An ortholog of TEOSINTE BRANCHED1 regulates inflorescence architecture and developmental rate in bread wheat in a dosage-dependent manner.
Abstract
The flowers of major cereals are arranged on reproductive branches known as spikelets, which group together to form an inflorescence. Diversity for inflorescence architecture has been exploited during domestication to increase crop yields, and genetic variation for this trait has potential to further boost grain production. Multiple genes that regulate inflorescence architecture have been identified by studying alleles that modify gene activity or dosage; however, little is known in wheat. Here, we show TEOSINTE BRANCHED1 (TB1) regulates inflorescence architecture in bread wheat (Triticum aestivum) by investigating lines that display a form of inflorescence branching known as “paired spikelets.” We show that TB1 interacts with FLOWERING LOCUS T1 and that increased dosage of TB1 alters inflorescence architecture and growth rate in a process that includes reduced expression of meristem identity genes, with allelic diversity for TB1 found to associate genetically with paired spikelet development in modern cultivars. We propose TB1 coordinates formation of axillary spikelets during the vegetative to floral transition and that alleles known to modify dosage or function of TB1 could help increase wheat yields.
INTRODUCTION
A grass inflorescence is a group of seed-producing flowers that are arranged on specialized branches, known as spikelets. A plant’s reproductive success is strongly influenced by the number and arrangement of spikelets and flowers that form on an inflorescence, known as inflorescence architecture, as this determines the maximum number of sites available for seed production. Consequently, diversity for inflorescence architecture has been important during the domestication of crop plants, as modifications that increase flower number have helped improve yields (Meyer and Purugganan, 2013; Zhang and Yuan, 2014). This diversity is often influenced genetically by alleles that alter the dosage of genes involved in spikelet and floret development (Doebley et al., 1997; Simons et al., 2006; Miura et al., 2010; Jiao et al., 2010; Yoshida et al., 2013; Houston et al., 2013; Zhu et al., 2013; Park et al., 2014; Greenwood et al., 2017; Debernardi et al., 2017; Soyk et al., 2017). An increased understanding of the genes that regulate spikelet and floret development, and selection of alleles that alter the activity of these genes, can therefore be used as a tool to increase crop yields (Miura et al., 2010; Jiao et al., 2010; Yoshida et al., 2013; Park et al., 2014; Wang et al., 2015; Soyk et al., 2017).
Bread wheat (Triticum aestivum) produces an inflorescence with a unique architecture among crops, forming single spikelets on opposite sides of a central rachis in an alternating phyllotaxy and a terminal spikelet at the apex, with each spikelet producing multiple florets. The number of spikelets that form on an inflorescence and the number of fertile florets in each spikelet are two major determinants of yield and are influenced by the rate of inflorescence development (Rawson, 1970; Miralles et al., 2000; Slafer, 2003; González et al., 2011). Despite the importance of these traits for grain production and the potential for improvements to contribute to yield increases required to feed the world’s growing population, very little is known about how they are regulated genetically (Reynolds et al., 2009; González-Navarro et al., 2015). Recent studies have investigated wheat lines with modified inflorescence architectures to identify genes that contribute to spikelet development, including our analysis of “paired spikelets” (Dobrovolskaya et al., 2015; Boden et al., 2015; Poursarebani et al., 2015). Paired spikelets are a form of supernumerary spikelets characterized by the formation of two precisely positioned spikelets at a given rachis node rather than the typical single spikelet (Boden et al., 2015; Sharman, 1944), which are distinct from other supernumerary spikelet structures including long lateral branches that form at the base of an inflorescence, and multirowed spikelets characterized by the formation of multiple spikelets at individual nodes (Dobrovolskaya et al., 2015; Poursarebani et al., 2015; Sharman, 1967). Paired spikelet development is regulated by multiple genes, with quantitative trait locus (QTL) analysis of a four parent multiparent advanced generation intercross (MAGIC) population detecting 18 QTLs that contribute to this trait (Boden et al., 2015). One of the QTLs detected in this study was underpinned by Photoperiod-1 (Ppd-1), a pseudo-response regulator gene that controls photoperiod response pathways in wheat (Beales et al., 2007). We found that Ppd-1 regulates inflorescence architecture by controlling the expression of the central regulator of flowering, FLOWERING LOCUS T1 (FT1), which modulates the strength of the flowering signal perceived in the developing inflorescence. Alleles of Ppd-1 and FT1 that produce a weak flowering signal facilitated paired spikelet production through reduced expression of genes that regulate spikelet development, relative to Ppd-1 and FT1 alleles that produce a strong flowering signal and suppress paired spikelet formation (Boden et al., 2015).
In this study, we investigated a pair of near-isogenic lines (NILs) developed from the MAGIC population to demonstrate that an ortholog of the maize (Zea mays) domestication gene, TEOSINTE BRANCHED1 (TB1), regulates wheat inflorescence architecture in a dosage-dependent manner. We show that TB1 protein interacts with FT1 and that increased dosage of TB1 promotes paired spikelet production and delays inflorescence growth by reducing expression of meristem identity genes during early developmental stages. We also identify variant alleles for TB1 on the B and D wheat genomes and show that these alleles are at least partially responsible for diversity of inflorescence architecture in modern wheat cultivars.
RESULTS
The Bread Wheat hb Line Displays Inflorescence and Plant Architecture Phenotypes
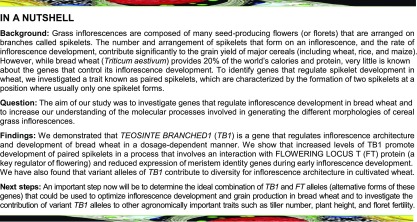
To identify genes that regulate inflorescence architecture in wheat, we investigated a pair of near-isogenic lines (NILs) derived from a single line of a four-parent MAGIC population for spring wheat (Huang et al., 2012). This line (MAGIC line 0053) was chosen because individuals within field plots displayed either wild-type inflorescences or inflorescences with multiple paired spikelets, suggesting that alleles for one or more gene(s) regulating spikelet architecture were segregating within this line. Seeds were independently selected from plants with wild-type and paired spikelet-producing inflorescences over multiple generations (Supplemental Figure 1) to produce true-breeding lines that formed either wild-type inflorescences or highly branched inflorescences with multiple paired spikelets (Figure 1A), which will henceforth be referred to as highly-branched (hb). In hb plants, 43.3% ± 3.8% of nodes on the inflorescence (aka rachis nodes) produced paired spikelets, with 28.7% ± 3.9% of all nodes containing fertile secondary spikelets (Figure 1B). Consistent with our previous analysis of this trait (Boden et al. 2015), paired spikelets were found most frequently at central nodes of the inflorescence, and secondary spikelets formed immediately adjacent to and directly below the primary spikelet during early developmental stages (Supplemental Figure 2).
Figure 1.
Inflorescence Architecture and Development and Tiller Number Are Modified in hb Plants.
(A) Wild-type (WT) and hb inflorescences (an image with secondary spikelets highlighted in blue is shown at the right). Bar = 1 cm.
(B) and (C) Frequency of total paired spikelets (TPS) and complete fertile paired spikelets (CPS) (B) and rachis node numbers of wild type (black) and hb (white) inflorescences (C).
(D) and (E) Flowering time of wild-type and hb plants, measured by days to head emergence (D) and total leaf number (E).
(F) Days until double ridge (since germination) for the wild type and hb.
(G) and (H) Inflorescence development phenotypes of wild-type (solid line) and hb (dashed line) plants, measured at intervals defined by leaf emergence. Inset in (H) is mature inflorescence length for the wild type and hb. Bar = 1 mm.
(I) and (J) Tiller numbers of wild-type and hb plants. Bar = 10 cm.
Data are mean ± se. n = 8 plants for (B) and (C), n = 10 for (D) to (F) and (J), n = 4 for (H), and n = 10 for (H) inset. All images are representatives of individuals used for quantification. **P < 0.01 and ***P < 0.001, compared with wild-type values.
As paired spikelet development is facilitated by genetic and environmental conditions that delay flowering and is associated with an increase in the number of positions (nodes) available for spikelet production, we tested if flowering time and rachis node numbers were affected in hb plants, relative to the wild type (Sharman, 1944; Boden et al., 2015). Under long-day photoperiods (16 h light/8 h dark), we observed no difference in the number of days until inflorescence emergence or the number of rachis nodes between the two genotypes, confirming that paired spikelet production is independent of flowering time in these lines (Figures 1C and 1D). However, we did notice a slight increase in the total leaf number of hb plants, relative to the wild type, suggesting that inflorescence development is delayed in hb plants (Figure 1E). To investigate this further, we analyzed the time required for shoot apical meristems (SAMs) of wild-type and hb lines to reach the double-ridge stage, which is the stage immediately after the vegetative-to-reproductive transition of the SAM occurs. We found that SAMs of wild-type plants reached the double-ridge stage earlier than hb plants, which explains the slight increase in leaf number of hb plants, relative to the wild type (Figures 1E and 1F).
Further investigation of inflorescence development and growth in hb plants, relative to wild-type plants, at intervals defined by leaf emergence from leaf 3 (L3) until leaf 8 (L8) revealed that there were also significant delays of inflorescence growth in hb plants at stages L5 and L6, which is when the terminal spikelet forms and spikelet primordia begin developing florets (Figures 1G and 1H). Growth of hb inflorescences was delayed dramatically at the L7 stage, relative to the wild type; the delay was allayed by L8, when no difference in inflorescence length was detected between the two genotypes (Figure 1H; Supplemental Figure 3). There was also no significant difference in the length of mature inflorescences between the two genotypes (inset of Figure 1H) nor was there a significant difference in leaf emergence or leaf elongation, suggesting that the inflorescence development and growth phenotypes are not the consequence of a delayed plastochron (Supplemental Figure 4). In addition to the inflorescence phenotypes, we found that hb plants produced fewer tillers than wild-type plants (Figures 1I and 1J). Dissection of leaf sheaths surrounding immature tillers showed that the hb plants produced tiller buds at each vegetative node that were smaller than those produced in wild-type plants (Supplemental Figure 5), suggesting that the reduced tiller numbers in the hb plants were due to suppressed outgrowth of the tiller buds rather than failure to develop tillers. Taken together, these results suggest that there is a pleiotropic effect on growth of lateral organs from the main shoot in hb plants, relative to the wild type.
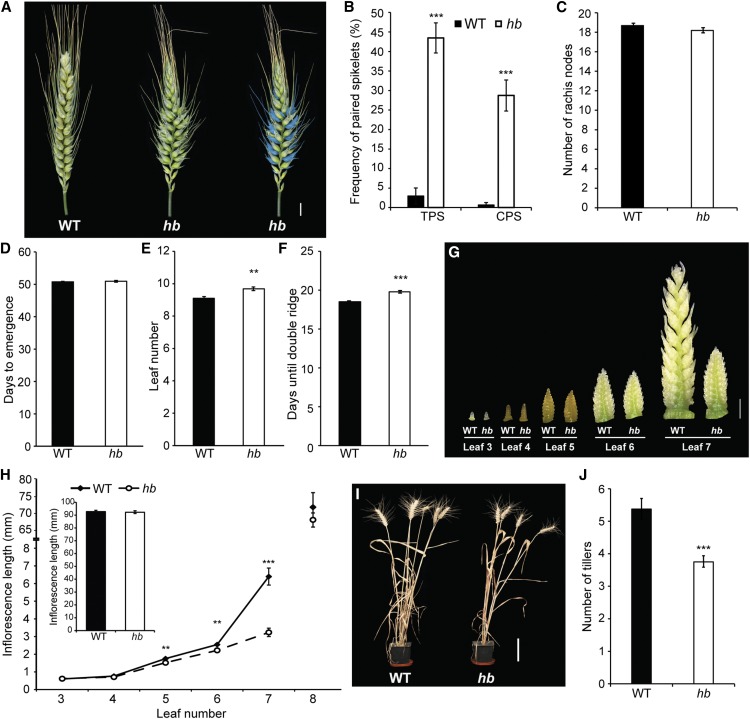
Chromosome 4D Is Duplicated in hb Lines
To identify the genetic factor(s) contributing to the inflorescence and plant architecture phenotypes of the hb NIL, we initially performed genotype analysis using the 90,000 single nucleotide polymorphism wheat array (Wang et al., 2014), which suggested there was a partial or complete duplication of chromosome 4D (data not shown). To investigate this further, we performed cytogenetic analysis of chromosomes from wild-type and hb plants, which showed that the hb line is tetrasomic for chromosome 4D, suggesting that increased dosage of chromosome 4D facilitates paired spikelet formation (Figures 2A and 2B). To test if regulation of paired spikelets by chromosome 4D dosage is a dominant or semidominant trait, we crossed the hb line to two spring wheat cultivars that do not produce paired spikelets: Sunstate and Pastor. Phenotype analysis showed that the first filial generation (F1) of offspring for both crosses produced paired spikelets (Supplemental Figure 6). As paired spikelet development was reduced in the F1 plants of the hb crosses, relative to the tetrasomic hb line, we conclude that this is a semidominant trait (Figure 1B; Supplemental Figure 6).
Figure 2.
Genetic Characterization and Expression Analysis Reveal Increased Dosage of TB-D1 in hb NILs.
(A) and (B) FISH analysis of wild-type (A) and hb (B) lines; copies of chromosome 4D are labeled.
(C) Schematic diagram of chromosome 4DS region in proximity to Rht-D1. Gene identities: DUF6-like gene, TRIAE_CS42_4DS_TGACv1_362748_AA1181880; Zn-finger protein, TRIAE_CS42_4DS_TGACv1_363130_AA1183390.
(D) ddPCR analysis of haploid gene copy number for TB-D1 in wild-type (black) and hb (white) lines.
(E) Relative expression of TB-A1, TB-B1, and TB-D1 in developing inflorescences of wild-type (solid bars) and hb (white bars) plants, at double ridge (DR; gray), leaf 5 (L5; blue), leaf 6 (L6; green), and leaf 7 (L7; magenta) developmental stages. Expression is relative to TB-B1 in the wild type.
(F) Expression analysis of TB-A1 (gray), TB-B1 (red), and TB-D1 (purple) in vegetative tissues of wild-type (solid bars) and hb (white bars) plants. Data of ddPCR and expression analyses are mean ± se of three to four biological replicates. *P < 0.05 and **P < 0.01, compared with wild-type values.
TB-D1 Expression Is Increased in Inflorescences and Tillers of hb Lines
Our previous analysis of QTLs that contribute to paired spikelet development within the four parent MAGIC population detected a QTL with a significant additive genetic effect on the short arm of chromosome 4D (4DS), which was detected using a marker positioned within the major Green Revolution gene Rht-D1 (Reduced height-D1; Rht-2) (Boden et al., 2015). Dominant dwarfing alleles for Rht-1 and Rht-2 (Rht-B1b and Rht-D1b, respectively) restrict plant growth and impede inflorescence development by reducing the plant’s sensitivity to the growth-promoting hormone gibberellin (Pearce et al., 2011; Hedden, 2003; Boden et al., 2014). Both the wild-type and hb lines contain the Rht-B1b (dwarfing) and Rht-D1a (wild-type) alleles, and we hypothesized that reduced sensitivity to gibberellin via increased dosage of Rht-D1a may facilitate paired spikelet development by restricting growth of spikelet meristems and the developing inflorescence. To test this hypothesis, we examined inflorescence architecture in a pair of NILs that contained either the wild-type Rht-D1a allele or the dwarfing Rht-D1b and in Rht-D1c (Rht10) lines that contain multiple copies of Rht-D1b (Pearce et al., 2011), relative to its NIL that contains a single copy. We did not detect paired spikelets in either of these genotypes, suggesting that increased dosage of Rht-D1 is not responsible for the modified inflorescence architecture of the hb line (Supplemental Figure 7).
Through our analysis of Rht-D1 and the surrounding genetic region underlying the QTL on 4D, we noticed that a wheat ortholog of an important maize domestication gene, TB1, is closely linked to Rht-D1 (Figure 2C). TB1 encodes a class II TCP (TEOSINTE BRANCHED1, CYCLOIDEA, PCF1) transcription factor, which is a member of a plant-specific family of transcription factors that bind to promoter regions of genes known to regulate developmental processes including branching, floral symmetry, leaf development, and germination (Doebley et al., 1997; Luo et al., 1996; Kosugi and Ohashi, 1997; Tatematsu et al., 2008). TB1 orthologs regulate tiller number and influence inflorescence architecture traits in maize, rice (Oryza sativa), and barley (Hordeum vulgare) (Doebley et al., 1995, 1997; Takeda et al., 2003; Studer et al., 2011; Ramsay et al., 2011), with the dominant Tb1 allele of domesticated maize restricting growth of lateral branches via increased gene expression, relative to the tb1 allele of the wild progenitor, teosinte (Zea mays ssp parviglumis) (Doebley et al. 1997); we therefore hypothesized that increased dosage of the wheat TB1 ortholog from the D genome (TB-D1) may be responsible for the paired spikelet and tiller phenotypes of hb plants.
Using digital droplet PCR (ddPCR), we confirmed that hb plants contain a haploid complement of two copies for TB-D1, and wild-type plants contain one copy (Figure 2D). We then compared expression levels for TB1 from the A (TB-A1), B (TB-B1), and D genomes in wild-type and hb plants at defined stages of inflorescence development and in vegetative tissues that included tiller buds, emerging tillers, leaves, roots, and germinating seedlings. We observed that TB-B1 and TB-D1 are expressed predominantly in developing inflorescences and tillers, relative to leaves, roots, and seedlings, and that TB-A1 expression is significantly lower than that of TB-B1 and TB-D1 (Figures 2E and 2F; P < 0.0001). Our analysis showed that TB-B1 and TB-D1 were expressed comparably in developing inflorescences and emerging tillers, and transcript levels for both genes were 4- to 10-fold higher in tiller buds (Supplemental Figure 8). We found that TB-D1 transcript levels were significantly higher in developing inflorescences of hb plants, relative to the wild type, at the double-ridge stage of development that is pivotal for paired spikelet formation (Boden et al., 2015) and during the L5–L7 developmental stages when inflorescence growth is delayed in hb plants (Figures 1G and 2E). We also detected a significant increase in TB-D1 transcripts in tiller buds and emerging tillers of hb plants, relative to the wild type, while no significant difference in TB-D1 transcript levels were detected between the two genotypes in roots, leaves, or germinating seedlings (Figure 2F). These results demonstrate that TB-D1 transcript levels are significantly higher in tissues of the hb plants that display architecture phenotypes, relative to wild-type plants, which was not observed for most other genes that are in proximity to TB-D1 on chromosome 4D (Supplemental Figure 9). These results are therefore supportive of increased TB-D1 expression promoting paired spikelet development and suppressing tiller outgrowth.
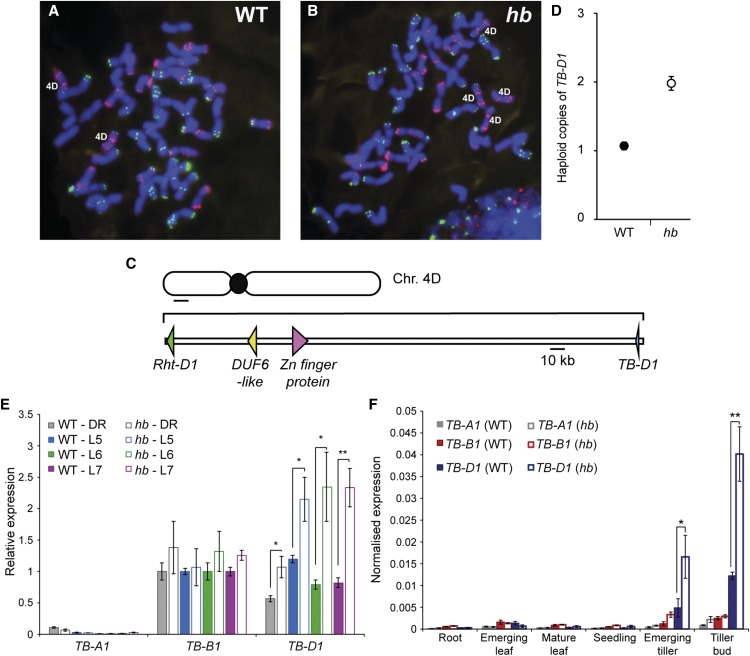
TB-D1 Regulates Inflorescence and Plant Architecture in a Dosage-Dependent Manner
To confirm genetically that TB-D1 is the gene responsible for the inflorescence and plant architecture phenotypes observed in hb plants, we crossed the hb line to a wheat TILLING mutant of cultivar Cadenza (Krasileva et al., 2017), line Cadenza1721, which contains a premature stop codon mutation (C>T, 49 bp of coding sequence) in TB-D1 and is henceforth referred to as tb-d1. As the crosses to Sunstate and Pastor showed that increased dosage of chromosome 4D is semidominant for paired spikelet production (Supplemental Figure 6), we hypothesized that F1 plants trisomic for chromosome 4D that contain two functional copies and one nonfunctional copy of TB-D1 (hb x tb-d1) would not produce paired spikelets and would produce more tillers, relative to F1 plants trisomic for chromosome 4D that contain three functional copies of TB-D1 (hb x Cadenza). Analysis of 29 F1 plants from the hb x Cadenza cross supported the results from the hb x Sunstate and hb x Pastor crosses, with F1 plants found to develop paired spikelets (6.34% ± 1.02% of nodes) (Figures 3A and 3C). Conversely, F1 plants of the hb x tb-d1 cross did not produce paired spikelets (45 plants) or formed rudimentary secondary spikelets at a very low frequency (one per inflorescence; five plants) (Figures 3A and 3C). F1 plants of the hb x tb-d1 cross also produced more tillers (3.86 ± 0.1) than the hb x Cadenza F1 plants (3.1 ± 0.125; P < 0.001) (Figures 3B and 3C). Based on these results, we conclude that TB-D1 is likely to be the gene responsible for the paired spikelet and tiller number phenotypes caused by increased dosage of chromosome 4D in the hb line.
Figure 3.
Increased Dosage of TB1 Regulates Paired Spikelet and Tiller Phenotypes in hb Plants.
(A) to (C) Inflorescence ([A] and [C]) and plant ([B] and [C]) architecture phenotypes of F1 offspring plants from crosses between hb and cv Cadenza (WT), hb, and the tb-d1 mutant line (Cadenza1721), with quantification (C) of frequency of nodes with secondary spikelets per inflorescence (given as total paired spikelets) and tillers per plant.
(D) to (F) Plant (D) and inflorescence (E) architecture phenotypes of hb and revertant lines, with quantification (F) of paired spikelets per inflorescence and tillers per plant. Secondary spikelets are highlighted in blue. n.d., not detected.
(G) KASP marker analysis of TB-B1 in hb (white) and revertant (green) lines, along with no template control (NTC; black).
Bars = 10 cm in (B) and (D) and 1 cm in (A) and (E). All images are representatives of individuals used for quantification. In (C), data are mean ± se of 29 F1 plants for hb x Cadenza and 50 plants for hb x tb-d1. *P < 0.05 and ***P < 0.001, compared with values for the F1 (hb x Cadenza) in (C) and to hb in (F).
This conclusion was further supported by analysis of plants from a revertant line that derived spontaneously from an individual hb plant, which no longer formed paired spikelets, produced more tillers, and were significantly taller than hb plants (Figures 3D to 3F; Supplemental Table 1). We hypothesized that the increased height of these plants was caused by loss of the dwarfing Rht-B1b allele and that the absence of paired spikelets and increased tiller numbers occurred through loss of the closely linked TB-B1, possibly through loss of chromosome 4B. Marker analysis confirmed that TB-B1 and Rht-B1b were indeed absent in the revertant lines (Figure 3G; Supplemental Figure 10). Taken together with the results from F1 plants of the hb x tb-d1 cross and the expression analysis, we conclude that increased dosage of TB1, with contributions from both the wheat D and B genomes, promotes paired spikelet development and reduces outgrowth of tillers in hb plants.
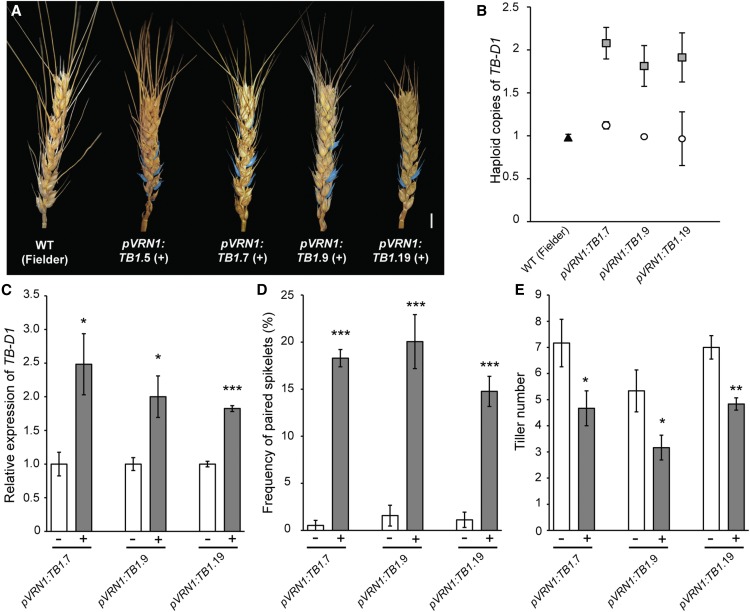
To further confirm genetically that increased dosage of TB-D1 promotes paired spikelet development in wheat, we generated transgenic lines that expressed TB-D1 using the well-characterized promoter of VERNALIZATION1 (VRN1) (Alonso-Peral et al., 2011) in the genetic background of Fielder, which forms paired spikelets at a very low frequency. VRN1 encodes a MADS box transcription factor that is expressed robustly in the developing inflorescence during the vegetative to reproductive transition, including the double-ridge stage when paired spikelets form (Yan et al., 2003; Alonso-Peral et al., 2011; Boden et al., 2015). We also confirmed that VRN1 is expressed in tiller buds and emerging tillers of wild-type plants (Supplemental Figure 11). The VRN1 promoter was used because initial attempts to overexpress TB-D1 using the Ubiquitin and Actin1 promoters failed to produce transgenic seedlings, and sequence for the TB-D1 promoter was not available. This experiment therefore increased the number of genomic copies for TB-D1 and promoted expression of the additional copies of TB-D1 within the inflorescence at stages pivotal for paired spikelet development. From this experiment, we identified 14 independent pVRN1:TB-D1 transgenic lines that formed paired spikelet-producing inflorescences (Figure 4; Supplemental Figure 12 and Supplemental Table 2). Genotype analysis of T1 transgenic lines identified lines that contained 1.5 or 2 haploid copies of TB-D1, while null transgenic plants and control Fielder plants contained 1 haploid copy of TB-D1 (Figure 4B; Supplemental Table 2). We selected three independent pVRN:TB1 lines to perform molecular and phenotypic analysis of T2 generation plants: pVRN:TB1.7, 9, and 19 (Figures 4C to 4E). RT-qPCR analysis showed that TB-D1 transcript levels were significantly higher in the developing inflorescences of each positive transgenic line, relative to their respective null transgenic siblings (Figure 4C). Phenotype analysis showed that inflorescences of the pVRN:TB1 transgenic lines formed secondary spikelets at 15 to 20% of the rachis nodes and produced fewer tillers than the null transgenic lines, which did not produce secondary spikelets (Figures 4D and 4E). These results therefore confirm our hypothesis that increased dosage of TB1 promotes paired spikelet development and reduces tiller outgrowth in wheat.
Figure 4.
Increased Dosage of TB-D1 Promotes Paired Spikelet Development and Reduces Tiller Number in Transgenic Lines.
(A) Inflorescences of pVRN1:TB1 transgenic plants (T1 generation), and the wild-type untransformed control (Fielder). Secondary spikelets are highlighted in blue. Bar = 1 cm.
(B) TB-D1 haploid gene copy number for pVRN1:TB1 transgenic plants (gray) and control (white, null transgenic; black, Fielder) lines.
(C) TB-D1 expression in developing inflorescences of pVRN1:TB1 transgenic lines (gray), relative to null siblings (white).
(D) and (E) Frequency of paired spikelets ([D]; given as total paired spikelets) and tiller number phenotypes (E) for pVRN1:TB1 transgenic lines (gray), relative to null siblings (white).
For (C), data are mean ± se of 4 biological replicates; for (D) and (E), data are mean ± se of 3 to 5 biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.001, compared with values for null siblings.
TB1 Interacts with FT1 and Expression of Meristem Identity Genes Is Reduced in Lines with Increased Dosage of TB-D1
To identify how increased TB1 dosage may promote paired spikelet development, we surveyed the literature for molecular interactions that involve TB1 orthologs and associate with genetic pathways known to influence inflorescence architecture. In Arabidopsis thaliana, rice, and wheat, the central integrator of flowering FT interacts with FLOWERING LOCUS D (FD; or FDL in wheat) and 14-3-3 proteins to form a floral activating complex, which induces expression of floral meristem identity genes (Abe et al., 2005; Wigge et al., 2005; Taoka et al., 2011; Li et al., 2015). It has also been shown that the Arabidopsis homolog of TB1, BRANCHED1 (BRC1) (Aguilar-Martínez et al., 2007), negatively regulates the vegetative-to-reproductive transition of axillary branches by interacting with FT and suppressing its ability to promote expression of floral meristem identity genes (Niwa et al., 2013). As we have shown previously that deletion of FT-B1 promotes paired spikelet development (Boden et al., 2015), we hypothesized that TB-D1 interacts with FT1 and that increased dosage of TB-D1 facilitates paired spikelet development by reducing the availability of FT1 to promote expression of spikelet meristem identity genes within the developing inflorescence.
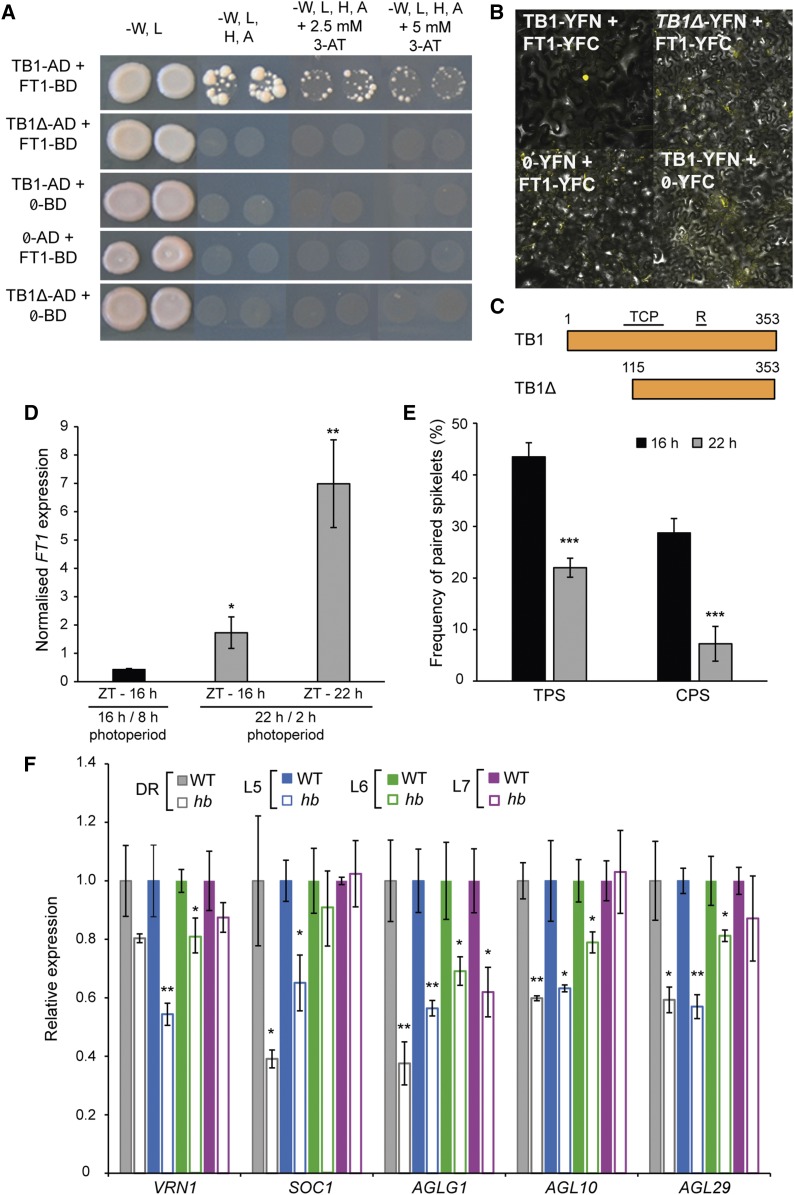
To test this hypothesis, we performed yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) analysis to determine if TB-D1 and FT1 proteins interact. Both the Y2H and BiFC analyses demonstrated that TB-D1 interacts directly with FT1, while no interaction was detected in control experiments, which included a truncated version of TB-D1 (TB1Δ) that lacks the first 115 amino acids of TB1, a region demonstrated previously to be essential for the interaction between BRC1 and FT from Arabidopsis (Figures 5A to 5C; Supplemental Figure 13) (Niwa et al., 2013). To test this interaction further and determine if the effect of increased TB-D1 levels could be suppressed by enhanced FT1 activity, we grew hb plants under extended long-day (LD) conditions (22-h/2-h photoperiod) and standard LD conditions (16 h/8 h). Under the extended LD conditions, FT1 transcript levels were significantly higher at dusk (Zeitgeber time [ZT] 22 h) and at the time point that would correspond to dusk in the standard LD photoperiod (ZT 16 h), relative to the levels at dusk under the standard LD conditions (ZT 16 h; Figure 5D).
Figure 5.
TB1 Interacts with FT1 and Modulates Expression of Meristem Identity Genes.
(A) Y2H assay between TB1 and FT1, including control reactions with truncated TB-D1 (TB1Δ) and empty vectors (Ø), grown on varying selective media.
(B) BiFC assay for TB1 and FT1, and control reactions including truncated TB-D1 (TB1Δ) and empty vectors (Ø).
(C) Schematic of full-length TB1 and truncated TB1 (TB1Δ) proteins used for Y2H and BiFC assays; TCP and R refer to domains of TB1 protein, and numbers indicate amino acid positions.
(D) and (E) Expression analysis of FT1 at dusk (D) and frequency of total paired spikelets (TPS) and complete fertile paired spikelets (CPS) in hb plants under standard LD photoperiods (black) and extended LD photoperiods (gray) (E).
(F) Expression of meristem identity genes in developing inflorescences of hb (white bars), relative to the wild type (solid bars), at double ridge (DR; gray), leaf 5 (L5; blue), leaf 6 (L6; green), and leaf 7 (L7; magenta) developmental stages. Data are mean ± se; n = 4 biological replicates for (D) and (F) and n = 10 biological replicates for (E). *P < 0.05, **P < 0.01, and ***P < 0.001, compared with standard LD values ([D] and [E]) and wild-type values (F).
We also found that paired spikelet development is significantly suppressed under the extended LD photoperiod, with reduced production of total paired spikelets and secondary spikelets with fertile florets, relative to plants grown under standard LD (Figure 5E). We then tested if increased dosage of TB-D1 affects expression of genes that regulate spikelet development by analyzing transcript levels of meristem identity genes from developing inflorescences of wild-type and hb plants (Figure 5F). These genes included VRN1, SOC1, AGAMOUS-LIKE GENE1, AGAMOUS-LIKE10 (AGL10), and AGL29, whose expression was found previously to be reduced in inflorescences of paired spikelet producing lines, including a mutant containing a deletion of FT-B1 (Boden et al., 2015). We found that transcript levels for all meristem identity genes were significantly reduced during early developmental stages in inflorescences of hb plants, relative to those of wild-type plants (except for VRN1 at the DR stage), which included stages pivotal for paired spikelet development (Figure 5F). This result was supported by analysis showing that transcript levels for meristem identity genes were significantly reduced in developing inflorescences of the pVRN1:TB1 transgenic plants, relative to null transgenic siblings (Supplemental Figure 14). At the leaf 7 developmental stage, transcript levels for VRN1, SOC1, AGL10, and AGL29 were not significantly different in wild-type and hb plants, which is consistent with the observed completion of spikelet and floret development in hb inflorescences at the L7 stage in preparation for growth between emergence of L7 and 8 (Figures 1G, 1H, and 5F). We confirmed that the changes in activity of meristem identity genes were not associated with reduced expression of FT1 in hb plants, relative to the wild type (Supplemental Figure 14). Based on these results, we conclude that TB1 interacts with FT1 and that increased dosage of TB1 promotes paired spikelet development and delays inflorescence growth between the L5 and L7 stages by reducing the expression of spikelet meristem identity genes.
Allelic Diversity for TB1 Regulates Inflorescence Architecture in Modern Wheat Cultivars
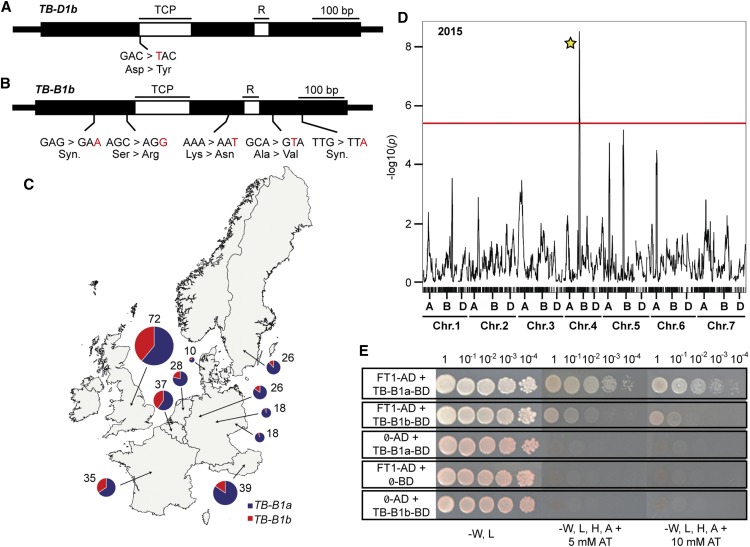
As TB1 contributes to important yield traits including inflorescence architecture and growth in other plants, we hypothesized that there are allelic variants for TB1 among modern wheat cultivars, which contribute to variation in spikelet development. To test this hypothesis and determine if a TB1 allele may be responsible for the paired spikelet QTL detected previously on chromosomes 4B and 4D (Boden et al., 2015), we sequenced TB-B1 and TB-D1 from the cultivars used to generate the four-parent spring wheat MAGIC population: Baxter, Chara, Westonia, and Yitpi. No sequence polymorphism was detected for TB-B1 among the four cultivars; however, one polymorphism was detected in the open reading frame of TB-D1 from Baxter, which is a nonsynonymous mutation (G-328 bp-T; D110Y) at the beginning of the conserved TCP domain (Figure 6A; Supplemental Figures 15 and 16). Interestingly, our previous analysis identified Baxter as the cultivar that contributes a positive genetic effect for the paired spikelet QTL on chromosome 4D that displayed LOD scores of 1.27 to 5.44 across five experiments, which were detected using the marker for Rht-D1 that is in proximity to TB-D1 (Boden et al., 2015). Taken together, these results suggest that the Baxter allele for TB-D1 (referred to as TB-D1b) may be responsible for the paired spikelet QTL detected on chromosome 4D.
Figure 6.
Allelic Variation for TB-B1 and TB-D1 Contributes to Inflorescence Architecture Diversity in Spring and Winter Wheat.
(A) and (B) Schematic diagrams of variant alleles for TB-D1 ([A]; TB-D1b) from spring wheat and TB-B1 ([B]; TB-B1b) from winter wheat, with positions of polymorphisms (indicated in red text) and molecular effect indicated (Syn., synonymous). TCP and R refer to genic regions that encode domains of the TB1 protein.
(C) Geographical distribution of TB-B1a (purple) and TB-B1b (red) alleles in European wheat cultivars of the GEDIFLUX collection.
(D) Genome-wide composite interval mapping scan displaying QTLs that contribute to paired spikelet development from the UK winter wheat MAGIC population from the 2015 trial, including a highly significant QTL on chromosome 4BS (indicated by yellow star). Red line indicates threshold of high significance (P < 0.01).
(E) Y2H assay between TB-B1 alleles (TB-B1a and TB-B1b) and FT1, including control reactions with empty vectors (Ø), grown on varying selective media. A 10-fold serial dilution was performed for each interaction experiment.
To gain further evidence of allelic variation for TB1, we surveyed 230 modern winter wheat cultivars for paired spikelet development over two growing seasons (Supplemental Figure 17, Supplemental Table 3, and Supplemental Data Set 1). We identified 96 cultivars that produced paired spikelets, including Abbot, Ambrosia, Einstein, Gladiator, and Hyperion, which formed secondary spikelets at 20 to 35% of rachis nodes (Supplemental Figure 17 and Supplemental Table 3). Digital droplet PCR and sequence analysis for TB-B1 and TB-D1 on a subset of these cultivars showed that there was no CNV for TB-B1 or TB-D1 (Supplemental Figure 18) and no sequence variant alleles for TB-D1. However, we did detect an allele for TB-B1 (referred to as TB-B1b) that contained five sequence polymorphisms in complete linkage disequilibrium, including two synonymous mutations (G-198 bp-A, G-912 bp-A) and three nonsynonymous mutations (C-309 bp-G, S103R; A-663 bp-T, K221N; C-812 bp-T, A271V) (Figure 6B; Supplemental Figures 19 and 20). KASP-based marker analysis showed that the TB-B1b allele was widely distributed across a geo-referenced panel of 264 European wheat varieties (the GEDIFLUX panel) and was observed most frequently in cultivars of Great Britain, Belgium, and France (Figure 6C; Supplemental Table 4 and Supplemental Data Set 2).
We also found that both the TB-B1a (wild type; reference sequence) and TB-B1b alleles were present among the founder genotypes used to generate the eight parent UK winter wheat MAGIC population, with Rialto, Robigus, and Soissons containing the TB-B1a allele and Alchemy, Brompton, Claire, Hereward, and Xi 19 containing the TB-B1b allele (Supplemental Table 4). We hypothesized, therefore, that phenotype and genetic analyses for paired spikelets within this MAGIC population would identify a QTL on chromosome 4B in the region proximal to TB-B1. Paired spikelet measurements were recorded for field-grown replicated trials over two successive years (643 lines with two replicates in 2015 and 493 lines with two replicates in 2016), with broad sense heritability estimated at 72.4% and 94.1% for 2015 and 2016, respectively. In both years, genetic mapping identified a highly significant QTL in a region of chromosome 4B that included TB-B1, which was bound by markers wsnp_BF482960B_Ta_1_4 (47.64 cM, chr 4B position 28,954,488 bp) and RAC875_c27536_611 (48.65 cM, chr 4B position 32,250,720 bp) and explained 6% and 7.5% of phenotypic variance (R2) in the 2015 and 2016 trials, respectively (Figure 6D; Supplemental Figure 21 and Supplemental Tables 5 and 6). Alleles donated by Rialto, Robigus, and Soissons were predicted to contribute positively toward the paired spikelet trait, suggesting that the TB-B1a allele facilitates paired spikelet development (Supplemental Table 6). This conclusion was supported by TB-B1-specific marker analysis of a subset of 212 MAGIC lines that formed either wild-type or paired spikelet-producing inflorescences, which confirmed the TB-B1a allele was associated genetically with paired spikelet development (McNemar’s Test, P < 0.05; Supplemental Table 7). To test if the nonsynonymous mutations of TB-B1b alter the function of TB-B1, we performed Y2H analysis to determine if TB-B1a and TB-B1b both interact with FT1. Both proteins interacted with FT1; however, analysis of serial dilutions indicated that the interaction between TB-B1a and FT1 was slightly stronger than the interaction between TB-B1b and FT1 (Figure 6E). These results show that variant TB1 alleles are present on the B and D genomes of winter and spring wheat, respectively, and that diversity for TB1 modulates inflorescence architecture in modern wheat cultivars.
DISCUSSION
Inflorescence architecture contributes significantly to seed production in plants and is a major determinant of crop yield. Seed production can be increased by developing inflorescences with more elaborate branching patterns, which can be achieved by breeding with alleles that modify the activity of genes involved in spikelet or floret development (Miura et al., 2010; Jiao et al., 2010; Yoshida et al., 2013; Bommert et al., 2013; Meyer and Purugganan, 2013; Zhu et al., 2013; Park et al., 2014; Poursarebani et al., 2015; Soyk et al., 2017). In this study, we have shown that increased dosage of TB1 promotes inflorescence branching in the form of paired spikelets and that allelic diversity for TB1 is associated with spikelet architecture traits in modern wheat cultivars.
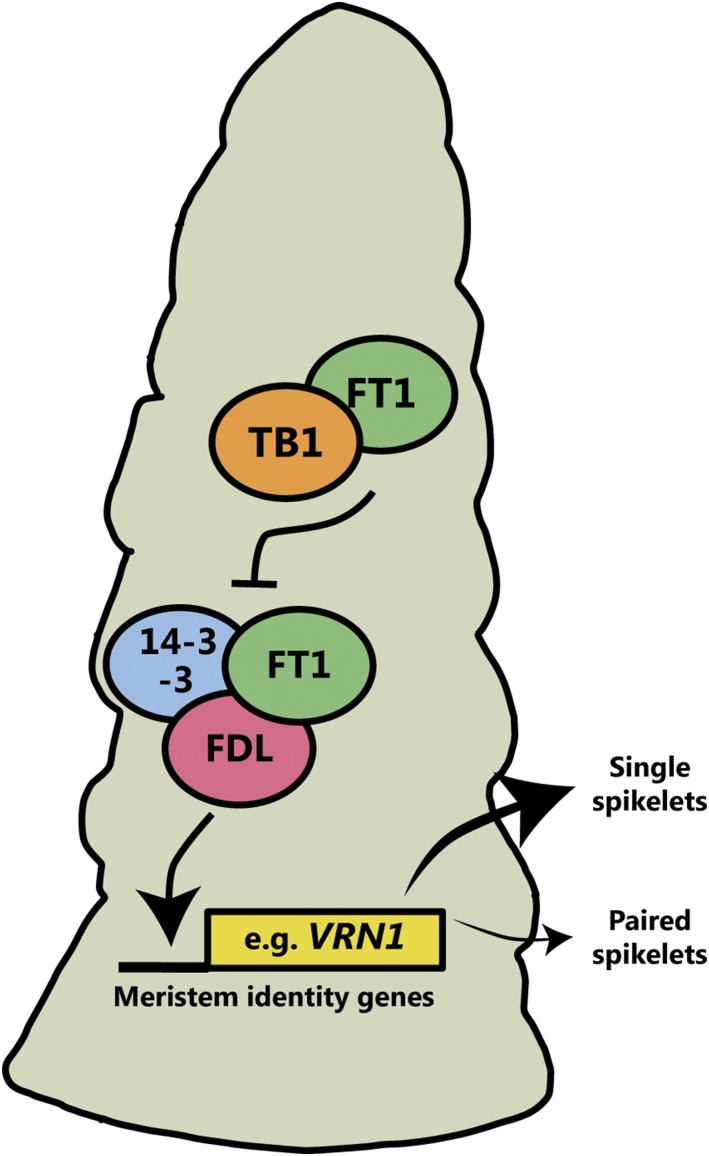
Our analysis of the near-isogenic and transgenic lines, and the F1 plants from the hb x tb-d1 cross, demonstrates that increased dosage of TB1 promotes paired spikelet development, which is associated with reduced expression of spikelet meristem identity genes. The reduced expression of the meristem identity genes is consistent with our previous analysis of paired spikelet-producing genotypes that have loss-of-function alleles for key flowering genes, Ppd-D1 and FT1 (Boden et al., 2015). Based on these results, and protein interaction experiments that show TB1 interacts with FT1, we propose a model for TB1-dependent regulation of inflorescence architecture (Figure 7), whereby increased dosage of TB1 facilitates paired spikelet development by reducing the availability of FT1 to form part of the floral activating complex and promote expression of spikelet meristem identity genes (Taoka et al., 2011; Boden et al., 2015; Li et al., 2015). This effect of increased TB1 dosage is consistent with studies of the Arabidopsis TB1 homolog, BRC1, which showed BRC1 is a negative regulator of flowering that interacts with FT and restricts the expression of floral meristem identity genes within axillary meristems (Niwa et al., 2013). We propose that this role for TB1 is conserved in wheat, where it helps regulate the development of axillary spikelet meristems from the inflorescence meristem during the vegetative to floral transition. At standard levels, TB1 facilitates formation of unbranched primary spikelets along the inflorescence; however, when TB1 levels are increased, the maturation of spikelet meristems is delayed, which results in development of a short branch that is limited to two spikelets (Figure 7). This effect of increased TB1 levels is consistent with studies performed in rice and tomato (Solanum lycopersicum), which have demonstrated that inflorescences with more elaborate branching patterns and increased yields can be developed by reducing the activity of flowering signals and extending the duration of meristem maturation (Park et al., 2012, 2014; Yoshida et al., 2013; Kyozuka et al., 2014; Wang et al., 2015; Lemmon et al., 2016; Soyk et al., 2017). An important area for future analysis will be to use wheat NILs that contain diverse alleles of TB1 and FT1 to confirm the TB1:FT1 interaction genetically and to examine the optimum combination of TB1 and FT1 alleles for improved inflorescence architecture. These NILs could also be used to investigate possible TB1-dependent regulation of inflorescence development and architecture that may occur independently of its interaction with FT1, possibly through regulation of meristem size or meristem dormancy.
Figure 7.
Model Explaining the Effect of Increased Dosage of TB1 on Spikelet Development in Wheat.
Increased levels of TB1 restrict the interaction of FT1 with other members of the floral activating complex (FDL and 14-3-3), which reduces expression of meristem identity genes (e.g., VRN1) and facilitates paired spikelet development. At standard levels of TB1, the floral activating complex promotes robust expression of meristem identity genes, and paired spikelet development is suppressed.
Our observation that increased dosage of TB1 promotes paired spikelet development and reduces tiller numbers in wheat suggests that there is a conserved role for this gene in regulating inflorescence and plant architecture of cereals. In maize, the Tb1 allele of domesticated cultivars is expressed at a higher level than the tb1 allele of teosinte, and it suppresses development and growth of axillary branches (Doebley et al., 1997; Hubbard et al., 2002). Consequently, the Tb1 allele modifies plant architecture by suppressing tiller development to produce a monoculm plant, which is consistent with observations in barley, rice, and wheat that show TB1 negatively regulates tiller outgrowth (Doebley et al., 1995, 1997; Hubbard et al., 2002; Takeda et al., 2003; Lewis et al., 2008; Ramsay et al., 2011). In regard to inflorescence architecture, the Tb1 allele contributes to development of maize ears with cupules that contain two spikelets, relative to the less active tb1 allele of teosinte that is associated with formation of single spikelet cupules (Doebley et al., 1995). Taken together with results shown here, these reports suggest that TB1 has a conserved role among cereals in regulating the rate at which an axillary meristem completes its development into a spikelet meristem, such that increased dosage of TB1 delays maturation of the spikelet meristem to promote formation of spikelet pairs rather than single spikelets. This conclusion is supported by a report of wheat plants overexpressing the maize TB1 gene that produced fewer tillers and formed inflorescences with an increased number and density of rachis nodes and many small spikelets containing incomplete florets, which is consistent with a description of paired spikelets (Lewis et al., 2008). This effect of TB1 dosage on spikelet architecture may also explain the six-rowed spikelet phenotype of barley lines that contain loss-of-function alleles of the barley TB1 ortholog, INTERMEDIUM-C (INT-C/HvTB1). Mutant alleles of HvTB1 facilitate development and outgrowth of fertile lateral spikelets, which are sterile in two-rowed barley that contain the wild-type allele of HvTB1 (Ramsay et al., 2011). Based on results presented here, the improved fertility of lateral spikelets in barley lines that carry loss-of-function mutant alleles of HvTB1 may be explained by reduced dormancy of the lateral spikelet primordia and associated with increased expression of meristem identity genes within the lateral spikelets. This hypothesis is supported by results from rice showing that an OsTB1 allele (SCM3) that confers increased TB1 expression is associated with increased dormancy of axillary meristems to form inflorescences with more primary and secondary panicle branches and that transcript levels of MADS box genes were altered by overexpression of OsTB1 (Yano et al., 2015). Interestingly, the maize Tb1 allele has been linked recently to negative transcriptional regulation of MADS box genes in the context of glume development, by suppressing transcription of the Squamosa Promoter Binding protein transcription factor, teosinte glume architecture1 (Studer et al., 2017). Taken together with the results shown here, these studies suggest that TB1 may act as a transcriptional and posttranslational regulator of genes that control agronomically important traits of inflorescence development, such as MADS box transcription factors. Further investigation of meristem identity gene expression in inflorescences of maize, rice, and barley NILs that contain variant alleles of Tb1, OsTB1, and INT-C, respectively, would determine if the TB1-dependent mechanism for regulating spikelet architecture is conserved among cereals.
In addition to the inflorescence architecture phenotypes shown here, an important observation from this study is that inflorescence growth was delayed in the hb NIL, relative to the wild type, between emergence of leaf 5 and leaf 7. Inflorescence growth rate contributes significantly to floret fertility and is therefore an important component of yield (Miralles et al., 2000; Slafer, 2003; Reynolds et al., 2009). Wheat plants typically produce 10 to 12 floret primordia per spikelet, yet only 2 to 4 of these primordia survive to produce grain; hence, optimization of floret fertility is a key objective for improving wheat yield (Reynolds et al., 2009; González et al., 2011; González-Navarro et al., 2015; Guo and Schnurbusch, 2015). The delayed inflorescence growth of the hb NIL, relative to the wild type, suggests that TB1 dosage influences the rate of inflorescence growth during early developmental stages. It remains to be determined whether this phenotype is directly linked to TB1 function or is a consequence of secondary spikelet development in these lines. However, this result, along with the rapid inflorescence growth that occurs between emergence of leaf 6 and 8, suggests that wheat inflorescences undergo a transition from a phase of axillary organ development to a period of growth in between the terminal spikelet and white anther stages. The timing of this transition correlates developmentally with the stage when the maximum number of viable floret primordia is observed (Guo and Schnurbusch, 2015), suggesting that this may be a point during development when the peak sink potential of the inflorescence is established prior to initiation of inflorescence growth. The conclusion that this growth phenotype is linked to events occurring within the developing inflorescence and is not caused by a delayed plastochron is supported by our analysis showing that leaf emergence and elongation rates were not significantly different between hb and wild-type plants. Comparative analysis of gene expression and metabolite levels in inflorescences of wild-type and hb NILs during these developmental stages will therefore be useful for identifying molecular pathways that influence the rate of inflorescence growth and floret fertility.
An important outcome of this research is the identification of variant alleles for TB1 on the B and D wheat genomes that regulate inflorescence architecture in modern wheat cultivars. We identified an alternate allele for TB-D1 in spring wheat that is a candidate gene for a spikelet architecture QTL detected previously on chromosome 4D (Boden et al., 2015). We also identified a variant allele for TB-B1 in winter wheat that contains five sequence polymorphisms (TB-B1b) and demonstrated that allelic diversity for TB-B1 contributes to the most significant paired spikelet QTL in the winter wheat MAGIC population. Y2H analysis was performed to investigate the comparative strength of the interaction between FT1 and TB-B1a versus FT1 and TB-B1b, which suggested that TB-B1a interacts more strongly with FT1 than does TB-B1b. Taken together with our genetic analysis of the two TB-B1 alleles, this result supports our proposed model that increased activity of TB1 promotes paired spikelet development by modulating the function of FT1 (Figure 7). An intriguing outcome from our analysis of multiple wheat cultivars is that many were found to produce paired spikelets, which is surprising given that paired spikelets are undesired within breeding programs because florets of the secondary spikelets often produce small grain that reduces the uniformity of grain size (Crop Committee Handbook, 2013). Taken together with our analysis of TB1, this phenomenon may be explained by alleles that promote paired spikelet development also contributing beneficial effects for important yield components of plant development, such as inflorescence growth, floret fertility, or tiller number. Interestingly, we observed that the TB-B1b allele appears more frequently in winter wheat cultivars of the UK, France, and Belgium, relative to other regions of Europe; this finding suggests that there may be an adaptive advantage for this allele under certain environmental conditions, which is consistent with the proposed role for tb1 regulating the plasticity of plant architecture traits of teosinte in response to changes in the local environment (Doebley et al., 1995). Further characterization of these alleles through generation of NILs, or development of CRISPR/Cas9 transgenic lines that target individual polymorphisms within the TB-B1b allele, will be important for determining the effect of these mutations on inflorescence and plant architecture. Nonetheless, these results show that variant TB1 alleles are present on the B and D genomes of winter and spring wheat, respectively, and that diversity for TB1 modulates inflorescence architecture in modern wheat cultivars.
In summary, we conclude that TB1 is a key regulator of inflorescence and plant architecture in wheat that interacts with FT1 and regulates axillary spikelet development in a dosage-dependent manner. Our demonstration that increased dosage of TB1 promotes paired spikelet development is consistent with reports from other crops showing that more elaborate inflorescence branching can be achieved using alleles that modify dosage of gene activity (Doebley et al., 1995; Miura et al., 2010; Jiao et al., 2010; Park et al., 2012, 2014; Yoshida et al., 2013; Bommert et al., 2013; Houston et al., 2013; Zhu et al., 2013; Wang et al., 2015; Soyk et al., 2017). In the case of TB1, these results point toward a conserved role for this gene regulating the determinacy and development of axillary spikelet meristems in cereals (Doebley et al., 1995). These results therefore highlight the potential for alleles that modify TB1 activity to increase the number of flowers available for seed production, which is important given our increasing need to identify genetic traits that will enhance yields of our staple crop plants (Reynolds et al., 2009; Fischer et al., 2014).
METHODS
Plant Materials and Growth Conditions
Hexaploid wheat (Triticum aestivum) used in this study included the following genotypes: MAGIC line 0053, from which the NILs termed “wild-type” and “hb” were derived (wild type refers to lines of MAGIC line 0053 that form regular inflorescences; Supplemental Figure 1); tall revertant NIL derived from the “hb” NIL; transgenic lines expressing TB-D1 using the VRN1 promoter generated in the cv Fielder genetic background (see details below); cultivars Sunstate, Pastor, and Cadenza, and the hexaploid wheat TILLING line, Cadenza1721 (tb-d1) (Krasileva et al., 2017); Rht NILs (BC6F3 NILs in the cv Paragon background, containing either the Rht-D1b dwarf allele from cv Alchemy, or the Rht-D1a allele from cv Paragon); Rht-D1c (aka Rht10) (Pearce et al. 2011); and multiple wheat cultivars listed in Supplemental Tables 3 and 4 and Supplemental Data Set 1.
Wild-type hb plants, transgenic lines, and the F1 plants of the hb crosses to Cadenza, tb-d1 (Cadenza1721), Sunstate, and Pastor that were used for phenotype analysis and gene expression studies were grown in controlled growth chambers under short-day (10 h light/14 h dark) or long-day (16 h light/8 h dark) photoperiods at 300 µmol/m2/s (using Plantastar 400-W HQI bulbs [Osram] and Maxim 60-W tungsten bulbs), with a day temperature of 20°C and a night temperature of 15°C. Extended photoperiod conditions of 22 h light/2 h dark, as used in speed-breeding (Watson et al., 2018), were used for the hb x Cadenza and hb x tb-d1 crosses and to analyze the effect of increased FT1 activity on paired spikelet development. Winter wheat varieties surveyed for paired spikelet phenotypes were grown at field sites of KWS based in Thriplow, UK (52°06'00.6"N, 0°05'29.1"E) in 1-m2 plots. Phenotype data for the winter wheat varieties were collected over two growing seasons (2015 and 2016).
The ‘NIAB Elite MAGIC’ bread wheat population has been previously described (Mackay et al., 2014), and F8 seed was sourced from NIAB, UK. The paired spikelet phenotype was assessed, following previously published protocols (Boden et al., 2015) but with fertile or infertile classifications for complete PS (resulting in 0–3 scores). PS phenotypes were scored in two field trials; both of which were undertaken at NIAB (52°13’19”N, 0°5′46”E). The first consisted of 643 progeny (2 replicates) and the 8 founders (2 replicates), sown in autumn 2014 and phenotyped at maturity in 2015 (the 2015 trial). The second trial used 493 progeny (2 replicates) and the 8 founders (≥4 replicates), sown in autumn 2015 and phenotyped at maturity in 2016 (the 2016 trial). Each replicate in both trials was sown as two adjacent rows of 1 m each within 1-m2 plots of six rows total.
Inflorescence Architecture Measurements
Rachis node and paired spikelet measurements were recorded for the inflorescences of the main stem and first tiller. Paired spikelet measurements for each plant have been normalized by calculating the frequency of rachis nodes that contain paired spikelets, represented as a percentage. Data shown for wild-type and hb plants is the average ± se of at least eight plants. Data shown for winter wheat varieties is the average ± se of four plants, and these varieties were scored in two growing seasons. Data shown for hb and tall revertant lines is the average ± se of at least three plants. Data shown for positive transgenic plants is average ± sd of three to four plants, and for null transgenic plants the data are average ± sd of two to four plants and average ± sd of 10 plants for cv Fielder.
Flowering Time Measurements
Developmental flowering time by total leaf number was determined by counting leaves of the main stem until emergence of the flag leaf. Heading date was measured as the number of days since germination when the inflorescence first emerged from the sheath on the main stem (Zadoks scale, Z = 47). Three independent flowering time experiments were performed, with 10 to 20 plants measured in each experiment.
Dissection of Inflorescences and Tillers
Developing inflorescences and immature tillers were isolated with a binocular dissecting microscope and then digitally photographed on a Leica MZ16 Stereo dissecting microscope with a Leica DFC420 color camera. Developing inflorescences were harvested at developmental stages determined by sequential emergence of leaves from germination. Lengths of inflorescences were determined at each developmental stage using Fiji software (Schindelin et al., 2012), with the embedded scale used as a reference. Two independent experiments were performed, with at least four inflorescences measured at each developmental stage. For tiller dissections, six to eight plants were examined of each genotype, with two independent experiments performed.
Paired Spikelet Distribution Analysis
Paired spikelet distribution analysis was performed as described previously (Boden et al., 2015), with 12 inflorescences from six plants (six main stems and six tiller inflorescences) collected and phenotyped from each of the wild-type and hb genotypes.
Scanning Electron Microscopy Analysis
Developing inflorescences were dissected from wild-type and hb plants, and immediately fixed in 100% ethanol, as described previously (Talbot and White, 2013). Samples were critical point dried using a Leica CPD300 automated critical point drier. Basal spikelets were dissected from dried tissue to expose secondary spikelets of central rachis nodes, and samples were mounted on aluminum stubs with double-sided sticky carbon and coated with gold (∼20 nm) using a sputter coater (Agar Scientific). Samples were then examined using a Zeiss Supra 55 scanning electron microscope (Carl Zeiss).
Fluorescence in Situ Hybridization Analysis
Lines were characterized by fluorescence in situ hybridization (FISH) as described previously (Zhang et al., 2004). Plasmid clone pSc119.2 contains highly repetitive sequences from rye (Secale cereale; Bedbrook et al., 1980), whereas clone pAs1 contains a repetitive sequence that belongs to the Afa family isolated from Aegilops tauschii (Rayburn and Gill, 1986). Using two clones simultaneously, all B and D genome chromosomes and chromosomes 1A, 4A, and 5A of hexaploid wheat can be identified (Mukai et al., 1993). One microgram of plasmid DNA of pSc119.2 and pAs1 was labeled with fluorescein-12-dUTP (fluoresces green) and rhodamine-5-dUTP (fluoresces red), respectively, using nick translation in accordance with the manufacturer’s protocol (Roche Diagnostics). The hybridization and posthybridization washes were conducted as described previously (Zhang et al., 2004). Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (Life Technologies) and pseudo-colored blue. Chromosome preparations were analyzed with a Zeiss Axio Imager epifluorescence microscope (Carl Zeiss Microimaging). Images were captured with a Retiga EXi CCD camera (QImaging) operated with Image-Pro Plus 7.0 software (Media Cybernetics) and processed with Photoshop version 8.0 software (Adobe Systems).
Leaf Emergence and Elongation Assays
Wild-type and hb plants were grown under LD conditions in a controlled growth chamber. Measurements of leaf emergence were recorded daily from leaf 2 onwards, determined by when the growing tip of the new leaf could be seen emerging from the sheath of the previous leaf. Measurements were recorded for leaf 2 to leaf 8. For leaf elongation assays, length of the emerging leaf blade was recorded daily until growth ceased, which coincided with emergence of the next leaf. Measurements were recorded from leaf 2 to leaf 5. Twelve biological replicates were used for each genotype per experiment, and two independent experiments were performed for each assay.
DNA Extractions and Sequence Analysis
Nucleotide sequences for TB-A1, TB-B1, and TB-D1 were obtained by BLAST search from the Ensembl Plants website, using the maize (Zea mays) TB1 protein sequence as a query sequence. Genomic DNA extractions were performed as described previously (Paterson et al., 1993). Clones of TB-B1 and TB-D1 were amplified using Phusion DNA polymerase (New England Biolabs) and the following oligonucleotides: TB-B1, ATGTTTCCTTTCTATGATTCCC, CATCCGGTTCTTTTCCCTAGT, and CGAGGGGAAGAAGCAGGTG, CACAAATAATCCATTGAACAAAGC; TB-D1, CTCTTCCACCCGCAGACAC, TCAGTAGGGCTGCGAGTTG. DNA fragments were sequenced using the Big-Dye Terminator Sequencing v3.1 Ready Reaction Kit (Perkin-Elmer, Applied BioSystems, Thermo Fischer Scientific) or with Mix2Seq Kit (Eurofins). Multiple nucleotide and protein sequence alignments of TB-A1, TB-B1, and TB-D1 were performed using MUSCLE. Pairwise nucleotide and protein sequence alignments of cv Chinese Spring reference sequences for TB-B1 and TB-D1 with alleles identified from other wheat varieties were performed using EMBOSS Needle, which were formatted in Microsoft Word. TB-B1 and TB-D1 alleles from wheat cultivars that contain sequence polymorphisms relative to the reference sequences from Chinese Spring were named TB-B1b and TB-D1b, following convention for naming alternate alleles for other wheat genes.
RNA Extraction and Expression Analysis
For experiments with wild-type and hb plants, RNA was extracted from tiller, root, young emerging leaf, mature leaf, and seedling tissue using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) and from developing inflorescences using the RNeasy Plant Mini Kit (Qiagen). “Tiller buds” refer to immature tillers that were transparent or light green in color for which no leaf blade had begun to emerge, while “emerging tillers” refers to young tillers where leaf blade tissue was beginning to emerge from the coleoptile-like tissue. RNA was extracted from developing inflorescences at the double-ridge stage and inflorescences harvested at complete emergence of leaf 5, leaf 6, and leaf 7. Analysis of FT1 expression was performed using RNA extracted from leaves of wild-type and hb plants, which were grown under a short-day photoperiod until the leaf 5 stage and then transferred to LD photoperiod for 7 d. For analysis of FT1 expression under the extended LD photoperiod conditions, hb leaf samples were collected at time points corresponding to dusk (i.e., at ZT 16 h for the standard LD photoperiod and ZT 22 h for the extended LD photoperiod), and at 16 h for the extended LD photoperiod to be used as a control time point in reference to dusk collection of the standard LD condition. For experiments with the pVRN1:TB1 transgenic lines, developing inflorescences were dissected at the spikelet primordium stage, with four inflorescences pooled for each biological replicate sample, and RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). Synthesis of cDNA and RT-qPCR were performed as described previously (Boden et al., 2014). Oligonucleotides for RT-qPCR analysis are provided in Supplemental Table 8. Expression of candidate genes in leaves, roots, tillers, and seedlings was normalized using RNA polymerase 15-kD subunit (TaRP15; Shaw et al., 2012) and with Traes_6DS_BE8B5E56D.1 (Borrill et al., 2016) in developing inflorescences. All RT-qPCR data points are the average of at least three biological replicates, with two technical replicates performed in each reaction.
Kompetitive Allele-Specific PCR Analysis
Oligonucleotides for Kompetitive allele-specific PCR (KASP) analysis were designed using Polymarker (http://polymarker.tgac.ac.uk; Ramirez-Gonzalez et al., 2015a) and contained the standard FAM or HEX compatible tails (FAM tail, 5′-GAAGGTGACCAAGTTCATGCT-3′; HEX tail, 5′-GAAGGTCGGAGTCAACGGATT-3′). Oligonucleotide sequences are provided in Supplemental Table 9. The KASP assay was performed as described previously (Ramirez-Gonzalez et al., 2015b), with the following modifications: Assays were set up as 2.4-μL reactions (1.8 μL template [10–20 ng of DNA] dried to assay plate, 1.18 μL of V4 2× Kaspar mix [LGC Group], 1.18 μL deionized water and 0.032 μL primer mix); and the PCR cycling included hotstart at 95°C for 15 min, followed by 10 touchdown cycles (95°C for 20 s; touchdown 65°C, −1°C per cycle, 25 s), followed by 40 cycles of amplification (95°C for 10 s; 57°C for 60 s).
ddPCR Reactions and Data Analysis
ddPCR was performed to determine gene copy numbers for TB-B1 and TB-D1, in reference to TaCONSTANS2 (TaCO2), which has a single copy on each of the three wheat genomes. Oligonucleotide sequences are provided in Supplemental Table 10. The ddPCR reaction mix included 10 μL of 2× QX200 ddPCR EvaGreen Supermix (Bio-Rad), 6 μL of genomic DNA template (2.5 ng/μL; 15 ng), 2 μL of mixed oligonucleotides (2 μM each), and 0.2 μL of HindIII-HF (New England Biolabs), in a 20-μL reaction volume. Droplets were generated in eight-well cartridges using the QX200 droplet generator (Bio-Rad). Water-in-oil emulsions were transferred to a 96-well plate and amplified in a C1000 Touch Thermal Cycler (Bio-Rad). Thermal cycling conditions were 7 min at 95°C; 40 cycles of a three-step thermal profile comprising of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C, with ramp rate 2.0°C/s. After cycling, each sample was incubated at 90°C for 5 min and then cooled to 4°C. Plates were then transferred to the QX200 droplet reader (Bio-Rad). Data acquisition and analysis were performed using QuantaSoft software (Bio-Rad) to quantify numbers of positive droplets containing amplicons and negative droplets without amplicons. Calculations for copy number of TB-B1 and TB-D1 were performed in Microsoft Excel. As oligonucleotides for TaCO2 annealed to sequence and produced amplicons from the A, B, and D genomes, gene copy numbers for TB-B1 and TB-D1 were determined by dividing the number of positive droplets for TB-B1 and TB-D1 by one-third of the number of positive droplets for TaCO2.
Construct Design and Wheat Transformation
The construct used to generate pVRN1:TB-D1 transgenic plants contained TB-D1 with the G328T SNP present in cv Baxter, wild-type, and hb lines, as well as synonymous mutations (C30G, G309T, and C1023G) that modified restriction sites to facilitate cloning and construct design. NcoI and NotI sites flanking the start and stop codon positions, respectively, were used to clone TB-D1 into the KR PRO 5 plasmid containing the VRN1 promoter (Alonso-Peral et al., 2011) and the kanamycin resistance gene for selection. The pVRN1:TB-D1 cassette was cloned into the vecBarII construct containing the Tn7 spectinomycin resistance gene and the BAR open reading frame under the regulatory control of the CaMV 35S promoter and the Agrobacterium tumefaciens octopine synthase terminator sequences.
Transgenic wheat plants were generated by Agrobacterium-mediated transformation of the hexaploid wheat cv Fielder, as described previously (Ishida et al., 2015; Richardson et al., 2014). Genomic DNA extractions were performed as described previously (Paterson et al., 1993), and genotype analysis of T1 transgenic lines was performed as described previously using qRT-PCR (Mieog et al., 2013; Boden et al., 2014) and the 2−ΔΔCT method to determine relative copy number for TB-D1. Amplicons of TB-D1 were amplified using oligonucleotides shown in Supplemental Table 10 and were normalized to Epsilon Cyclase-A1 (oligonucleotides in Supplemental Table 10), which is present once on the “A” genome of wheat (Mieog et al., 2013). Normalized levels of TB-D1 for each transgenic plant were then normalized to those identified for cv Fielder to determine relative copy numbers of TB-D1 for each transgenic line. Data for RT-qPCR are the average of two technical replicates, with 100 ng of template DNA per reaction. Reactions were performed using the Bio-Rad CFX384 real-time system.
Y2H Analysis
Y2H assays were performed at 28°C in the yeast strain AH109 (Clontech) using the cotransformation technique (Egea-Cortines et al., 1999). Open reading frames of full-length TB-D1, TB-B1a, TB-B1b, and FT1, and a truncated version of TB-D1 (TB1Δ) that encodes a protein lacking the first 114 amino acids of the full-length protein. The TB-D1 allele used here was the allele from Baxter, as this is the allele found in the wild-type and hb lines. These open reading frames were cloned into the Gateway vector GAL4 system (pGADT7 and pGBKT7; Clontech) passing through pDONR207 (Life Technologies, Thermo Fischer Scientific). Strength of protein interactions was tested on selective yeast synthetic dropout medium (YSD) lacking leucine (L), tryptophan (W), adenine (A), and histidine (H), supplemented with different concentrations of 3-aminotriazole (2.5, 5, and 10 mM). Images were taken 7 d after plating on the selective media.
BiFC Analysis
Open reading frames of full-length TB-D1 and FT1, and truncated TB1 (TB1Δ) in pDONR207 (Life Technologies) generated for the Y2H assay, were each cloned into pYFPN43 and pYFPC43. pEAQ-HT vector (Sainsbury et al., 2009), which expresses the silencing suppressor p19 of tomato bushy stunt virus, was added to the vector combination. BiFC was performed in Nicotiana benthamiana as described previously (Belda-Palazón et al., 2012). Images were taken with a Zeiss LSM780 confocal microscope 72 h after infiltration.
Genetic Analysis of NIAB Elite MAGIC Population
Trial entries were arrayed in an incomplete block design using the software Block Designs U (http://www.expdesigns.co.uk). Paired spikelet phenotype data were analyzed using GenStat (16th edition; VSNi) with a generalized linear mixed model employing a Poisson distribution with a logarithmic link function (Bolker et al., 2009) providing best linear unbiased estimate values. Broad-sense heritability for the replicated trials was calculated as per Equation 1 (Tenesa and Haley, 2013), where 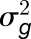 is the genotypic variance and
is the genotypic variance and 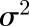 is the residual error variance with
is the residual error variance with 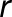 replicates.
replicates.
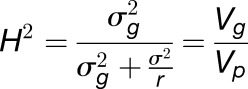 |
(1) |
In total, 643 F5 lines from the NIAB Elite MAGIC wheat population (Mackay et al., 2014) were genotyped using a wheat Illumina 90k iSelect array designed by Wang et al. (2014) generating 18,601 polymorphic markers mapped to 7369 unique genetic map locations (Gardner et al., 2016). Using these markers, a high confidence subset of identity by descent (IBD) segments were calculated in a three-point probabilistic manner (Huang and George, 2011) for their parental contributions over the 7369 uniquely mapped markers. Founder probabilities were assigned with a 33% threshold, as identity-by-descent relationships among the founders complicate direct observation of true parental origin. The resulting founder haplotype probabilities were used for composite interval mapping analysis.
QTL analysis was performed using an IBD composite interval mapping (IBD-CIM) method (Milner et al., 2016). IBD-CIM was performed using the mpIM function of the R package mpMap (Huang and George, 2011). Primarily, a linear model was fitted for estimation of the QTL fixed effects for every founder at each hypothetical QTL position (with the founder “Xi19” arbitrarily fixed as the reference haplotype). Simple interval mapping was run and then the inclusion of marker covariates was determined by a forward selection process in an Akaike information criterion-dependent manner, up to a maximum of five covariates. To determine significance thresholds, a null distribution is formed with the same variance as the trial’s score data and the significance threshold is then determined empirically. The proportion of phenotypic variance explained by any given QTL, accounting for all other QTL, is the square of the semipartial correlation coefficient (Milner et al., 2016). QTL nomenclature follows recommended rules, as listed in the GrainGenes catalog (https://wheat.pw.usda.gov/ggpages/wgc/98/Intro.htm).
Genetic markers of interest were aligned to the unannotated IWGSC RefSeq v1.0 reference wheat genome sequence (cv Chinese Spring 42; www.wheatgenome.org/) using BLASTn with a maximum E-value threshold of 0.1 (Altschul et al., 1990). Annotation within target regions was undertaken using GMap (Wu and Watanabe, 2005) to map high-confidence TGAC v1 gene models (Clavijo et al., 2017) (cv Chinese Spring 42, Earlham Institute) to IWGSC RefSeq v1.0 using 95% minimum identity and 80% minimum coverage thresholds. The highest scoring BLASTx hit for the resulting gene models in wheat from the SwissProt (The UniProt Consortium, 2017) database were identified with a maximum E-value threshold of one (Supplemental Table 7).
Statistical Analysis
Differences between treatments were tested by two-tailed Student’s t test. Results in figures are shown as means ± se, except for Figures 2H and 2I, which show the mean ± sd because some null transgenic lines contained only two individuals. For the TB-B1-specific marker analysis of 212 UK winter wheat MAGIC lines, a McNemar’s test was performed using 1 degree of freedom.
Accession Numbers
Accession numbers for all genes investigated in this study are provided in Supplemental Tables 8 and 10.
Supplemental Data
Supplemental Figure 1. Schematic outlining pedigree of wild-type and hb lines from MAGIC line 0053.
Supplemental Figure 2. Paired spikelet development on hb inflorescences.
Supplemental Figure 3. Wild-type and hb inflorescences at the leaf 8 developmental stage.
Supplemental Figure 4. Leaf emergence and leaf elongation rates for wild-type and hb plants.
Supplemental Figure 5. Immature tillers of wild-type and hb plants.
Supplemental Figure 6. Inflorescence architecture phenotypes of F1 offspring from hb crosses.
Supplemental Figure 7. Inflorescence architecture phenotypes of Rht-D1 NILs.
Supplemental Figure 8. Expression analysis of TB-B1 and TB-D1 in tillers and developing Inflorescences.
Supplemental Figure 9. Expression analysis of genes from chromosome group 4 that are in proximity to Rht-1 and TB1.
Supplemental Figure 10. KASP marker analysis of Rht-B1 in hb and revertant lines.
Supplemental Figure 11. Expression analysis of VRN1.
Supplemental Figure 12. Inflorescence architecture phenotypes of pVRN1:TB1 transgenic plants.
Supplemental Figure 13. TEOSINTE BRANCHED1 interacts with FLOWERING LOCUS T1.
Supplemental Figure 14. Expression analysis of meristem identity genes and FLOWERING LOCUS T1.
Supplemental Figure 15. Alignment of coding nucleotide sequence for TB-D1a and TB-D1b alleles.
Supplemental Figure 16. Alignment of predicted amino acid sequence for TB-D1a and TB-D1b.
Supplemental Figure 17. Paired spikelet development in modern winter wheat cultivars.
Supplemental Figure 18. Analysis of copy number variation for TB-D1 and TB-B1 in winter wheat.
Supplemental Figure 19. Alignment of coding nucleotide sequence for TB-B1a and TB-B1b alleles.
Supplemental Figure 20. Alignment of amino acid sequence for TB-B1a and TB-B1b.
Supplemental Figure 21. Chromosome-wide CIM scans displaying the 4B QTL that contributes to paired spikelet development in the UK wheat MAGIC population.
Supplemental Table 1. Phenotype information for hb revertant lines, relative to hb.
Supplemental Table 2. Phenotype information for T1 transgenic pVRN1:TB1 lines, with controls.
Supplemental Table 3. Proportion of rachis nodes with paired spikelets in subset of modern wheat cultivars.
Supplemental Table 4. TB-B1 alleles of modern winter wheat cultivars investigated in this study.
Supplemental Table 5. Summary of paired spikelet QTLs identified in the NIAB Elite MAGIC population using IBD-CIM.
Supplemental Table 6. Predicted effects of seven founder alleles for QTL on chromosome 4B, relative to Xi 19.
Supplemental Table 7. TB-B1-specific KASP marker analysis of a subset of 212 NIAB MAGIC lines.
Supplemental Table 8. Oligonucleotide sequences used in RT-qPCR assays.
Supplemental Table 9. Oligonucleotide sequences used in KASP marker assays.
Supplemental Table 10. Oligonucleotide sequences used in digital droplet PCR assays.
Supplemental Data Set 1. Paired spikelet phenotypes for modern winter wheat cultivars.
Supplemental Data Set 2. Summary of TB-B1 allele genotypes for GEDIFLUX population.
Acknowledgments
We acknowledge the Biotechnology and Biological Sciences Research Council (BBS/E/J/000C0677, BB/P016855/1, BB/M011666/1, and BB/N00518X/1), the Royal Society (UF150081), International Wheat Yield Partnership, and a CSIRO O.C.E. Fellowship for funding this research. We also acknowledge Grant Calder, Elaine Barclay, Sara Simonini, Simon Griffiths, Richard Goram, Anna Backhaus (all JIC), Matthew Moscou and Inma Hernandez-Pinzon (TSL), and Beatrice Corsi (NIAB) for technical assistance; Ben Trevaskis (CSIRO) for the VRN1 promoter; Jacob Lage and Nicholas Bird (KWS) for access to field trials of modern wheat cultivars; and Keith Gardner (NIAB) for genetic mapping guidance. We thank the Wheat Genetic Improvement Network for access to the GEDIFLUX collection. We thank the International Wheat Genome Sequencing Consortium for prepublication access to the wheat genome RefSeq v0.4 and v1.0 that was used to identify genes located in the chromosome 4D region.
AUTHOR CONTRIBUTIONS
S.A.B., L.E.D., J.R.G., and S.M.S. designed the research. S.A.B. and L.E.D. performed phenotype and genotype analyses of wild-type and hb plants, modern wheat cultivars, and MAGIC lines, and conducted expression analyses. S.A.B. and J.R.G. performed crosses using the hb line. J.R.G. analyzed transgenic plants and performed expression analysis. S.B. performed Y2H and BiFC assays. P.Z. performed FISH experiments. J.C. and G.M. collected MAGIC phenotype data and performed QTL analysis. S.M.S., K.R., and C.C. performed initial genotype analysis and characterization of MAGIC line 0053. S.A.B., L.E.D., and J.R.G. wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. [DOI] [PubMed] [Google Scholar]
- Aguilar-Martínez J.A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Peral M.M., Oliver S.N., Casao M.C., Greenup A.A., Trevaskis B. (2011). The promoter of the cereal VERNALIZATION1 gene is sufficient for transcriptional induction by prolonged cold. PLoS One 6: e29456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Beales J., Turner A., Griffiths S., Snape J.W., Laurie D.A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115: 721–733. [DOI] [PubMed] [Google Scholar]
- Bedbrook J.R., Jones J., O’Dell M., Thompson R.D., Flavell R.B. (1980). A molecular description of telometic heterochromatin in Secale species. Cell 19: 545–560. [DOI] [PubMed] [Google Scholar]
- Belda-Palazón B., Ruiz L., Martí E., Tárraga S., Tiburcio A.F., Culiáñez F., Farràs R., Carrasco P., Ferrando A. (2012). Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS One 7: e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden S.A., Cavanagh C., Cullis B.R., Ramm K., Greenwood J., Jean Finnegan E., Trevaskis B., Swain S.M. (2015). Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat. Plants 1: 14016. [DOI] [PubMed] [Google Scholar]
- Boden S.A., Weiss D., Ross J.J., Davies N.W., Trevaskis B., Chandler P.M., Swain S.M. (2014). EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 26: 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolker B.M., Brooks M.E., Clark C.J., Geange S.W., Poulsen J.R., Stevens M.H.H., White J.-S.S. (2009). Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol. Evol. (Amst.) 24: 127–135. [DOI] [PubMed] [Google Scholar]
- Bommert P., Nagasawa N.S., Jackson D. (2013). Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 45: 334–337. [DOI] [PubMed] [Google Scholar]
- Borrill P., Ramirez-Gonzalez R., Uauy C. (2016). expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol. 170: 2172–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo B.J., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crop Committee Handbook (2013). Crop Committee Handbook. (Kenilworth, UK: Agriculture and Horticulture Development Board Cereals and Oilseeds).
- Debernardi J.M., Lin H., Chuck G., Faris J.D., Dubcovsky J. (2017). microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144: 1966–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaya O., et al. (2015). FRIZZY PANICLE drives supernumerary spikelets in bread wheat. Plant Physiol. 167: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Gustus C. (1995). teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386: 485–488. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Byerlee D., Edmeades G. (2014). Crop Yields and Global Food Security. (Canberra, Australia: Australian Centre of International Agricultural Research; ). [Google Scholar]
- Gardner K.A., Wittern L.M., Mackay I.J. (2016). A highly recombined, high-density, eight-founder wheat MAGIC map reveals extensive segregation distortion and genomic locations of introgression segments. Plant Biotechnol. J. 14: 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González F.G., Miralles D.J., Slafer G.A. (2011). Wheat floret survival as related to pre-anthesis spike growth. J. Exp. Bot. 62: 4889–4901. [DOI] [PubMed] [Google Scholar]
- González-Navarro O.E., Griffiths S., Molero G., Reynolds M.P., Slafer G.A. (2015). Dynamics of floret development determining differences in spike fertility in an elite population of wheat. Field Crops Res. 172: 21–31. [Google Scholar]
- Greenwood J.R., Finnegan E.J., Watanabe N., Trevaskis B., Swain S.M. (2017). New alleles of the wheat domestication geneQreveal multiple roles in growth and reproductive development. Development 144: 1959–1965. [DOI] [PubMed] [Google Scholar]
- Guo Z., Schnurbusch T. (2015). Variation of floret fertility in hexaploid wheat revealed by tiller removal. J. Exp. Bot. 66: 5945–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. (2003). The genes of the Green Revolution. Trends Genet. 19: 5–9. [DOI] [PubMed] [Google Scholar]
- Houston K., et al. (2013). Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc. Natl. Acad. Sci. USA 110: 16675–16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B.E., George A.W. (2011). R/mpMap: a computational platform for the genetic analysis of multiparent recombinant inbred lines. Bioinformatics 27: 727–729. [DOI] [PubMed] [Google Scholar]
- Huang B.E., George A.W., Forrest K.L., Kilian A., Hayden M.J., Morell M.K., Cavanagh C.R. (2012). A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 10: 826–839. [DOI] [PubMed] [Google Scholar]
- Hubbard L., McSteen P., Doebley J., Hake S. (2002). Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162: 1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y., Tsunashima M., Hiei Y., Komari T.. 2015. Wheat transformation. In Agrobacterium Protocols: Methods in Molecular Biology, K. Wang, ed (New York: Springer), pp. 189–198. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., Qian Q., Li J. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544. [DOI] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva K.V., et al. (2017). Uncovering the hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA 114: E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J., Tokunaga H., Yoshida A. (2014). Control of grass inflorescence form by the fine-tuning of meristem phase change. Curr. Opin. Plant Biol. 17: 110–115. [DOI] [PubMed] [Google Scholar]
- Lemmon Z.H., Park S.J., Jiang K., Van Eck J., Schatz M.C., Lippman Z.B. (2016). The evolution of inflorescence diversity in the nightshades and heterochrony during meristem maturation. Genome Res. 26: 1676–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.M., Mackintosh C.A., Shin S., Gilding E., Kravchenko S., Baldridge G., Zeyen R., Muehlbauer G.J. (2008). Overexpression of the maize Teosinte Branched1 gene in wheat suppresses tiller development. Plant Cell Rep. 27: 1217–1225. [DOI] [PubMed] [Google Scholar]
- Li C., Lin H., Dubcovsky J. (2015). Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 84: 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799. [DOI] [PubMed] [Google Scholar]
- Mackay I.J., Bansept-Basler P., Barber T., Bentley A.R., Cockram J., Gosman N., Greenland A.J., Horsnell R., Howells R., O’Sullivan D.M., Rose G.A., Howell P.J. (2014). An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 (Bethesda) 4: 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R.S., Purugganan M.D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14: 840–852. [DOI] [PubMed] [Google Scholar]
- Mieog J.C., Howitt C.A., Ral J.-P. (2013). Fast-tracking development of homozygous transgenic cereal lines using a simple and highly flexible real-time PCR assay. BMC Plant Biol. 13: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner S.G., Maccaferri M., Huang B.E., Mantovani P., Massi A., Frascaroli E., Tuberosa R., Salvi S. (2016). A multiparental cross population for mapping QTL for agronomic traits in durum wheat (Triticum turgidum ssp. durum). Plant Biotechnol. J. 14: 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles D., Richards R.A., Slafer G.A. (2000). Duration of the stem elongation period influences the number of fertile florets in wheat and barley. Aust. J. Plant Physiol. 27: 931–940. [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.-J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Mukai Y., Nakahara Y., Yamamoto M. (1993). Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36: 489–494. [DOI] [PubMed] [Google Scholar]
- Niwa M., Daimon Y., Kurotani K., Higo A., Pruneda-Paz J.L., Breton G., Mitsuda N., Kay S.A., Ohme-Takagi M., Endo M., Araki T. (2013). BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25: 1228–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Schatz M.C., Lippman Z.B. (2012). Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. USA 109: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Tal L., Yichie Y., Gar O., Zamir D., Eshed Y., Lippman Z.B. (2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46: 1337–1342. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., Brubaker C.L., Wendel J.F. (1993). A rapid method for extraction of cotton (Gossypium ssp.) genomic DNA suitable for RFLP and PCR analysis. Plant Mol. Biol. Report. 11: 122–127. [Google Scholar]
- Pearce S., et al. (2011). Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiol. 157: 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poursarebani N., et al. (2015). The genetic basis of composite spike form in barley and ‘Miracle-Wheat’. Genetics 201: 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Gonzalez R.H., Uauy C., Caccamo M. (2015a). PolyMarker: a fast polyploid primer design pipeline. Bioinformatics 31: 2038–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Gonzalez R.H., Segovia V., Bird N., Fenwick P., Holdgate S., Berry S., Jack P., Caccamo M., Uauy C. (2015b). RNA-Seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat. Plant Biotechnol. J. 13: 613–624. [DOI] [PubMed] [Google Scholar]
- Ramsay L., et al. (2011). INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 43: 169–172. [DOI] [PubMed] [Google Scholar]
- Rawson H.M. (1970). Spikelet number, its control and relation to yield per ear in wheat. Aust. J. Biol. Sci. 23: 1–15. [Google Scholar]
- Rayburn A.L., Gill B.S. (1986). Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant Mol. Biol. Report. 4: 102–109. [Google Scholar]
- Reynolds M., Foulkes M.J., Slafer G.A., Berry P., Parry M.A.J., Snape J.W., Angus W.J. (2009). Raising yield potential in wheat. J. Exp. Bot. 60: 1899–1918. [DOI] [PubMed] [Google Scholar]
- Richardson T., Thistleton J., Higgins T.J., Howitt C., Ayliffe M. (2014). Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell Tissue Organ Cult. 119: 647–659. [Google Scholar]
- Sainsbury F., Thuenemann E.C., Lomonossoff G.P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7: 682–693. [DOI] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharman B.C. (1944). Branched heads in wheat and wheat hybrids. Nature 153: 497–498. [Google Scholar]
- Sharman B.C. (1967). Interpretation of the morphology of various naturally occurring abnormalities of the inflorescence of wheat (Triticum). Can. J. Bot. 45: 2073–2080. [Google Scholar]
- Shaw L.M., Turner A.S., Laurie D.A. (2012). The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J. 71: 71–84. [DOI] [PubMed] [Google Scholar]
- Simons K.J., Fellers J.P., Trick H.N., Zhang Z., Tai Y.-S., Gill B.S., Faris J.D. (2006). Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slafer G.A. (2003). Genetic basis of yield as viewed from a crop physiologist’s perspective. Ann. Appl. Biol. 142: 117–128. [Google Scholar]
- Soyk S., et al. (2017). Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 169: 1142–1155.e12. [DOI] [PubMed] [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J., Doebley J. (2011). Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43: 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A.J., Wang H., Doebley J.F. (2017). Selection during maize domestication targeted a gene network controlling plant and inflorescence architecture. Genetics 207: 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Suwa Y., Suzuki M., Kitano H., Ueguchi-Tanaka M., Ashikari M., Matsuoka M., Ueguchi C. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33: 513–520. [DOI] [PubMed] [Google Scholar]
- Talbot M.J., White R.G. (2013). Methanol fixation of plant tissue for scanning electron microscopy improves preservation of tissue morphology and dimensions. Plant Methods 9: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335. [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Nakabayashi K., Kamiya Y., Nambara E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53: 42–52. [DOI] [PubMed] [Google Scholar]
- Tenesa A., Haley C.S. (2013). The heritability of human disease: estimation, uses and abuses. Nat. Rev. Genet. 14: 139–149. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45: D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sun S., Jin J., Fu D., Yang X., Weng X., Xu C., Li X., Xiao J., Zhang Q. (2015). Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA 112: 15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., et al. ; International Wheat Genome Sequencing Consortium (2014). Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 12: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4: 23–29. [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wu T.D., Watanabe C.K. (2005). GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. [DOI] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., Dubcovsky J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100: 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., et al. (2015). Isolation of a novel lodging resistance QTL gene involved in strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol. Plant 8: 303–314. [DOI] [PubMed] [Google Scholar]
- Yoshida A., et al. (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. USA 110: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Yuan Z. (2014). Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 65: 553–578. [DOI] [PubMed] [Google Scholar]
- Zhang P., Li W., Fellers J., Friebe B., Gill B.S. (2004). BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma 112: 288–299. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Tan L., Fu Y., Liu F., Cai H., Xie D., Wu F., Wu J., Matsumoto T., Sun C. (2013). Genetic control of inflorescence architecture during rice domestication. Nat. Commun. 4: 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]