersensitive induced reaction (HIR2) | IV.3.a | X | X | MGNLFCCVLV | C14-gNLFccVLVK | 2 | |
| AT5G63030.1 | Glutaredoxin-C1 | III.1.a | X | X | X | MGSMFSGNRM | C14-gSmFSGNR | 2 |
| AT2G48150.1 | Glutathione peroxidase 4 (GPX4) | IV.2 | X | X | X | MGASASVPER | C14-gASASVPER | 2 |
| AT3G63080.1 | Glutathione peroxidase 5 (GPX5) | IV.1.a | X | X | X | MGASSSSSVS | C14-gASSSSSVSEK | 2 |
| AT1G69880.1 | Thioredoxin H-type 8 (TrxH8) | X | X | MGANVSTPDQ | C14-gANVSTPDQR | 2 | ||
| AT3G08710.1 | Thioredoxin H-type 9 (TrxH9) | IV.1.b | X | X | MGSCVSKGKG | C14-gSCVSK | 2 | |
| AT5G06260.1 | TLD-domain containing nucleolar protein | X | X | MGNSNSSSVD | C14-gNSNSSSVDHR | 2 | ||
| AT2G45380.1 | TLD-domain unknown function | X | K.GNSNSSSVDHR.F | C14-gNSNSSSVDHR | 98 | |||
| AT1G70490.1, AT3G62290.1, AT1G10630.1 | ADP-ribosylation factor (ARFA1D); (ARFA1E); (ARFA1F) | II.1 | X | X | MGLSFAKLFS | C14-gLSFAKLFSRLFAK | 2 | |
| X | X | MGLSFGKLFS | C14-gLSFGKLFSKLFAK | 2 | ||||
| X | X | MGLSFAKLFS | C14-gLSFAK | 2 | ||||
| AT4G35230.1 | IV.1.b | X | X | MGCCQSLFSG | C14-gccQSLFSGDNPLGK | 2 | ||
| AT5G46570.1 | BR-signaling kinase 2 (BSK2) | IV.1.b | X | X | X | MGCLHSKTAN | C14-gcLHSK | 2 |
| AT4G00710.1 | BR-signaling kinase 3 (BSK3) | IV.1.b | X | X | X | MGGQCSSLSC | C14-gGQcSSLSccR | 2 |
| AT5G41260.1 | BR-signaling kinase 8 (BSK8) | IV.1.b | X | X | MGCEVSKLSA | C14-gCEVSK (possible ambiguity) | 2 | |
| AT1G63500.1 | BR-signaling kinase 7 (BSK7) | IV.1.b | X | X | MGCEVSKLCA | 2 | ||
| AT3G06270.1 | Protein phosphatase 2C family protein (PP2C -like) | IV.1.b | X | X | MGCVQCKCCS | C14-gcVQcK | 2 | |
| AT1G03590.1 | Protein phosphatase 2C family protein (PP2C6) | IV.1.b | X | MGGCISKTSW | C14-gGCISK | 2 | ||
| AT1G16220.1 | Protein phosphatase 2C family protein (PP2C -like) | X | X | MGLCHSKIDK | C14-gLCHSK | 2 | ||
| AT4G03415.1 | Protein phosphatase PP2C52 | IV.1.b | X | X | MGGCVSTSSK | C14-gGcVSTSSK | 2 | |
| AT1G14370.1 | Protein kinase (KIN1) | IV.1.b | X | MGNCLDSSAK | C14-gNcLDSSAK | 2 | ||
| AT2G02800.1 | Protein kinase (KIN2) | IV.3.b | X | MGNCLDSSAK | C14-gNCLDSSAK | 2 | ||
| AT5G11410.1 | Protein kinase superfamily | X | MGNCLKPLKE | C14-gNcLKPLK | 2 | |||
| AT2G20140.1, AT4G29040.1 | 26S protease regulatory complex subunit (RPT2b, RPT2a) | IV.3.b | X | X | X | MGQGPSGGLN | C14-gQGPSGGLNR | 2 |
| X | X | MGQGPSGGLN | C14-gQGPSGGLNR | 2 | ||||
| AT3G09770.1 | Ubiquitin E3 ligase (LOG2) | X | X | MGNISSSGGE | C14-gNISSSGGEGR | 2 | ||
| AT4G17615.1 | Calcineurin B-like protein 1 (CBL1) | IV.1.b | X | X | MGCFHSKAAK | C14-gCFHSK | 2 | |
| AT5G47100.1 | Calcineurin B-like protein 9 (CBL9) | IV.1.b | X | X | X | MGCFHSTAAR | C14-gcFHSTAAR | 2 |
| AT5G04870.1 | Calcium-dependent protein kinase 1 (CPK1) | IV.1.b | X | X | X | MGNTCVGPSR | C14-gNTcVGPSR | 2 |
| AT4G35310.1 | Calcium-dependent protein kinase 5 (CPK5) | IV.1.b | X | X | MGNSCRGSFK | C14-gNSCR | 2 | |
| AT2G17290.1 | Calcium-dependent protein kinase 6 (CPK6) | IV.1.b | X | X | MGNSCRGSFK | C14-gNSCR | 2 | |
| AT5G12480.1 | Calcium-dependent protein kinase 7 (CPK7) | IV.1.b | X | X | X | MGNCCGNPSS | C14-gNccGNPSSATNQSK | 2 |
| AT5G19450.1 | Calcium-dependent protein kinase 8 (CPK8) | IV.1.b | X | X | X | MGNCCASPGS | C14-gNccASPGSETGSK | 2 |
| AT3G20410.1 | Calcium-dependent protein kinase 9 (CPK9) | IV.1.b | X | X | X | MGNCFAKNHG | C14-gNcFAK | 2 |
| AT1G18890.1 | Calcium-dependent protein kinase 10 (CPK10) | IV.1.b | X | X | X | MGNCNACVRP | C14-gNcNAcVRPDSK | 2 |
| AT3G51850.1 | Calcium-dependent protein kinase 13 (CPK13) | IV.1.b | X | X | MGNCCRSPAA | C14-gNCCR | 2 | |
| AT4G21940.1 | Calcium-dependent protein kinase 15 (CPK15) | IV.1.b | X | X | MGCFSSKHRN | C14-gCFSSK | 2 | |
| AT4G04720.1 | Calcium-dependent protein kinase 21 (CPK21) | IV.1.b | X | X | MGCFSSKHRK | C14-gCFSSK | 2 | |
| AT1G74740.1 | Calcium-dependent protein kinase 30 (CPK30) | IV.1.b | X | X | X | MGNCIACVKF | C14-gNcIAcVK | 2 |
| AT3G57530.1 | Calcium-dependent protein kinase 32 (CPK32) | IV.1.b | X | X | X | MGNCCGTAGS | C14-gNccGTAGSLAQNDNKPK | 2 |
| AT1G50700.1 | Calcium-dependent protein kinase 33 (CPK33) | IV.1.b | X | X | MGNCLAKKYG | C14-gNCLAK | 2 | |
| AT1G64850.1 | Calcium-binding EF hand family protein (Ca EF) | II.2 | X | X | MGQVFNKLRG | C14-gQVFNK | 2 | |
| AT1G49580.1 | CDPK-related kinase 8 (CRK8) | IV.1.b | X | X | MGGCTSKPST | C14-gGCTSK | 2 | |
| AT3G50530.1 | CDPK-related kinase 5 (CRK5) | IV.1.b | X | X | X | MGLCTSKPNS | C14-gLcTSKPNSSNSDQTPAR | 2 |
| AT5G24430.1 | CDPK-related kinase 4 (CRK4) | IV.3.a | X | X | MGHCYSRNIS | C14-gHCYSR | 2 | |
| AT2G46700.1 | CDPK-related kinase 3 (CRK3) | IV.1.b | X | X | MGQCYGKVNQ | C14-gQCYGK | 2 | |
| AT4G39320.1 | Unknown function | IV.1.b | X | X | MGNCALKPKV | C14-gNcAIKPK | 2 | |
| AT3G23280.1 | RING E3 Ligase (XBAT35) | X | X | MGQQQSKGEL | C14-gQQQSK | 2 | ||
| AT1G14060.1 | Unknown function | II.1 | X | X | MGITSSTDPK | C14-gITSSTDPK | 2 | |
| AT5G07910.1 | Leucine-rich repeat family protein (LRR-like) | IV.1.b | X | X | X | MGCCASNTAG | C14-gccASNTAGGSK | 2 |
| AT3G27930.1 | Unknown function | I.2 | X | X | X | MGNALVKKEP | C14-gNAIVK | 2 |
| AT5G66290.1 | Unknown function | III.1.a | X | X | MGASDSTLLG | C14-gASDSTLLGSQENR | 2 | |
| AT5G39590.1 | Unknown function | III.1.b | X | X | X | MGASSSTDDK | C14-gASSSTDDKESSEK | 2 |
| AT1G73390.1 | Unknown function | IV.1.b | X | X | MGCFASRPND | C14-gcFASRPNDTGGNR | 2 | |
| AT1G52320.2 | Unknown function | IV.1.b | X | X | MGCAQSKIEN | C14-gCAQSK | 2 | |
| AT1G32190.1 | α/β-Hydrolases superfamily | IV.3.b | X | X | X | MGCMFSHLAA | C14-gcmFSHLAAK | 2 |
| AT5G14390.1 | α/β-Hydrolases superfamily | IV.1.b | X | MGGVTSSVAA | C14-gGVTSSVAAK | 2 | ||
| AT3G01690.1 | α/β-Hydrolases superfamily | X | MGGVTSSVAA | C14-gGVTSSVAAK | 2 | |||
| AT1G66900.1 | α/β-Hydrolases superfamily | X | MGGVTSSIAA | C14-gGVTSSIAAK | 2 | |||
| AT4G24760.1 | α/β-Hydrolases superfamily | X | MGGVTSSMAA | C14-gGVTSSmAAK | 2 | |||
| AT3G30380.1 | α/β-Hydrolases superfamily | X | MGAVTSSMAA | C14-gAVTSSmAAK | 2 | |||
| AT1G13610.1 | α/β-Hydrolases superfamily | X | MGIATSTMAA | C14-gIATSTmAAK | 2 | |||
| AT5G23395.1 | Cox19-like CHCH family protein (MIA40) | X | MGQAQSDENS | C14-gQAQSDENSIPTTTTTNTPPPSANSPR | 2 | |||
| AT3G27210.1 | Unknown function | X | X | MGSSSSSSLN | C14-gSSSSSSLNNSPIR | 2 | ||
| AT1G04200.1 | Unknown function | X | X | MGGVPSTPRK | C14-gGVPSTPR | 2 | ||
| AT2G44260.1 | Unknown function (DUF946 domain) | X | X | X | MGNCLSTSDP | C14-gNcLSTSDPSHEDVSK | 2 | |
| AT1G53400.1 | Unknown function (Ubiquitin domain) | X | X | X | MGCAGSTQSQ | C14-gcAGSTQSQADGSVK | 2 | |
Protein accession number (TAIR v.10). Proteins identified ambiguously are indicated as protein groups.
Protein name based on PPDB annotation.
Cluster number.
Predicted myristoylome (Martinez et al., 2008).
Identification of MYRed peptide by LTQ-Orbitrap (Velos) (left column) and by Triple-TOF (right column).
N-terminal protein sequence (10 amino acids).
Sequence of identified N-terminal MYRed peptides. Identified modified amino acid is indicated in lowercase. Myristoyl moiety is indicated as C14.
Position of the MYRed Gly.
For the majority of NMT targets, the myristoyl moiety is added on Gly2 after cotranslational removal of the first Met (Giglione et al., 2015). Several posttranslational MYR events have also been identified in animal cells (Martin et al., 2011; Peng et al., 2016). While the majority of MYRed peptides identified here contained a myristoyl moiety at Gly2, we also identified a myristoyl moiety at Gly98 [K.(MYR)gNSNSSSVDHR.F] of AT2G45380, a protein of unknown function. An identical sequence (MYRgNSNSSSVDHR) was identified at the N terminus of another protein of unknown function (AT5G06260) with a bona fide N-terminal semitryptic peptide carrying a myristoyl group at Gly2. Both peptides were identified in independent runs, suggesting that, despite their shared sequence, MYR can occur in a posttranslational or cotranslational manner, at least for these two proteins.
Proximal Putative Secondary Acylation Sites or Polybasic Regions Correlate with the MYRed Proteome Distributions in the PM and DRM Fractions
To visualize relative enrichment of the identified myristoylome in subcellular fractions, the distributions of predicted and directly identified MYRed proteins were plotted as a function of protein abundance and the number of MYRed proteins showing maximal accumulation in a given fraction (Figure 5). This double quantification avoided possible bias from a few abundant proteins versus higher numbers of proteins of lower abundance. The greater number of predicted MYRed proteins for which no MYRed peptide was identified (159; Figure 3) showed 50% less total protein abundance than the less numerous set of proteins identified as MYRed (72; Figure 5A), confirming that the identification of MYRed peptides was hampered by the overall lower abundance of such proteins. Of note, a global comparison of relative abundance of the identified MYRed proteins versus the entire identified protein data set revealed that MYRed proteins belonged to low abundance categories (Supplemental Figure 4).
Figure 5.
Influence of PAL and PBR Secondary Targeting Signals on the Localization of Identified MYRed Proteins.
(A) The relative abundance of MS-identified MYRed proteins (i.e., with MYR detected or predicted) in different subcellular fractions was quantified based on ∑(Narea) (solid gray and red bars) or the number of proteins showing the maximum expression in a given fraction (gray and red hatched bars). Identified myristoylome is the pool of 231 MS-identified proteins for which the N-terminal MYRed peptide was directly detected (72 proteins) plus those predicted to undergo MYR (159 proteins; see Figure 3). Detected MYRed peptides belong to the set of 72 for which MYRed peptide was directly detected alone (red bars).
(B) Percentage of identified MYRed proteins containing a second targeting signal (PBR, PAL, both PAL and PBR, or no second signal).
(C) Relative abundance of identified MYRed proteins either containing or not containing a second targeting signal in the analyzed cellular fractions as in (A).
(D) Venn diagram representing the overlap between the identified protein sets: proteins identified and predicted as MYRed (232 proteins, MYR+), proteins experimentally determined as PAL by affinity purification (Hemsley et al., 2013) (492 proteins, PAL+), and identified proteins nonpredicted to be MYRed but carrying a Gly2 (692 proteins, MGxx+).
The observed MYRed and predicted MYRed proteins were mainly enriched in the PM and DRM fractions (Figure 5A). Although this suggests that MYR is required for PM/DRM interactions, it does not demonstrate that it is sufficient for PM/DRM targeting. Indeed, both the observed and predicted MYRed protein sets were heterogeneous in terms of the presence of a second targeting signal such as an S-acylation (PAL) and/or PBRs. The current predicted Arabidopsis myristoylome includes ∼37% of MYRed proteins with at least one putative PAL site close to the MYR site (i.e., within amino acids 2 to 6), 20% with a polybasic track, and ∼43% not displaying any second signal (Martinez et al., 2008). Within our entire set of identified MGxx proteins undergoing MYR, a similar percentage of proteins contained PBRs (19%) as a unique second targeting signal (Figure 5B, first column). However, proteins carrying a putative PAL or both PAL and PBR sites represented 53% of the identified myristoylome (versus only 37% in the predicted Arabidopsis myristoylome). The same tendency was observed when only the 72 identified MYRed proteins for which we could retrieve the N-terminal myristoyl moiety were considered: Only 8% of proteins carried a PBR, while 58% carried a PAL or PAL and PBR (Figure 5B, second column). Such PAL site enrichment agrees with the suggested role of PAL as an important second PM targeting signal for MYRed proteins. When the presence or otherwise of the second signal was considered, the abundance of identified MYRed proteins without a second signal was widely distributed across all fractions (Figure 5C). However, a clear shift in protein abundance toward PM/DRM fractions was observed when a putative second targeting signal such as PBR, PAL, or both were present (Figure 5C).
These protein localization data were further used to explore the influence of MYR/PAL/PBR signals on subcellular protein localization interplay. Three protein subsets were considered (Figure 5D): (1) all proteins identified as MYRed (214+18 proteins, noted as MYR+); (2) proteins previously determined as PALed by affinity purification (Hemsley et al., 2013) (492 proteins, noted as PAL+); and (3) identified proteins not predicted to be myristoylated but carrying the main requirement for a putative MYR (i.e., a Gly2, 692 proteins, MGxx+). There was relatively little overlap between these subsets; only 22 and 35 proteins were identified in the overlapping MYR+ and the MGxx+ sets, respectively, suggesting that the majority of in vivo identified MYRed proteins displaying both MYR and PAL sites (22) were retrieved in PM and DRM fractions (Figure 5C). Only three other proteins with a potentially double acylation, HIR1, HIR3, and HIR4, were characterized in the µZ and one, CRK2, in the Golgi fraction (Supplemental Data Set 1.1). Although this covers only a fraction of our complete data set, the distribution agrees with previous studies supporting a preferential PM localization of MYRed proteins with adjacent PAL sites (Traverso et al., 2013b).
Frequency Distribution of Cys Residues in All Identified Predicted MYRed Proteins
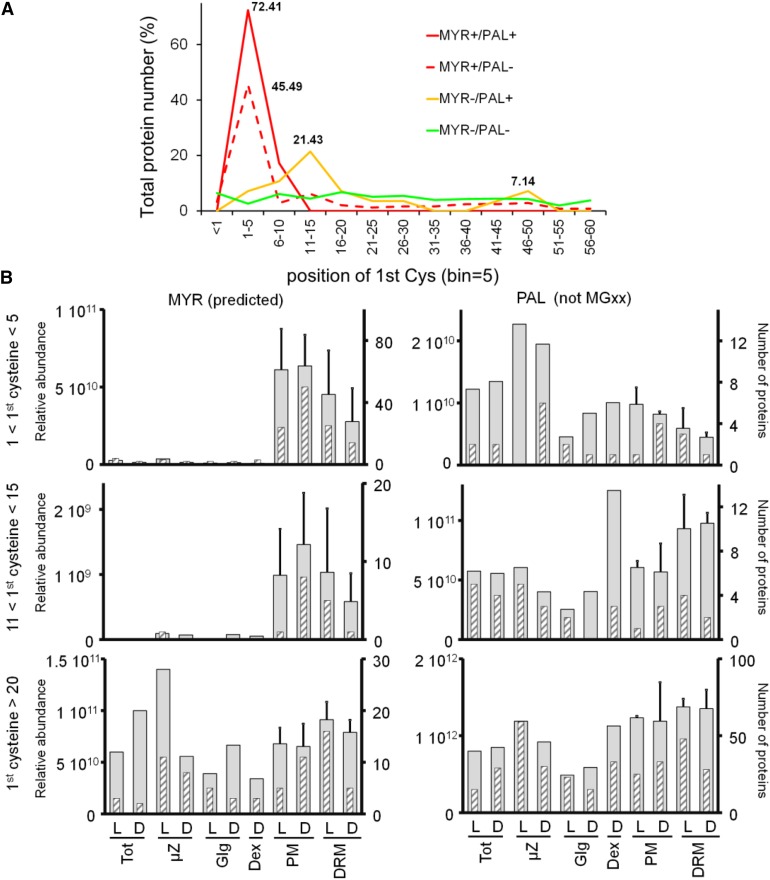
Within the predicted MYRed protein set, positions 3 to 6 frequently featured putative palmitoylable Cys residues. Indeed, there was striking enrichment (72%) of Cys residues at positions 1 to 5 for all proteins predicted to undergo MYR (Figure 6A). This Cys enrichment was not observed in any other protein data set (Hemsley et al., 2013). For proteins that were putatively only palmitoylable but displaying an N-Gly (MYR-/PAL+ set), limited but significant Cys enrichment was observed at positions 11-15 (21%) and overlapping the end of the Cys position peak observed for the MYR+/ PAL+ set. The Cys enrichment at positions 4-5 and 11-15 was clearly linked to proteins with preferential PM/DRM localization (Figure 6B). In fact, the MYR+/PAL+ set distribution with Cys residues at positions 4-5 or 11-15 was quasi-exclusively concentrated in PM/DRM fractions. This was in contrast to MYRed proteins with the first Cys residue situated beyond position 20, which were present in all analyzed fractions.
Figure 6.
Frequency Distribution of the First N-Terminal Cys Position in Relation to Subcellular Localization.
(A) Frequency distribution of the first Cys position is shown for the following subsets of identified proteins: MYRed (observed and predicted) and PALed (MYR+/PAL+, red line), MYRed but not PALed (MYR+/PAL-, dashed red line), not predicted to be MYRed but carrying a Gly2 (MGxx set) and PALed (MYR-/PAL+, yellow line), not predicted to be MYRed but carrying a Gly2 and not PALed (MYR-/PAL-, green line). Percentages for peaks of frequency distributions are indicated.
(B) Protein subcellular localization based on relative abundance expressed as ∑(Narea) (gray large bars) or based on the number of proteins showing the maximum expression in a given fraction (gray hatched thin bars) were plotted for MYRed predicted or PALed-only identified subsets as a function of the position of the first Cys in the protein sequence.
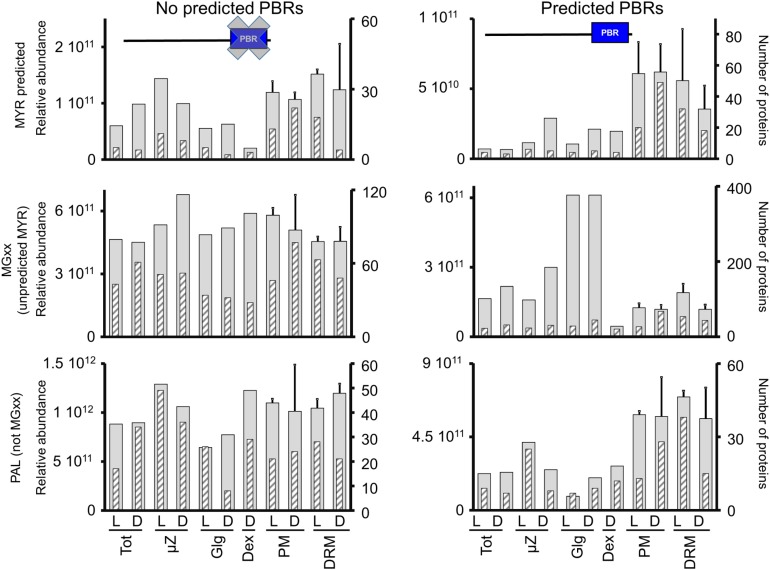
Within the experimentally observed PALed proteome (Hemsley et al., 2013) excluding all putative MYRed proteins (i.e., within the MYR-/PAL+ protein set), the position of the first Cys residue in the proximity of the N terminus had no impact on subcellular localization. This suggests that in the absence of a MYR signal, PALed-only proteins are targeted to a variety of subcellular compartments (including PM/DRM) and that the position of palmitoylable residues is not related to a specific protein localization. Compared with the MYR+/PAL+ set, PALed-only or MYRed-only proteins accumulated in almost all fractions. However, of those, a number of proteins showed more pronounced PM/DRM enrichment compared with the MYRed set, suggesting that additional PM targeting signals exist. To investigate this hypothesis, we analyzed the subcellular localization of putatively MYRed or PALed proteins as a function of the presence of close PBRs predicted on the basis of short, condensed amino acid stretches (Martinez et al., 2008), and additional identification of these regions was performed on our MYRed and non-MYRed predicted experimental data set and verified PALed-only proteins (Hemsley et al., 2013). The relative distribution in subcellular fractions of proteins carrying a putative PBR or for which no PBR was predicted is shown in Figure 7. Within the MYRed set, a higher specificity for PM/DRM localization was observed for proteins carrying a putative PBR. Similarly, within the PAL-only protein set, clear PM/DRM clustering was observed in the presence of a putative PBR. By contrast, no preferential accumulation in a specific fraction was observed in the pool of non-MYRed proteins as a function of the presence or absence of a putative PBR (Figure 7). Polybasic tracks might act as second PM/DRM targeting signal but only in combination with MYR and/or PAL.
Figure 7.
Influence of Predicted PBRs on Protein Subcellular Localization.
Protein subcellular localization, taking in account the presence or absence of predicted PBRs, was plotted based on relative abundance expressed as ∑(Narea) (gray, large bars) or based on the number of proteins identified with maximal accumulation in a given fraction (gray hatched thin bars) for MYRed-predicted proteins, MYRed not predicted but carrying a Gly2, and for experimentally determined PALed proteins not carrying a Gly2 (PAL+/MGxx-).
Characterization and Distribution of Protein Lipidation Enzymes
Of the protein lipidation catalysts (Supplemental Data Set 1.2), NMT1 was present and highly abundant in most fractions and enriched in the Dex fraction, most likely indicating endomembrane localization in addition to the cytosolic localization previously reported in plants and animals (Pierre et al., 2007; Dudek et al., 2015). NMT1 accumulated identically in dark and light samples. Ten of twenty-four predicted palmitoyl-acyl-transferases (PATs) were also identified, of which nine were localized to the PM and/or DRM fractions, as reported previously (Batistic, 2012) and in Supplemental Data Set 1.2. Each PAT accumulated between 0.5- to 5-fold less than NMT1 with the exception of PAT24 (Tip1), which accumulated one order of magnitude more than NMT1 in the Dex fractions.
Interestingly, the farnesyltransferase PFT/PGGT α subunit, an enzyme responsible for prenylation, and many RAB geranylgeranyl transferases were identified in the DRMD sample (Supplemental Data Set 1.2). The most abundant RAB geranylgeranyl transferases Rab-GGTβ1 and GGTβ2 were also found in µZD, whereas FPS1 geranylgeranyl transferase was identified with maximum accumulation in Dex. A geranylgeranyl is added after proteolytic maturation of the C terminus by a specific CaaX domain processing protease Ste24 (AT4G01320.1) that was identified in Dex fractions, in agreement with its predicted localization to the endoplasmic reticulum (ER; Bracha et al., 2002).
Effect of Light/Dark on MYRed Protein Distribution in Different Compartments
It is known that MYRed proteins can dynamically relocalize in response to specific signals, including light signaling (Assmann, 2002; Giammaria et al., 2011; Yin et al., 2017), particularly when associated with a reversible second signal (Turnbull and Hemsley, 2017). Some of the entire set of identified proteins and a few proteins in the MGxx pool not predicted to undergo MYR differentially accumulated in PM and DRM under dark and light growth conditions (Supplemental Figure 5). Although this differential accumulation was much more evident for proteins found in the DRM fractions, the analysis of the identified myristoylome in PM or DRM under light and dark growth conditions showed that there were no significant changes in protein abundance for the majority of these proteins (Supplemental Figure 5).
DISCUSSION
Over 90% of the predicted Arabidopsis myristoylome still awaits direct confirmation, and few in vivo studies have successfully characterized this subproteome, particularly in plants (Hemsley, 2015). Here, we adopted a multifaceted proteomics approach in Arabidopsis to produce the largest in vivo MYRed protein data set to date. This approach was based on (1) protein enrichment via membrane fractionation, (2) high-resolution separation of hydrophobic peptides, and (3) development of an MS data-dependent acquisition method targeting MYRed peptides. We also determined the subcellular distribution and accumulation of many proteins, including most of the predicted MYRed proteins in PM and DRM fractions in response to light or dark. Furthermore, the endogenous N-terminal myristoyl moiety was identified on 72 distinct proteins implicated in a variety of cellular functions including calcium signaling, redox regulation, pathogen responses, and metabolism.
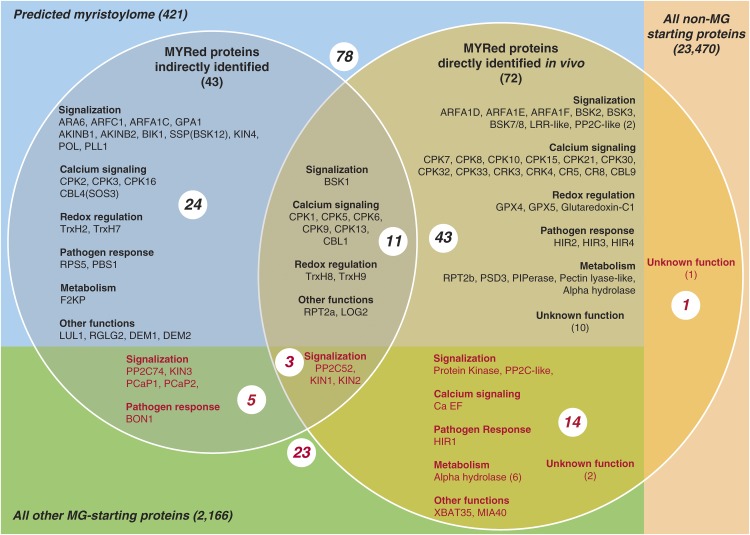
Previous indirect approaches have suggested MYR in 42 Arabidopsis proteins (Gagne and Clark, 2010; Ishitani et al., 2000; Ueda et al., 2001; Lu and Hrabak, 2002; Boisson et al., 2003; Dammann et al., 2003; Pierre et al., 2007; Yin et al., 2007; Batistic et al., 2008; Benetka et al., 2008; Nagasaki et al., 2008; Bayer et al., 2009; Kato et al., 2010; Li et al., 2010; Mehlmer et al., 2010; Meng et al., 2010; Yamauchi et al., 2010; Burr et al., 2011; Held et al., 2011; Stael et al., 2011; Pratelli et al., 2012; Takemoto et al., 2012; Tsugama et al., 2012a, 2012b; Feng et al., 2013; Lu and Hrabak, 2013; Traverso et al., 2013b; Kang et al., 2015), among which eight were not included in the predicted myristoylome made of 421 proteins (Figure 8). The endogenous MYR moiety of 11 of the 42 aforementioned proteins was confirmed. Moreover, the MYR tag was identified on three of the previously identified proteins not included in the predicted myristoylome (P2C52, KIN1, and KIN2), and an additional 43 in vivo MYRed proteins were identified and predicted to undergo MYR. Of note, we also highlighted the in vivo MYR of 18 proteins not previously predicted to undergo MYR, highlighting that the current predicted Arabidopsis myristoylome remains incomplete. Our consolidated report of 101 MYRed proteins and the cellular abundance of almost half (214) of the actual predicted myristoylome (421) represents the most comprehensive MYR data set for any organism reported to date.
Figure 8.
Relationship between Various Sets of Putative and Identified MYRed Proteins in Arabidopsis.
Proteins identified in this study are shown in the right circle (72). Those identified indirectly in other studies (43) are in the left circle. The intersection between the two data sets is indicated (14 proteins). Proteins proposed to be part of the myristoylome through bioinformatics analysis (421) are in the blue rectangle (78). Those not yet predicted (23) are in red. The sets of Arabidopsis proteins starting with MGxx correspond to the green and blue rectangles. All others not starting with MGxx are in the right light pink rectangle. Only one identification corresponds to this set.
The Majority of MYRed Proteins Localize to the PM and Are Involved in Calcium Signaling and Pathogen Responses
The most important protein family undergoing MYR is related to calcium signaling, as MYR occurred in 13/34 calcium-dependent protein kinases (CPKs; for review, see Boudsocq and Sheen, 2013). Twenty-seven of 30 (88%) identified CPKs carried a Gly2, 23 were previously predicted to be MYRed, and 8 were indirectly shown to undergo this lipidation. Our targeted approach revealed the myristoyl moiety on CPK1, 5, 6, 9, and 13 and on eight additional CPKs (CPK7, 8, 10, 15, 21, 30, 32, and 33), mostly in the PM fraction, and a few (CPK15, 21, 30, 32, and 33) in lesser amounts in the DRM fraction, consistent with previous reports showing PM/DRM localization of several CPKs (Dammann et al., 2003; Mehlmer et al., 2010; Hemsley et al., 2013). Since (1) 80% of the identified CPKs (25/30) carried at least one Cys at positions 3, 4, or 5, with several already shown to belong to the PAL proteome (Hemsley et al., 2013), and (2) most directly identified MYRed CPKs also displaying PBRs, we suggest that these secondary signals directly contribute to PM localization in association with MYR. One interesting case is CPK1, with previous studies showing that LeCPK1 specifically localizes to the PM in an N-terminal Gly-dependent manner (Rutschmann et al., 2002), unlike Arabidopsis CPK1, which localizes to peroxisomes and lipid bodies (Dammann et al., 2003; Coca and San Segundo, 2010). In our analysis, CPK1 was most abundant in the PM/DRM in agreement with LeCPK1, but a non-negligible amount was also found in the Tot fraction and to a minor extent in µZ fractions. However, we relied only on endogenous proteins and not on overexpressed, GFP-fused constructs. Thus, although we cannot rule out that Arabidopsis CPK1 is not targeted to peroxisomes, the relative accumulation in this compartment was less than in PM/DRM, suggesting diverse, compartment-specific roles for this protein.
Another interesting category of MYRed proteins was the receptor-like cytoplasmic kinases such as those of the RLCK-XII subfamily, known as brassinosteroid (BR) signaling kinases (BSK). BSK1 plays a role not only in plant growth and BR signaling but also in mediating flg22-dependent pattern-triggered immunity (Shi et al., 2013). In this latter case, membrane localization, most likely due to MYR, is necessary for BSK1 to function as a positive regulator of pattern-triggered immunity signaling. BSK1 and BSK12 were the only RLCK-XII subfamily BSKs for which MYR was previously indirectly shown to occur. The characterization of unequivocal MYR for other members of the family, their identical PM membrane localization, palmitoylable Cys, and partial overlapping functions for plant growth and BR signaling (Kong et al., 2012; Sreeramulu et al., 2013) suggest that additional MYRed BSKs might contribute to plant immunity. Of note, a myristoyl moiety was also identified in a number of proteins involved in general pathogen responses (Figure 8, Table 1).
MYR also occurred on the calcineurin B-like (CBL) Ca2+ sensor proteins CBL1 and CBL9; MYR of CBL1 has previously been reported (Batistic et al., 2008). Both CBLs carry putative PAL sites near the N-terminal Gly2 (Cys3), and CBL9 has also been identified in PAL-enriched proteomes in Arabidopsis (Hemsley et al., 2013). Both proteins were localized to PM fractions, whereas both interacting targets were found in the DRM. Interestingly, the nonmyristoylable CBL2 (lacking Gly2) previously identified in a PAL-enriched proteome (Hemsley et al., 2013) was identified in the microsomal fraction. In agreement with our findings, the triple PAL of CBL2 creates a tonoplast targeting signal (Batistič et al., 2012), confirming that PAL alone can relocalize proteins to compartments other than the PM/DRM.
Finally, we also identified 13 leucine-rich repeat (LRR) domain proteins in PM/DRM fractions, only two of which were previously described to undergo MYR and PAL: PBS1 and RPS5 (Takemoto et al., 2012; Qi et al., 2014). Transient expression of the N-terminal part of PBS1 and RPS5 or of the corresponding mutated versions affecting their MYR and PAL sites in Arabidopsis or Nicotiana benthamiana suggested that these acylations are necessary for PM localization (Takemoto et al., 2012; Qi et al., 2014). We retrieved PBS1 in both the PM and DRM fractions, whereas RPS5 was mainly found in the DRM. These data are consistent with MYR playing a prominent role in pathogen responses.
Alternative Localization of MYRed Proteins in Non-PM Compartments
Here, 188/214 (88%) of the predicted MYRed proteins targeted the PM/DRM, the others mostly located in the ER/Golgi fraction. For instance, there was a broader distribution of HIR1, 2, 3, and 4 in the µZ and PM/DRM fractions. Interestingly, HIR was not part of the predicted myristoylome and because HIR1, 2, 3, and 4 are in the palmitoylome (Hemsley et al., 2013), these findings emphasize the reversibility of the PAL modification and that two pools of the same protein display different N-terminal acylations that guide their distribution in distinct compartments.
MYR also occurred at the N terminus of a calcium binding EF protein, which did not display an apparent second signal and was mainly found in the TotL and µZ fractions, consistent with previous studies suggesting that MYR mostly targets endomembrane system proteins (Traverso et al., 2013b). A similar pattern was observed for all identified small GTPase ARFs displaying a predicted MYR.
An unforeseen myristoyl moiety was recovered at the N-terminal Gly of two interesting proteins (XBAT35 and MIA40) for which the contribution of the lipid modification seems to be different. XBAT35 is a structurally related ankyrin repeat-containing RING E3 ligase intricately involved in ethylene-mediated responses via ubiquitin-protein degradation (Carvalho et al., 2012). The corresponding gene undergoes alternative splicing to produce two proteins with and without a nuclear localization signal. Although we could not discriminate between the two isoforms, the most abundant protein predominantly accumulated in the DEX fraction, suggesting nuclear localization. Nuclear localization of predicted MYRed proteins has previously been observed in different organisms. For instance, another RING-type ubiquitin ligase McCPN1 in Mesembryanthemum crystallinum was involved in AGO4 degradation in response to salt stress, had a myristoylable N-terminal Gly, and localized to the nucleus in addition to the PM and cytoplasm (Li et al., 2014). It is tempting to speculate that MYR of these ligases is involved in ubiquitin-proteasome-controlled proteolysis of specific substrates via regulation of the localization of the MYRed ligase. This agrees with previous findings showing that MYR of proteasome subunit Rpt2 reduces the nuclear export of the proteasome in yeast and plays an additional role in protein quality control within the nucleus (Kimura et al., 2016).
MIA40 is one of two components of the mitochondrial intermembrane space (IMS) disulfide relay system, which ensures import of Cys-containing proteins into the IMS (Herrmann and Riemer, 2012). Arabidopsis MIA40 resides in the IMS and peroxisomes and is thought to be responsible for importing and trapping the chaperone for Mn-superoxide dismutase (CCS1), Cu/Zn superoxide dismutase 1 (CSD1), and Shewanella-like protein phosphatase 2 (SLP2) in the IMS and the Cu/Zn CSD3 in peroxisomes (Carrie et al., 2010; Uhrig et al., 2017). Peroxisome localization has only been observed using GFP-MIA40-N terminus fusion constructs that block MYR. Moreover, in yeast, MIA40 is anchored to the IM via an N-terminal hydrophobic stretch that is not conserved in plants (Herrmann and Riemer, 2012). Here, MIA40 was mainly found in the µZ fraction, which is the most enriched fraction in mitochondria, suggesting that MYR could overcome the evolutionary absence of the yeast N-terminal hydrophobic stretch.
Thus, although to a much lesser extent than the PM/DRM, MYR also directly contributes to dedicated functions in other compartments including the ER/Golgi, mitochondria, peroxisome, and nucleus. The distribution of the same protein between the PM/DRM and ER/Golgi appears to depend on partial PAL of the nearby Cys.
Posttranslational MYR in Plants
Most MYR moieties are added to an N-terminal Gly after cotranslational removal of the N-terminal Met. In animals, non-N-terminal MYR affects the pro-apoptotic BID protein, in which an internal Gly residue is MYRed following caspase cleavage. This leads to relocalization of the proteolytic apoptotic BID fragment to mitochondria (Zha et al., 2000). Surprisingly, we observed posttranslational MYR in a protein of unknown function (AT2G45380), with a MYR moiety identified at Gly98 [K.g(MYR)NSNSSSVDHR.F]. An identical MYR-gNSNSSSVDHR sequence was identified at the N terminus of a TLD domain-containing nucleolar protein with a bona fide N terminus peptide carrying a Gly2 MYR. Both peptides were identified in independent runs suggesting that, despite their shared sequence, MYR could occur in a post- and cotranslational manner. A pBLAST search (Supplemental Figure 6) revealed four Arabidopsis proteins sharing a highly conserved 50-amino acid stretch starting with the conserved GN(S/L)NSSSVDH MYR consensus sequence. The position of the conserved stretch was either the N terminus (position 2 for AT5G60260), classically observed for myristoylated proteins, or was situated downstream of a 98- or 48-amino acid N-terminal extension in AT2G45380 and AT4G34070, respectively. Both AT5G60260 and AT2G45380 proteins were highly expressed in PM/DRM fractions, while AT4G34070 was identified with low scores in the µZ fraction. The fourth member of this protein family (AT2G44850), carrying a shorter N-terminal extension (23 amino acids) and an S27/L substitution within the N-terminal end of the conserved 50-amino acid stretch, escaped MS identification. Interestingly, AT5G06260 and AT4G34070 are related to the calcium binding EF-hand protein family. We hypothesize that these isoforms have undergone functional specialization, with at least two members undergoing MYR in a co- and posttranslational manner, respectively, directing the proteins to the PM/DRM. In contrast, AT4G34070, despite having an internal myristoylatable sequence, should not undergo posttranslational MYR like AT2G45380, taking into account its chloroplast localization (i.e., MYR occurs only in the cytosol). In the case of AT2G44850, the Ser-to-Leu substitution within the MYRed sequence is unlikely to inhibit MYR, and the sequence is still positively predicted by TermiNator3. It is possible that the N terminus of AT2G44850 undergoes proteolytic cleavage in response to specific stress signals that expose the N-terminal Gly to posttranslational modification. Such identification of MYR on internal Gly residues provides evidence of posttranslational MYR events for the first time in plants.
The predominance of signaling and pathogen-related functions suggests that the myristoylome might act as a dynamic proteome in response to abiotic or biotic stresses. Future work will involve characterizing this myristoylome dynamicity under different stress conditions.
METHODS
All solvent and chemicals were purchased from Sigma-Aldrich and Bio-Rad unless otherwise stated.
Arabidopsis Cell Suspension Cultures
Arabidopsis thaliana Col-0 cell suspension cultures were grown in Jouanneau and Péaud-Lenoël (JPL; Jouanneau and Péaud-Lenoël, 1967) medium in either continuous light (70 µE s−1 m−2) or continuous dark at 23°C and at 90 rpm rotation. Cells were subcultured at 9-d intervals. For subcellular fractionation, cells were harvested during exponential growth phase five days after 1/10 dilution in fresh JPL medium.
Purification of PM and DRM Fractions
Microsomal, PM, and DRM fractions were prepared using as a starting material 500 mL cultured Arabidopsis cells per biological replicate. In total, four biological replicates were prepared from light and dark grow cells for MS analysis. Three best preparations were chosen for further analysis (as indicated below in MS section). Briefly, as described by Marmagne et al. (2006), PMs were purified from the microsomal pellet (100,000g) over a two-phase dextran and polyethylene glycol (PEG) system using 6.4% dextran, 6.4% PEG, and 5 mM phosphate buffer (KH2PO4/K2HPO4), pH 7.8 (Marmagne et al., 2006). Protein concentrations were determined by the Bradford assay (Bradford, 1976). The dextran fraction was diluted 10-fold in 10 mM HEPES, pH 8.0, and 5 mM MgCl2 (buffer A) and pelleted at 120,000g for 20 min. The procedure was repeated twice to remove soluble contaminants. PEG fractions were precipitated by addition of phosphate buffer, and the precipitated PM were recovered by centrifugation and washed twice with buffer A. Enriched DRM fractions were prepared from PM fractions treated with Triton X-100 for 30 min at 4°C at a 1:15 protein to detergent ratio on a rotary wheel. DRM fractions were pelleted at 120,000g for 20 min. After discarding the supernatant, the DRM-enriched pellets were washed twice with buffer A by centrifugation at 120,000g for 20 min.
Purification of Golgi-Enriched Fractions
The preparation of Golgi-enriched fractions was performed as described (Graham, 2001).
Immunological Methods
Rabbit polyclonal antibodies raised against cytosolic sucrose synthase (SPS), H+-ATPase, and RbcL were produced at Agrisera and used at 1:5000 dilution for immunoblot analysis as previously described (Adam et al., 2011).
Protein Identification and Analysis by LC-MS/MS
For all analyzed fractions, 150 µg of protein was resuspended in 1× Laemmli buffer and denatured at 95°C for 1 min. Insoluble fractions were removed by centrifugation (10,000g for 5 min at 4°C). Proteins were separated by SDS-PAGE, and the whole gel lane was cut into eight gel bands followed by a tryptic in-gel digestion including reduction and alkylation (Shevchenko et al., 1996). Peptides were resuspended in 20 μL nLC buffer B (0.1 formic acid [FA] and 5% acetonitrile [ACN]). Tryptic peptide mixtures were analyzed by LC-MS/MS using the nano-flow Proxeon Thermo LC and Orbitrap Velos (Thermo Fisher Scientific). Peptides were separated with Easy-nLC (Thermo Fisher Scientific) and loaded on a guard column (NS-MP-10; Nanoseparation) followed by separation on a Nikkyo Technos analytical column (NTCC-360/100-5-153) using 40-min gradients with 0.1% FA in water (solvent C) and 0.1% FA in ACN (solvent B) at a flow rate of 300 nL/min. The gradients were optimized to allow a maximum time elution at high mobile phase (0→5 min/10→40% B, 6→27 min/40→60% B, 28→29 min/60→80%, B and 29→35 min/80% B). Each sample analysis was followed by a blank injection and 30 min run to reduce carryover. The survey scan was acquired by Fourier-transform MS scanning 400 to 2000 D at the maximal 100,000 resolution in the Orbitrap using internal calibration. This was followed by top 20 data-dependent tandem MS scans acquired in the LTQ with a dynamic exclusion set at 30 s. When no inclusion list acquisition method was used, the MS/MS triggering option “Nth most intense ions” in the data-dependent settings panel was used. When MYR inclusion lists were used, the option “Nth most intense from list” button was deactivated. Predicted masses of MYRed peptides for MYR inclusions lists were generated based on in silico tryptic digestion of the Arabidopsis MGxx protein set using N-terminal peptides with fixed MYR-N-ter modification and combinatory occurrence of variable methionine oxidation and carbamidomethylation. The generated list contained ∼4000 masses, so, due to limitations of the Xcalibur software (Thermo Fisher Scientific), the obtained inclusion list was split into two lists and used in sequential runs. All inclusion lists from data-dependent acquisitions were applied to a given sample in a sequence of three successive analyses as follows: no-inclusion/inclusion1/inclusion2 to ensure a qualitative comparison reference for the identification of MYR-N-ter peptides. Spectral data with parent ion signal higher than 1 count and with an S/N ratio higher than 1.5 were extracted using Proteome Discoverer v1.3 (Thermo Fisher Scientific) and were searched against the Arabidopsis genome (TAIR v10; http://www.arabidopsis.org/) supplemented with chloroplast and mitochondria genomes using Mascot v2.4 (Matrix Science). Variable MYR-N-ter, methionine oxidation, and fixed Cys carbamidomethylation were used. A minimal ion score threshold of 25 yielded a peptide false discovery rate (FDR) below 1%, with peptide false positive rate calculated as: 2 × (decoy_hits)/total_hits. The quantitative proteomics pipeline used Mascot for protein identification and label-free quantification based on MS precursor areas using Proteome Discoverer (Supplemental Figure 2). The relative quantification allowed us to determine the three best independent biological replicates (out of four) for PM and DRM. The selection of the best biological replicates was based on enrichment of cell compartment markers (data extracted from PPDB, http://ppdb.tc.cornell.edu/; Sun et al., 2009) and compared with previously published PM (Marmagne et al., 2007) and DRM (Kierszniowska et al., 2009; Minami et al., 2009) proteomes. Approximately 2.4 million MS/MS spectra were acquired, and ∼1.3 million spectra were assigned to peptide sequences that passed quality filters (FDR <1%) and assigned to 9256 protein accessions. After removing ambiguous gene models and grouping ambiguously identified protein families, the entire identified protein set regrouped 8837 different proteins.
Quantification and Statistical Analysis
Label-free quantitative data were extracted using Proteome Discoverer v.1.3 (Thermo Fisher Scientific). Quantification was based on the integration of precursor peak areas. The settings used were to avoid automatic protein grouping based on shared peptides as those often generate large protein groups (>50 members) with no obvious biological relevance. To increase accuracy and avoid overquantification of protein groups, we developed an in-house bioinformatics pipeline that allowed processing of raw quantification data from PD1.3. Considering differences in protein complexity versus abundance between different cell subproteomes analyzed in this study, sums of precursor areas obtained per protein were normalized per sample and adjusted to the total ion count for each biological replicate (Friso et al., 2011; Renna et al., 2013), especially for PM biological replicates with very high total ion counts. Relative protein amounts were expressed as the sum of normalized precursor areas averaged between biological replicates. Quantification confidence cutoffs were estimated by plotting the correlation between protein frequency distribution and the coefficient of variation (CV) between biological replicates. The cutoff for quantification confidence was set as >70% of proteins with a CV <50%, which translated into a protein abundance cutoff of108 Σ(Narea). Proteins with low numbers of spectral counts (<5) and/or low precursor areas (<108) were considered to be of low confidence for quantification (Supplemental Data Set 1.3). Functional annotations and localizations were obtained from the Plant Proteomics database (http://ppdb.tc.cornell.edu/dbsearch/subproteome.aspx) (Sun et al., 2009) and TAIR (http://www.arabidopsis.org/). To functionally assign proteins to cellular functions, we used the MapManBin functional classification (Thimm et al., 2004). Protein expression profiles through unsupervised hierarchical clustering were performed as described (Majeran et al., 2012).
Accession Numbers
Mass spectrometry proteomics data are deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al., 2013), with the data set identifier PXD006850. Additional accession numbers are in Table 1.
Supplemental Data
Supplemental Figure 1. Purification of subcellular Arabidopsis membrane proteomes.
Supplemental Figure 2. Experimental and quantitative proteomics work flow.
Supplemental Figure 3. Relative enrichment of cell compartment marker proteins in analyzed fractions.
Supplemental Figure 4. Frequency distribution of relative protein abundance of MYRed proteins versus other proteins.
Supplemental Figure 5. Light versus dark accumulation of identified MYRed proteins in PM and DRM fractions.
Supplemental Figure 6. Homology alignment of four members of the TLD-domain unknown proteins carrying a highly conserved N-terminal region including a myristoylatable peptide.
Supplemental Data Set 1.1. Identified MYRed proteins including proteins from predicted myristoylome and proteins directly identified with a myristoyl moiety.
Supplemental Data Set 1.2. Relative abundance and localization of enzymes involved in protein lipidations.
Supplemental Data Set 1.3. Summary of all identifications in MS analysis with functional annotations, cluster analysis, and MS relevant information.
Acknowledgments
This work was supported by Agence Nationale de la Recheche Grant PALMYR PROT (ANR-10-BLAN-1611), by Labex Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS), and by ARC Grant SFI2011120111203841. We thank W. Bienvenut and D. Cornu for assistance with MS analysis, M. Boudsocq for help with CDPK family annotations, and P. Millares for developing the in-house parsing algorithm for PD1.3. This work used the facilities of the SICaPS platform at I2BC (Gif).
AUTHOR CONTRIBUTIONS
C.G. and T.M. conceived and supervised the project. W.M. designed and performed biochemistry purifications. W.M. and J.-P.L.C. performed mass spectrometry analysis. J.-P.L.C. developed the in-house parsing algorithm for PD1.3. T.M. designed and carried out predictive MYR in silico analysis. L.P. designed and performed hierarchical clustering. W.M., T.M., and C.G. wrote the manuscript. All authors discussed the results and contributed to the final version of the manuscript.
References
- Adam Z., Frottin F., Espagne C., Meinnel T., Giglione C. (2011). Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis. Plant Cell 23: 3745–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann S.M. (2002). Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 14 (suppl.): S355–S373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O. (2012). Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family. Plant Physiol. 160: 1597–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O., Sorek N., Schültke S., Yalovsky S., Kudla J. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20: 1346–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O., Rehers M., Akerman A., Schlücking K., Steinhorst L., Yalovsky S., Kudla J. (2012). S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res. 22: 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488. [DOI] [PubMed] [Google Scholar]
- Benetka W., Mehlmer N., Maurer-Stroh S., Sammer M., Koranda M., Neumüller R., Betschinger J., Knoblich J.A., Teige M., Eisenhaber F. (2008). Experimental testing of predicted myristoylation targets involved in asymmetric cell division and calcium-dependent signalling. Cell Cycle 7: 3709–3719. [DOI] [PubMed] [Google Scholar]
- Bhatnagar R.S., Gordon J.I. (1997). Understanding covalent modifications of proteins by lipids: where cell biology and biophysics mingle. Trends Cell Biol. 7: 14–20. [DOI] [PubMed] [Google Scholar]
- Boisson B., Meinnel T. (2003). A continuous assay of myristoyl-CoA:protein N-myristoyltransferase for proteomic analysis. Anal. Biochem. 322: 116–123. [DOI] [PubMed] [Google Scholar]
- Boisson B., Giglione C., Meinnel T. (2003). Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 278: 43418–43429. [DOI] [PubMed] [Google Scholar]
- Bologna G., Yvon C., Duvaud S., Veuthey A.L. (2004). N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics 4: 1626–1632. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Sheen J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P.C., Schwizer S., Hind S.R., Kraus C.M., De la Torre Diaz S., He B., Martin G.B. (2016). Detecting N-myristoylation and S-acylation of host and pathogen proteins in plants using click chemistry. Plant Methods 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha K., Lavy M., Yalovsky S. (2002). The Arabidopsis AtSTE24 is a CAAX protease with broad substrate specificity. J. Biol. Chem. 277: 29856–29864. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Breiman A., Fieulaine S., Meinnel T., Giglione C. (2016). The intriguing realm of protein biogenesis: Facing the green co-translational protein maturation networks. Biochim. Biophys. Acta 1864: 531–550. [DOI] [PubMed] [Google Scholar]
- Burr C.A., Leslie M.E., Orlowski S.K., Chen I., Wright C.E., Daniels M.J., Liljegren S.J. (2011). CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol. 156: 1837–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C., Giraud E., Duncan O., Xu L., Wang Y., Huang S., Clifton R., Murcha M., Filipovska A., Rackham O., Vrielink A., Whelan J. (2010). Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J. Biol. Chem. 285: 36138–36148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S.D., Saraiva R., Maia T.M., Abreu I.A., Duque P. (2012). XBAT35, a novel Arabidopsis RING E3 ligase exhibiting dual targeting of its splice isoforms, is involved in ethylene-mediated regulation of apical hook curvature. Mol. Plant 5: 1295–1309. [DOI] [PubMed] [Google Scholar]
- Chen T.F., Yoder J.D., Hruby D.E. (2004). Mass spectrometry analysis of synthetically myristoylated peptides. Eur. J. Mass Spectrom. (Chichester) 10: 501–508. [DOI] [PubMed] [Google Scholar]
- Coca M., San Segundo B. (2010). AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 63: 526–540. [DOI] [PubMed] [Google Scholar]
- Dammann C., Ichida A., Hong B., Romanowsky S.M., Hrabak E.M., Harmon A.C., Pickard B.G., Harper J.F. (2003). Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 132: 1840–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J.S., Andriotis V.M., Wu A.J., Rathjen J.P. (2006). Tomato Pto encodes a functional N-myristoylation motif that is required for signal transduction in Nicotiana benthamiana. Plant J. 45: 31–45. [DOI] [PubMed] [Google Scholar]
- Dudek E., Millott R., Liu W.X., Beauchamp E., Berthiaume L.G., Michalak M. (2015). N-Myristoyltransferase 1 interacts with calnexin at the endoplasmic reticulum. Biochem. Biophys. Res. Commun. 468: 889–893. [DOI] [PubMed] [Google Scholar]
- Feng X., Shi W., Wang X., Running M.P. (2013). In vitro myristoylation assay of Arabidopsis proteins. Methods Mol. Biol. 1043: 135–139. [DOI] [PubMed] [Google Scholar]
- Friso G., Olinares P.D., van Wijk K.J. (2011). The workflow for quantitative proteome analysis of chloroplast development and differentiation, chloroplast mutants, and protein interactions by spectral counting. Methods Mol. Biol. 775: 265–282. [DOI] [PubMed] [Google Scholar]
- Gagne J.M., Clark S.E. (2010). The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell 22: 729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammaria V., Grandellis C., Bachmann S., Gargantini P.R., Feingold S.E., Bryan G., Ulloa R.M. (2011). StCDPK2 expression and activity reveal a highly responsive potato calcium-dependent protein kinase involved in light signalling. Planta 233: 593–609. [DOI] [PubMed] [Google Scholar]
- Giglione C., Fieulaine S., Meinnel T. (2015). N-terminal protein modifications: Bringing back into play the ribosome. Biochimie 114: 134–146. [DOI] [PubMed] [Google Scholar]
- Glover C.J., Hartman K.D., Felsted R.L. (1997). Human N-myristoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction. J. Biol. Chem. 272: 28680–28689. [DOI] [PubMed] [Google Scholar]
- Graham J.M. (2001). Isolation of Golgi membranes from tissues and cells by differential and density gradient centrifugation. Curr. Protoc. Cell Biol. 10: 3.9.1–3.9.24. [DOI] [PubMed] [Google Scholar]
- Hannoush R.N. (2015). Synthetic protein lipidation. Curr. Opin. Chem. Biol. 28: 39–46. [DOI] [PubMed] [Google Scholar]
- Held K., Pascaud F., Eckert C., Gajdanowicz P., Hashimoto K., Corratgé-Faillie C., Offenborn J.N., Lacombe B., Dreyer I., Thibaud J.B., Kudla J. (2011). Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 21: 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P.A. (2015). The importance of lipid modified proteins in plants. New Phytol. 205: 476–489. [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Weimar T., Lilley K.S., Dupree P., Grierson C.S. (2013). A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol. 197: 805–814. [DOI] [PubMed] [Google Scholar]
- Herrmann J.M., Riemer J. (2012). Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J. Biol. Chem. 287: 4426–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Liu J., Halfter U., Kim C.S., Shi W., Zhu J.K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanneau J.P., Péaud-Lenoël C. (1967). Croissance et synthèse des protéines de suspensions cellulaires de tabac sensibles à la kinétine. Physiol. Plant. 20: 834–850. [Google Scholar]
- Jurgens G. (2004). Membrane trafficking in plants. Annu. Rev. Cell Dev. Biol. 20: 481–504. [DOI] [PubMed] [Google Scholar]
- Kang G.H., Son S., Cho Y.H., Yoo S.D. (2015). Regulatory role of BOTRYTIS INDUCED KINASE1 in ETHYLENE INSENSITIVE3-dependent gene expression in Arabidopsis. Plant Cell Rep. 34: 1605–1614. [DOI] [PubMed] [Google Scholar]
- Kato M., Nagasaki-Takeuchi N., Ide Y., Maeshima M. (2010). An Arabidopsis hydrophilic Ca2+-binding protein with a PEVK-rich domain, PCaP2, is associated with the plasma membrane and interacts with calmodulin and phosphatidylinositol phosphates. Plant Cell Physiol. 51: 366–379. [DOI] [PubMed] [Google Scholar]
- Kierszniowska S., Seiwert B., Schulze W.X. (2009). Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Mol. Cell. Proteomics 8: 612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Kurata Y., Nakabayashi J., Kagawa H., Hirano H. (2016). N-Myristoylation of the Rpt2 subunit of the yeast 26S proteasome is implicated in the subcellular compartment-specific protein quality control system. J. Proteomics 130: 33–41. [DOI] [PubMed] [Google Scholar]
- Kong X., Pan J., Cai G., Li D. (2012). Recent insights into brassinosteroid signaling in plants: its dual control of plant immunity and stomatal development. Mol. Plant 5: 1179–1181. [DOI] [PubMed] [Google Scholar]
- Lanyon-Hogg T., Faronato M., Serwa R.A., Tate E.W. (2017). Dynamic protein acylation: new substrates, mechanisms, and drug targets. Trends Biochem. Sci. 42: 566–581. [DOI] [PubMed] [Google Scholar]
- Li C.H., Chiang C.P., Yang J.Y., Ma C.J., Chen Y.C., Yen H.E. (2014). RING-type ubiquitin ligase McCPN1 catalyzes UBC8-dependent protein ubiquitination and interacts with Argonaute 4 in halophyte ice plant. Plant Physiol. Biochem. 80: 211–219. [DOI] [PubMed] [Google Scholar]
- Li Y., Gou M., Sun Q., Hua J. (2010). Requirement of calcium binding, myristoylation, and protein-protein interaction for the Copine BON1 function in Arabidopsis. J. Biol. Chem. 285: 29884–29891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Hrabak E.M. (2002). An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol. 128: 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Hrabak E.M. (2013). The myristoylated amino-terminus of an Arabidopsis calcium-dependent protein kinase mediates plasma membrane localization. Plant Mol. Biol. 82: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., Watkins K.P., Barkan A., van Wijk K.J. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 158: 156–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmagne A., Salvi D., Rolland N., Ephritikhine G., Joyard J., Barbier-Brygoo H. (2006). Purification and fractionation of membranes for proteomic analyses. Methods Mol. Biol. 323: 403–420. [DOI] [PubMed] [Google Scholar]
- Marmagne A., Ferro M., Meinnel T., Bruley C., Kuhn L., Garin J., Barbier-Brygoo H., Ephritikhine G. (2007). A high content in lipid-modified peripheral proteins and integral receptor kinases features in the arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 6: 1980–1996. [DOI] [PubMed] [Google Scholar]
- Martin D.D., Beauchamp E., Berthiaume L.G. (2011). Post-translational myristoylation: Fat matters in cellular life and death. Biochimie 93: 18–31. [DOI] [PubMed] [Google Scholar]
- Martinez A., Traverso J.A., Valot B., Ferro M., Espagne C., Ephritikhine G., Zivy M., Giglione C., Meinnel T. (2008). Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8: 2809–2831. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S., Eisenhaber B., Eisenhaber F. (2002a). N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 317: 541–557. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S., Eisenhaber B., Eisenhaber F. (2002b). N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J. Mol. Biol. 317: 523–540. [DOI] [PubMed] [Google Scholar]
- Mehlmer N., Wurzinger B., Stael S., Hofmann-Rodrigues D., Csaszar E., Pfister B., Bayer R., Teige M. (2010). The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 63: 484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Wong J.H., Feldman L.J., Lemaux P.G., Buchanan B.B. (2010). A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc. Natl. Acad. Sci. USA 107: 3900–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami A., Fujiwara M., Furuto A., Fukao Y., Yamashita T., Kamo M., Kawamura Y., Uemura M. (2009). Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol. 50: 341–359. [DOI] [PubMed] [Google Scholar]
- Morel J., Claverol S., Mongrand S., Furt F., Fromentin J., Bessoule J.J., Blein J.P., Simon-Plas F. (2006). Proteomics of plant detergent-resistant membranes. Mol. Cell. Proteomics 5: 1396–1411. [DOI] [PubMed] [Google Scholar]
- Nagasaki N., Tomioka R., Maeshima M. (2008). A hydrophilic cation-binding protein of Arabidopsis thaliana, AtPCaP1, is localized to plasma membrane via N-myristoylation and interacts with calmodulin and the phosphatidylinositol phosphates PtdIns(3,4,5)P(3) and PtdIns(3,5)P(2). FEBS J. 275: 2267–2282. [DOI] [PubMed] [Google Scholar]
- Peitzsch R.M., McLaughlin S. (1993). Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry 32: 10436–10443. [DOI] [PubMed] [Google Scholar]
- Peng T., Thinon E., Hang H.C. (2016). Proteomic analysis of fatty-acylated proteins. Curr. Opin. Chem. Biol. 30: 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre M., Traverso J.A., Boisson B., Domenichini S., Bouchez D., Giglione C., Meinnel T. (2007). N-myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell 19: 2804–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R., Guerra D.D., Yu S., Wogulis M., Kraft E., Frommer W.B., Callis J., Pilot G. (2012). The ubiquitin E3 ligase LOSS OF GDU2 is required for GLUTAMINE DUMPER1-induced amino acid secretion in Arabidopsis. Plant Physiol. 158: 1628–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D., Dubiella U., Kim S.H., Sloss D.I., Dowen R.H., Dixon J.E., Innes R.W. (2014). Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on s-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol. 164: 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renna L., Stefano G., Majeran W., Micalella C., Meinnel T., Giglione C., Brandizzi F. (2013). Golgi traffic and integrity depend on N-myristoyl transferase-1 in Arabidopsis. Plant Cell 25: 1756–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running M.P. (2014). The role of lipid post-translational modification in plant developmental processes. Front. Plant Sci. 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann F., Stalder U., Piotrowski M., Oecking C., Schaller A. (2002). LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 129: 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858. [DOI] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., Zhao T., Katagiri F., Tang D. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvius J.R., l’Heureux F. (1994). Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry 33: 3014–3022. [DOI] [PubMed] [Google Scholar]
- Sreeramulu S., Mostizky Y., Sunitha S., Shani E., Nahum H., Salomon D., Hayun L.B., Gruetter C., Rauh D., Ori N., Sessa G. (2013). BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74: 905–919. [DOI] [PubMed] [Google Scholar]
- Stael S., Bayer R.G., Mehlmer N., Teige M. (2011). Protein N-acylation overrides differing targeting signals. FEBS Lett. 585: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D., van Wijk K.J. (2009). PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37: D969–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D., Rafiqi M., Hurley U., Lawrence G.J., Bernoux M., Hardham A.R., Ellis J.G., Dodds P.N., Jones D.A. (2012). N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol. Plant Microbe Interact. 25: 379–392. [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Traverso J.A., Giglione C., Meinnel T. (2013a). High-throughput profiling of N-myristoylation substrate specificity across species including pathogens. Proteomics 13: 25–36. [DOI] [PubMed] [Google Scholar]
- Traverso J.A., Micalella C., Martinez A., Brown S.C., Satiat-Jeunemaître B., Meinnel T., Giglione C. (2013b). Roles of N-terminal fatty acid acylations in membrane compartment partitioning: Arabidopsis h-type thioredoxins as a case study. Plant Cell 25: 1056–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugama D., Liu S., Takano T. (2012a). A putative myristoylated 2C-type protein phosphatase, PP2C74, interacts with SnRK1 in Arabidopsis. FEBS Lett. 586: 693–698. [DOI] [PubMed] [Google Scholar]
- Tsugama D., Liu H., Liu S., Takano T. (2012b). Arabidopsis heterotrimeric G protein β subunit interacts with a plasma membrane 2C-type protein phosphatase, PP2C52. Biochim. Biophys. Acta 1823: 2254–2260. [DOI] [PubMed] [Google Scholar]
- Turnbull D., Hemsley P.A. (2017). Fats and function: protein lipid modifications in plant cell signalling. Curr. Opin. Plant Biol. 40: 63–70. [DOI] [PubMed] [Google Scholar]
- Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20: 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig R.G., Labandera A.M., Tang L.Y., Sieben N.A., Goudreault M., Yeung E., Gingras A.C., Samuel M.A., Moorhead G.B. (2017). Activation of mitochondrial protein phosphatase SLP2 by MIA40 regulates seed germination. Plant Physiol. 173: 956–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J.A., et al. (2013). The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41: D1063–D1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox C., Hu J.S., Olson E.N. (1987). Acylation of proteins with myristic acid occurs cotranslationally. Science 238: 1275–1278. [DOI] [PubMed] [Google Scholar]
- Witte C.P., Keinath N., Dubiella U., Demoulière R., Seal A., Romeis T. (2010). Tobacco calcium-dependent protein kinases are differentially phosphorylated in vivo as part of a kinase cascade that regulates stress response. J. Biol. Chem. 285: 9740–9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi S., Fusada N., Hayashi H., Utsumi T., Uozumi N., Endo Y., Tozawa Y. (2010). The consensus motif for N-myristoylation of plant proteins in a wheat germ cell-free translation system. FEBS J. 277: 3596–3607. [DOI] [PubMed] [Google Scholar]
- Yin C., Karim S., Zhang H., Aronsson H. (2017). Arabidopsis RabF1 (ARA6) is involved in salt stress and dark-induced senescence (DIS). Int. J. Mol. Sci. 18: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X.J., et al. (2007). Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19: 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha J., Weiler S., Oh K.J., Wei M.C., Korsmeyer S.J. (2000). Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science 290: 1761–1765. [DOI] [PubMed] [Google Scholar]