Pseudomonas syringae activates autophagy in an effector-dependent manner to degrade proteasomes for enhanced pathogenicity, whereas NBR1-dependent autophagic processes counteract disease progression.
Abstract
Autophagy and the ubiquitin-proteasome system (UPS) are two major protein degradation pathways implicated in the response to microbial infections in eukaryotes. In animals, the contribution of autophagy and the UPS to antibacterial immunity is well documented and several bacteria have evolved measures to target and exploit these systems to the benefit of infection. In plants, the UPS has been established as a hub for immune responses and is targeted by bacteria to enhance virulence. However, the role of autophagy during plant-bacterial interactions is less understood. Here, we have identified both pro- and antibacterial functions of autophagy mechanisms upon infection of Arabidopsis thaliana with virulent Pseudomonas syringae pv tomato DC3000 (Pst). We show that Pst activates autophagy in a type III effector (T3E)-dependent manner and stimulates the autophagic removal of proteasomes (proteaphagy) to support bacterial proliferation. We further identify the T3E Hrp outer protein M1 (HopM1) as a principle mediator of autophagy-inducing activities during infection. In contrast to the probacterial effects of Pst-induced proteaphagy, NEIGHBOR OF BRCA1-dependent selective autophagy counteracts disease progression and limits the formation of HopM1-mediated water-soaked lesions. Together, we demonstrate that distinct autophagy pathways contribute to host immunity and bacterial pathogenesis during Pst infection and provide evidence for an intimate crosstalk between proteasome and autophagy in plant-bacterial interactions.
INTRODUCTION
Autophagy is a major degradation and recycling pathway in eukaryotes that directs the bulk or selective engulfment of cytoplasmic content into double membrane vesicles, termed autophagosomes, for subsequent turnover (Klionsky and Codogno, 2013). The sequential steps of autophagosome formation and delivery to lytic compartments (i.e., vacuole or lysosome) rely on a complex set of membrane trafficking and fusion events and involve the coordinated action of conserved autophagy-related (ATG) proteins (Yin et al., 2016; Reggiori and Ungermann, 2017; Yu et al., 2017). For instance, two ubiquitin-like conjugation pathways produce ATG12-ATG5-ATG16 complexes and lipidated ATG8 proteins required for the expansion and sealing of the isolation membrane (or phagophore) around the nearby cellular cargo (Mizushima and Komatsu, 2011). In addition, membrane-anchored ATG8 acts as an important docking site for selective autophagy receptors that deliver a multitude of substrates to the growing autophagosome, including single or aggregated proteins, entire organelles, and invading microbes (Zaffagnini and Martens, 2016). In plants, NEIGHBOR OF BRCA1 (NBR1) is the best characterized cargo receptor and functions in the degradation of polyubiquitinated protein aggregates (aggrephagy) as well as viral components and particles (xenophagy) (Svenning et al., 2011; Zhou et al., 2013; Hafrén et al., 2017, 2018). Recent findings also revealed that the ubiquitin-binding proteasome subunit REGULATORY PARTICLE NON-ATPASE SUBUNIT10 (RPN10) acts as a specific autophagy receptor for the degradation of proteasomes (proteaphagy) in response to chemical or genetic proteasome inhibition (Marshall et al., 2015). This interplay between both major cellular degradation pathways appears to be conserved in other eukaryotes as malfunctioning proteasomes are also degraded in yeast and mammals, albeit via different cargo receptors (Cohen-Kaplan et al., 2016; Marshall et al., 2016).
Altered expression of ATG and cargo receptor genes has been widely explored to dissect the functions and mechanisms of autophagy processes. These studies have established important roles for autophagy in cellular homeostasis, development, metabolism, and stress adaptation in various eukaryotic organisms (Boya et al., 2013; Klionsky and Codogno, 2013). In addition, autophagy is induced in response to a wide range of pathogens and contributes to various aspects of adaptive and innate immunity during animal infections (Levine et al., 2011; Gomes and Dikic, 2014). In turn, several intracellular viruses and bacteria have evolved measures to suppress and evade antimicrobial autophagy or even hijack autophagic processes for enhanced pathogenicity (Dong and Levine, 2013; Mostowy, 2013). In plants, autophagy was initially ascribed to the regulation of the hypersensitive response as part of effector-triggered immunity against avirulent oomycete, viral, and bacterial pathogens (Liu et al., 2005; Hofius et al., 2009; Kwon et al., 2013; Han et al., 2015). Subsequently, autophagy was shown to be involved in basal resistance and the control of disease-associated cell death upon infection with necrotrophic fungi (Lai et al., 2011; Lenz et al., 2011; Li et al., 2016). The identification of an ATG8-interacting oomycete effector that antagonizes the NBR1 autophagy receptor further indicated an important role of selective autophagy in defense responses (Dagdas et al., 2016). In support of this notion, NBR1 was also found to function in antiviral immunity by targeting the viral capsid protein and particles of Cauliflower mosaic virus (CaMV) for xenophagic degradation (Hafrén et al., 2017). However, NBR1-independent bulk autophagy promotes host survival during CaMV infection and thus serves as a proviral pathway by extending the time span for particle production and potential vector transmission (Hafrén et al., 2017).
Despite recent advances in the understanding of autophagy during compatible interactions of plants with oomycetes, fungi, and viruses (Zhou et al., 2014; Hofius et al., 2017), the functions of bulk and selective autophagy pathways upon virulent bacterial infection remained unclear. Autophagy-deficient Arabidopsis thaliana mutants such as atg5 were previously shown to display enhanced resistance to infection with Pseudomonas syringae pv tomato DC3000 (Pst). This phenotype has been linked to a potential, as yet unknown role of autophagy in the negative regulation of salicylic acid (SA) levels and defense signaling (Yoshimoto et al., 2009; Lenz et al., 2011). Notably, Pst secretes several type III effector (T3E) proteins to suppress proteasome activity and block SA signaling (Üstün et al., 2016), probably mediated by the impaired turnover of the defense regulator NONEXPRESSOR OF PATHOGENESIS-RELATED1 (NPR1), as seen during Xanthomonas campestris infection (Üstün et al., 2013; Üstün and Börnke, 2015). These findings imply a potential crosstalk between autophagy and proteasome functions during bacterial infections, the specifics of which are not yet known.
In this study, we investigated whether and how Pst alters autophagy responses to interfere with host immunity and enhance pathogenicity in Arabidopsis. Our results show that Pst induces autophagy and increases autophagic flux upon delivery of the T3E HopM1. We further demonstrate that HopM1 stimulates proteaphagy, thus providing the mechanistic basis for Pst-mediated inhibition of proteasome function that suppresses immune responses in plants. While Pst-induced autophagy promotes bacterial proliferation, NBR1-mediated selective autophagic processes seem to counteract it by suppressing the formation of HopM1-induced water-soaked lesions. Our data indicate that distinct selective autophagy pathways operate simultaneously during virulent P. syringae infection and have opposing functions in plant immunity and bacterial pathogenesis.
RESULTS
Autophagy Is Required for Pst-Induced Proteasome Inhibition
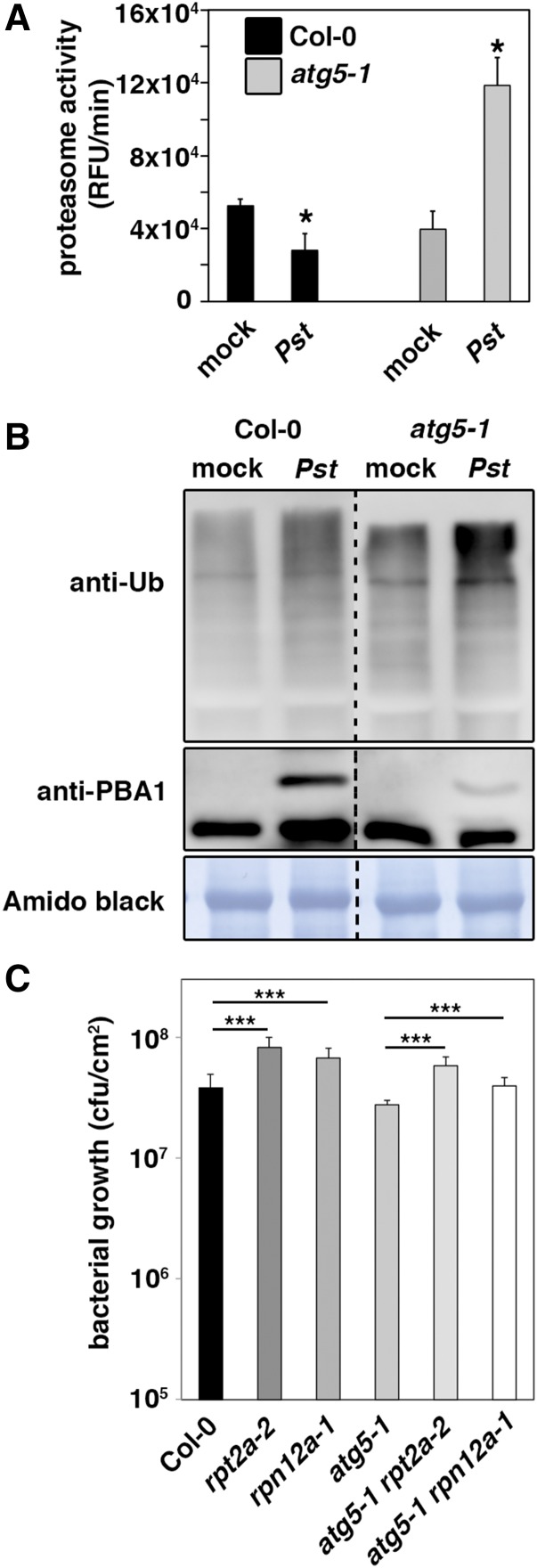
To analyze the role of autophagy and its potential interplay with the ubiquitin-proteasome system during bacterial infection, we first assayed whether the previously observed impact of Pst on proteasome function (Üstün et al., 2016) is altered in Arabidopsis mutants defective in the core autophagy machinery. As expected, Pst infection inhibited proteasome activity in Col-0 wild-type plants, determined by cleavage of a fluorogenic peptide (Suc-LLVY-AMC) via the 20S proteasome. This suppressive effect was reversed in atg5-1 (Figure 1A), atg2-1, and atg7-2 null mutant plants (Supplemental Figure 1A), leading to a higher proteasome activity upon infection compared with Col-0. Notably, loss of function of NPR1 or the SA biosynthetic gene SALICYLIC ACID INDUCTION DEFICIENT2 (SID2) in atg5-1 npr1-1 and atg5-1 sid2 double mutants did not change the proteasome response to Pst infection compared with atg5-1, indicating that the phenotype occurs independently of SA levels and SA-mediated defense signaling (Supplemental Figure 1B). We then analyzed the accumulation of the proteasome subunit PBA1, whose posttranslational cleavage at the N terminus is blocked upon chemical or bacterial suppression of proteasome activity and maturation (Book et al., 2010; Üstün et al., 2016). The unprocessed form of PBA1 was still detectable in the atg5-1 mutant but accumulated to a substantially lower amount compared with Col-0 (Figure 1B), supporting the observed inability of Pst to reduce proteasome activity in the autophagy-deficient background. Furthermore, accumulation of ubiquitinated proteins during Pst infection was increased in atg5 relative to Col-0 (Figure 1B), indicating that Pst stimulates autophagic degradation of ubiquitinated target proteins. Finally, the enhanced resistance phenotype of atg5 plants to Pst infection (Lenz et al., 2011) was reverted by introducing the proteasome subunit loss-of-function mutations rpt2a-2 and rpn12a-1 (Kurepa et al., 2008) (Figure 1C). Collectively, these results show that Pst-induced proteasome inactivation to enhance virulence (Üstün et al., 2016) is dependent on a functional autophagy pathway.
Figure 1.
Pst-Induced Proteasome Suppression Is Dependent on Autophagy.
(A) Proteasome activity in leaves of Col-0 and atg5-1 plants upon infection with either Pst wild-type bacteria or mock. Leaf samples were taken 1 d postinoculation (dpi), and the relative proteasome activity was determined. Bars represent means ± sd (n = 3 biological replicates). MgCl2 infiltration served as the mock control. Asterisks indicate statistical significance (*P < 0.05) determined by Student’s t test (compared with mock control). The experiment was repeated at least three times with similar results.
(B) Immunoblot analysis of the 20S proteasome subunit PBA1 and ubiquitinated proteins in Col-0 and atg5-1 leaves upon infection with Pst and mock. Total proteins were extracted from infiltrated leaves at 1 dpi, separated by SDS-PAGE, and probed with specific anti-PBA1 and anti-Ub antibodies. Mock-infected plants served as control, and amido black staining verified equal protein loading. Immunoblot analysis was reproduced at least three times with comparable results.
(C) Growth of Pst in 4- to 5-week-old Col-0, rpt2a-2, rpn12a-1, atg5-1, atg5-1 rpt2a-2, and atg5-1 rpn12a-1 plants 3 d after syringe infiltration at OD600 = 0.0001. Bars represent means ± sd (n = 4 biological replicates), and asterisks indicate statistical significance (***P < 0.001) determined by Student’s t test. The resistance assay was done twice with comparable results.
Pst-Induced Autophagy Promotes Plant Susceptibility
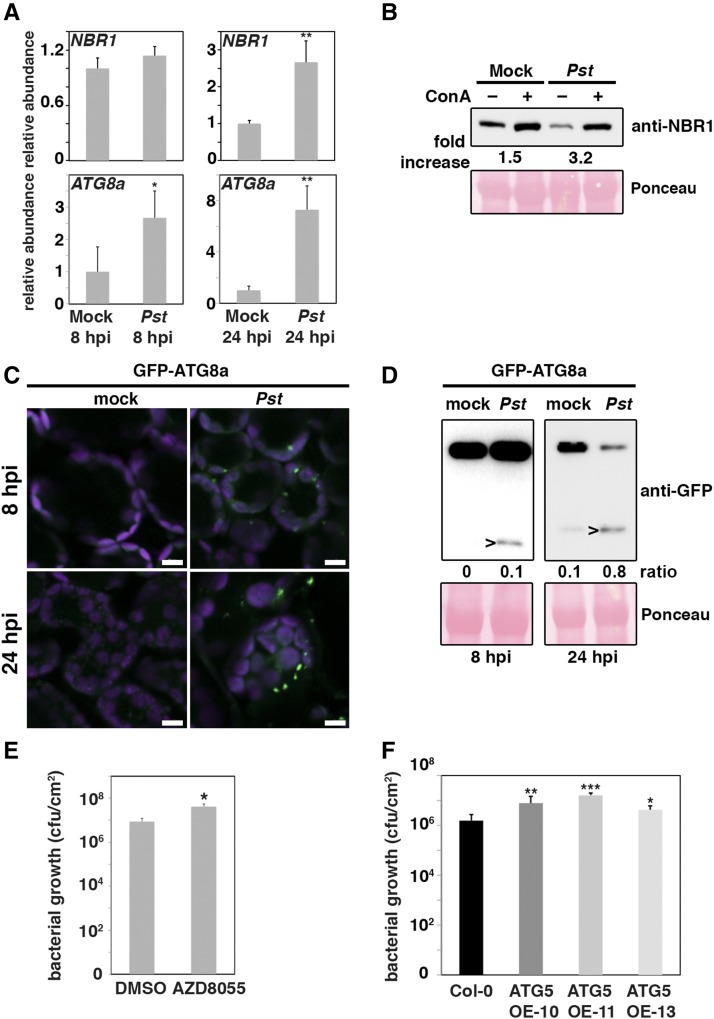
The requirement of autophagy for Pst-induced proteasome suppression in Arabidopsis prompted us to investigate whether autophagy is induced during Pst infection. We first monitored the expression of the autophagy core gene ATG8a and the selective autophagy receptor NBR1, which represent well-established markers for the autophagy response (Thompson et al., 2005; Zhou et al., 2013; Hafrén et al., 2017). Transcript levels of both genes were increased upon bacterial infection (Figure 2A), with ATG8a showing an earlier accumulation compared with NBR1. Importantly, the steady state level of NBR1 protein appeared diminished during Pst infection, suggesting enhanced NBR1 turnover (Figure 2B). Indeed, stabilization of NBR1 by treatment with concanamycin A (ConA), an inhibitor of vacuolar acidification and hence autophagic body degradation, verified Pst-induced enhancement of autophagic flux (Svenning et al., 2011; Minina et al., 2013). We then infected a transgenic line expressing GFP-ATG8a and found that Pst stimulated the formation of GFP-labeled autophagosomal structures compared with the noninfected control (Figure 2C). In addition, we monitored by immunoblotting the release of free GFP from the GFP-ATG8a fusion protein, which serves as marker for vacuolar turnover of autophagic bodies (Chung et al., 2010). Detection of an increased free GFP/GFP-ATG8a ratio at early and particularly at later stages of infection (24 h postinoculation [hpi]) compared with the mock control further indicated an enhanced autophagy activity and flux upon Pst challenge (Figure 2D).
Figure 2.
Pst Activates Autophagy to Enhance Plant Susceptibility.
(A) RT-qPCR analysis of ATG8a and NBR1 transcript levels in Col-0 upon challenge with Pst at 8 and 24 hpi compared with mock-infected plants. Values represent means ± sd (n = 4) relative to mock control and were normalized to PP2A. Statistical significance (*P < 0.05 and **P < 0.01) was revealed by Student’s t test. The experiment was repeated at least twice with similar results.
(B) Immunoblot analysis of NBR1 protein levels in 14-d-old Col-0 seedlings. Seedlings were treated with DMSO (−) or ConA at 10 hpi for additional 14 h. Numbers correspond to the fold increase of NBR1 in ConA-treated samples in comparison to the respective DMSO control. Ponceau S staining served as a loading control and for normalization of NBR1 signal intensities. The experiment was repeated at least three times with similar results.
(C) Confocal microscopy detection of GFP-ATG8a-labeled puncta in stably expressing Col-0 seedlings at 8 and 24 hpi with Pst compared with mock treatment. Images represent single confocal planes from abaxial epidermal cells and were acquired with identical microscope settings. The experiment was repeated at least three times with similar results. Bars = 10 μm.
(D) Immunoblot analysis of GFP-ATG8a processing in transgenic Col-0 plants at 8 and 24 hpi with Pst compared with mock treatment. Total proteins were extracted from infiltrated leaves and probed with anti-GFP antibody. Arrowheads indicate free GFP at 25 kD and Ponceau S staining of RbcL served as a loading control and for normalization of signal intensities. The ratio between free GFP and GFP-ATG8a is indicated. Immunoblot analysis was repeated three times with equal results.
(E) Bacterial growth in 5-week-old Col-0 plants in the presence or absence of the TOR inhibitor AZD8055 (final concentration 15 μM). Leaves were syringe-infiltrated with Pst at OD600 = 0.0001, and colony-forming units were determined at 3 dpi. Bars represent means ± sd (n = 3). Asterisks indicate statistical significance (**P < 0.01) determined by Student’s t test in comparison to Col-0. The experiment was repeated twice with similar results.
(F) Bacterial growth in 6-week-old Col-0 and ATG5 overexpression (OE) lines 10, 11, and 13. Leaves were spray-infiltrated with Pst at OD600 = 0.1, and colony-forming units were determined at 3 dpi. Bars represent means ± sd (n = 4). Asterisks indicate statistical significance (***P < 0.001, **P < 0.01, and *P < 0.05) determined by Student’s t test in comparison to Col-0.
To assess the relevance of stimulated autophagy during infection, we determined bacterial growth in plants treated with AZD8055, an inhibitor of the negative autophagy regulator TARGET OF RAPAMYCIN (TOR), which is known to promote autophagic flux (Liu and Bassham, 2010; Pu et al., 2017; Supplemental Figure 2). Pst growth was slightly but significantly enhanced by AZD8055-mediated autophagy activation in Col-0 plants (Figure 2E), whereas inhibitor treatment did not affect bacterial proliferation in atg5-1 (Supplemental Figure 3). To further exclude autophagy-independent TOR signaling effects on the altered resistance response, we analyzed bacterial growth in ATG5 overexpressing Arabidopsis plants that show constitutive upregulation of autophagy (Minina et al., 2018). Consistent with the results from the AZD8055 inhibitor treatment, we observed significantly increased proliferation of Pst in independent transgenic lines compared with the wild-type control (Figure 2F). Together, these data show that Pst-induced autophagy promotes bacterial susceptibility in Arabidopsis.
Pst Effector Proteins Drive Autophagy Induction
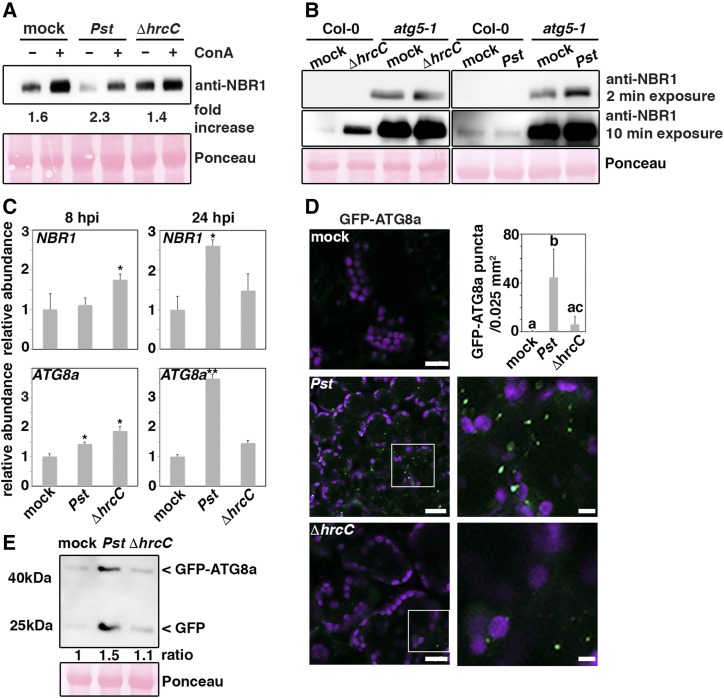
We next addressed whether Pst triggers autophagy through the activity of T3E proteins. T3Es are delivered into the plant cell to suppress pathogen-associated molecular pattern (PAMP)-triggered immunity and to manipulate other cellular functions for enhanced virulence (Macho, 2016). We therefore analyzed the autophagy responses to nonpathogenic PstΔhrcC, a mutant strain defective in the T3 secretion system and thus incapable of effector translocation. In contrast to wild-type Pst, NBR1 protein levels were slightly increased upon infection with PstΔhrcC and showed only a minor response to ConA treatment comparable to that of the mock control (Figure 3A). In addition, PstΔhrc infection did not further increase NBR1 accumulation in the atg5 mutant background as detected for the wild-type strain (Figure 3B). Transcriptional upregulation of ATG8a upon infection with PstΔhrcC was reduced compared with Pst, while NBR1 expression was increased only at early infection time points, possibly explaining the elevated NBR1 protein levels at steady state (Figure 3C). Furthermore, autophagosomal structures labeled with GFP-ATG8a were only slightly enhanced (Figure 3D), and accumulation of the GFP-ATG8a fusion protein as well as free GFP release was hardly affected by PstΔhrcC in comparison to the mock control (Figure 3E). This minor autophagic response upon PstΔhrcC challenge suggests that autophagy is stimulated by the delivery of T3Es. Indeed, treatment with the bacterial PAMP flg22 or heat-inactivated wild-type Pst did not induce the formation of GFP-ATG8a puncta to similar levels as seen upon Pst infection (Supplemental Figure 4). Together, these results indicate that Pst-induced enhancement of autophagy is mediated by T3Es and is part of a bacterial virulence strategy.
Figure 3.
Autophagy Activation Is Mediated by the Secretion of T3Es.
(A) Immunoblot analysis of NBR1 protein levels in 14-d-old Col-0 seedlings upon flood inoculation with Pst and PstΔhrcC strains compared with the mock-infected control. Seedlings were treated with DMSO (–) or ConA (+) at 10 hpi for an additional 14 h. Numbers correspond to the fold increase of NBR1 in ConA-treated samples in comparison to the respective DMSO control. Ponceau staining was used as loading control and quantified for normalization of NBR1 signal intensities. The experiment was repeated at least three times with similar results.
(B) Immunoblot analysis of NBR1 protein levels in Col-0 and atg5-1 backgrounds upon infection with Pst and PstΔhrcC. Infiltrated leaves were sampled at 1 dpi and subjected to immunoblot analysis using an anti-NBR1 antibody. Ponceau S staining of RbcL served as a loading control. Immunoblot analysis was repeated twice with similar results.
(C) RT-qPCR analysis of NBR1 and ATG8a transcript levels in Col-0 plants upon challenge with Pst and PstΔhrcC at 8 and 24 hpi compared with mock-infected plants. Values represent mean ± sd (n = 4) relative to mock control and were normalized to PP2A. Statistical significance (*P < 0.05 and **P < 0.01) was revealed by Student’s t test compared the Col-0 mock control. The analysis was repeated twice with similar results.
(D) Detection of GFP-ATG8a-labeled autophagosomal structures in transgenic Col-0 seedlings infected with Pst and PstΔhrcC strains compared with mock control. Images represent single confocal planes from abaxial epidermal cells and were taken with identical microscope settings (bars = 10 μm). Images in the right panel show magnifications of boxed areas (bars = 5 μm). GFP-ATG8a puncta were quantified in areas of 0.025 mm2 from mock-, Pst-, and PstΔhrcC-infected leaves (upper right panel) and are presented as means ± sd (n = 8 independent areas). Different letters indicate statistically significant differences (P < 0.05) as determined by one-way ANOVA. The experiment was repeated at least three times with similar results.
(E) Immunoblot analysis of GFP-ATG8a processing in transgenic Col-0 plants at 8 hpi with Pst and PstΔhrcC compared with mock treatment. Total proteins were extracted from infiltrated leaves and probed with an anti-GFP antibody. The ratio between free GFP and GFP-ATG8a is indicated. Ponceau S staining of RbcL served as a loading control. Immunoblot analysis was repeated twice with similar results.
The Pst Effector HopM1 Mediates Autophagy Activation
To identify potential T3Es that are responsible for autophagy modulation, we utilized a quantitative autophagy assay in the Nicotiana benthamiana system. This assay is based on the Agrobacterium tumefaciens-mediated transient expression of Renilla luciferase (RLUC) fused to ATG8a together with free Firefly luciferase (FLUC). While RLUC-ATG8a is sequestered into autophagosomes and targeted for vacuolar degradation, cytoplasmic FLUC serves as an internal reference for protein expression. The readout of autophagic flux is derived from the ratio of RLUC-ATG8a to FLUC levels and compared in the presence or absence of the coexpressed effector protein.
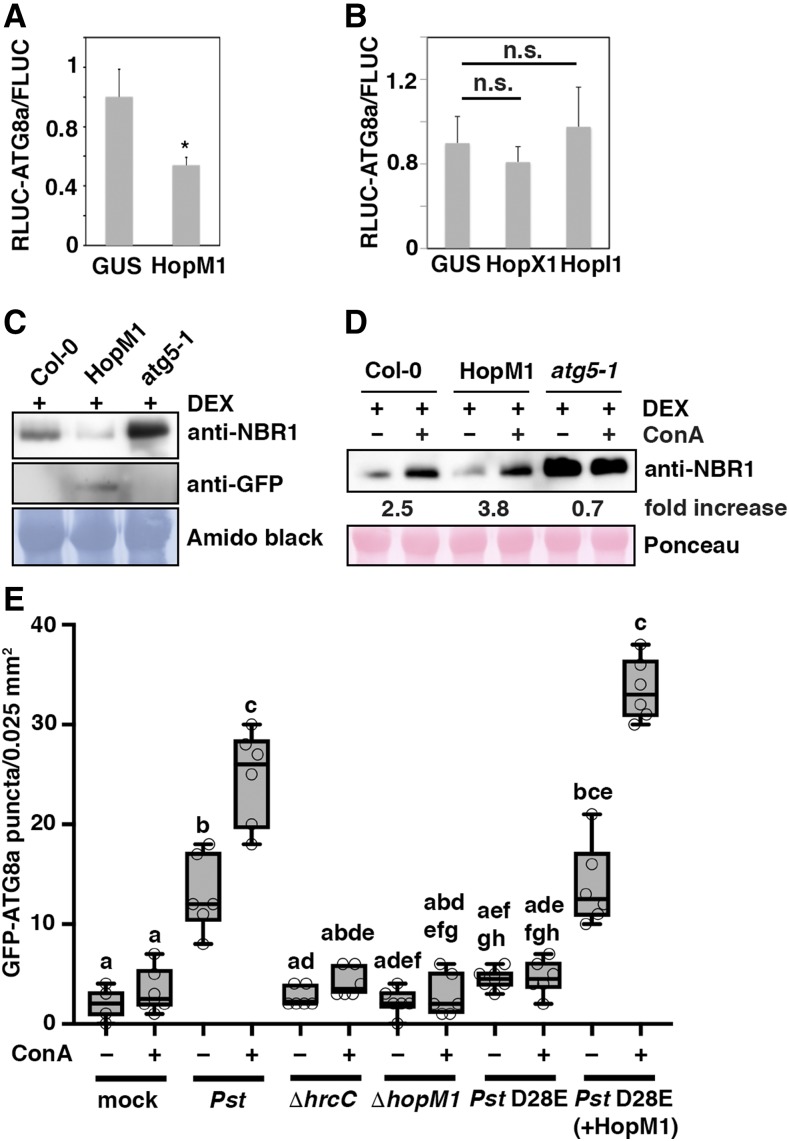
Based on our observation that Pst-induced proteasome suppression requires autophagy, we speculated that effector proteins previously identified as suppressors of proteasome activity (Üstün et al., 2016) might equally well be autophagy modulators. We therefore selected HopM1 as a candidate, given that it has the strongest proteasome inhibiting capacity (Üstün et al., 2016) and suppresses SA-dependent defense responses (DebRoy et al., 2004). Measurement of luciferase activities in N. benthamiana leaves upon coexpression of HopM1 and the autophagy reporter constructs revealed a significant decrease in the RLUC-ATG8a/FLUC ratio, indicating increased autophagy activity (Figure 4A). In contrast, the T3Es HopX1 and Hopl1, capable of suppressing SA-mediated defenses (Jelenska et al., 2007; Gimenez-Ibanez et al., 2014) but not proteasome activity (Üstün et al., 2016), did not alter the RLUC-ATG8a/FLUC ratio (Figure 4B). Expression of all T3Es used in the quantitative autophagy assay was verified by immunoblot analysis (Supplemental Figure 5). This finding indicated that the modulation of autophagy is not a general feature of T3E interfering with SA signaling.
Figure 4.
Induction of Autophagy Is Dependent on the Delivery of the T3E HopM1.
(A) and (B) T3E-mediated influence on autophagic flux determined by the quantitative dual-luciferase assay. RLUC-ATG8a and FLUC constructs were coexpressed with HopM1 (A) or HopX1 and HopI1 (B) in N. benthamiana leaves in comparison to the GUS control, and Renilla and Firefly luciferase activities were simultaneously measured in leaf extracts at 48 h postinfiltration using the dual-luciferase system. Values represent the mean ratio of RLUC-ATG8a and FLUC activities and error bars show sd (n = 3). The significantly decreased ratio (*P < 0.05, Student’s t test) in the presence of HopM1 indicates increased autophagic flux due to enhanced turnover of vacuole-targeted RLUC-ATG8a protein. n.s., not significant.
(C) Immunoblot analysis of NBR1 protein levels in Col-0, transgenic DEX:HopM1-GFP, and atg5-1 seedlings upon treatment with dexamethasone (DEX; 30 μM) using an anti-NBR1 antibody. Induced expression of HopM1-GFP was validated with an anti-GFP antibody. The experiment was repeated twice with similar results.
(D) Immunoblot analysis of NBR1 protein levels in Col-0, transgenic DEX:HopM1-GFP, and atg5-1 seedlings upon treatment with DEX (30 μM) in combination with either DMSO (–) or ConA (+) for 14 h. Numbers correspond to the fold increase of NBR1 in ConA-treated samples in comparison to the respective DMSO control. Ponceau staining was used as loading control and for normalization of NBR1 signal intensities. The experiment was repeated three times with similar results.
(E) GFP-ATG8a-labeled autophagosomal structures were quantified from plants infected with the indicated bacterial strains 6 hpi in the presence or absence of ConA. Puncta were calculated in single planes of independent areas (n = 6 per condition) using ImageJ. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles and data points are plotted as open circles. Different letters indicate statistically significant differences (P < 0.05) as determined by one-way ANOVA. The experiment was repeated twice with similar results.
To verify the effect of HopM1 on autophagy levels, we used a dexamethasone-inducible HopM1-GFP transgenic Arabidopsis line (Nomura et al., 2006, 2011). HopM1 expression resulted in decreased NBR1 protein accumulation at steady state (Figure 4C) that could be reverted to control level in the presence of ConA, thus confirming enhanced autophagy flux (Figure 4D). To verify that HopM1 also activates autophagic degradation in the infection context, we took advantage of a Pst strain that lacks 28 T3Es (Pst D28E) and can be engineered for delivery of single effectors (Cunnac et al., 2011). Pst D28E containing HopM1 increased the formation of GFP-ATG8a-labeled autophagosomal structures to a similar extent as the wild-type Pst strain in comparison to PstΔhrcC, PstΔhopM1, PstD28E, and mock controls (Figure 4E; Supplemental Figure 6). The dramatic rise in GFP-ATG8a puncta upon ConA treatment of HopM1-delivering PstD28E strongly substantiates the finding that HopM1 activates autophagic flux and suggests that this T3E is the major contributor to the autophagy-inducing capacity of Pst.
Pst Induces Proteaphagy during Infection
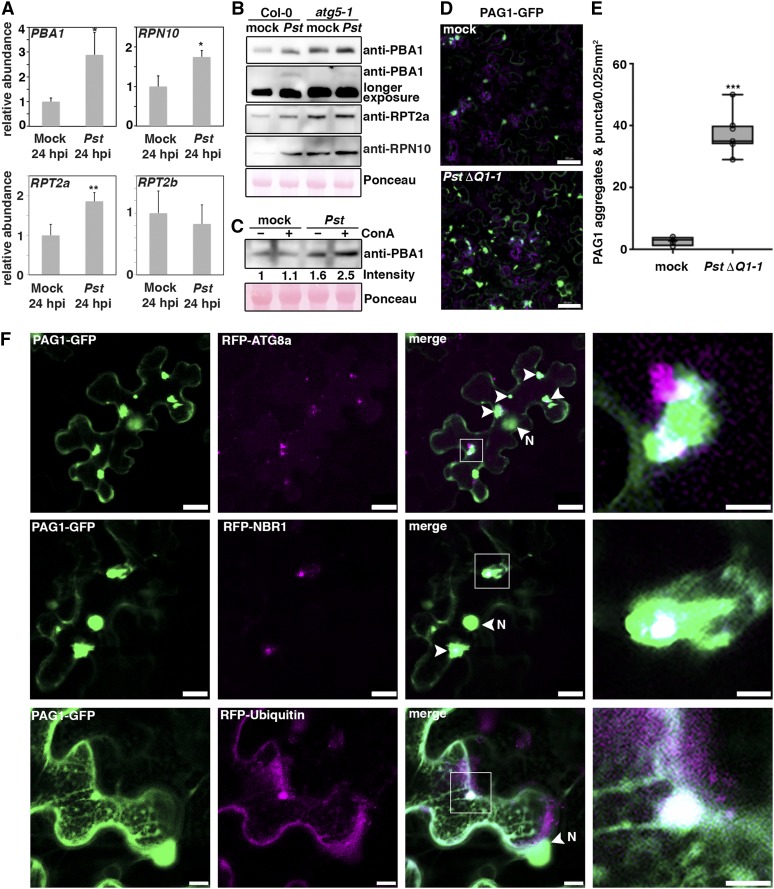
Autophagy has recently been shown to mediate the turnover of proteasomes during nutrient starvation and in response to chemical or genetic proteasome inhibition (Marshall et al., 2015). Since Pst suppresses proteasome activity and activates autophagy in Arabidopsis, we analyzed whether proteaphagy is induced during bacterial infection. Similar to treatment with the proteasome inhibitor MG132 (Gladman et al., 2016), Pst infection led to transcriptional upregulation of proteasome subunits at 8 and 24 hpi (Figure 5A; Supplemental Figure 7). Transcript levels of PBA1, RPT2a, and the proteaphagy receptor RPN10 were significantly increased, while expression of RPT2b, which is nonresponsive to proteasome inhibition (Gladman et al., 2016), remained unaltered during Pst infection (Figure 5A). Consistent with the transcriptional response, levels of Arabidopsis proteasome subunits were also elevated in Col-0 upon challenge with Pst (Figure 5B), but not with PstΔhrcC (Supplemental Figure 8), substantiating the T3E dependency of the phenotype. Notably, ConA treatment further increased PBA1 levels upon infection compared with mock control (Figure 5C), indicating that the transcriptional upregulation of proteasome subunits masks their autophagic degradation both in response to Pst infection and chemical proteasome inhibition (Marshall et al., 2015). Together, these results reveal that Pst activates proteaphagy to degrade proteasomes.
Figure 5.
P. syringae Infection Triggers Proteaphagy.
(A) RT-qPCR analysis of PBA1, RPN10, RPT2a, and RPT2b transcript levels upon challenge of Col-0 plants with Pst at 24 hpi compared with mock-infected plants. Values represent mean ± sd (n = 4) relative to the mock control and were normalized to PP2A. Statistical significance (*P < 0.05 and **P < 0.01) was revealed by Student’s t test (compared with mock control). The experiment was repeated twice with similar results.
(B) Immunoblot analysis of PBA1, RPT2a, and RPN10 protein levels in Col-0 and atg5-1 at 1 dpi with Pst. Proteins were extracted from leaves at 1 dpi and probed with specific antibodies. Ponceau S staining of RbcL served as a loading control. Immunoblots were repeated twice with similar results.
(C) Immunoblot analysis of PBA1 in the absence (–) and presence (+) of ConA during Pst infection. Proteins were extracted from leaves at 6 hpi and probed with specific antibodies. Numbers correspond to the relative signal intensities compared with the mock control without ConA. Ponceau S staining of RbcL was used as loading control and quantified for normalization of PBA1 signals. The experiment was repeated twice with similar results.
(D) Confocal analysis of transiently expressed PAG1-GFP in N. benthamiana at 6 hpi with PstΔhopQ1-1 compared with mock control. Images represent single confocal planes from abaxial epidermal cells and were acquired with identical microscope settings. The experiment was repeated at least three times with similar results. Bars = 50 μm.
(E) Quantification of PAG1-GFP aggregates in mock- and PstΔhopQ1-1-infected leaves analyzed in (C). The number of aggregates were calculated from single planes of independent leaf areas of 0.025 mm2 (n = 9) using ImageJ. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles and data points are plotted as open circles. Asterisks indicate statistical significance (***P < 0.001) determined by Student’s t test in comparison with the control.
(F) Colocalization analysis of PAG1-GFP with RFP-ATG8a, RFP-NBR1, or RFP-Ub in N. benthamiana leaves. After 2 d of transient expression, plants were infected with PstΔhopQ1-1 to induce PAG1-GFP aggregates, and imaging was done at 4 to 6 hpi (bars = 20 μm). Arrows indicate areas of partial colocalization, and N marks the nucleus that also shows GFP fluorescence. Images in the right column are magnifications of boxed areas (bars = 5 μm). The experiment was repeated three times with similar results.
To test whether Pst-induced reduction of proteasome activity results in autophagic clearance of proteasomes, we applied a cell biological approach in the N. benthamiana system to demonstrate the association of the proteasome with autophagy proteins during infection. For this purpose, we transiently expressed the GFP-tagged proteasome core particle subunit PAG1, which has previously been verified as a reliable proteasome reporter (Marshall et al., 2015). Subsequently, leaves were infected with a T3E HopQ1-1-deficient Pst strain (ΔhopQ1-1) that is fully virulent in N. benthamiana and also capable of inhibiting proteasome activity (Cunnac et al., 2011; Misas-Villamil et al., 2017). PstΔhopQ1-1 induced the formation of PAG1-GFP puncta and aggregates (Figures 5D and 5E) in a manner comparable to MG132 treatment (Supplemental Figure 9). Pst-induced aggregation of PAG1 also proved to be dependent on T3E secretion, as it was not observed upon infection with PstΔhrcC (Supplemental Figure 9), in agreement with the gene expression response of proteasome subunits at transcript and protein level. Importantly, we found colocalization of PAG1-GFP with RFP-ATG8a, RFP-NBR1, RFP-Ubiquitin (Figure 5F), and RFP-tagged ATG8e and ATG8g isoforms (Supplemental Figure 10). These results suggest that the Pst-induced degradation of proteasomes is linked to the autophagy pathway.
Finally, we tested whether loss of the proteaphagy receptor RPN10 impacts Pst-induced proteasome malfunction in Arabidopsis. Similar to other autophagy mutants, proteasome suppression by Pst was abolished in the weak rpn10-1 mutant allele (Smalle et al., 2003) that lacks the ubiquitin-interacting motifs required for binding of ubiquitylated proteasomes and ATG8 (Supplemental Figure 11A). Bacterial growth was elevated in rpn10-1 (Supplemental Figure 11B) similar to other proteasome subunit mutants compared with the wild type (Figure 1B; Üstün et al., 2016), suggesting that the function of RPN10 in proteaphagy is masked by additional pleiotropic functions as ubiquitin receptor in targeting substrates to the proteasome (Smalle et al., 2003).
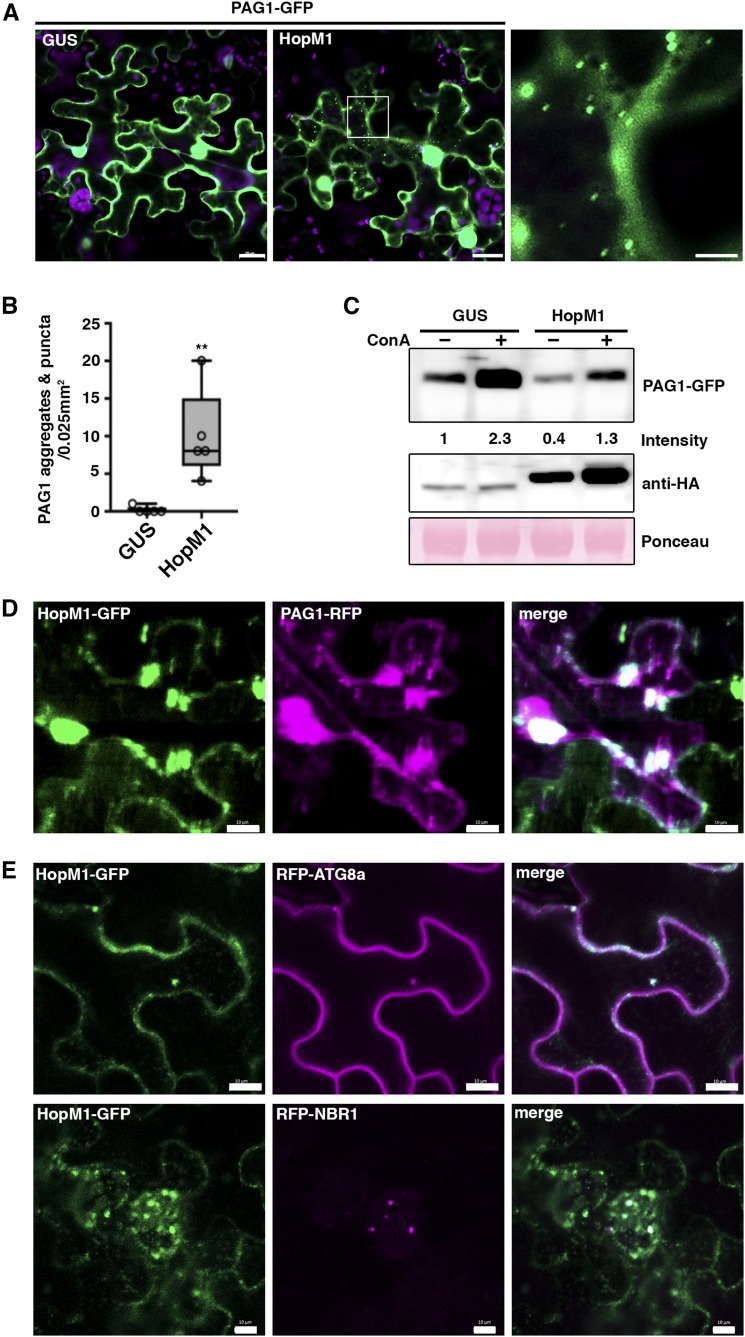
HopM1 Induces Proteaphagy
As HopM1 contributes to both proteasome suppression (Üstün et al., 2016) and autophagy induction (Figure 4), we then tested whether the T3E-dependent activation of proteaphagy involves HopM1. Both transient expression of HopM1 via agroinfiltration as well as T3SS-dependent delivery by PstD28E (+HopM1) triggered the formation of PAG1-GFP labeled punctate structures and aggregates in N. benthamiana leaves (Figures 6A and 6B; Supplemental Figure 12). In addition, we found that the protein abundance of PAG1-GFP was diminished in the presence HopM1 (Figure 6C). ConA treatment reverted this HopM1-induced effect but also stabilized PAG1-GFP at control conditions. These results suggest that proteasomes are turned over by autophagy at basal levels and that proteaphagy is further stimulated in response to HopM1 activity. Confocal microscopy verified colocalization of transiently expressed HopM1 with PAG1-GFP aggregates (Figure 6D) as well as association with the autophagy markers ATG8a and NBR1 (Figure 6E). Furthermore, HopM1-induced PAG1-GFP aggregates and punctuate structures colocalize with NBR1 and ATG8e (Supplemental Figure 13). These observations link HopM1 to the proteasome and autophagy machinery, further supporting HopM1-induced activation of proteaphagy. From these data we conclude that Pst reduces proteasome activity and triggers proteaphagy via the activity of the T3E HopM1, thereby impairing proteasome function.
Figure 6.
HopM1 Induces Proteaphagy.
(A) Coexpression of PAG1-GFP with GUS or HopM1 in N. benthamiana leaves. Images were taken 2 d postinfiltration (bars = 20 μm). The right image shows a magnification of the boxed area (bar = 5 μm). The experiment was repeated three times with similar results.
(B) Quantification of PAG1-GFP aggregates in N. benthamiana leaves coexpressing PAG1-GFP with GUS or HopM1. The number of aggregates were calculated from single planes of independent leaf areas of 0.025 mm2 (n = 5) using ImageJ. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles and data points are plotted as open circles. Asterisks indicate statistical significance (**P < 0.01) determined by Student’s t test in comparison with the control.
(C) Immunoblot analysis of PAG1-GFP upon transient expression of HA-tagged HopM1 and the GUS control in the presence (+) or absence (–) of ConA. The constructs were transiently expressed in N. benthamiana leaves for 48 h and samples were taken after treatment with DMSO or ConA for an additional 6 h. Extracted proteins were probed with anti-GFP and anti-HA antibodies, and numbers correspond to the relative PAG1-GFP signal intensities compared with the GUS control (–ConA). Ponceau S staining of RbcL was used as loading control and quantified for normalization of PBA1 signals. The analysis was repeated twice with similar results.
(D) Colocalization analysis of PAG1-RFP with HopM1-GFP in N. benthamiana leaves. Imaging was performed 2 d after transient expression and images represent single confocal planes from abaxial epidermal cells (bars = 20 μm). The experiment was repeated twice with similar results.
(E) Colocalization analysis of HopM1-GFP with RFP-ATG8a and RFP-NBR1 in N. benthamiana leaves. Imaging was performed 2 d after transient expression (bars = 10 μm) and the experiment was repeated twice with similar results.
NBR1 Counteracts HopM1-Triggered Water-Soaked Lesions and Bacterial Pathogenicity
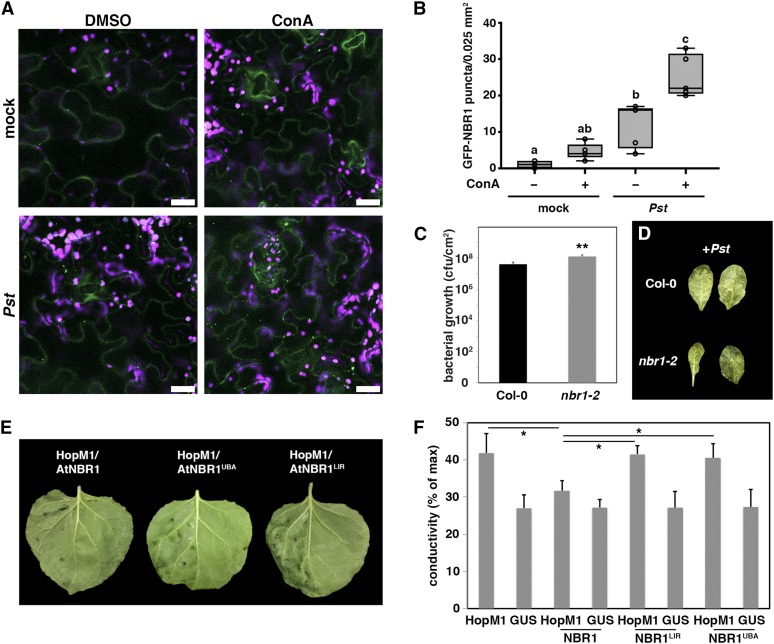
Based on our initial observation that Pst infection causes enhanced NBR1 flux and association of NBR1 with aggregated proteasomes and HopM1, we wondered whether NBR1-mediated selective autophagy influences host immunity and bacterial pathogenesis. The induction of NBR1-dependent selective processes by Pst was further analyzed using a transgenic Arabidopsis line expressing GFP-NBR1. Compared with the noninfected control, GFP-NBR1-labeled punctate structures were more abundant upon infection and increased substantially upon ConA treatment (Figures 7A and 7B). Additionally, NBR1 was found in immunoprecipitates of GFP-ATG8a but not of free GFP from Pst-infected plants (Supplemental Figure 14), further supporting the notion that NBR1-mediated selective autophagy is activated upon Pst challenge. To assess the biological relevance of NBR1-dependent processes during infection, we determined the Pst resistance response in Arabidopsis upon loss of NBR1 function. Intriguingly, nbr1-2 plants supported significantly more bacterial growth and displayed more pronounced disease progression and water-soaked lesions compared with wild-type plants (Figures 7C and 7D).
Figure 7.
NBR1 Counteracts P. syringae Infection.
(A) Detection of GFP-NBR1 labeled structures in transgenic Col-0 plants upon mock and Pst infection in the presence (ConA) and absence (DMSO) of ConA. Images represent single confocal planes from abaxial epidermal cells and were taken at 8 hpi with identical microscope settings (bars = 20 μm). This experiment was repeated three times with similar results.
(B) GFP-NBR1 puncta were quantified in single planes of independent areas (n = 6) using ImageJ. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles and data points are plotted as open circles. Different letters indicate statistically significant differences (P < 0.05) as determined by one-way ANOVA.
(C) Bacterial growth in leaves of Col-0 and nbr1-2 plants infected with Pst. Leaves were syringe infiltrated with a bacterial suspension at OD600 = 0.0001, and bacterial multiplication was determined at 3 dpi. Bars represent means ± sd (n = 4). Asterisks indicate statistical significance determined by Student’s t test (**P < 0.01) in comparison to Col-0. This experiment was repeated three times with similar results.
(D) Phenotypes of Pst-infected leaves of Col-0 and nbr1-2 at 3 dpi.
(E) Phenotype of N. benthamiana leaves expressing HopM1 together with Arabidopsis NBR1 wild-type (AtNBR1) or NBR1 variants mutated in the ATG8/LC3-interacting motif (AIM/LIR) and main ubiquitin binding (UBA2) domain (AtNBR1LIR and AtNBR1UBA) at 2 dpi. Only the left part of the leaves was infiltrated with agrobacteria expressing the respective constructs. The experiment was repeated at least three times with similar results.
(F) Quantification of HopM1-triggered water-soaking by ion leakage measurements. The indicated constructs were transiently expressed in N. benthamiana and samples were taken at 48 hpi. Bars represent the means ± sd calculated from four leaf discs per treatment with four replicates within an experiment. Asterisks indicate significant differences (*P < 0.05) based on Student’s t test.
Recent data have shown that water-soaking is mediated by HopM1 and plays an active role in establishing an aqueous apoplast for bacterial pathogenesis (Xin et al., 2016). We therefore tested whether NBR1-mediated autophagy antagonizes HopM1-induced water-soaking in N. benthamiana leaves. Indeed, transient expression of NBR1 reduced the appearance of HopM1-triggered water-soaked lesions and the extent of electrolyte leakage from leaf tissue, which was dependent on the ATG8/LC3-interacting motif (AIM/LIR) and main ubiquitin binding domain (UBA2) of NBR1 (Figures 7E and 7F). The observed phenotypes were not due to lower HopM1 protein levels, as HopM1 abundance remained largely unchanged upon coexpression of wild-type and mutant forms of NBR1 (Supplemental Figure 15).
Based on these data, we speculate that NBR1 is involved in the autophagic degradation of an as yet unknown factor required for HopM1-induced water-soaking. In agreement with this notion, nbr1-2 mutants hyperaccumulate ubiquitinated proteins to higher levels than in Col-0 upon Pst infection (Supplemental Figure 16), suggesting that NBR1 might contribute to the turnover of ubiquitinated substrates during infection.
Together, these findings reveal that NBR1-mediated selective autophagy plays a role in antibacterial immunity and thus acts contrary to NBR1-independent proteaphagy in bacterial pathogenesis.
DISCUSSION
Autophagy is well recognized as an intrinsic pathway with both anti- and promicrobial functions during viral and bacterial infections in animals (Levine et al., 2011; Mostowy, 2013; Paul and Münz, 2016). Recent evidence indicates that xenophagic mechanisms also target viruses and their components in plants and that some viruses evolved sophisticated strategies to escape or hijack autophagy processes for enhanced pathogenicity (Derrien et al., 2012; Hafrén et al., 2017, 2018; Haxim et al., 2017). Plant pathogenic bacteria differ from their animal counterparts and viral pathogens in the exclusive extracellular proliferation, but they exhibit a similar capacity to manipulate the intracellular machinery via secreted effector proteins. While autophagy is well known to contribute to the effector-triggered hypersensitive response upon avirulent P. syringae infection (Hofius et al., 2009; Kwon et al., 2013; Coll et al., 2014), the role of autophagic processes during virulent bacterial infections has remained to be clarified.
In this study, we demonstrate that Pst delivers the T3E HopM1 to enhance autophagic flux and activate proteaphagy, thus leading to the suppression of proteasome function and promotion of virulence. By contrast, the selective cargo receptor NBR1 counteracts HopM1-induced water-soaking and suppresses bacterial proliferation. Hence, our data establish the phytopathogenic bacterium P. syringae as a seminal example of the integration of distinct autophagy pathways into host immunity and bacterial pathogenesis. In addition, we provide evidence for the intimate crosstalk between autophagy and the proteasome during plant-pathogen interactions.
Probacterial Role of Autophagy during P. syringae Infection
Several pathogenic bacteria in animals inhibit autophagy to escape their elimination by xenophagy (e.g., Salmonella, Shigella, Legionella, and Mycobacterium) (Shpilka and Elazar, 2012; Mostowy, 2013), whereas others seem to exploit autophagy components and structures to promote their replication and survival (e.g., Listeria, Staphylococcus, and Brucella) (Mostowy, 2013). Our findings clearly support a probacterial function of T3E-triggered autophagy during virulent P. syringae infection in plants. In particular, the significant promotion of bacterial growth upon autophagy enhancement by AZD8055 treatment or ATG5 overexpression is in accordance with previous observations that transgenic plants expressing a constitutively active form of the positive autophagy regulator RabG3B are more susceptible (Kwon et al., 2013), and autophagy-deficient mutants more resistant (Lenz et al. 2011) to Pst infection.
Our data provide several lines of evidence for a novel mechanistic link between Pst-induced probacterial autophagy and inactivation of proteasome function: (1) Autophagy-defective atg mutants are insensitive to Pst-induced proteasome inhibition, which is not caused by altered SA levels and signaling; (2) enhanced bacterial resistance in atg mutants is abolished by mutations in proteasome subunits; and (3) proteaphagy is induced by the autophagy-stimulating T3E HopM1. Because proteaphagy is activated upon chemical proteasome inhibition (Marshall et al., 2015), and some effectors from Xanthomonas ssp and P. syringae pv lachrymans interact directly with the proteasome to block its activity (Üstün et al., 2013, 2014; Üstün and Börnke, 2015), we anticipate that HopM1-mediated interference with the proteasome leads to proteaphagy activation and suppression of proteasome function during Pst infection.
In support of this model (illustrated in Figure 8), HopM1 was previously shown to inhibit proteasome activity and associate with proteasome subunits (Üstün et al., 2016). In addition, the presence of the unprocessed form of PBA1 in the atg5 background upon Pst infection confirms direct effector targeting of the proteasome independent of autophagy activation. We also observed several hallmarks of chemical proteasome inhibition during bacterial infection (Marshall et al., 2015), including increased transcript and protein levels of multiple proteasome subunits as well as proteasome aggregation. However, in contrast to Pst infection, MG132 treatment is still sufficient to inactivate the proteasome in atg mutants (Marshall et al., 2016), suggesting different modes of HopM1 and MG132 action. Indeed, HopM1 is mainly associated with the regulatory particle of the 26S proteasome (Üstün et al., 2016), while MG132 targets its catalytic subunits (Bibo-Verdugo et al., 2017). Hence, an efficient block of proteasome activity during Pst infection requires a functional proteaphagy pathway involving the core autophagy machinery (ATG2, ATG5, and ATG7) and cargo receptor (RPN10). Intriguingly, proteasome activity appeared rather elevated in the atg mutants. This observation might give at least an indirect explanation why autophagy-deficient mutants are more resistant to Pst infection (Lenz et al., 2011). Enhanced proteasome activity was demonstrated to correlate with activated PAMP-triggered immune responses and resistance toward nonpathogenic bacteria (Üstün et al., 2016).
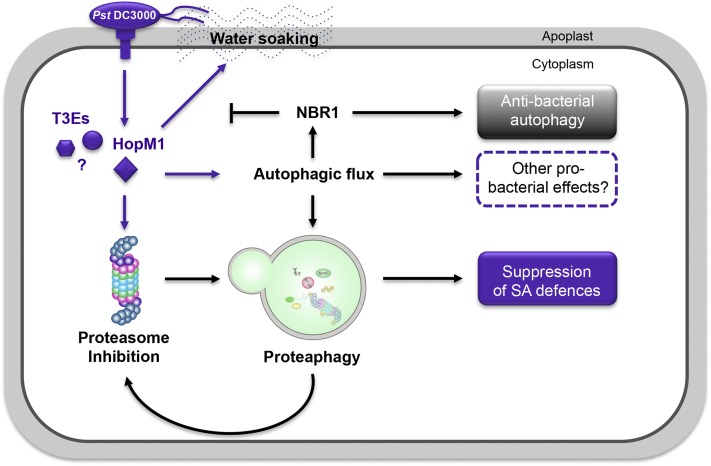
Figure 8.
A Working Model Showing the Pro- and Antibacterial Roles of Autophagy during Pst Infection.
Pst DC3000 delivers a large repertoire of T3Es into the cytoplasm to alter host functions. HopM1-mediated inhibition of the proteasome (Üstün et al., 2016) activates proteaphagy required to efficiently block proteasome function during infection, thus resulting in suppression of SA-dependent defense responses. HopM1 also contributes significantly to the overall increase in autophagy activity and flux in the host cell, which probably supports the effective removal of inactivated proteasomes and serves additional probacterial functions. The concurrent stimulation of NBR1-dependent autophagy, however, counteracts HopM1-mediated water-soaking and creation of an aqueous apoplast, thereby dampening bacterial virulence and proliferation. Arrows indicate activation of a process and T-bars inhibition.
The strong overall increase in cellular autophagy levels upon Pst infection and HopM1 delivery in Arabidopsis still implies the possibility that HopM1 and potentially additional T3Es stimulate bulk and selective autophagy pathways by other as yet unknown mechanisms. Notably, HopM1 has been localized to the trans-Golgi network/early endosomes (TGN/EEs) where it exerts virulence function (Nomura et al., 2006). In general, there is substantial evidence for the extensive overlap and crosstalk between autophagy and endosomal trafficking pathways in eukaryotes including plants (Teh and Hofius, 2014; Kalinowska and Isono, 2017; Noda, 2017) and the TGN/EE has recently been implicated in the regulation of autophagosome formation in mammalian cells (Guo et al., 2012; Mattera et al., 2017). Thus, it is tempting to speculate that HopM1 might target host proteins and associated processes at the TGN, resulting in autophagy induction or promotion. Such primarily enhanced autophagic activities may result in concurrent turnover of proteasomes, resembling the situation of starvation-induced proteaphagy (Marshall et al., 2015). Which host targets are mainly responsible for the effector-triggered autophagy enhancement and proteaphagy induction will be an important subject of future research.
Besides selective proteaphagy to suppress proteasome-dependent host defense, the stimulation of bulk autophagy may have additional benefits for the hemibiotrophic bacterium. One possibility is related to the survival function of autophagy in suppressing stress- and disease-associated cell death (Üstün et al., 2017). Increased plant viability and life span by autophagy activation has been demonstrated to serve the production of CaMV particles and, thus, potential vector transmission (Hafrén et al., 2017). Hence, Pst-induced autophagy may prolong the biotrophic phase during infection, thereby supporting bacterial survival and proliferation. Alternatively, effector-triggered autophagy might also be involved in the recycling and rerouting of nutrients in favor of the pathogen during the establishment of infection or upon transition from the biotrophic to necrotrophic phase, as speculated previously for the hemibiotrophic bacterium Ralstonia solanacearum (Popa et al., 2016) and the oomycete Phytophthora infestans (Dagdas et al., 2016).
Antibacterial Role of Autophagy during P. syringae Infection
While bulk autophagy and proteaphagy contribute to bacterial pathogenicity, NBR1-dependent selective autophagy seems to restrict bacterial infection. NBR1 proteins in plants are structurally and functionally related to the mammalian autophagic cargo receptors NBR1 and p62/sequestosome-1 (SQSTM1) that either act alone or cooperate in targeting mostly ubiquitinated substrates to the growing phagophore (Svenning et al., 2011). Unlike NBR1 homologs, p62 proteins are confined to metaozans, but Arabidopsis NBR1 and the N. benthamiana homolog Joka2 share their ability of self-oligomerization and aggregation (Svenning et al., 2011; Zientara-Rytter and Sirko, 2014). Thus, plant NBR1 proteins are regarded as functional hybrids of mammalian NBR1 and p62 and have been assigned a general role in targeting polyubiquitinated protein aggregates for autophagic degradation, e.g., in abiotic stress conditions (Svenning et al., 2011; Zhou et al., 2013). Recently, NBR1-mediated xenophagy of nonassembled viral capsid protein and particles has been linked to antiviral immunity against CaMV infection (Hafrén et al., 2017). Intriguingly, the RXLR effector PexRD54 counteracts the function of Joka2 in host defense during P. infestans infection of N. benthamiana (Dagdas et al., 2016). A model was proposed in which Joka2 facilitates the removal of plant or pathogen proteins negatively regulating immunity. PexRD54 might hijack this pathway to selectively eliminate defense compounds or to reallocate nutrients to the infection structures (i.e., haustoria) (Dagdas et al., 2016). Our finding of NBR1-mediated suppression of HopM1-induced water-soaking together with the enhanced susceptibility of nbr1-2 mutants to Pst infection supports the conserved integration of NBR1-dependent processes in plant immunity against different pathogens with a (hemi)-biotrophic lifestyle. Bacterial infection increases the amount of ubiquitinated proteins due to the reduction of proteasome activity (Üstün et al., 2016) and autophagy activation. The increased abundance of polyubiquitinated proteins in nbr1-2 plants in response to Pst infection indicates that NBR1 contributes to the turnover of these proteins, which is possibly required for activation, or maintenance, of antibacterial defenses. Given that transient overexpression of NBR1 reduces the establishment of water-soaked lesion, we speculate that NBR1 is involved in the selective degradation of negative immune regulators and/or substrates required for HopM1-triggered water-soaking. Although we find partial association of HopM1-labeled structures with NBR1 and ATG8, there is currently no substantial evidence suggesting that NBR1-mediated selective autophagy may be involved in the direct targeting of bacterial effectors. However, similar to P. infestans, distinct effectors and NBR1 may coincide at the same cellular destinations due to their competition in modulating similar host targets and processes.
In conclusion, we propose that Pst has evolved an effector-based strategy to exploit bulk autophagy and selective proteaphagy for enhanced pathogenicity, whereas NBR1-dependent selective autophagy contributes to host immunity and restricts bacterial proliferation (Figure 8). Our findings indicate that autophagy pathways with opposing pro- and antimicrobial functions act in parallel during Pst infection, which is a likely consequence of the long-lasting coevolution of plants and their associated bacterial pathogens.
METHODS
Plant Material and Growth Conditions
Wild-type plants were Arabidopsis thaliana ecotype Columbia (Col-0). Loss-of-function mutants atg5-1, atg2-1, atg7-2, nbr1-2, rpn10-1, rpn12a-1, rpt2a-2, atg5-1 npr1-1, atg5-1 sid2-2, and atg5-1 nbr1-2, as well as the GFP-ATG8a, GFP-NBR1, and DEX:HopM1-GFP transgenic lines have been described previously (Smalle et al., 2002, 2003; Kurepa et al., 2008; Hofius et al., 2009; Yoshimoto et al., 2009; Nomura et al., 2011; Zhou et al., 2013; Munch et al., 2014; Hafrén et al., 2017). Autophagy- and proteasome-defective double mutants were obtained by crossing and genotyping was performed by PCR using the gene-specific primers listed in Supplemental Table 1. Arabidopsis plants were grown on soil for infection experiments under short-day conditions (8/16-h light/dark cycles) in a growth chamber or for maintenance and crossings under long-day conditions (16/8-h light/dark cycles) in a growth room with light intensity of 150 μE from a Osram FQ 24W/840 HQ Constant Lumilux Cool White light source, 21°C, and 70% relative humidity, respectively. Nicotiana benthamiana plants were grown under the same long-day conditions for transient expression assays. Sterile plants were cultivated in vitro on half-strength Murashige and Skoog (MS) medium with a 16-h photoperiod at 150 μE/m2s and 21°C.
Bacterial Infection and Resistance Assay
Pseudomonas syringae pv tomato DC3000 wild-type and derivatives [PstΔhrcC, PstΔhopM1, PstD28E, PstD28E (+HopM1) and PstΔhopQ1-1] were grown in King’s B medium with appropriate antibiotics (i.e., rifampicin) at 28°C. All strains were kind gifts from Alan Collmer (Cornell University). For growth assays, bacterial suspensions with an OD600 of 0.0001 were syringe-infiltrated into three fully expanded leaves per soil-grown plant. At 3 d postinoculation, leaf discs from infiltrated leaf areas were homogenized using two technical replicates (leaf discs from the same plant) each from four independent plants, and leaf extracts were plated in appropriate dilutions on solid King’s B medium. Colony-forming units were determined after plate incubation for 1 to 2 d at 28°C. Bacterial infections in seedlings were performed as described (Ishiga et al., 2011) with slight modifications. In brief, 10- to 14-d-old seedlings were flood-inoculated for 3 min with a bacterial suspension at an OD600 of 0.1 containing 0.025% Silwet L-77. For NBR1 flux assays and immunoblot analysis, seedlings were harvested 10 h after inoculation. Bacterial infections for RT-qPCR analysis, proteasome activity measurements, confocal microscopy, and immunoblot analysis were performed with a bacterial suspension at an OD600 of between 0.1 and 0.2 to provide fast and robust responses.
Plasmid Construction
For transient expression experiments, the coding region of the proteasome subunit PAG1 was cloned into pENTR/D-TOPO and subsequently recombined into pGWB505 (Nakagawa et al., 2007), while the coding region of ubiquitin (UBQ11) was similarly cloned into pENTR/D-TOPO and then recombined into pUBN-DEST-RFP (Grefen et al., 2010). The RFP-ATG8a, RFP-NBR1, HopM1-HA, and HopI1-HA constructs were described previously (Hafrén et al., 2017; Üstün et al., 2016). All binary plasmids were transformed into Agrobacterium tumefaciens strain C58C1 and infiltration of N. benthamiana was done at the four- to six-leaf stage. For the quantitative autophagy assay, the coding sequences of the luciferases from Photinus pyralis (Firefly luciferase [FLUC]) and Renilla reniformis (Renilla luciferase [RLUC]) were amplified from the pRD400-35S:FLUC and pRD400-35S:RLUC vectors (Eskelin et al., 2011) using FLUC_fwd/FLUC_rev and RLUC_fwd/RLUC_rev as primer pairs, respectively (Supplemental Table 1). The FLUC sequence was then introduced into pMDC32 (Curtis and Grossniklaus, 2003) between the double 35S promoter and the Gateway cassette using KpnI/AscI restriction sites resulting in the pMDC32:FLUC vector. To produce a pMDC32 vector with kanamycin resistance for expression of RLUC, the hygromycin B resistance gene of pMDC32 was replaced by the NPTII gene. RLUC was then introduced between the double 35S promoter and the Gateway cassette of the modified pMDC32 vector using the same KpnI/AscI restriction sites.
Transient Expression in N. benthamiana by Agrobacterium-Mediated Leaf Infiltration
Agrobacterium strains harboring the indicated plasmids were grown overnight in liquid LB media at 28°C with appropriate antibiotics and harvested by centrifugation at 3000g for 5 min. Bacterial pellets were washed once in double-distilled water, resuspended in infiltration buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 150 μM acetosyringone), and incubated for 2 h at room temperature without shaking. The OD600 of each culture was adjusted such that approximately equal numbers of bacteria from cultures expressing different transgenes were mixed and infiltrated into N. benthamiana leaves using a needleless syringe.
Confocal Microscopy
Live-cell images were acquired from abaxial leaf epidermal cells using a Zeiss LSM 780 microscope. Excitation/detection parameters for GFP and RFP were 488 nm/490 to 552 nm and 561 nm/569 to 652 nm, respectively, and sequential scanning mode was used for colocalization of both fluorophores. Inhibitor treatment was performed by either syringe infiltration of mature leaves or incubation of seedlings in half-strength MS with 0.5 μM ConA for 6 or 14 h prior to confocal analysis. Confocal images were processed with ZEN (version 2011) and ImageJ (version 1.48v) software. Quantification of GFP-ATG8a-labeled autophagosomal structures was done on single planes that were converted to eight-bit grayscale and then counted for GFP-ATG8 puncta either manually or by the Particle Analyzer function of ImageJ.
Immunoprecipitation
GFP pull-down assays were performed as previously described (Dagdas et al., 2016) with minor modifications. Approximately 2.5 g of Arabidopsis leaf material was ground to a fine powder in liquid nitrogen and homogenized in 5 mL of extraction buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM DTT, 1 mM EDTA, 2% [w/v] polyvinylpolypyrrolidone, 1% [v/v] protease inhibitor cocktail [Sigma], and 0.1% [v/v] Nonidet P-40). Insoluble debris was pelleted by centrifugation for 20 min with 2000g at 4°C. Immunoprecipitation was performed by adding 30 μL of GFP-Trap coupled to agarose beads (ChromoTek) and samples were incubated for 2 h at 4°C with continual rotation. Beads were subsequently washed five times with Tris-buffered saline containing 0.1% (v/v) Nonidet P-40, and immunoprecipitates were eluted with 30 μL of 2× SDS loading buffer at 70°C for 10 min.
Immunoblot Analysis
Proteins were extracted in 100 mM Tris (pH 7.5) containing 2% SDS, boiled for 10 min in SDS loading buffer, and cleared by centrifugation. The protein extracts were then separated by SDS-PAGE, transferred to PVDF membranes (Amersham, GE Healthcare), blocked with 5% skimmed milk in PBS, and incubated with primary antibodies anti-NBR1 (Svenning et al., 2011), anti-ATG8a (Yoshimoto et al., 2004), anti-ubiquitin (Agrisera), anti-GFP (Clontech and Santa Cruz), anti-PBA1 (Enzo Life Science), anti-RPT2a (Enzo Life Science), and anti-RPN10 (Lin et al., 2011) primary antibodies using 1:2000 dilutions in PBS containing 0.1% Tween 20. This was followed by incubation with horseradish peroxidase-conjugated secondary antibodies diluted 1:10,000 in PBS containing 0.1% Tween 20. The immunoreaction was developed using an ECL Prime Kit (GE Healthcare) and detected with a LAS-3000 Luminescent Image Analyzer (Fujifilm).
RT-qPCR
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen), and on-column DNA digestion was performed with DNase I (Qiagen). First-strand cDNA was synthesized from 1 μg of total RNA using the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative PCR analysis was done with Maxima SYBR Green/Fluorescein qPCR Master Mix (Thermo Fisher Scientific) using the CFX Connect Real-Time PCR detection system (Bio-Rad) with the gene-specific primers listed in Supplemental Table 1. Normalization was done using PP2A (AT1G69960).
Proteasome Activity Measurement
Proteasome activity was determined as described previously (Üstün et al., 2016; Üstün and Börnke, 2017).
Dual Luciferase Assay
The dual luciferase reporter assay was performed according to the manufacturer’s instructions (Dual-Luciferase Reporter Assay System; Promega) with slight modifications. Briefly, four leaf discs were homogenized in 200 μL lysis buffer and cleared by centrifugation. For detection and measurement of the Firefly luciferase activity, 40 μL of the luciferase assay reagent was added to 5 μL of plant extracts. To measure Renilla luciferase activity, 40 μL of the Stop and Glo reagent was added to the mixture. The measurement was performed in a plate reader.
Ion Leakage Measurement
For quantification of water-soaking in HopM1-expressing N. benthamiana leaves, electrolyte leakage assays were done essentially as described (Üstün et al., 2012). In brief, four leaf discs (0.9 cm in diameter) per biological replicate were sampled at 48 h post-agroinfiltration, placed into a reaction tube with 8 mL double-distilled water, and incubated over night at 4°C under constant rotation. Conductivity of the bath solution was measured with a conductometer (EcoScan Hand-held Series; Eutech Instruments), and values were expressed as the percentage of maximum values obtained by subsequent boiling of the samples for 30 min.
Data Analysis and Presentation
Data are presented as mean ± sd. Statistical significance was analyzed by Student’s t test (*P < 0.05, **P < 0.01, and P*** < 0.001) or one-way ANOVA (P < 0.05). The number of biological replicates (n) is given in the figure legends.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Information Resource or GenBank/EMBL databases under the following accession numbers: ATG2 (AT3G19190), ATG5 (AT5G17290), ATG7 (AT5G45900), NPR1 (AT1G64280), SID2 (AT1G74710), NBR1 (AT4G24690), ATG8a (AT4G21980), PBA1 (AT4G31300), RPN10 (AT4G38630), RPT2a (AT4G29040), RPT2b (AT2G20140), RPN12a (AT1G64520); PAG1 (AT2G27020), Ubiquitin/UBQ11 (AT4G05050), PP2A (AT1G69960), HopM1 (NP_791202), HopI1 (NP_794511), and HopX1 (NP_808672).
Supplemental Data
Supplemental Figure 1. Pst-mediated inhibition of proteasome function is dependent on a functional autophagy pathway and independent of the SA pathway.
Supplemental Figure 2. TOR inhibitor AZD8055 induces autophagy.
Supplemental Figure 3. AZD8055 promotion of bacterial growth is dependent on a functional autophagy pathway.
Supplemental Figure 4. Kinetics of GFP-ATG8a accumulation upon challenge with Pst and bacterial PAMPs.
Supplemental Figure 5. Protein expression analysis of T3Es used in the luciferase-based autophagy assay in N. benthamiana leaves.
Supplemental Figure 6. HopM1 induces accumulation of GFP-ATG8a puncta upon delivery of an “effectorless” Pst strain.
Supplemental Figure 7. Transcriptional induction of proteasome subunit genes in response to Pst infection.
Supplemental Figure 8. Proteasome subunit levels do not change in response to PstΔhrcC.
Supplemental Figure 9. PAG1-GFP localization upon chemical proteasome inhibition and PstΔhrcC infection.
Supplemental Figure 10. Colocalization of PAG1-GFP with RFP-ATG8e and RFP-ATG8g in N. benthamiana leaves.
Supplemental Figure 11. Proteasome activity and bacterial growth in rpn10 mutant.
Supplemental Figure 12. PAG1-GFP aggregates and punctuate structures are induced upon delivery of HopM1 by PstD28E.
Supplemental Figure 13. Colocalization of PAG1-GFP puncta/aggregates with ATG8 and NBR1 upon transient expression of HopM1 in N. benthamiana.
Supplemental Figure 14. Coimmunoprecipitation of GFP-ATG8a and NBR1 upon Pst infection.
Supplemental Figure 15. Protein abundance of HopM1 upon coexpression with wild-type NBR1 or UBA2 and LIR mutant variants.
Supplemental Figure 16. Hyperaccumulation of ubiquitinated proteins during Pst infection in nbr1-2.
Supplemental Table 1. Oligonucleotides.
Acknowledgments
We thank Sheng-Yang He (Michigan State University, East Lansing, MI) for the DEX:HopM1-GFP construct and transgenic Arabidopsis lines, Alan Collmer (Cornell University, Ithaca, NY) for providing the P. syringae strains used in this study, and Hongyong Fu (Academia Sinica, Taiwan) for the anti-RPN10 antibody. We also thank Kristiina Mäkinen (University of Helsinki, Finland) for providing pRD400-35S:FLUC and pRD400-35S:RLUC vectors. This research was funded by grants from the Swedish University of Agricultural Sciences, the Knut and Alice Wallenberg Foundation, the Carl Tryggers Foundation, and the Swedish Research Council VR to D.H., the Swedish Research Council FORMAS to D.H. and A.H., the Swedish Research Council VR and Olle Engkvist Foundation to P.V.B., and a fellowship from the Federation of European Biochemical Societies to S.Ü. R.S.M. and R.D.V. were supported by a grant from the U.S. Department of Energy Office of Science, Office of Basic Energy Science, Chemical Sciences, Geosciences and Biosciences Division (DE-FG02-88ER13968).
AUTHOR CONTRIBUTIONS
S.Ü. and D.H. designed experiments. S.Ü., A.H., and Q.L. performed experiments. S.Ü., A.H., Q.L., R.S.M., R.D.V., and D.H. analyzed the data. R.S.M., E.A.M., P.V.B., and R.D.V. contributed novel experimental material. S.Ü. and D.H. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Bibo-Verdugo B., Jiang Z., Caffrey C.R., O’Donoghue A.J. (2017). Targeting proteasomes in infectious organisms to combat disease. FEBS J. 284: 1503–1517. [DOI] [PubMed] [Google Scholar]
- Book A.J., Gladman N.P., Lee S.S., Scalf M., Smith L.M., Vierstra R.D. (2010). Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. J. Biol. Chem. 285: 25554–25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P., Reggiori F., Codogno P. (2013). Emerging regulation and functions of autophagy. Nat. Cell Biol. 15: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493. [DOI] [PubMed] [Google Scholar]
- Cohen-Kaplan V., Livneh I., Avni N., Fabre B., Ziv T., Kwon Y.T., Ciechanover A. (2016). p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. USA 113: E7490–E7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N.S., Smidler A., Puigvert M., Popa C., Valls M., Dangl J.L. (2014). The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. 21: 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S., Chakravarthy S., Kvitko B.H., Russell A.B., Martin G.B., Collmer A. (2011). Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 108: 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas Y.F., et al. (2016). An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5: e10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S., Thilmony R., Kwack Y.B., Nomura K., He S.Y. (2004). A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA 101: 9927–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 109: 15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Levine B. (2013). Autophagy and viruses: adversaries or allies? J. Innate Immun. 5: 480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelin K., Hafrén A., Rantalainen K.I., Mäkinen K. (2011). Potyviral VPg enhances viral RNA Translation and inhibits reporter mRNA translation in planta. J. Virol. 85: 9210–9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Boter M., Fernandez-Barbero G., Chini A., Rathjen J.P., Solano R. (2014). The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 12: e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladman N.P., Marshall R.S., Lee K.H., Vierstra R.D. (2016). The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. Plant Cell 28: 1279–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes L.C., Dikic I. (2014). Autophagy in antimicrobial immunity. Mol. Cell 54: 224–233. [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Guo Y., Chang C., Huang R., Liu B., Bao L., Liu W. (2012). AP1 is essential for generation of autophagosomes from the trans-Golgi network. J. Cell Sci. 125: 1706–1715. [DOI] [PubMed] [Google Scholar]
- Hafrén A., Macia J.L., Love A.J., Milner J.J., Drucker M., Hofius D. (2017). Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 114: E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén A., Ustun S., Hochmuth A., Svenning S., Johansen T., Hofius D. (2018). Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 176: 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Wang Y., Zheng X., Jia Q., Zhao J., Bai F., Hong Y., Liu Y. (2015). Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27: 1316–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxim Y., et al. (2017). Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 6: e23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Li L., Hafrén A., Coll N.S. (2017). Autophagy as an emerging arena for plant-pathogen interactions. Curr. Opin. Plant Biol. 38: 117–123. [DOI] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783. [DOI] [PubMed] [Google Scholar]
- Ishiga Y., Ishiga T., Uppalapati S.R., Mysore K.S. (2011). Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions. Plant Methods 7: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J., Yao N., Vinatzer B.A., Wright C.M., Brodsky J.L., Greenberg J.T. (2007). A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17: 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowska K., Isono E. (2017). All roads lead to the vacuole-autophagic transport as part of the endomembrane trafficking network in plants. J. Exp. Bot. 19: doi/10.1093/jxb/erx395. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Codogno P. (2013). The mechanism and physiological function of macroautophagy. J. Innate Immun. 5: 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J., Toh-E A., Smalle J.A. (2008). 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 53: 102–114. [DOI] [PubMed] [Google Scholar]
- Kwon S.I., Cho H.J., Kim S.R., Park O.K. (2013). The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 161: 1722–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Wang F., Zheng Z., Fan B., Chen Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66: 953–968. [DOI] [PubMed] [Google Scholar]
- Lenz H.D., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66: 818–830. [DOI] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H.W. (2011). Autophagy in immunity and inflammation. Nature 469: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kabbage M., Liu W., Dickman M.B. (2016). Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell 28: 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.L., Sung S.C., Tsai H.L., Yu T.T., Radjacommare R., Usharani R., Fatimababy A.S., Lin H.Y., Wang Y.Y., Fu H. (2011). The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. Plant Cell 23: 2754–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2010). TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577. [DOI] [PubMed] [Google Scholar]
- Macho A.P. (2016). Subversion of plant cellular functions by bacterial type-III effectors: beyond suppression of immunity. New Phytol. 210: 51–57. [DOI] [PubMed] [Google Scholar]
- Marshall R.S., McLoughlin F., Vierstra R.D. (2016). Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Reports 16: 1717–1732. [DOI] [PubMed] [Google Scholar]
- Marshall R.S., Li F., Gemperline D.C., Book A.J., Vierstra R.D. (2015). Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/Ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 58: 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R., Park S.Y., De Pace R., Guardia C.M., Bonifacino J.S. (2017). AP-4 mediates export of ATG9A from thetrans-Golgi network to promote autophagosome formation. Proc. Natl. Acad. Sci. USA 114: E10697–E10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E.A., et al. (2018). Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot., doi/10.1093/jxb/ery010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E.A., Filonova L.H., Fukada K., Savenkov E.I., Gogvadze V., Clapham D., Sanchez-Vera V., Suarez M.F., Zhivotovsky B., Daniel G., Smertenko A., Bozhkov P.V. (2013). Autophagy and metacaspase determine the mode of cell death in plants. J. Cell Biol. 203: 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misas-Villamil J.C., van der Burgh A.M., Grosse-Holz F., Bach-Pages M., Kovács J., Kaschani F., Schilasky S., Emon A.E., Ruben M., Kaiser M., Overkleeft H.S., van der Hoorn R.A. (2017). Subunit-selective proteasome activity profiling uncovers uncoupled proteasome subunit activities during bacterial infections. Plant J. 90: 418–430. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Komatsu M. (2011). Autophagy: renovation of cells and tissues. Cell 147: 728–741. [DOI] [PubMed] [Google Scholar]
- Mostowy S. (2013). Autophagy and bacterial clearance: a not so clear picture. Cell. Microbiol. 15: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch D., Rodriguez E., Bressendorff S., Park O.K., Hofius D., Petersen M. (2014). Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 10: 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Noda T. (2017). Autophagy in the context of the cellular membrane-trafficking system: the enigma of Atg9 vesicles. Biochem. Soc. Trans. 45: 1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223. [DOI] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y.N., Imboden L.A., Chang J.H., He S.Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P., Münz C. (2016). Autophagy and mammalian viruses: roles in immune response, viral replication, and beyond. Adv. Virus Res. 95: 149–195. [DOI] [PubMed] [Google Scholar]
- Popa C., Li L., Gil S., Tatjer L., Hashii K., Tabuchi M., Coll N.S., Ariño J., Valls M. (2016). The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Sci. Rep. 6: 27058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y., Luo X., Bassham D.C. (2017). TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front. Plant Sci. 8: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Ungermann C. (2017). Autophagosome maturation and fusion. J. Mol. Biol. 429: 486–496. [DOI] [PubMed] [Google Scholar]
- Shpilka T., Elazar Z. (2012). Essential role for the mammalian ATG8 isoform LC3C in xenophagy. Mol. Cell 48: 325–326. [DOI] [PubMed] [Google Scholar]
- Smalle J., Kurepa J., Yang P., Babiychuk E., Kushnir S., Durski A., Vierstra R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J., Kurepa J., Yang P., Emborg T.J., Babiychuk E., Kushnir S., Vierstra R.D. (2003). The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh O.K., Hofius D. (2014). Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J. Exp. Bot. 65: 1297–1312. [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S., Börnke F. (2015). The Xanthomonas campestris type III effector XopJ proteolytically degrades proteasome subunit RPT6. Plant Physiol. 168: 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S., Hafrén A., Hofius D. (2017). Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 40: 122–130. [DOI] [PubMed] [Google Scholar]
- Üstün S., König P., Guttman D.S., Börnke F. (2014). HopZ4 from Pseudomonas syringae, a member of the HopZ type III effector family from the YopJ superfamily, inhibits the proteasome in plants. Mol. Plant Microbe Interact. 27: 611–623. [DOI] [PubMed] [Google Scholar]
- Üstün S., Börnke F. (2017). Ubiquitin proteasome activity measurement in total plant extracts. Bio Protoc. 7: e2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S., Bartetzko V., Börnke F. (2013). The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic-acid mediated plant defence. PLoS Pathog. 9: e1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S., Sheikh A., Gimenez-Ibanez S., Jones A., Ntoukakis V., Börnke F. (2016). The proteasome acts as a hub for plant immunity and is targeted by Pseudomonas type III effectors. Plant Physiol. 172: 1941–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S., Müller P., Palmisano R., Hensel M., Börnke F. (2012). SseF, a type III effector protein from the mammalian pathogen Salmonella enterica, requires resistance-gene-mediated signalling to activate cell death in the model plant Nicotiana benthamiana. New Phytol. 194: 1046–1060. [DOI] [PubMed] [Google Scholar]
- Xin X.F., Nomura K., Aung K., Velásquez A.C., Yao J., Boutrot F., Chang J.H., Zipfel C., He S.Y. (2016). Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539: 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Pascual C., Klionsky D.J. (2016). Autophagy: machinery and regulation. Microb. Cell 3: 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Chen Y., Tooze S.A. (2017). Autophagy pathway: Cellular and molecular mechanisms. Autophagy 1–9: doi/10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G., Martens S. (2016). Mechanisms of selective autophagy. J. Mol. Biol. 428: 1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Yu J.Q., Chen Z. (2014). The perplexing role of autophagy in plant innate immune responses. Mol. Plant Pathol. 15: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang J., Cheng Y., Chi Y.J., Fan B., Yu J.Q., Chen Z. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K., Sirko A. (2014). Significant role of PB1 and UBA domains in multimerization of Joka2, a selective autophagy cargo receptor from tobacco. Front. Plant Sci. 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]