Maize ZmMAC1 protein delivered to the apoplast by the smut pathogen Ustilago maydis cell-autonomously corrects both somatic cell division and differentiation defects in Zmmac1-1 mutant anthers.
Abstract
Small proteins are crucial signals during development, host defense, and physiology. The highly spatiotemporal restricted functions of signaling proteins remain challenging to study in planta. The several month span required to assess transgene expression, particularly in flowers, combined with the uncertainties from transgene position effects and ubiquitous or overexpression, makes monitoring of spatiotemporally restricted signaling proteins lengthy and difficult. This situation could be rectified with a transient assay in which protein deployment is tightly controlled spatially and temporally in planta to assess protein functions, timing, and cellular targets as well as to facilitate rapid mutagenesis to define functional protein domains. In maize (Zea mays), secreted ZmMAC1 (MULTIPLE ARCHESPORIAL CELLS1) was proposed to trigger somatic niche formation during anther development by participating in a ligand-receptor module. Inspired by Homer’s Trojan horse myth, we engineered a protein delivery system that exploits the secretory capabilities of the maize smut fungus Ustilago maydis, to allow protein delivery to individual cells in certain cell layers at precise time points. Pathogen-supplied ZmMAC1 cell-autonomously corrected both somatic cell division and differentiation defects in mutant Zmmac1-1 anthers. These results suggest that exploiting host-pathogen interactions may become a generally useful method for targeting host proteins to cell and tissue types to clarify cellular autonomy and to analyze steps in cell responses.
INTRODUCTION
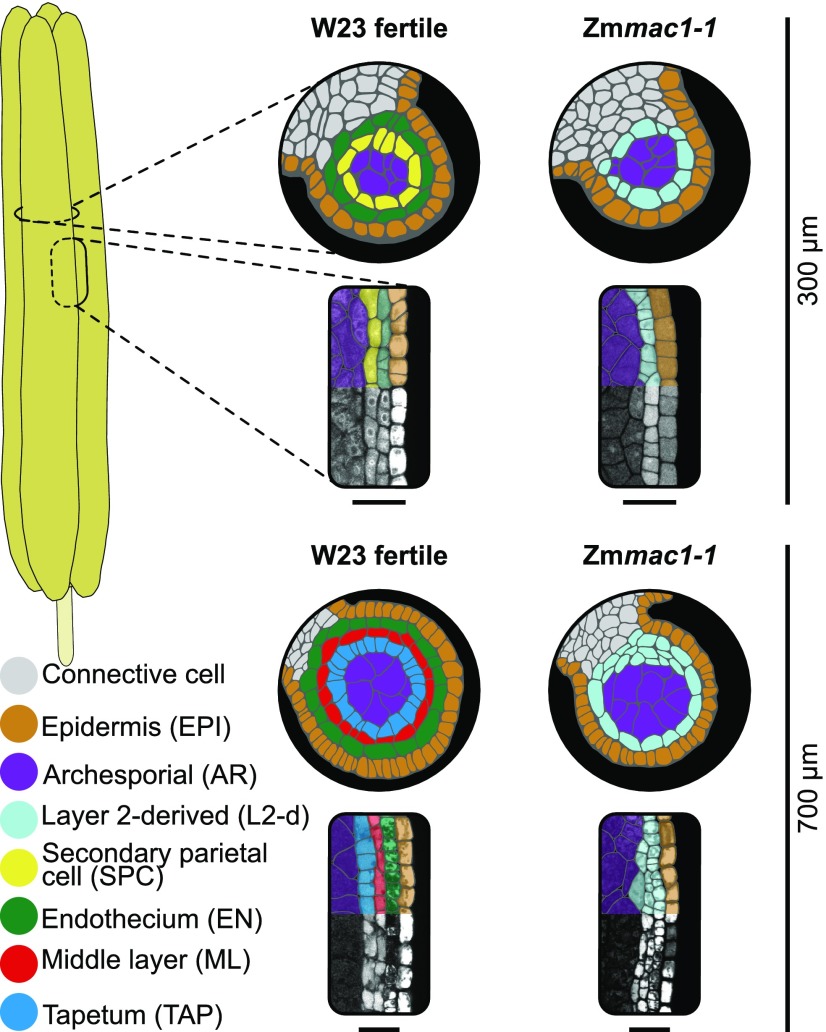
In contrast to animals, plants lack a germline and must switch from vegetative to reproductive growth. In angiosperms, shoot apical meristems redifferentiate into inflorescence meristems and then generate somatic floral organs within which a few cells become specified for meiosis. These processes depend on small proteins such as CLV3 (CLAVATA3) (Clark et al., 1995) and MSCA1 (MALE STERILE CONVERTED ANTHER1) (Kelliher and Walbot, 2012). Stamens, the male reproductive organ, consist of a thin filament subtending four-lobed anthers. Anther lobes initially contain pluripotent cells, and a key step in reproductive development is specification of premeiotic and somatic niche cells (Kelliher and Walbot, 2012). The somatic niche contains four distinct cell types: the outer epidermis and three internal layers, the endothecium (EN), middle layer (ML), and tapetum (TAP).
In maize (Zea mays), anther development is highly correlated with anther length permitting accurate staging of developmental events. At lobe inception, layer 2-derived (L2-d) cells are pluripotent, then growth-induced hypoxia triggers premeiotic archesporial (AR) cell differentiation at 150-µm anther length (Kelliher and Walbot, 2012). At 180 to 280 µm of anther length, L2-d cells that are surrounding the central AR undergo a periclinal division generating two layers, EN and secondary parietal cells (SPC). This division step establishes the somatic niche (Figure 1). By 500 µm, the SPC start to divide periclinally, forming two new layers within lobes, the ML and TAP, a process completed by ∼700 µm to yield the final premeiotic lobe anatomy (Figure 1) (Kelliher and Walbot, 2011). At this point, each somatic layer (EN, ML, and TAP) is only a single cell thick, has a distinctive morphology, and all layers are required to support the centrally located AR cells for successful completion of meiosis (Figure 1).
Figure 1.
The Zmmac1-1 Mutant Has an Early Premeiotic Anther Phenotype.
Colorized tracings of transverse and longitudinal confocal images of W23 fertile and Zmmac1-1 lobes at two stages of premeiotic development. Bars = 40 µm.
In Arabidopsis thaliana and rice (Oryza sativa), somatic cell identity has been scored shortly before meiosis and is maintained through a ligand:receptor module involving a secreted cysteine-rich protein and a leucine-rich repeat receptor-like kinase (LRR-RK) partner: for Arabidopsis AtTPD1 (TAPETUM DETERMINANT1):AtEMS1/EXS (EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS) (Zhao et al., 2002; Jia et al., 2008) and for rice OsTDL1A (TPD1-like1A):OsMSP1 (MULTIPLE SPOROCYTE1) (Nonomura et al., 2003; Zhao et al., 2008). The rice and Arabidopsis gene pairs are epistatic, and their proteins interact biochemically in yeast (Jia et al., 2008; Zhao et al., 2008). Recently, it was proposed that those ligands undergo cleavage into smaller peptides to be functionally active (Huang et al., 2016; Yang et al., 2016). The maize ligand homolog ZmMAC1 (MULTIPLE ARCHESPORIAL CELLS1) is expressed very early, in just-specified AR cells (Wang et al., 2012a), and was found to be secreted in a transient assay in plant cells (Wang et al., 2012a). The Zmmac1-1 mutant shares the key phenotype of extra AR cells in both ovules and anthers paralleling rice and Arabidopsis ligand mutants (Sheridan et al., 1996, 1999); accompanying AR overproliferation, somatic cell specification fails and the L2-d cells proliferate slowly (Figure 1). These initial somatic phenotypes have been evaluated earlier in maize anthers compared with rice or Arabidopsis (Walbot and Egger, 2016). To date, it is unknown whether ZmMAC1 is further processed, if it acts as a receptor ligand, and if so, what its receptor partner is. Furthermore, despite analysis in all three species, many questions remain concerning the precise timing, location, and function of the ligand in vivo.
To answer these questions, both genetic and direct biochemical evidence is required. In maize, this is handicapped by laborious and costly plant transformation. Studies using external application of ZmMAC1, as was done for other small proteins (Huffaker et al., 2011; Haruta et al., 2014), is problematic because the protein needs to reach subepidermal layers in anthers that are surrounded by glumes, palea, and lemma. Also, the correct timing of protein delivery to L2-d cells cannot be guaranteed by this method. Neither overexpression using transgenic plants nor external application of ZmMAC1 could permit comparison of recipient and nonrecipient cells within an anther, which is needed to clarify key developmental questions, such as determining whether ZmMAC1 is a cell-autonomous signal. In a metaphorical sense, these challenges are the same the ancient Greeks faced when trying to conquer Troy with its unscalable city walls, as described in Homer’s Odyssey. To this end, the Greeks gifted Troy with a wooden horse with soldiers hidden inside. The so-called Trojan horse (TH) was rolled into the city of Troy, and at night the soldiers emerged and opened the city gate for the Greek army who then conquered the city. Inspired by this ancient trick, we aimed to engineer a molecular TH exploiting the secretory machinery of the pathogenic smut fungus Ustilago maydis. U. maydis can infect all aerial maize organs; during infection, it secretes an array of small proteins to tamper with host defenses and redirect host metabolism (Doehlemann et al., 2008a, 2008b). As for many other pathogens, U. maydis is readily transformed and promoters active during specific stages of host infection are known (Kämper et al., 2006; Doehlemann et al., 2009, 2011a; Wahl et al., 2010). Leader sequences for conventional and unconventional secretion are defined allowing control of posttranslational modifications (Stock et al., 2012). Because most U. maydis effectors are small, cysteine-rich proteins, the secretory machinery should be well primed to secrete correctly folded ZmMAC1 (Kämper et al., 2006; Doehlemann et al., 2009, 2011b).
In this study, we show that U. maydis can be used as a TH to deliver ZmMAC1 protein to individual maize cells at a defined time. Using the TH strategy, we demonstrated that secreted ZmMAC1is a cell-autonomous signal that triggers L2-d periclinal division to establish two new cell types. Furthermore, our results suggest that ZmMAC1 does not need to undergo processing into smaller peptides to be bioactive. To assess the applicability of our findings on ZmMAC1 to other plant ligands, such as OsTDL1A and AtTPD1, we provide evidence that ZmMAC1 functions as a ligand for the LRR-RK ZmMSP1, a homolog of AtEMS1/ EXS and OsMSP1. These functional insights into a conserved developmental signaling module critical for flowering plant reproduction highlight the advantages of TH-delivered protein assays over conventional genetic complementation. The TH strategy of exploiting a pathogen should be generalizable to any host-pathogen interaction in which the pathogen is accessible to genetic modifications and uses secreted proteins to modify the host.
RESULTS
U. maydis Secretes ZmMAC1 Protein into the Anther Apoplast
To experimentally determine the impact of ZmMAC1 on cell division and whether it acts on individual cells or regionally within anthers, in vivo delivery of spatially and temporally restricted ZmMAC1 protein is required. To also overcome difficulties with maize transformation, we aimed to establish a protein delivery system based on the maize smut fungi U. maydis. We generated the U. maydis strain SG200PUmpit2::SpUmpit2-ZmMac1-mCherry-Ha (SG200Zmmac1) to secrete ZmMAC1, fused to mCherry and an HA epitope (Figure 2A). Fungal secretion of fusion protein occurs into the apoplastic space surrounding infected maize cells—this biotrophic interaction zone is a narrow space between the fungal and maize plasma membranes—recapitulating the juxtaposition between AR and neighboring L2-d cells (Figures 2B to 2E). Detection of fluorescence was limited to a narrow part of the apoplastic space surrounding the hyphae growing within an invagination of the plant plasma membrane of an infected cell, indicating that individual infected maize cells receive the ZmMAC1 secreted by SG200Zmmac1 (Figures 2B to 2E). Because SG200Zmmac1 was identical in pathogenicity to its progenitor strain SG200 (Kämper et al., 2006) (Supplemental Figure 1), we assumed that SG200Zmmac1 follows the same time line of infection as SG200, which requires 1 to 2 days after injection to reach subepidermal cells (Doehlemann et al., 2008b)
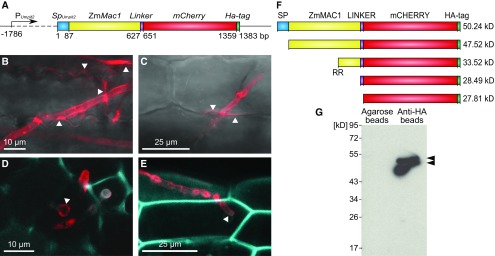
Figure 2.
U. maydis Strain SG200Zmmac1 Secretes Full-Length ZmMAC1 Fusion Protein into the Maize Apoplast.
(A) Schematic overview of the Trojan horse construct integrated into the genome of U. maydis strain SG200 to obtain strain SG200PUmpit2::SpUmpit2-ZmMac1-mCherry-Ha (SG200Zmmac1). The ZmMac1 coding cDNA sequence was fused to mCherry and an HA-tag. To guarantee successful fungal secretion, the coding region was fused at the N terminus with the signal peptide coding region of the U. maydis effector pit2, which targets ZmMAC1 to the maize apoplast (Doehlemann et al., 2011a). Because Umpit2 is highly expressed in planta 2 to 3 dpi, its promoter was used to drive SpUmpit2ZmMac1-mCherry-Ha expression (Doehlemann et al., 2011a).
(B) to (E) Secretion of the ZmMAC1-fusion protein monitored by confocal imaging.
(B) and (C) U. maydis hyphae infecting seedling leaf epidermal cells. SG200Zmmac1 hyphae within epidermal cells of maize seedling leaves. The fusion protein (red) localized around the hyphae and spreads from the fungal hyphae into the apoplast of contiguous cells at sites of cell-to-cell passage (indicated by white arrowheads).
(D) and (E) Transverse section of an anther lobe (D) and anther epidermal cells (E) infected with SG200Zmmac1. The fungal hyphae are growing intracellularly, while the ZmMAC1-fusion protein (red) is secreted and surrounds the fungal hyphae (indicated by white arrowheads). To visualize maize cell walls using autofluorescence (blue), an excitation of 405 nm and detection at 435 to 480 nm were used.
(F) Schematic overview of different ZmMAC1 protein cleavage versions including predicted molecular weight. RR refers to a predicted protease site (Supplemental Figure 2).
(G) Immunodetection by anti-HA serum of ZmMAC1-fusion protein pulled down from SG200Zmmac1 infected spikelet extract with anti-HA or unconjugated agarose matrix after extraction in the presence of protease inhibitors and after formaldehyde incubation of spikelets.
Previously, it was shown that high apoplastic protease activity in tomato (Solanum lycopersicum) results in cleavage of affinity-tagged effectors from Cladosporium fulvum (van Esse et al., 2006). In maize, there are pathogen-activated apoplastic proteases; however, analysis of apoplastic fluid after compatible U. maydis infection showed no increased cysteine protease activity (van der Linde et al., 2012; Mueller et al., 2013). Several plant peptide hormones are reported to undergo processing by proteases (Srivastava et al., 2008, 2009). The ZmMAC1 homologs in Arabidopsis and rice were recently proposed to be further processed by cleavage into smaller peptides (Huang et al., 2016; Yang et al., 2016). In Arabidopsis, a dibasic motif, consisting of amino acids K135 and R136, was suggested as a protease processing site (Huang et al., 2016). This dibasic motif is also present in both OsTDL1A and ZmMAC1 (Figure 2F; Supplemental Figure 2). Taking these previous reports into account, we aimed to test if ZmMAC1 undergoes processing in addition to signal peptide cleavage in U. maydis or in the plant apoplast after secretion. We analyzed protein extracts from SG200Zmmac1-infected spikelets (each spikelet contains two flowers and six anthers) by pull-downs, immunoblotting, and immunodetection with anti-HA antibodies (Figure 2G). To capture ZmMAC1-fusion protein variants produced in vivo and exclude proteolysis during extraction, spikelets were treated with formaldehyde prior to extraction to block protease activity. In extracts from spikelets infected with SG200Zmmac1, two bands were detected after pull-down using anti-HA agarose beads (Figure 2G). These match the molecular mass of unsecreted (50 kD) protein and the secreted fusion protein (48 kD) (Figures 2F and 2G). When the same extract was incubated with agarose beads (no anti-HA), no bands were visualized, indicating the specificity of the anti-HA beads and serum (Figure 2G). Taken together, these results show that U. maydis is capable of secreting tagged ZmMAC1 in a highly spatially restricted way into the maize apoplast (Figure 3A). Our studies further suggested that neither in planta nor in U. maydis did processing beyond cleavage of the secretion signal occur.
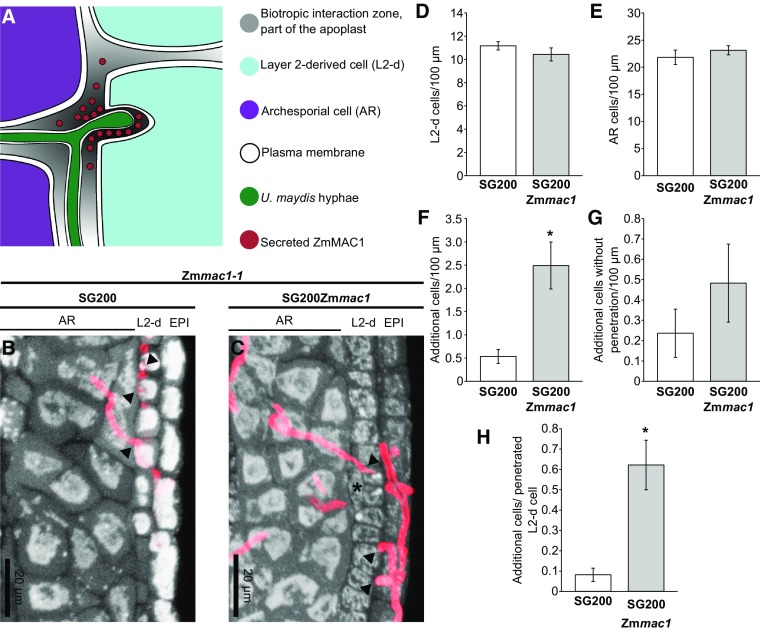
Figure 3.
Cellular Impact of Local ZmMAC1 Secretion by the Trojan Horse.
(A) Schematic view at the cellular level of an Zmmac1-1 anther infected by SG200PUmpit2::SpUmpit2-Zmmac1-mCherry-Ha (SG200Zmmac1). The fungal hyphae secrete the ZmMAC1 fusion protein into the biotrophic interaction zone that is part of the plant apoplast.
(B) and (C) Longitudinal images of Zmmac1-1 anthers infected with U. maydis strain SG200 (SG200) (B) or SG200Zmmac1 (C). False colors showing anther cells in gray and U. maydis hyphae are in red. Additional cells are marked with an asterisk. Plant cells were stained using propidium iodide. U. maydis hyphae were stained with WGA Alexa Fluor 488.
(D) to (G) Cell counts of L2-d (D), archesporial (E), additional cells (F), and additional cells without penetration (G) of Zmmac1-1 anthers infected with SG200 (white) or TH (gray) per 100 µm.
(H) Cells counts of additional cells next to a U. maydis penetrated L2-d cell. Cell counts were performed on 23 to 24 anther sections from a total of 17 anthers, collected in three independent biological replicates (Supplemental Data Sets 1 and 2). Asterisk indicates P values < 0.05 (by Student’s t test); error bars represent se.
ZmMAC1, a Cell-Autonomous Signal, Induces Periclinal Division of L2-d Cells
Typically, maize anther L2-d cells require ∼1 to 2 d after ZmMAC1 perception to complete the periclinal division that establishes the EN and SPC cell types in 180- to 500-µm anthers (Kelliher and Walbot, 2011, 2012; Wang et al., 2012a). To analyze the impact of ZmMAC1 fusion protein delivered to the L2-d and AR cells present in 120- to 300-µm anthers, Zmmac1-1 mutant tassels were infected with either SG200Zmmac1 or control SG200 fungal strains when they had 50- to 125-µm anthers; these immature anthers are several days younger than the stage when normal L2-d cells complete bilayer formation. Three days postinfection (dpi) 400- to 700-µm anthers were dissected for confocal imaging (Figures 3B and 3C). In all imaged Zmmac1-1 mutant anthers, the three morphologically distinct lobe cell layers, epidermis, L2-d, and AR, were infected by U. maydis (Figure 3B). In the SG200Zmmac1-infected anthers, additional cells were readily observed between AR cells and the L2-d layer where SG200Zmmac1 hyphae contacted individual L2-d cells (Figure 3C). The ectopic cells are morphologically more similar to L2-d and somatic niche cells than to AR cells. Therefore, these ectopic cells constitute a bilayered pair derived from the single L2-d layer and required periclinal division of an L2-d progenitor. Quantification of anther cell numbers showed that overall AR and L2-d cell numbers are not perturbed by infection with SG200Zmmac1 compared with anthers infected with the progenitor strain SG200 (Figures 3D and 3E; Supplemental Data Sets 1 and 2). However, there is a statistically significant increase in additional paired cells after SG200Zmmac1 infections (Figure 3F).
When examined more closely, we found no significant addition of cells in areas lacking direct U. maydis penetration (Figure 3G; Supplemental Data Sets 1 and 2). By contrast, cells penetrated in SG200Zmmac1 infected anthers showed a highly significant increase in periclinal division compared with SG200 infected cells (Figure 3H; Supplemental Data Sets 1 and 2). Collectively, half of the L2-d cells penetrated by SG200Zmmac1 hyphae had conducted a periclinal division, generating a bilayer (Figure 3H; Supplemental Data Sets 1 and 2). These results suggest that the TH-delivered ZmMAC1 fusion protein is functional and that apoplastic ZmMAC1 stimulates periclinal division of L2-d cells. Furthermore, this action is apparently cell-autonomous because the fusion protein showed very restricted apoplastic movement (Figures 2B and 2E) and infected cells that divided periclinally did not appear to influence uninfected neighbors.
To verify that ZmMAC1 secretion is essential for periclinal division of L2-d cells, we performed cell count experiments with control strain SG200PUmpit2::ZmMac1-mCherry-Ha (SG200Zmmac1 no SP) that expresses ZmMAC1 in the fungal cytosol (Supplemental Figures 3A to 3D and Supplemental Data Set 3). Compared with the progenitor strain SG200, infection of SG200Zmmac1 no SP showed no significant differences in anther lobe cell counts or any increase in paired somatic cells (Supplemental Figures 3E and 3F and Supplemental Data Set 3).
ZmMAC1 Determines Cell Fate Specification
The first cytological distinction between Zmmac1-1 and fertile anthers is bilayer formation by L2-d periclinal division (Figures 1 and 4A). Molecularly, these somatic niche cells exhibit an altered transcriptome compared with undifferentiated progenitors or AR (Zhang et al., 2014). To determine if TH delivery of ZmMAC1 protein results not only in cytological but also molecular cell differentiation in Zmmac1-1, gene expression was determined using RNA sequencing (Figures 4B to 4G). First, expression of U. maydis genes was analyzed as a proxy for equal infection among samples and replicates (Figure 4B). Maize expression patterns of fertile anthers infected with SG200, a control to show normal somatic niche and AR cell progression, or Zmmac1-1 sterile anthers infected with SG200Zmmac1 were compared with Zmmac1-1 sterile anthers infected with SG200, a negative control lacking the somatic niche and overproducing AR cells (Figures 4C to 4G). Previous publications determined sets of pluripotency, somatic niche, photosynthesis, AR-specific, and meiosis marker genes (Kelliher and Walbot, 2014; Zhang et al., 2014). Comparison of these early anther development markers for the positive control, or Zmmac1-1 anthers infected with SG200Zmmac1 against the negative control, resulted in distinct expression patterns. As expected, expression of pluripotency markers differed in Zmmac1-1 anthers, compared with fertile anthers, because the L2-d cells in Zmmac1-1 mutants fail to exit from pluripotency (Figure 4C). Expression of these genes in Zmmac1-1 infected with SG200Zmmac1 was comparable to the negative control (Figure 4C). Somatic niche marker genes in Zmmac1-1 infected with SG200Zmmac1 show an intermediate expression between positive and negative control (Figure 4D). Chloroplasts are a unique feature of normal EN cells (Figures 1 and 4A); enhanced expression of photosynthesis-related genes are markers for chloroplast differentiation (Zhang et al., 2014; Murphy et al., 2015). Infection of Zmmac1-1 anthers with SG200Zmmac1 led to an overcomplementation of photosynthesis-related gene expression (Figure 4E). Zmmac1-1 produces an excess of AR cells resulting in a different expression pattern of AR-associated and meiotic marker genes (Kelliher and Walbot, 2014) (Figure 4E). Expression of these marker genes in fertile anthers infected with SG200 showed weak correlation with Zmmac1-1 infected with SG200 (Figures 4F and 4G). The same holds true for Zmmac1-1 infected with SG200Zmmac1, yet the linear regression differed substantially from the positive control (Figures 4F and 4G). qRT-PCR analysis on somatic niche and photosynthesis genes confirmed the same trend detected in RNA sequencing (Supplemental Figure 4).
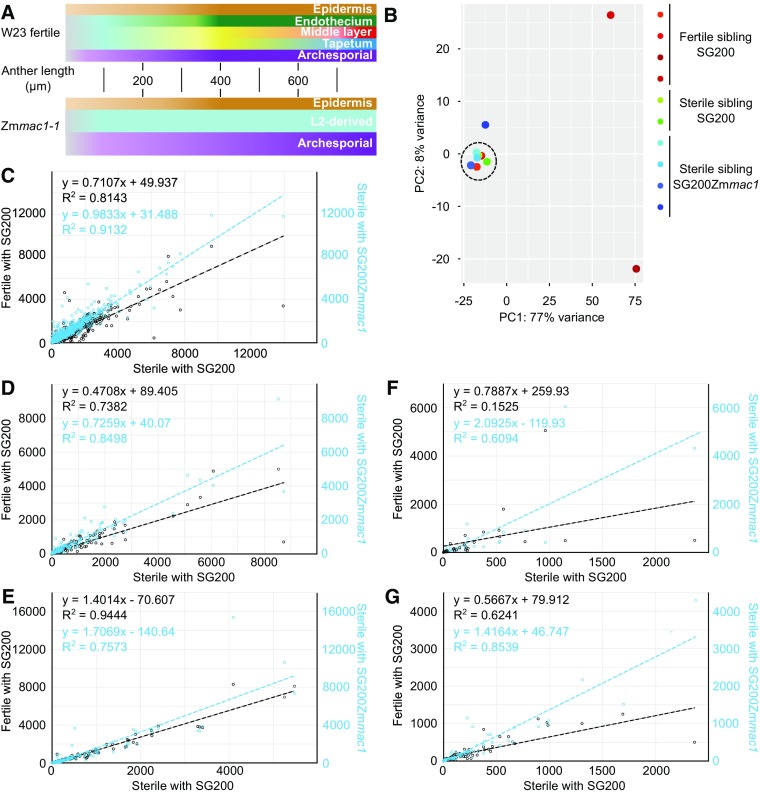
Figure 4.
Molecular Impact of the Trojan Horse on Somatic Cell Fate in Zmmac1-1 Anthers.
(A) Schematic comparison of cell fate specification progression in W23 fertile and Zmmac1-1 anthers up to 800 μm. Lighter hues indicate fate specification and darker hues mature cell types.
(B) to (G) RNA sequencing analysis of U. maydis genes or maize genes in fertile anthers infected with SG200 and Zmmac1-1 anthers infected with SG200 or SG200PUmpit2::SpUmpit2-Zmmac1-mCherry-Ha (SG200Zmmac1).
(B) Principal component analysis of U. maydis gene expression revealed differential infections between samples. Samples with comparable infection are highlighted by a dashed circle and used for the analysis shown in (C) to (G).
(C) to (G) Expression (normalized abundances) comparison by linear regression of pluripotency markers ([C]; Zhang et al., 2014), somatic niche markers ([D]; Kelliher and Walbot, 2014), photosynthesis-associated genes ([E]; Zhang et al., 2014; Murphy et al., 2015), archesporial marker genes ([F]; Kelliher and Walbot, 2014), and meiosis-associated genes ([G]; Kelliher and Walbot, 2014).
In summary, additional cells generated by periclinal division after infection with the TH most closely resembled somatic niche cells, and the transcriptome pattern of anthers was more similar to a fertile control in multiple molecular marker gene sets. In particular, EN cells were present, based on the available markers tested. Therefore, ZmMAC1 is a direct signal for periclinal division of L2-d cells. In addition, it may also regulate EN, SPC, and AR cell fate specification and differentiation.
ZmMAC1 and ZmMSP1, a Conserved Ligand-Receptor Pair
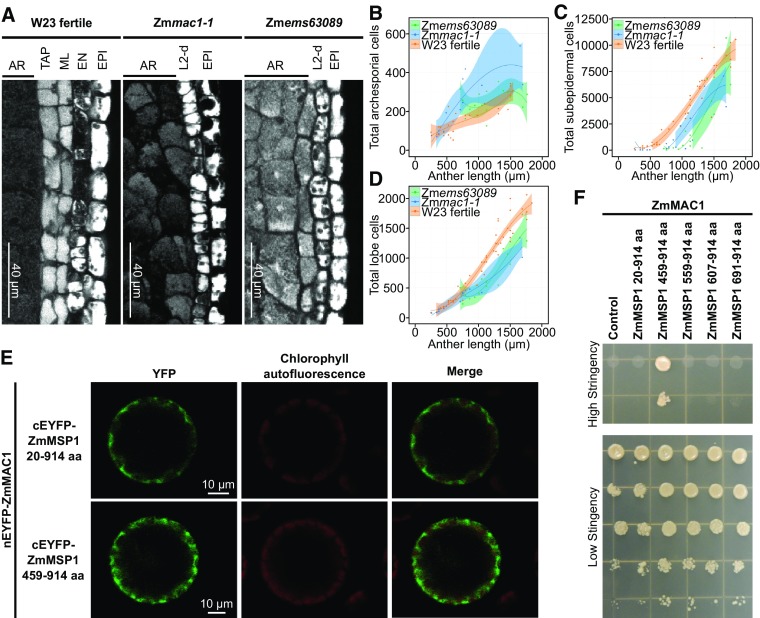
ZmMAC1 homologs in Arabidopsis and rice were reported to be recognized by LRR-RKs to promote anther cell fate differentiation (Huang et al., 2016; Yang et al., 2016). Is this mechanism conserved in maize, i.e., does ZmMAC1 also partner with an LRR-RK in a developmental signaling module? Analysis of the maize genome indicated three candidate LRR-RKs homologous to OsMSP1 and AtEMS1: ZmMSP1 (GRMZM2G447447), ZmMSP2 (GRMZM2G306771), and ZmMSP3 (GRMZM2G107484) (Supplemental Figure 5A and Supplemental Data Set 4). ZmMsp1 and ZmMsp3 exhibit high expression in tassels containing anthers at the developmental stages that require ZmMAC1 (Supplemental Figure 5B). We found that the mutation in the recently described, male sterile maize mutant Zmems63089 (Timofejeva et al., 2013; Egger and Walbot, 2016) maps to a chromosomal region containing ZmMsp1 (Supplemental Figure 6A). Further cloning and sequencing of Zmems63089 identified a G→A substitution in ZmMsp1 resulting in an A→T amino acid substitution, in a conserved region adjacent to the ZmMSP1 DFG domain, which controls kinase activity (Treiber and Shah, 2013) (Supplemental Figures 6B and 7A). To test if this amino acid substitution influences LRR-RK activity, this mutation was introduced into the Arabidopsis AtEMS1 receptor gene, encoding a ZmMSP1 homolog. This mutated allele failed to restore fertility to Arabidopsis Atems1 mutant plants (Supplemental Figure 7B), while the native AtEMS1 restored fertility (Supplemental Figure 7B). Therefore, the Zmems63089 mutation defines a critical amino acid for receptor activity. By confocal microscopy, both Zmmac1-1 and Zmems63089 mutants retain the L2-d layer at 700 µm, unlike fertile anthers (Figure 5A). Closer examination using cell counts at different anther stages (Figures 5B to 5D) showed an excess of AR (Figure 5B) cells and a significantly decreased number of both subepidermal cells (Figure 5C) and total lobe cells (Figure 5D) in both mutants compared with W23 anther lobes (Timofejeva et al., 2013; Egger and Walbot, 2016). Direct comparison of Zmmac1-1 and Zmems63089 found no significant difference in subepidermal cell and total lobe cell numbers between 700 and 1700 μm (Figures 5B to 5D). We concluded that these mutants share the same cellular proliferation defects. Based on the phenotypic identity to Zmmac1-1, we propose that the Zmems63089 mutant identifies ZmMsp1 as the ZmMAC1 receptor gene. To test direct interaction of ZmMAC1 and ZmMSP1 a bimolecular fluorescence complementation (BiFC) approach in Arabidopsis protoplasts was performed, indicating that ZmMAC1 interacts with the LRR domain of ZmMSP1 (Figure 5E). Additionally, interaction of one ZmMSP1 fragment (amino acids 459–914) was detected in yeast two-hybrid assays (Figure 5F). This fragment partially spans the corresponding sequences of ZmMSP1 homologs in Arabidopsis and rice that were previously reported to interact with the corresponding ligands by yeast two-hybrid analysis (Supplemental Figure 7C) (Jia et al., 2008; Yang et al., 2016).
Figure 5.
Genetic Identification and Characterization of the LRR-RK ZmMSP1.
(A) Microscopy images of longitudinal sections of W23 fertile, Zmmac1-1, and Zmems63089 lobes stained with propidium iodide. EPI, epidermis cells.
(B) to (D) Plots of archesporial (B), L2-d or SPC +ML+ TAP (C), and total cells (D), per lobe in W23 fertile (orange) (Egger and Walbot, 2016), Zmems63089 (green) (Egger and Walbot, 2016), and Zmmac1-1 (blue). Data sets (dots) were fit to a third degree polynomial (line), and the shaded region is the 95% confidence interval (Egger and Walbot, 2016). Zmmac1-1 cell counts were performed on 21 anthers from two individual plants.
(E) BiFC assays of ZmMAC1 and ZmMSP1 fragments in Arabidopsis protoplasts. Part of the ZmMAC1 coding region was fused to nEYFP and cEFYP was fused to fragments of ZmMSP1. The resulting constructs were expressed in Arabidopsis protoplasts showing fluorescence complementation (left panel, EYFP shown in green) as a result of interaction. Intactness of protoplasts was visualized by chlorophyll autofluorescence (middle panel, red).
(F) Interaction study of ZmMAC1 and ZmMSP1 by yeast-two-hybrid assay. ZmMAC1 (bait) was tested in combination with the extracellular domain of ZmMSP1 (amino acids 20–914), different fragments of the leucine-rich repeat domain of ZmMSP1 (amino acids are indicated in the Figure) or the empty prey vector (control). Yeast growth was monitored in a dilution series on high stringency (for interaction) and medium stringency media (for growth control).
DISCUSSION
Small proteins serve as signaling components in fundamental developmental processes in plant organs. In shoots and flowers, CLV3 controls the number of stem cells (Clark et al., 1995) and MSCA1 is required to establish meiotically competent cells in the anther (Li et al., 2009; Kelliher and Walbot, 2012), root meristem development is regulated by perception of ROOT GROWTH FACTOR (Matsuzaki et al., 2010), Casparian strip formation is controlled by CASPARIAN STRIP INTEGRITY FACTOR1 (CIF1) and CIF2 (Doblas et al., 2017; Nakayama et al., 2017), and RAPID ALKALINIZATION FACTOR suppresses cell elongation of the primary root (Haruta et al., 2014). The specific functions and interactions of these small proteins are a result of their spatiotemporal pattern in the plant (Breiden and Simon, 2016). Such small signaling proteins can be difficult to study because overexpression or external application do not provide the requisite temporal and spatial resolution within tissues or at individual cells.
Despite intense biochemical and heterologous expression studies of the ZmMAC1 homologs in rice and Arabidopsis, questions about the precise timing and function in planta remain open. To address in vivo functions of ZmMAC1 more precisely, we established the TH approach that exploits microbial pathogen genetics and secretion machinery to deliver small host proteins in situ. For maize, we deployed the maize smut fungus U. maydis to secrete ZmMAC1 into the anther apoplast. Our results suggest that the TH allows direct comparison of cells receiving the ZmMAC1 protein to adjacent cells that lack the biotrophic interface with U. maydis because the fusion protein accumulated exclusively in the apoplast of infected cells. Furthermore, this approach overcomes the bottleneck of maize transformation, promoter choice, position effects on transgene expression, gene silencing, and other difficulties with assessment of protein functions.
Using the TH approach, we found that secreted ZmMAC1 serves as a signal for the periclinal division of L2-d cells, as additional cells constituting a bilayer after periclinal division were readily observed without an increased number of L2-d cells. This confirms previous work based on microscopy observations of Zmmac1-1 in which it was proposed that ZmMAC1 specifically promotes this periclinal division rather than being a general regulator of L2-d cell proliferation (Wang et al., 2012a). Periclinal divisions occurred nearly exclusively at sites where SG200Zmmac1 hyphae had infected a L2-d cell, and in combination with the observed highly local secretion of ZmMAC1 by SG200Zmmac1, we conclude that only L2-d cells that directly receive ZmMAC1 undergo periclinal division and that ZmMAC1 function is cell-autonomous. This conclusion is further supported by the absence of L2-d periclinal division when ZmMAC1 is expressed in U. maydis but lacking the signal peptide for fungal secretion.
Using double mutant analyses of Zmmac1-1 and Zmocl4 (outer cell layer4), Wang et al. (2012a) proposed a balanced circuit that regulates L2-d periclinal division, in which AR secreted ZmMAC1 promotes divisions of L2-d subepidermal cells, and then the EP generates a suppressor that acts on the outer layer after this division to inhibit further division. In this context, a cell-autonomous function of ZmMAC1 might be favorable in terms of counterbalancing an EP-provided repressor to restrict over proliferation. At the molecular level, cell-type and cell fate marker gene expression becomes more similar to fertile anthers in Zmmac1-1 after cells received ZmMAC1, indicating that ZmMAC1 also regulates cell fate specification. In line with this model, ectopic expression of AtTPD1 in anther wall cells in the knockout mutant background induces periclinal division of subepidermal layers and expression of tapetal marker genes in Arabidopsis (Huang et al., 2016).
Interestingly, the recently described Zmems63089 mutant (Timofejeva et al., 2013; Egger and Walbot, 2016) shares key developmental cellular defects with Zmmac1-1. The point mutation in ZmMSP1 in the Zmems63089 allele is critical for kinase activity. Interaction was detected in BiFC and yeast two-hybrid assays between ZmMAC1 and a fragment of ZmMSP1, indicating that ZmMSP1, a homolog of AtEMS and OsMSP1, could be the corresponding receptor for ZmMAC1. Hence, our data support the idea that ZmMAC1, AtTPD, and OsTDL1A serve as ligands in an angiosperm conserved ligand-receptor pair for somatic niche formation in anthers (Huang et al., 2016; Yang et al., 2016). Although the module is conserved, there may be minor variations in terms of spatiotemporal function. For example, knockout mutants in rice, Arabidopsis, and maize show slight phenotypic variations. Furthermore, contrary to the data in rice and Arabidopsis, we did not observe cleavage of ZmMAC1 in planta and full-length secreted ZmMAC1 induced periclinal division and cell fate specification of L2-d cells. This is in line with previously reported immune detection results using an anti-ZmMAC1 antibody (Wang et al., 2012a). More evidence that ZmMAC1 does not require cleavage at the dibasic motif could be generated in future studies by analysis of dibasic motif mutant variants using the TH approach. Also, the roles of the two additional homologs LRR-RKs ZmMSP2 and ZmMSP3 merit future analysis. These could represent neofunctionalized gene duplications or could function as coreceptors as described for BRI1-ASSOCIATED KINASE1 (Chinchilla et al., 2007; Schulze et al., 2010). ZmMac1 is expressed during all stages of premeiotic and meiotic anther development (Wang et al., 2012a) and might be a signal at successive developmental stages of somatic cell layers adjacent to the AR. Future studies could explore potential roles of ZmMAC1 and the ZmMSP gene family in signaling to the AR cells, including coordinating meiotic start.
Considering the data from rice, Arabidopsis, and maize, we propose that ZmMAC1 and its homologs in other plants function as a cell fate specification signal perceived by LRR-RKs in pluripotent cells and potentially at later developmental stages. Furthermore, ZmMAC1 functions as a cell-autonomous signal that must be perceived by each L2-d cell individually. These results clarify the timing, cellular target, and results of ligand secretion to specify the somatic niche during maize anther development.
To date, a variety of transient RNA or DNA delivery systems are available for plants, e.g., virus-induced overexpression or Agrobacterium tumefaciens infiltration assays. However, it is often not clear which cells were transformed within the tissue, and most of these assays are only available for leaves. Stable transformation, on the other hand, can target expression in all plant tissues if promoters are known, yet not all plants are accessible and transformation processes are often costly and time-consuming. The data and insights presented here highlight the advantages of TH-delivered protein assays over conventional genetic complementation and transient assays delivering DNA or RNA. Diverse animal and plant pathogens (bacteria, fungal, and others) deploy secreted proteins, so-called effectors, to tamper with host defenses (Hueck, 1998; Laliberté and Carruthers, 2008; Hewitson et al., 2009; Kim et al., 2016). These effectors are often small, cysteine-rich proteins (Rep, 2005; Kamoun, 2006) and are thus structurally similar to cysteine-rich plant peptide hormones, sometimes even mimicking these endogenous signaling molecules (Ali et al., 2015), making correct folding within the TH likely. More broadly, the pathogenic arsenal of effector proteins spans numerous protein classes, promising that diverse protein classes should be accessible by the TH approach. In many cases, the pathogen secretion process is well studied and the requirements for conventional and unconventional secretion are well defined (Table 1). Different secretion pathways give the opportunity to control and if necessary to avoid foreign posttranslational modifications. Filamentous fungi harbor several homologs to glycosylation-specific genes studied from other organisms, possibly offering the opportunity to test posttranslationally modified proteins (Rosi-Marshall et al., 2007). Pathogens are often genetically tractable and promoters active at specific stages in the infection cycle are available, facilitating engineering of pathogens as Trojan horse delivery vehicles (Table 1); nowadays, some plant pathogens are even used as expression systems in biotechnology because of their exceptional protein folding and secretion capabilities (Royer et al., 1995; Sarkari et al., 2016). Therefore, we predict that the TH strategy of exploiting a pathogen to study secreted host proteins should be generalizable to a variety of host-pathogen interactions. In the particular case of U. maydis, it is important to recognize that it redirects host anther development into a tumor pathway, with gross overproliferation of some cell types (Gao et al., 2013). The perturbation of normal development is not observed until day 5-6 following infection; consequently, the TH can be successfully used prior to this point to assess host responses to an engineered secreted protein.
Table 1. Examples of Transformable Plant Pathogens, Secretion Systems, and Their Hosts.
| Pathogen (Reference for Transformation) | Kingdom | Secretion System | Host |
|---|---|---|---|
| Agrobacterium tumefaciens | Bacterium | Type IV (Vergunst et al., 2005) and type VI (Ma et al., 2014) secretion system | Variety of monocots and dicots |
| Botrytis cinerea (Noda et al., 2007) | Fungi | Signal peptide dependent (ten Have et al., 1998) | Over 200 different species |
| Cladosporium fulvum (Harling et al., 1988) | Fungi | Signal peptide dependent (Joosten et al., 1994) | S. lycopersicum |
| Colletotrichum graminicola (Amnuaykanjanasin and Epstein, 2006) | Fungi | Signal peptide dependent (Vargas et al., 2016) | Z. mays, Triticum aestivum |
| Fusarium oxysporum f. sp Lycopersici (Kistler and Benny, 1988) | Fungi | Signal peptide dependent (Rep et al., 2005) | S. lycopersicum |
| Leptosphaeria maculans (Gout et al., 2006) | Fungi | Signal peptide dependent (Gout et al., 2006) | Brassica napus |
| Magnaporthe oryzae (Talbot et al., 1993) | Fungi | Signal peptide dependent and biotrophic interfacial complex (Giraldo et al., 2013) | O. sativa, T. aestivum, Hordeum vulgare |
| Phytophthora infestans (Avrova et al., 2008) | Oomycete | Signal peptide dependent and BFA-insensitive (Wang et al., 2017) | Solanum tuberosum, S. lycopersicum, other Solanaceae |
| Pseudomonas syringae pathovars (Wendt-Potthoff et al., 1992) | Bacterium | Type III secretion system (He et al., 1993) and type VI (Haapalainen et al., 2012) | Variety of monocots and dicots |
| Sporisorium reilianum (Schulz et al., 1990) | Fungi | Signal peptide dependent (Schirawski et al., 2010) | Z. mays, Sorghum bicolor |
| U. hordei (Ali and Bakkeren, 2011) | Fungi | Signal peptide dependent (Doehlemann et al., 2009) | H. vulgare |
| U. maydis (Kämper et al., 2006) | Fungi | Signal peptide dependent (Kämper et al., 2006), unconventional secretion (Stock et al., 2012) | Z. mays |
| Verticillium dahlia (Wang et al., 2016) | Fungi | Signal peptide dependent and unconventional secretion (Chen et al., 2016) | Medicago sativus |
| Cynara scolymus | |||
| Capsicum annuum | |||
| Brassica oleracea var. capitate | |||
| Brassica oleracea var. botrytis | |||
| Gossypium hirsutum | |||
| Solanum melongena | |||
| Lactuca sativa | |||
| S. tuberosum | |||
| Citrullus vulgaris Schrader | |||
| Zymoseptoria tritici (Zwiers and De Waard, 2001) | Fungi | Signal peptide dependent (Motteram et al., 2009) | T. aestivum, T. turgidum ssp durum |
The TH approach, illustrated here with the ZmMAC1 ligand, may in future studies also offer a rapid method for identifying and evaluating protein motif requirements for efficacy and for interaction with partners in the normal cellular and developmental context. We also note that many of these small proteins are encoded by unannotated genes, reflecting the arbitrary 100-amino acid minimum applied in automated genome annotation (Schnable et al., 2009). Presently, ∼18,000 peptides are estimated to be encoded by the Arabidopsis genome (Breiden and Simon, 2016). In an initial attempt at functional analysis, 473 small proteins identified by proteomics and in silico were constitutively overexpressed in Arabidopsis. Forty-nine lines had visible phenotypes including nine lines overexpressing proteins with signal peptides (Hanada et al., 2013). In previous studies, we identified 424 proteins below 20 kD by mass spectrometry in meiotic maize anthers and 226 proteins smaller than 20 kD in premeiotic anthers; only 19 proteins were common in these two data sets (Wang et al., 2012b; Zhang et al., 2014). For the dozens of proteins with presumptive secretion signals, it is a daunting challenge to conduct traditional genetic analysis and expression in transgenic maize. Therefore, the TH strategy provides a tool to test the biological impact and to elucidate the as yet undiscovered roles of small proteins.
METHODS
Abbreviations, loci, and references for all plant mutants, genes, and proteins are summarized in Supplemental Data Set 5.
Plants Used in This Study and Growth Conditions
Maize (Zea mays) plants used in this study were grown under greenhouse conditions with 14 h/28°C day, 10 h/22°C night, or under field conditions at Stanford University. Ustilago maydis disease ratings were performed on maize cultivar Early Golden Bantam or W23 seedlings and on tassels of Gaspe Flint (indicated in the figure legend). For all Zmmac1-1 experiments, a stock segregating 1:1 for mac1-1//mac1-1 male sterile:mac1-1//+ fertile (Wang et al., 2012a) introgressed to 87.5% into inbred W23 was used. Genotyping of Zmmac1-1 plants was performed by PCR with primers 3F, 3R, and 5R (Supplemental Data Set 6; Wang et al., 2012a) after genomic DNA extraction.
The maize mutant Zmems63089 was generated by Jay Hollick (Ohio State University) by treating inbred A619 pollen with EMS and then crossing pollen onto inbred A632 ears; after self-pollination of the M1 plants, a family segregating for male sterility was identified (Timofejeva et al., 2013). For mapping and positional cloning of Zmems63089, families segregating for the Zmems63089 mutant in the hybrid A619/A632 background were used.
Seeds of Arabidopsis thaliana segregating for the recessive mutant Atems1 (Zhao et al., 2002) generated by maize transposon Ds mutagenesis were kindly provided by D. Zhao (University of Wisconsin-Milwaukee). Seeds were sown on selective media with kanamycin (50 µg/mL). Resistant plants were grown in Biolan soil mixed with vermiculite (7:3) under 16 h/23°C day and 8 h/19°C night conditions in a greenhouse. Seed of Arabidopsis ecotype Col-0 were grown in Biolan soil mixed with vermiculite (7:3) under 16 h/23°C day and 8 h/19°C night conditions in a greenhouse.
Genetic Mapping and Positional Cloning
A mapping population was generated by crossing Zmems63089 mutant plants of A619/A632 background with fertile plants of A632. The first-generation plants were self-pollinated and M2 (F2) plants were scored for sterility. DNA pools of 23 sterile and 21 fertile plants were used for genetic mapping of Zmems63089 via quantitative bulked segregant analysis. This mapping strategy utilizes Sequenom-based SNP assays (Liu et al., 2010). The whole length of candidate genes was sequenced in sterile and fertile plants of the same family as well as in both parental inbred lines and searched for mutations.
Ustilago maydis Strains Used in This Study
To obtain strain SG200PUmpit2::SpUmpit2-ZmMac1-mCherry-Ha (SG200Zmmac1), PCR products of p123-Ppit2-pit2-mCherry-Ha (Mueller et al., 2013) and ZmMac1 (for primers used, see Supplemental Table 5) were digested with NcoI and XbaI and then ligated. The resulting construct (p123-PUmpit2-SpUmpit2-Zmmac1-mCherry-Ha) was integrated into the ip-locus of U. maydis strain SG200 (Kämper et al., 2006).
For construction of strain SG200PUmpit2::ZmMac1-mCherry-Ha (SG200Zmmac1 no SP), inverse PCR was performed on p123-PUmpit2-SpUmpit2-Zmmac1-mCherry-Ha (for primers used, see Supplemental Data Set 6) and the resulting product was circularized by ligation. This plasmid was integrated into the ip-locus of U. maydis strain SG200 (Kämper et al., 2006). The resulting strain and the progenitor strain SG200 (Kämper et al., 2006) were used as controls in plant experiments.
Plant Infections and Disease Ratings
U. maydis strains SG200 (Kämper et al., 2006) and SG200Zmmac1 were grown in axenic culture to an OD600 of between 0.8 and 1.0 and harvested by centrifugation at 3000g, RT for 10 min (Kämper et al., 2006). For disease ratings, cells were resuspended in water to an OD600 of 1.0. For all other experiments, fungal cells were resuspended to an OD600 of 3.0 in water. Tassel and seedling infections as well as disease ratings were performed as described previously (Kämper et al., 2006; Schilling et al., 2014; Redkar et al., 2015). Based on published studies describing U. maydis infection patterns and the role of ZmMAC1 in anther development (Doehlemann et al., 2008b; Wang et al., 2012a), tassels of Zmmac1-1 and its fertile siblings were infected when anthers had a length of between 50 and 125 μm.
RNA Sequencing and Data Analysis
For whole-transcriptome analysis, anther samples containing 50 or more anthers between 450 and 600 µm were collected. Anthers were dissected at 3 dpi with U. maydis. Total RNA for RNA sequencing libraries was isolated using the PureLink Plant RNA Reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. Total RNA quality and quantity were accessed using an RNA 6000 Pico Kit (Agilent) and Qubit RNA BR Assay Kit (Thermo Fisher Scientific), respectively. An aliquot of 500 ng of total RNA was treated with DNase I (New England Biolabs) and then cleaned with RNA Clean and Concentrator-5 (Zymo Research). An aliquot containing 200 ng of treated RNA was used for library construction. The TruSeq Stranded Total RNA with RiboZero-Plant Kit (Illumina) was used for library construction following the manufacturer’s instructions. Libraries were sequenced in an Illumina HiSeq 2500 instrument with single-end mode with 101-bp reads. All of the sequencing runs were performed at the University of Delaware Sequencing and Genotyping Center in Newark, DE.
RNA sequencing libraries were trimmed using Trimmomatic version 0.32 (Bolger et al., 2014). The reads were then mapped to version 3 of the maize genome (Kersey et al., 2016) as well as version 2 of the U. maydis genome (Kämper et al., 2006) using TopHat version 2.0.12 (Trapnell et al., 2009). Transcripts for each genome were assembled from RNA sequencing libraries using the Cufflinks package (Trapnell et al., 2010). Upon completion, raw gene expression abundances were attained from featureCounts version 1.5.0 (Liao et al., 2014) to generate count tables which were then imported into the R statistical environment (R Development Core Team, 2011). Using the U. maydis data first, we normalized the transcript abundances using Deseq2 (Love et al., 2014) and removed reads with fewer than one read to create principal component analysis (PCA) graphs to identify the libraries with the same amount of U. maydis infection. With these date, we were able to remove three libraries (two fertile and one SG200) from the maize differential expression analysis. For the remaining libraries, we used Deseq2 (Love et al., 2014) again to normalize the assembled maize transcripts with more than one read and identify the final differentially expressed genes between three sample types.
RT-qPCR
For RT-qPCR experiments, RNA and cDNA were prepared as described (Zhang et al., 2014) from pools of 50 anthers of three size classes (300–400 µm, 400–500 µm, and 500–600 µm) or from 0.5- to 2.0-cm tassels. All anthers were dissected 3 dpi for both U. maydis strain SG200 and SG200Zmmac1 infections. Primers used for quantification of maize gene transcript levels are summarized in Supplemental Data Set 5. The amplification conditions used and calculation of expression relative to the ZmCyanase control gene were performed as previously described (van der Linde et al., 2011). Experiments were performed in two independent biological replicates in four technical repetitions.
Staining of Anthers, Imaging, and Cell Counts
For cell counts of TH infection, Zmmac1-1 anthers were stained with propidium iodide and WGA-Alexa Fluor 488 as described previously (Doehlemann et al., 2009; Kelliher and Walbot, 2011). Imaging was performed on a Leica SP8 scanning confocal microscope with an AOBS system and HyD SMD detectors using a 63× water objective (Leica Biosystems). The white light laser was used at an excitation of 488 nm for WGA Alexa Fluor 488 and 535 nm for propidium iodide. Emission of propidium iodide was collected between 590 and 640 nm and emission of WGA Alexa Fluor 488 between 500 and 540 nm. Images of whole lobes were taken in 0.5-μm stacks, and images were analyzed in Volocity (Perkin-Elmer). One longitudinal column of L2-d cells on the outer arc of the lobe (facing away from the connective tissue) was selected for analysis; the total number of AR cells and additional cells per penetration were counted in a 20 stack (total 10 μm thick) merged longitudinal lobe image. Sample sizes were chosen based on data in previous studies in which these sample sizes were sufficient to detect statistically significant, nonvisible differences between mutant and fertile sibling cell numbers at similar stages of anther development (Kelliher and Walbot, 2012; Gao et al., 2013; Egger and Walbot, 2016). Student’s t test was used for statistical analysis and P values < 0.05 are indicated by asterisks in figures.
Secretion of SPUmPit2-ZmMAC1-mCHEERY-HA in vivo was observed using an excitation of 561 nm (mCherry) and detection at 580 to 630 nm. Autofluorescence of anther cell wall material was excited at 405 nm and detected at 415 to 460 nm. All images were taken as described above with the SP8 system and analyzed in Volocity.
Imaging and cell counts for comparison of Zmmac1-1 to previously published data for Zmems63089 and W23 fertile anthers were performed as described previously (Egger and Walbot, 2016).
Pull-Down of ZmMAC1 Fusion Protein from TH-Infected Spikelets
For pull-downs followed by immunoblotting and immune detection, spikelets from a 1:1 Zmmac1-1 segregating population containing 150- to 1000-μm anthers 3 dpi by SG200Zmmac1 were fixed by vacuum infiltration in PBS containing 1% formaldehyde for 10 min followed by vacuum infiltration in 1 M glycine for 5 min prior to extraction. For total protein extraction, spikelets were ground to powder in liquid N2 and mixed in a ratio of 1:2 (v/v) with extraction buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM EDTA, 1% IGEPAL CA-630, 0.5 mM DTT, 1% [v/v] plant protease inhibitor cocktail, 0.2 mM E-64, and 4 mM Pefabloc [all Sigma-Aldrich]) and incubated with constant inversion for 1 h at 4°C followed by centrifugation at 2500g for 5 min. Supernatant protein concentration was determined by a Bradford assay (Bio-Rad) and adjusted to a final concentration of 1.8 mg/mL. For precleaning, 36 mg of total protein was incubated with 400 μL uncoupled agarose matrix (equilibrated with three matrix volumes of 0.5× extraction buffer without DTT) at 4°C for 1 h with gentle agitation followed by centrifugation at 2500g for 10 min at 4°C. Then, 18 mg of precleaned total protein was mixed with 100 μL anti-HA-affinity matrix (Roche) and 18 mg precleaned total protein was mixed with 100 μL uncoupled agarose matrix; both matrices were equilibrated with three matrix volumes of 0.5× extraction buffer without DTT. The samples were incubated overnight with constant agitation at 4°C, and then the matrix was collected by centrifugation for 30 s at 500g at 4°C and washed three times with 200 μL 0.5× extraction buffer without DTT. For protein elution, each matrix was mixed with 120 μL of 2× NuPAGE LDS sample buffer (Invitrogen) and incubated at 65°C for 45 min. After elution, DTT (Sigma-Aldrich) was added to a final concentration of 100 mM. Then, 40 μL of eluate was separated by SDS-PAGE and analyzed by immunoblotting followed by immunodetection with anti-HA antibody according to standard protocols (Doehlemann et al., 2009).
Complementation Analysis in Arabidopsis
A QuickChange II XL site-directed mutagenesis kit (Agilent Technologies) was used for whole-plasmid mutagenesis. The entire Gateway pENTR/D-TOPO vector harboring the 1740-bp AtEMS1/EXS promoter and full-length AtEMS1/EXS cDNA (kindly provided by D. Zhao, University of Wisconsin-Milwaukee) was amplified with PfuUltra high-fidelity DNA polymerase using the complementary primers At105 and At106 (Supplemental Data Set 6) with a designed G→A or C→T mutation in the middle. The parental methylated DNA template was digested by DpnI endonuclease. The presence of the G→A mutation in the plasmid was confirmed by sequencing. Both the wild-type and mutated fragments (AtEMS1/EXS and AtEMS1/EXS G→A) were transferred from entry vectors to Gateway binary vector pGWB16 via LR reaction with LR clonase (Invitrogen) following the manufacturer’s protocol. pGWB16 vector was kindly provided by T. Nakagawa (Shimane University, Matsue, Japan) (Nakagawa et al., 2007). The resulting constructs (pGWB16-AtEMS1/EXS and pGWB16- AtEMS1/EXS G→A) were introduced into Agrobacterium tumefaciens strain GV3101, and Arabidopsis plants heterozygous for the Atems1 mutation (Zhao et al., 2002) were transformed with Agrobacterium by the floral dip method (Clough and Bent, 1998). Arabidopsis plants were genotyped using three primers simultaneously (At109, At125, and At126; Supplemental Data Set 6): one associated with the Ds transposon and two associated with the AtEMS1/EXS genomic DNA upstream and downstream of the Ds insertion site. Seeds of T1 transformants were screened with 50 µg/mL hygromycin. Transformed plants were genotyped again using a Ds-specific primer (At109; Supplemental Data Set 6), an AtEMS1/EXS gene-specific primer binding upstream of the Ds transposon insertion (At197; Supplemental Data Set 6), and the primer binding genomic DNA downstream of AtEMS1/EXS (At198; Supplemental Data Set 6) to distinguish the endogenous AtEMS1/EXS wild-type allele from the wild-type transgene introduced by pGW16-AtEMS1/EXS. The presence of a mutated or wild-type transgene was verified with the AtEMS1/EXS gene-specific primer binding the region encoding the kinase domain (At123; Supplemental Data Set 5) and the pGWB16 vector-specific primer (At195; Supplemental Data Set 6).
Yeast Two-Hybrid Assay
ZmMac1, ZmMsp1, or fragments of ZmMsp1 were amplified by PCR (for primers, see Supplemental Data Set 6) and cloned into pENTR/D-TOPO (Invitrogen). Via LR reaction (Invitrogen) fragments were transferred into pGBKT7gw or pGADT7gw. The resulting plasmids, pGBKT7gwZmmac1, pGADT7gwZmmsp1, or pGADT7gw, containing fragments of ZmMsp1 were cotransformed in Saccharomyces cerevisiae strain AH109. Selection and interaction studies were performed according to Mueller et al. (2013).
BiFC in Arabidopsis Protoplasts
Fragments encoding the maize truncated ZmMSP1 or ZmMAC1 proteins were amplified from cDNA with gene-specific primers (for primers, see Supplemental Data Set 6) using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific). The PCR fragments were cloned into pENTR/D-TOPO vector (Invitrogen) and after sequencing subcloned into Gateway pSAT vectors (Citovsky et al., 2006) using the Gateway LR recombinase II enzyme mix (Invitrogen). This resulted in BiFC constructs, where nEYFP (the N-terminal part of EYFP) was fused to MAC1 (without the signal peptide), while cEYFP (the C-terminal part of EYFP) was fused to fragments of MSP1 (459–914 amino acids or 20–914 amino acids).
Protoplast isolation from the leaves of 4-week-old Arabidopsis Col-0 ecotype and protoplast transfection for the transient expression of both MSP1 and MAC1 proteins were performed as previously described (Hansen and van Ooijen, 2016).
Accession Numbers
Sequence data from major genes in this article can be found at www.maizegdb.org using version 3 of the reference genome (Maize B73 RefGen-v3) under the following accession numbers: ZmMac1, GRMZM2G027522; ZmMsp1, GRMZM2G447447; ZmMsp2, GRMZM2G306771; and ZmMsp3, GRMZM2G107484. All sequence data with accession numbers used in this article are summarized in Supplemental Data Set 5. All RNA sequencing data have been deposited into the Gene Expression Omnibus under the accession code GSE109644.
Supplemental Data
Supplemental Figure 1. Pathogenicity assays of U. maydis strains SG200 and SG200Zmmac1.
Supplemental Figure 2. A potential dibasic cleavage motif is conserved among ligands.
Supplemental Figure 3. Characterization of strain SG200PUmpit2::ZmmMac1-mCherry-Ha.
Supplemental Figure 4. Molecular impact of the Trojan horse on cell fate in Zmmac1-1 anthers.
Supplemental Figure 5. Receptor-ligand modules governing somatic fate setting in Arabidopsis, rice, and maize.
Supplemental Figure 6. Mapping of Zmems63089.
Supplemental Figure 7. Conservation of structure and function in the ligand-receptor module regulating somatic niche formation in anthers.
Supplemental Data Set 1. Cell counts of Zmmac1-1 anthers lobes infected with U. maydis strain SG200.
Supplemental Data Set 2. Cell counts of Zmmac1-1 anther lobes infected with U. maydis strain SG200PUmpit2::SpUmpit2-ZmMac1-mCherry-Ha (SG200Zmmac1).
Supplemental Data Set 3. Cell counts of Zmmac1-1 anthers lobes infected with U. maydis strain SG200PUmpit2::ZmMac1-mCherry-Ha (SG200Zmmac1 no SP).
Supplemental Data Set 4. Alignment of leucine-rich repeat receptor-like kinase from Arabidopsis, rice, and maize.
Supplemental Data Set 5. Gene, protein, and mutant abbreviations in alphabetical order.
Supplemental Data Set 6. Oligonucleotides used in this study.
Acknowledgments
This work was supported by the National Science Foundation (Award IOS13-39229). K.v.d.L. was supported by the National Academy of Sciences-Leopoldina (Germany). L.T. and B.I. were supported by Grant IUT19-3 from the Estonian Ministry of Education and Research. We thank Darren Morrow, Sidae Lee, Jorgen Holm, Mary Beth Mudgett, Heather Cartwright, and the Carnegie Department of Plant Biology Imaging facility for advice and technical support. Vectors pGBKT7gw and pGADT7gw, and S. cerevisiae strain AH109 were a kind gift from Mary Beth Mudgett and Jung-Gun Kim.
AUTHOR CONTRIBUTIONS
K.v.d.L., L.T., R.L.E., B.I., G.D., B.M., and V.W. designed the experiments. K.v.d.L., L.T., R.L.E., B.I., and C.T. performed research. K.v.d.L., L.T., R.L.E., B.I., and R.H. analyzed data. K.v.d.L., L.T., and V.W. wrote the manuscript with input from all authors.
References
- Ali S., Bakkeren G. (2011). Introduction of large DNA inserts into the barley pathogenic fungus, Ustilago hordei, via recombined binary BAC vectors and Agrobacterium-mediated transformation. Curr. Genet. 57: 63–73. [DOI] [PubMed] [Google Scholar]
- Ali S., Magne M., Chen S., Côté O., Stare B.G., Obradovic N., Jamshaid L., Wang X., Bélair G., Moffett P. (2015). Analysis of putative apoplastic effectors from the nematode, Globodera rostochiensis, and identification of an expansin-like protein that can induce and suppress host defenses. PLoS One 10: e0115042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amnuaykanjanasin A., Epstein L. (2006). A class Vb chitin synthase in Colletotrichum graminicola is localized in the growing tips of multiple cell types, in nascent septa, and during septum conversion to an end wall after hyphal breakage. Protoplasma 227: 155–164. [DOI] [PubMed] [Google Scholar]
- Avrova A.O., Boevink P.C., Young V., Grenville-Briggs L.J., van West P., Birch P.R., Whisson S.C. (2008). A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell. Microbiol. 10: 2271–2284. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiden M., Simon R. (2016). Q&A: How does peptide signaling direct plant development? BMC Biol. 14: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.Y., Xiao H.L., Gui Y.J., Zhang D.D., Li L., Bao Y.M., Dai X.F. (2016). Characterization of the Verticillium dahliae exoproteome involves in pathogenicity from cotton-containing medium. Front. Microbiol. 7: 1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Lee L.Y., Vyas S., Glick E., Chen M.H., Vainstein A., Gafni Y., Gelvin S.B., Tzfira T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362: 1120–1131. [DOI] [PubMed] [Google Scholar]
- Clark S.E., Running M.P., Meyerowitz E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067. [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Doblas V.G., Smakowska-Luzan E., Fujita S., Alassimone J., Barberon M., Madalinski M., Belkhadir Y., Geldner N. (2017). Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Reissmann S., Assmann D., Fleckenstein M., Kahmann R. (2011a). Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis-induced tumour formation. Mol. Microbiol. 81: 751–766. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Reissmann S., Assmann D., Fleckenstein M., Kahmann R. (2011b). Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis-induced tumour formation. Mol. Microbiol. 81: 751–766. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Vraneš M., de Vries R.P., Kämper J., Kahmann R. (2008a). Establishment of compatibility in the Ustilago maydis/maize pathosystem. J. Plant Physiol. 165: 29–40. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., van der Linde K., Assmann D., Schwammbach D., Hof A., Mohanty A., Jackson D., Kahmann R. (2009). Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5: e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Horst R.J., Voll L.M., Usadel B., Poree F., Stitt M., Pons-Kühnemann J., Sonnewald U., Kahmann R., Kämper J. (2008b). Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 56: 181–195. [DOI] [PubMed] [Google Scholar]
- Egger R.L., Walbot V. (2016). A framework for evaluating developmental defects at the cellular level: An example from ten maize anther mutants using morphological and molecular data. Dev. Biol. 419: 26–40. [DOI] [PubMed] [Google Scholar]
- Gao L., Kelliher T., Nguyen L., Walbot V. (2013). Ustilago maydis reprograms cell proliferation in maize anthers. Plant J. 75: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo M.C., Dagdas Y.F., Gupta Y.K., Mentlak T.A., Yi M., Martinez-Rocha A.L., Saitoh H., Terauchi R., Talbot N.J., Valent B. (2013). Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 4: 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout L., Fudal I., Kuhn M.L., Blaise F., Eckert M., Cattolico L., Balesdent M.H., Rouxel T. (2006). Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol. Microbiol. 60: 67–80. [DOI] [PubMed] [Google Scholar]
- Haapalainen M., Mosorin H., Dorati F., Wu R.F., Roine E., Taira S., Nissinen R., Mattinen L., Jackson R., Pirhonen M., Lin N.C. (2012). Hcp2, a secreted protein of the phytopathogen Pseudomonas syringae pv. tomato DC3000, is required for fitness for competition against bacteria and yeasts. J. Bacteriol. 194: 4810–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., et al. (2013). Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc. Natl. Acad. Sci. USA 110: 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L.L., van Ooijen G. (2016). Rapid analysis of circadian phenotypes in Arabidopsis protoplasts transfected with a luminescent clock reporter. J. Vis. Exp. 115: doi/10.3791/54586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harling R., Kenyon L., Lewis B.G., Oliver R.P., Turner J.G., Coddington A. (1988). Conditions for efficient isolation and regeneration of protoplasts from Fulvia fulva. J. Phytopathol. 122: 143–146. [Google Scholar]
- Haruta M., Sabat G., Stecker K., Minkoff B.B., Sussman M.R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S.Y., Huang H.C., Collmer A. (1993). Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73: 1255–1266. [DOI] [PubMed] [Google Scholar]
- Hewitson J.P., Grainger J.R., Maizels R.M. (2009). Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol. Biochem. Parasitol. 167: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhang T., Linstroth L., Tillman Z., Otegui M.S., Owen H.A., Zhao D. (2016). Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet. 12: e1006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck C.J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62: 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Dafoe N.J., Schmelz E.A. (2011). ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 155: 1325–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Liu X., Owen H.A., Zhao D. (2008). Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc. Natl. Acad. Sci. USA 105: 2220–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten M.H., Cozijnsen T.J., De Wit P.J. (1994). Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367: 384–386. [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44: 41–60. [DOI] [PubMed] [Google Scholar]
- Kämper J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101. [DOI] [PubMed] [Google Scholar]
- Kelliher T., Walbot V. (2011). Emergence and patterning of the five cell types of the Zea mays anther locule. Dev. Biol. 350: 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T., Walbot V. (2012). Hypoxia triggers meiotic fate acquisition in maize. Science 337: 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T., Walbot V. (2014). Maize germinal cell initials accommodate hypoxia and precociously express meiotic genes. Plant J. 77: 639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P.J., et al. (2016). Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 44: D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.T., Jeon J., Choi J., Cheong K., Song H., Choi G., Kang S., Lee Y.H. (2016). Kingdom-Wide Analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front. Plant Sci. 7: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler H.C., Benny U.K. (1988). Genetic transformation of the fungal plant wilt pathogen, Fusarium oxysporum. Curr. Genet. 13: 145–149. [Google Scholar]
- Laliberté J., Carruthers V.B. (2008). Host cell manipulation by the human pathogen Toxoplasma gondii. Cell. Mol. Life Sci. 65: 1900–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lauri A., Ziemann M., Busch A., Bhave M., Zachgo S. (2009). Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Liu S., Chen H.D., Makarevitch I., Shirmer R., Emrich S.J., Dietrich C.R., Barbazuk W.B., Springer N.M., Schnable P.S. (2010). High-throughput genetic mapping of mutants via quantitative single nucleotide polymorphism typing. Genetics 184: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.S., Hachani A., Lin J.S., Filloux A., Lai E.M. (2014). Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16: 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067. [DOI] [PubMed] [Google Scholar]
- Motteram J., Küfner I., Deller S., Brunner F., Hammond-Kosack K.E., Nürnberger T., Rudd J.J. (2009). Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain-containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 22: 790–799. [DOI] [PubMed] [Google Scholar]
- Mueller A.N., Ziemann S., Treitschke S., Aßmann D., Doehlemann G. (2013). Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9: e1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.M., Egger R.L., Walbot V. (2015). Chloroplasts in anther endothecium of Zea mays (Poaceae). Am. J. Bot. 102: 1931–1937. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakayama T., Shinohara H., Tanaka M., Baba K., Ogawa-Ohnishi M., Matsubayashi Y. (2017). A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284–286. [DOI] [PubMed] [Google Scholar]
- Noda J., Brito N., Espino J.J., González C. (2007). Methodological improvements in the expression of foreign genes and in gene replacement in the phytopathogenic fungus Botrytis cinerea. Mol. Plant Pathol. 8: 811–816. [DOI] [PubMed] [Google Scholar]
- Nonomura K., Miyoshi K., Eiguchi M., Suzuki T., Miyao A., Hirochika H., Kurata N. (2003). The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing).
- Redkar A., Hoser R., Schilling L., Zechmann B., Krzymowska M., Walbot V., Doehlemann G. (2015). A secreted effector protein of Ustilago maydis guides maize leaf cells to form tumors. Plant Cell 27: 1332–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M. (2005). Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 253: 19–27. [DOI] [PubMed] [Google Scholar]
- Rep M., Meijer M., Houterman P.M., van der Does H.C., Cornelissen B.J.C. (2005). Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Mol. Plant Microbe Interact. 18: 15–23. [DOI] [PubMed] [Google Scholar]
- Rosi-Marshall E.J., Tank J.L., Royer T.V., Whiles M.R., Evans-White M., Chambers C., Griffiths N.A., Pokelsek J., Stephen M.L. (2007). Toxins in transgenic crop byproducts may affect headwater stream ecosystems. Proc. Natl. Acad. Sci. USA 104: 16204–16208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer J.C., et al. (1995). Fusarium graminearum A 3/5 as a novel host for heterologous protein production. Biotechnology (N.Y.) 13: 1479–1483. [DOI] [PubMed] [Google Scholar]
- Sarkari P., Feldbrügge M., Schipper K. (2016). The corn smut fungus Ustilago maydis as an alternative expression system for biopharmaceuticals. In Gene Expression Systems in Fungi: Advancements and Applications, Schmoll M. and Dattenböck C., eds (Cham, Switzerland: Springer International Publishing; ), pp. 183–200. [Google Scholar]
- Schilling L., Matei A., Redkar A., Walbot V., Doehlemann G. (2014). Virulence of the maize smut Ustilago maydis is shaped by organ-specific effectors. Mol. Plant Pathol. 15: 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J., et al. (2010). Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330: 1546–1548. [DOI] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schulz B., Banuett F., Dahl M., Schlesinger R., Schäfer W., Martin T., Herskowitz I., Kahmann R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60: 295–306. [DOI] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A.K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285: 9444–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan W.F., Golubeva E.A., Abrhamova L.I., Golubovskaya I.N. (1999). The mac1 mutation alters the developmental fate of the hypodermal cells and their cellular progeny in the maize anther. Genetics 153: 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan W.F., Avalkina N.A., Shamrov I.I., Batygina T.B., Golubovskaya I.N. (1996). The mac1 gene: controlling the commitment to the meiotic pathway in maize. Genetics 142: 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R., Liu J.-X., Howell S.H. (2008). Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 56: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R., Liu J.X., Guo H., Yin Y., Howell S.H. (2009). Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 59: 930–939. [DOI] [PubMed] [Google Scholar]
- Stock J., Sarkari P., Kreibich S., Brefort T., Feldbrügge M., Schipper K. (2012). Applying unconventional secretion of the endochitinase Cts1 to export heterologous proteins in Ustilago maydis. J. Biotechnol. 161: 80–91. [DOI] [PubMed] [Google Scholar]
- Talbot N.J., Ebbole D.J., Hamer J.E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5: 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Have A., Mulder W., Visser J., van Kan J.A. (1998). The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant Microbe Interact. 11: 1009–1016. [DOI] [PubMed] [Google Scholar]
- Timofejeva L., Skibbe D.S., Lee S., Golubovskaya I., Wang R., Harper L., Walbot V., Cande W.Z. (2013). Cytological characterization and allelism testing of anther developmental mutants identified in a screen of maize male sterile lines. G3 (Bethesda) 3: 231–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber D.K., Shah N.P. (2013). Ins and outs of kinase DFG motifs. Chem. Biol. 20: 745–746. [DOI] [PubMed] [Google Scholar]
- van der Linde K., Kastner C., Kumlehn J., Kahmann R., Doehlemann G. (2011). Systemic virus-induced gene silencing allows functional characterization of maize genes during biotrophic interaction with Ustilago maydis. New Phytol. 189: 471–483. [DOI] [PubMed] [Google Scholar]
- van der Linde K., Hemetsberger C., Kastner C., Kaschani F., van der Hoorn R.A., Kumlehn J., Doehlemann G. (2012). A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell 24: 1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse H.P., Thomma B.P., van ’t Klooster J.W., de Wit P.J. (2006). Affinity-tags are removed from Cladosporium fulvum effector proteins expressed in the tomato leaf apoplast. J. Exp. Bot. 57: 599–608. [DOI] [PubMed] [Google Scholar]
- Vargas W.A., Sanz-Martín J.M., Rech G.E., Armijos-Jaramillo V.D., Rivera L.P., Echeverria M.M., Díaz-Mínguez J.M., Thon M.R., Sukno S.A. (2016). A fungal effector with host nuclear localization and DNA-binding properties is required for maize anthracnose development. Mol. Plant Microbe Interact. 29: 83–95. [DOI] [PubMed] [Google Scholar]
- Vergunst A.C., van Lier M.C., den Dulk-Ras A., Stüve T.A., Ouwehand A., Hooykaas P.J. (2005). Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. USA 102: 832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl R., Wippel K., Goos S., Kämper J., Sauer N. (2010). A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biol. 8: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V., Egger R.L. (2016). Pre-meiotic anther development: Cell fate specification and differentiation. Annu. Rev. Plant Biol. 67: 365–395. [DOI] [PubMed] [Google Scholar]
- Wang C.J., Nan G.L., Kelliher T., Timofejeva L., Vernoud V., Golubovskaya I.N., Harper L., Egger R., Walbot V., Cande W.Z. (2012a). Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 139: 2594–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Adams C.M., Fernandes J.F., Egger R.L., Walbot V. (2012b). A low molecular weight proteome comparison of fertile and male sterile 8 anthers of Zea mays. Plant Biotechnol. J. 10: 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xing H., Hua C., Guo H.S., Zhang J. (2016). An improved single-step cloning strategy simplifies the Agrobacterium tumefaciens-mediated transformation (ATMT)-based gene-disruption method for Verticillium dahliae. Phytopathology 106: 645–652. [DOI] [PubMed] [Google Scholar]
- Wang S., Boevink P.C., Welsh L., Zhang R., Whisson S.C., Birch P.R.J. (2017). Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol. 216: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt-Potthoff K., Niepold F., Backhaus H. (1992). High-efficiency electro-transformation of the plant pathogen Pseudomonas syringae pv. syringae R32. J. Microbiol. Methods 16: 33–37. [Google Scholar]
- Yang L., Qian X., Chen M., Fei Q., Meyers B.C., Liang W., Zhang D. (2016). Regulatory role of a receptor-like kinase in specifying anther cell identity. Plant Physiol. 171: 2085–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Egger R.L., Kelliher T., Morrow D., Fernandes J., Nan G.L., Walbot V. (2014). Transcriptomes and proteomes define gene expression progression in pre-meiotic maize anthers. G3 (Bethesda) 4: 993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D.Z., Wang G.F., Speal B., Ma H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16: 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., de Palma J., Oane R., Gamuyao R., Luo M., Chaudhury A., Hervé P., Xue Q., Bennett J. (2008). OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J. 54: 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiers L.H., De Waard M.A. (2001). Efficient Agrobacterium tumefaciens-mediated gene disruption in the phytopathogen Mycosphaerella graminicola. Curr. Genet. 39: 388–393. [DOI] [PubMed] [Google Scholar]