RecQ DNA helicases are important genome surveillance proteins that are found in meristematic tissues, where they function in plant development, DNA repair, and gene targeting in the moss Physcomitrella patens.
Abstract
RecQ DNA helicases are genome surveillance proteins found in all kingdoms of life. They are characterized best in humans, as mutations in RecQ genes lead to developmental abnormalities and diseases. To better understand RecQ functions in plants we concentrated on Arabidopsis thaliana and Physcomitrella patens, the model species predominantly used for studies on DNA repair and gene targeting. Phylogenetic analysis of the six P. patens RecQ genes revealed their orthologs in humans and plants. Because Arabidopsis and P. patens differ in their RecQ4 and RecQ6 genes, reporter and deletion moss mutants were generated and gene functions studied in reciprocal cross-species and cross-kingdom approaches. Both proteins can be found in meristematic moss tissues, although at low levels and with distinct expression patterns. PpRecQ4 is involved in embryogenesis and in subsequent development as demonstrated by sterility of ΔPpRecQ4 mutants and by morphological aberrations. Additionally, ΔPpRecQ4 displays an increased sensitivity to DNA damages and an increased rate of gene targeting. Therefore, we conclude that PpRecQ4 acts as a repressor of recombination. In contrast, PpRecQ6 is not obviously important for moss development or DNA repair but does function as a potent enhancer of gene targeting.
INTRODUCTION
RecQ helicases are genome surveillance proteins that are found in all kingdoms of life. They are best investigated in humans as mutations in each of the five HsRecQ genes result in developmental abnormalities and cancer predispositions and various other diseases (e.g., Bloom, Werner, and Rothmund-Thomson syndromes; Croteau et al., 2014). Animals, algae, and plants harbor four to seven RecQs, but fungi have only one to three different RecQs (Hartung and Puchta, 2006). Their phylogenetic relationship is unsolved but we can assign orthologs to the best investigated eukaryotic RecQ, the Bloom syndrome protein (Blm helicase), named RecQ4 in plants, Sgs1 (small growth suppressor 1) in Saccharomyces cerevisiae, and Rqh1 (RecQ homolog 1) in Schizosaccharomyces pombe. Two Arabidopsis thaliana RecQ helicases, RecQ4A and RecQ4B, are functional and phylogenetic orthologs of Blm and Sgs1 (Bagherieh-Najjar et al., 2005; Hartung and Puchta, 2006; Hartung et al., 2007). All sequenced members of the Brassicaceae encode these two RecQ4 genes, whereas all other plants possess only one RecQ4 (Phytozome v12.0; www.phytozome.net). Therefore, RecQ4A and RecQ4B probably originated from the duplication of a single RecQ4 after the separation of the last common ancestor of Carica papaya and Arabidopsis ∼70 million years ago (Proost et al., 2011). The ancestral RecQ4 is present in early diverging land plants, e.g., the moss Physcomitrella patens and the lycophyte Selaginella moellendorfii, and is therefore regarded as the original Blm/Sgs1 ortholog (Hartung et al., 2007). Intriguingly, mutations in AtRecQ4A and AtRecQ4B result in different phenotypes: While Atrecq4a exhibits enhanced sensitivity toward genotoxins and increased homologous recombination (HR) in somatic cells, Atrecq4b shows slightly decreased HR frequencies (Bagherieh-Najjar et al., 2005; Hartung et al., 2006, 2007). One explanation of these different phenotypes is a split of function of the ancestral RecQ4 protein after gene duplication. Alternatively, the positive role of AtRecQ4B in HR is a neofunctionalization after duplication. Thus, although the widespread appearance of RecQs suggests their essential functions, these are unclear for plant RecQs. Furthermore, inference of RecQ functions in plants is mainly based on a single species, which may represent an exception rather than the rule, as exemplified by the duplicated RecQ4 with opposing effects of the two paralogs. Many plants outside the Brassicales encode an additional RecQ protein, which was named OsRecQ886 in rice (Oryza sativa), based upon its number of amino acids. It shows low identity to other RecQ proteins, covering only the helicase domain. Therefore, it was proposed that it is of ancient origin and was lost during evolution in the Brassicales/Malvales (Hartung and Puchta, 2006) which is supported by the fact that P. patens also encodes a RecQ886 homolog. As protein lengths vary between species, we propose renaming RecQ886 to RecQ6. Saotome et al. (2006) showed that OsRecQ886/OsRecQ6 is expressed mainly in proliferating cells and may be involved in the repair of alkyl DNA lesions, oxidative DNA damage, and DNA double-strand breaks (DSBs). However, there is no functional characterization based on Osrecq6 mutants available.
To gain a deeper understanding of plant RecQ functions, we undertook an evolutionary approach and concentrated on the species predominantly used in the analysis of DNA recombination, Arabidopsis and P. patens, a haploid-dominant plant with outstandingly efficient HR in somatic cells (Reski, 1998a). First, we identified six RecQ genes in P. patens based on the recent version of its genome (Lang et al., 2018) and analyzed their phylogeny in comparison to different land plants. Additionally, we identified the orthologous relations between human and plant RecQ proteins. Second, we analyzed the expression of the PpRecQ genes in P. patens wild type and studied experimentally the tissue-specific distribution of PpRecQ4 and PpRecQ6 in reporter lines. Third, we generated ΔPpRecQ4 and ΔPpRecQ6 mutants by targeted gene disruption, a technique well established in this organism (Strepp et al., 1998). Fourth, we performed cross-species complementation experiments in Atrecq4a and ΔPpRecQ4 mutant backgrounds with the respective orthologs from the other species. Therefore, we cloned the PpRecQ4 cDNA and transferred it into the previously described Atrecq4a mutant of Arabidopsis (Hartung et al., 2007). Vice versa, AtRecQ4A and AtRecQ4B cDNAs were expressed in ΔPpRecQ4. Additionally, we tested if a cross-kingdom complementation of the ΔPpRecQ4 phenotype by the human Blm cDNA is possible.
RESULTS
Phylogeny of Plant RecQ Proteins
To resolve the relationship of plant RecQs, we used the RecQ orthologs of P. patens, Arabidopsis, and Homo sapiens as a reference set to query the proteomes of 60 Viridiplantae, S. cerevisiae and S. pombe (Supplemental Data Set 1). We identified candidates by searching with the Hidden Markov Models (HMMs) of these reference sequences from Panther (Thomas et al., 2003) and applied strict filtering based on Pfam (Finn et al., 2016) domain composition to obtain 360 RecQ orthologs. Multiple sequence alignments and subsequent construction of phylogenetic trees revealed six clusters corresponding to the six RecQ subfamilies (Figure 1; Supplemental Figures 1 to 7). Interestingly, each of the five human RecQs aligned basally to an individual cluster. Among all six, only orthologs in the clusters RecQ2, RecQ4, and RecQ5 were identified in all major clades of Viridiplantae (Supplemental Figures 2, 4, and 5), whereas RecQ4 is the only cluster with additional orthologs in yeasts. This subfamily comprises the highest amount of paralogs including a putative duplication in the last common ancestor of Brassicaceae. The clustering also suggests that RecQsim originated within the clade of RecQ6. Alternatively, RecQsim was present in the last common ancestor of all land plants and lost in the lineage including Marchantia polymorpha.
Figure 1.
Cladogram of the RecQ Family.
Midpoint-rooted maximum-likelihood tree based on a multiple sequence alignment of RecQ CDS sequences identified in the Viridiplantae, H. sapiens, S. cerevisiae, and S. pombe. The cladogram was calculated with RAxML using the GTRGAMMA nucleotide substitution model and 1000 rapid bootstrap samples. Tip labels show the corresponding species abbreviation for each sequence. The six colored clades highlight the RecQ subfamilies.
According to all genomic and transcriptomic data available (Rensing et al., 2008; Zimmer et al., 2013; Lang et al., 2018; Hiss et al., 2014; Matasci et al., 2014; Ortiz-Ramírez et al., 2016) P. patens does not encode RecQ1 and RecQ3. Our analysis also indicated that the second moss in our data set, Sphagnum fallax, does not harbor these two genes, yet the liverwort M. polymorpha does (Supplemental Figures 1 and 3). In contrast, P. patens possesses a duplicated RecQsim, whereas S. fallax harbors one single ortholog and M. polymorpha none (Supplemental Figure 7). To gain more insight into other mosses, we searched this clade in the 1KP database (Matasci et al., 2014). While no orthologs for AtRecQ1 were identified, orthologs for AtRecQ3 are expressed in a number of mosses including the class of Bryopsida, but neither in P. patens nor in Physcomitrium spec., the closest related species (Beike et al., 2014) present in this database (Supplemental Data Set 2). The properties of the six PpRecQ genes are shown in Supplemental Figure 8.
P. patens RecQ4 and RecQ6: Gene Expression Patterns and Tissue-Specific Protein Accumulation
The expression of all six P. patens RecQs was analyzed in silico in publicly available transcriptomic databases on microarray experiments (Hiss et al., 2014, Ortiz-Ramírez et al., 2016; Supplemental Figures 9A and 9B) and found to be low in all tissues and developmental stages and conditions. Even upon induction of DSBs, no significant changes in the expression of the PpRecQs are reported (Kamisugi et al., 2016). Subsequently, we concentrated on RecQ4 and RecQ6 because Arabidopsis encodes a second RecQ4 but no RecQ6, suggesting that this difference might account for the dramatic difference in gene targeting (GT) efficiency between the two plant species. To assay protein distribution and abundance in planta, GUS reporter lines for PpRecQ4 and PpRecQ6 were established. Knockin constructs were generated in order to insert the coding sequence of GUS in frame at the end of the coding sequence of the endogenous gene via HR, replacing the original stop codon, thereby creating a PpRecQ4:GUS (Supplemental Figure 10A) or PpRecQ6:GUS fusion (Supplemental Figure 10B). The fused transcripts were detected in five PpRecQ4:GUS (Supplemental Figure 10A) and two PpRecQ6:GUS lines (Supplemental Figure 10B). All subsequent analyses were performed using these lines, which showed consistent results and did not deviate in their morphology or development from P. patens wild type.
PpRecQ4 and PpRecQ6 proteins accumulate in meristematic cells of buds and young gametophores (Figures 2A and 2J). Additionally, both proteins are detected throughout young gametangia, female archegonia including the egg cell, and in male antheridia (Figures 2B to 2F and 2K to 2P). Upon development of the sporophyte after fertilization of the egg cell, the GUS signal disappears in PpRecQ4:GUS (Figures 2G and 2H). It remains present in PpRecQ6:GUS sporophytes but exclusively restricted to the seta (Figures 2P and 2Q). Neither PpRecQ4 nor PpRecQ6 is detected in spores or protonema (Figures 2I and 2R). Additionally, PpRecQ6 accumulates at side branch initials (Figures 2K and 2L) and partially upper parts of rhizoids (Figure 2P). Treatment of protonema, young gametophores, or gametophores carrying gametangia with bleomycin did not affect the pattern or strength of GUS accumulation compared with untreated controls.
Figure 2.
Tissue-Specific Localization of PpRecQ4 and PpRecQ6 Visualized by GUS Staining of Reporter Lines.
The GUS signal is detected in specific tissues throughout the life cycle by expression of the GUS-tagged endogenous PpRecQ4 ([A] to [I]) or PpRecQ6 ([J] to [R]) by fusion of the gene with the GUS coding sequence by insertion directly in front of the native stop codon.
(A) and (J) Young gametophores: arrows point to the GUS signal at the apex. Bars = 50 µm.
(B) and (K) Single gametophore prepared from a colony with GUS signal at the apex in gametangia highlighted by the box in (B) and arrow in (K). Bars = 1 mm.
(C) and (L) Close-up (C) of apex highlighted by box in (B) (bar = 200 µm) and close-up (L) of the basal region of PpRecQ6:GUS with stained initials for side branches highlighted with the box in (K) (bar = 100 µm).
(D) and (M) The developing antheridia ([D], left; [M], top) are completely stained, while in mature antheridia before release of sperm cells with the tip cell still intact ([D], right, [M], bottom), no GUS signal was detected. Bars = 25 µm.
(E) and (N) Young archegonia with closed neck were completely stained. Bars = 25 µm.
(F) and (O) Archegonia ready for fertilization with an opened neck showed accumulation of the GUS signal in the lower part; arrows point to the egg cell. Bars = 25 µm.
(G) No GUS signal was detected in a developing PpRecQ4:GUS embryo and the surrounding tissue of the archegonium. Bar = 100 µm.
(H) and (P) Gametophores carrying spore capsules. While the spore capsules of PpRecQ6:GUS did not show a signal, the foot of the sporophyte (highlighted by box) and the rhizoids showed GUS staining (box in [P]). Bars = 1 mm.
(Q) Close-up of the PpRecQ6:GUS sporophyte foot. Bar = 50 µm.
(I) and (R) Germinating spores after 5 d did not show any GUS staining. Bars = 50 µm.
Rescue of the Arabidopsis recq4a Mutant by P. patens RecQ4
We cloned the full-length PpRecQ4 cDNA into pKD1 containing either the promoter sequence of AtRecQ4A or AtRecQ4B (Hajdukiewicz et al., 1994). Using the floral dip method, the respective constructs and controls were transferred into Atrecq4a (Hartung et al., 2007). Controls included the empty vector, Col-0 wild type and the IC9C line that contains an HR reporter for visualization of somatic intermolecular recombination (Molinier et al., 2004). Several single locus lines were obtained and showed 3:1 segregation on gentamycin selection medium.
To analyze whether these lines show a reversion of the Atrecq4a phenotype of sensitivity to the DNA damaging agents cisplatin and/or methyl methanesulfonate (MMS), we subjected them to several assays in liquid germination medium. Two analyzed Atrecq4a lines containing PpRecQ4 driven by the AtRecQ4B promoter showed full rescue of the sensitivity phenotype treated with either cisplatin (Figure 3A) or MMS (Figure 3B). In contrast, different lines containing PpRecQ4 driven by the AtRecQ4A promoter exhibited a variable degree of complementation (Figure 3) depending on methylation of the transgene promoter, an effect revealed by bisulfite sequencing of the putative promoter region upstream of the ATG to the 3′-untranslated region (UTR) of the preceding gene (Supplemental Figure 11).
Figure 3.
Plant Growth Assay Using Different Concentrations of Cisplatin and MMS with Complementation Lines Containing PpRecQ4 Controlled by the 4A or 4B Promoter of Arabidopsis RecQ4 in recq4A Mutant Background.
Fresh weight of 10 plantlets per line at each cisplatin or MMS concentration was measured and put into relation to the untreated plants of the same line. Each assay was performed at least five times, and the mean values including sd are shown. The color code of the lines is as follows: Three wild-type lines containing either no new construct, the empty vector, or the PpRecQ4 construct under the control of the AtRecQ4B promoter are in green, two fully complementing lines containing PpRecQ4 under control of the AtRecQ4B promoter are depicted in blue, and three only partially complementing lines containing PpRecQ4 under control of the AtRecQ4A promoter are shown in lilac. The original Atrecq4A mutant of Arabidopsis and the same mutant containing the empty vector are shown in red and orange, respectively.
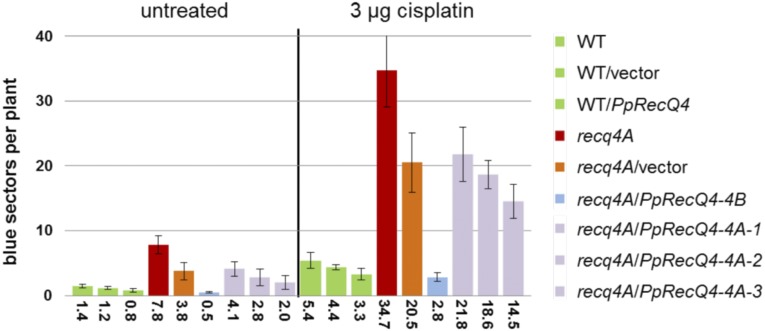
Another phenotype described for Atrecq4a is its enhanced HR frequency (Hartung et al., 2007). Therefore, we tested these lines in an HR assay as described by Hartung et al. (2007). In short, in this assay an interrupted GUS gene is repaired by recombination between the sister chromatids or the homologous chromosomes. After staining, each recombination event results in a blue sector of the leaf. Thus, the number of spots indicates the HR frequency (Molinier et al., 2004). The lines expressing PpRecQ4 driven by the AtRecQ4B promoter exhibited full rescue. Again, the more or less methylated lines containing PpRecQ4 under control of the AtRecQ4A promoter showed variable degrees of rescue but never full complementation (Figure 4). Together, these experiments reveal an evolutionary conservation of RecQ4 function in DNA repair and regulation of HR between a moss and a flowering plant, representing an evolutionary distance of ∼450 million years (Lang et al., 2008).
Figure 4.
Homologous Recombination Assay without or with Cisplatin Treatment in Complementation Lines Containing PpRecQ4 Controlled by the AtRecQ4A or AtRecQ4B Promoter in the recq4A Mutant Background.
The capability of HR in Arabidopsis lines carrying different complementation constructs is shown. On the y axis the number of blue sectors is shown as a scale, and on the x axis the number of blue sectors is shown as average resulting from several experiments. The color code of the lines is as follows: Three wild-type lines containing either no new construct, the empty vector, or the PpRecQ4 construct under the control of the AtRecQ4B promoter are in green, two fully complementing lines containing PpRecQ4 under control of the AtRecQ4B promoter are depicted in blue, and three only partially complementing lines containing PpRecQ4 under control of the AtRecQ4A promoter are shown in lilac. The original Atrecq4A mutant of Arabidopsis and the same mutant containing the empty vector are shown in red and orange, respectively.
Disruption of PpRecQ4 and PpRecQ6 and Cross-Species Complementation of ΔPpRecQ4 by AtRecQ4A
Because the expression of PpRecQ4 rescues Atrecq4a, we investigated the function of PpRecQ4 and PpRecQ6 by generating moss knockout mutants. For ΔPpRecQ4, we tested if complementation is possible by AtRecQ4A or AtRecQ4B. The targeting strategy (Supplemental Figure 12A) to obtain ΔPpRecQ4 deletion mutants was based on the gene model Pp3c2_1760V3.1. A 4.3-kb-long fragment was amplified from gDNA and the central part of this fragment comprising exons 15 to 22 was replaced by the nptII cassette. After transformation, selection, and regeneration (Hohe et al., 2004) initial screening was done by leaflet PCR (Schween et al., 2002). Seven ΔPpRecQ4 lines were validated by RT-PCR as they showed no PpRecQ4 transcript (Supplemental Figure 12B). DNA gel blot analysis revealed single copy integrations of the targeting construct in four of these lines (Supplemental Figure 12C) and flow cytometry (Schween et al., 2003a) confirmed their haploidy. For all further experiments two of these lines were used (ΔPpRecQ4-1 and ΔPpRecQ4-2).
Subsequently, we tested if cross-species complementation is also possible the other way by expressing AtRecQ4A or AtRecQ4B in ΔPpRecQ4. Therefore, the complete AtRecQ4A coding sequence (CDS) of 3567 bp was amplified from pPZP221-K-RecQ4A (Schröpfer et al., 2014) and cloned into two different P. patens vectors with the transgene under the control of the strong Actin5 promoter from moss (PpAct5; Büttner-Mainik et al., 2011), additionally containing a hygromycin (hpt) marker. The expression of AtRecQQ4A was detected in five lines (Supplemental Figure 13A). As none of them showed an integration into the PIG1 locus (Okano et al., 2009), we used the PTA2 vector (Kubo et al., 2013; Mueller and Reski, 2015). In five plants, expression of AtRecQ4A was verified (Supplemental Figure 13B). Subsequently, expression levels were quantified via RT-qPCR. Two haploid lines with the highest AtRecQ4A expression were subsequently analyzed (AtRecQ4A_ΔPpRecQ4-1#1 with targeted integration, ∼1000 transcripts per 50 ng RNA; AtRecQ4A_ΔPpRecQ4-1#2 with untargeted integration, ∼2000 transcripts per 50 ng RNA; Supplemental Figure 13C).
ΔPpRecQ6 mutants were designed on the basis of the gene model Pp3c12_1610V3.1 (Supplemental Figure 14A). A 3-kb genomic fragment was amplified and a central part of this amplicon comprising exon 16 to the first bases of intron 18 was released and replaced by the nptII cassette. After transformation, selection and regeneration screening were done via leaflet PCR. Nine putative ΔPpRecQ6 mutants were validated as they showed no PpRecQ6 transcript (Supplemental Figure 14B). According to DNA gel blot analysis, three plants showed less than 10 integrations of the construct (Supplemental Figure 14C) and flow cytometry confirmed their haploidy. Two of them with different integration patterns were subsequently analyzed with a qPCR-based method (Noy-Malka et al., 2014; ΔPpRecQ6-1 with 9 to 10 integrations and ΔPpRecQ6-2 with 5 integrations but additional 8 integrations of the nptII cassette).
Effects of ΔPpRecQ4 and ΔPpRecQ6 and Expression of AtRecQ4A in ΔPpRecQ4 on Morphology and Development
Targeted knockout of PpRecQ4 severely affected development and morphology (Figure 5). In ΔPpRecQ4, protonema biomass gain was significantly reduced to around 70%, while there were no significant changes in ΔPpRecQ6 during the period of 18 d (dry weight at day 18: wild type 525.5 mg/L, ΔPpRecQ4-1 362.2 mg/L, and ΔPpRecQ6-2 443.3 mg/L; t test, P ≤ 0.001; Supplemental Figure 15A). While protoplast regeneration was unchanged with respect to filament size and branching pattern during the first 2 weeks, bud formation was delayed in ΔPpRecQ6 and even more in ΔPpRecQ4 (Supplemental Figure 15B). Caulonema is a distinct protonemal cell type mediating expansion of colonies and bud formation (Reski, 1998b). The ability for caulonema formation is unchanged in ΔPpRecQ4. Nevertheless, colony sizes and gametophores differ: ΔPpRecQ4 colonies were smaller with a reduced number of gametophores compared with the wild type and ΔPpRecQ6 (Figure 5A). ΔPpRecQ4 colonies were spreading less and thus grew in a more compact shape. Length of ΔPpRecQ4 gametophores was significantly reduced (average length 1.2 mm ± 0.4, n = 104) compared with the wild type (2.3 mm ± 0.7, n = 241) (Figure 5B; Supplemental Figure 15C).
Figure 5.
Morphology of Colonies, Gametophores, and Leaves Altered in ΔPpRecQ4 Compared with Wild-Type and AtRecQ4A_ΔPpRecQ4 Plants.
(A) Colonies of wild type, ΔPpRecQ4-1 (ΔRecQ4), ΔPpRecQ6-1 (ΔRecQ6), and AtRecQ4A_ΔPpRecQ4-1#2 (AtRecQ4A_ΔRecQ4) were grown from protonema spot inocula on solid medium for 26 d. Bars = 2 mm.
(B) Single gametophores of wild type, ΔPpRecQ4-1 (ΔRecQ4), ΔPpRecQ6-1 (ΔRecQ6), and AtRecQ4A_ΔPpRecQ4-1#1 (AtRecQ4A_ΔRecQ4) have been prepared from colonies grown from protonema spot inocula on solid medium for two months. Bars = 2 mm.
(C) Single leaves prepared from gametophores grown on solid medium for 2 months of wild type (wild type), ΔPpRecQ6-1 (ΔRecQ6), ΔPpRecQ4-1 (ΔRecQ4), and AtRecQ4A_ΔPpRecQ4-1#1 (AtRecQ4A_ΔRecQ4). Bars = 0.5 mm.
Expression of AtRecQ4A restored the ΔPpRecQ4 phenotype to wild-type-like regarding colony morphology, gametophore length, as well as leaf shape and size. The average gametophore length in AtRecQ4A_ΔPpRecQ4 (AtRecQ4A_ΔPpRecQ4-1#1 2.1 ± 0.7, n = 88) did not differ significantly (unpaired t test, P ≤ 0.0001) from the wild type in contrast to ΔPpRecQ4 (Supplemental Figure 15C). Additionally, compared with the wild type and ΔPpRecQ6, leaf morphology is altered (Figure 5C). In the wild type (n = 96), the leaves from more than 95% of all gametophores showed an undisturbed shape, while in ΔPpRecQ4 (n = 137) in all single gametophores analyzed, on average three leaves showed an aberrant shape, like a bifurcated tip, protuberances, or indentations at the leaf margins (Figure 5C). Leaves detached from gametophores, not including the basal four or developing leaves at the apex resulting in roughly 20 leaves analyzed per gametophore, were measured in length and width, revealing a shorter length in ΔPpRecQ4 (Supplemental Figure 15D). Taken together, these experiments reveal an evolutionary conservation of PpRecQ4 and AtRecQ4A in the control of development.
Previous studies of P. patens mutants affected in genes involved in DNA repair revealed a mutator phenotype (Trouiller et al., 2006; Schaefer et al., 2010; Kamisugi et al., 2012; Charlot et al., 2014). In order to evaluate whether loss of PpRecQ4 influences the genetic stability, spontaneous mutation frequency of the APT reporter was recorded according to Trouiller et al. (2006). Non-sense mutations in this gene confer resistance to 2-fluoroadenine (2-FA). In total, 3.2 × 106 protoplast-derived colonies for ΔPpRecQ4 were tested on medium containing 10 µM 2-FA. As mutants were not detected (Supplemental Figure 16A), loss of PpRecQ4 does not lead to a significant mutator phenotype. Additionally, ΔPpRecQ4 and ΔPpRecQ6 gametophores were propagated every month by transfer of single gametophores to new plates over a period of 1 year (12 cycles of vegetative propagation). Alterations in growth or morphology were not detected for either ΔPpRecQ4 (Supplemental Figure 16B) or ΔPpRecQ6 (Supplemental Figure 16C). Hence, both tests did not provide evidence for genome instability in the mutants.
Upon conditions inducing sporophyte formation (Hohe et al., 2002), gametangia were simultaneously developing on the wild type, ΔPpRecQ4, and ΔPpRecQ6. No morphological differences were detected for archegonia or antheridia, from which the mature spermatozoids were visualized according to Horst and Reski (2017). However, in contrast to the wild type with an average of 3.5 sporophytes per colony, no sporophytes developed in ΔPpRecQ4 and only three sporophytes formed on 98 ΔPpRecQ6 colonies (Table 1). Spores derived from these capsules (Supplemental Figure 17A) were viable but even when their germination was delayed, gametophores of F1 plants developed as in the wild type (Supplemental Figure 17B). Upon sporophyte induction of F1 plants, the reduction in the number of sporophytes was consistent in ΔPpRecQ6 (ΔPpRecQ6-2 with 2 spore capsules on 91 colonies) compared with the wild type (115 spore capsules on 116 colonies) (Table 1). The impairment of sporophyte formation of ΔPpRecQ4 was not restored upon the expression of AtRecQ4A (Table 1), indicating a specific function of PpRecQ4 in sexual reproduction of P. patens.
Table 1. Sporophyte Formation Is Completely Abolished in ΔPpRecQ4 and Severely Impaired in ΔPpRecQ6.
| No. Capsules | No. Colonies | Ø No. Capsules/Colony | |
|---|---|---|---|
| Wild type | 422 | 122 | 3.46 |
| ΔRecQ4 | 0 | 84 | 0 |
| ΔRecQ6 | 3 | 98 | 0.03 |
| F1 wild type | 115 | 116 | 0.99 |
| F1 ΔRecQ6 | 2 | 91 | 0.02 |
| AtRecQ4A_ΔPpRecQ4 | 0 | 26 | 0 |
Numbers of spore capsules observed for the different genotypes. Wild type, 122 colonies, 422 capsules; ΔRecQ4, ΔPpRecQ4-1 40 colonies without capsules, ΔPpRecQ4-2 44 colonies without capsules; ΔRecQ6, ΔPpRecQ6-1 52 colonies, 3 capsules; ΔPpRecQ6-2, 45 colonies without capsules; F1 wild type, wild-type clones regenerated from germinated single spores 116 colonies, 115 capsules; F1 ΔRecQ6, ΔPpRecQ6-1 clones regenerated from germinated single spores, 91 colonies, 2 capsules; AtRecQ4A_ΔRecQ4, AtRecQ4A_ΔPpRecQ4-1#2, 26 colonies without capsules.
PpRecQ4, but Not PpRecQ6, Is Involved in DNA Repair
To assay sensitivity to the induction of DNA damages, we applied UV stress to the wild type and ΔPpRecQ4. Sensitivity of ΔPpRecQ4 and wild-type strains to UV-B was investigated using a protoplast survival assay (Trouiller et al., 2006) and revealed an increased sensitivity of ΔPpRecQ4. While regeneration after irradiation with 60 mJ UV-B is ∼75% in the wild type, it is less than 10% in ΔPpRecQ4, which is even lower than for the wild type irradiated with 180 mJ UV-B (Supplemental Figure 18).
To assay the sensitivity of ΔPpRecQ4 and ΔPpRecQ6 toward the induction of DSBs, protonemata were treated with bleomycin, directly inducing DSBs (Favaudon, 1982). Alternatively, cisplatin was used, inducing intrastrand cross-links leading to DSBs upon replication if not repaired (Chu, 1994). After washing the treated or control material, spot inocula were placed on solid medium and the degree of damage was assayed by survival of the filaments and finally colony size (Figure 6). The wild type and ΔPpRecQ6 were mostly unaffected after treatment with 50 µg/mL bleomycin, whereas ΔPpRecQ4 suffered, indicated by a low number of newly developing filaments. After treatment with a higher concentration (200 µg/mL), which was lethal for ΔPpRecQ4, the wild type and ΔPpRecQ6 were also severely affected (Figure 6A). The same dose-dependent effect was observed upon treatment with cisplatin leading to severe damage in ΔPpRecQ4 already at a low concentration and affecting the wild type and ΔPpRecQ6 at higher concentrations only (Figure 6B). Upon expression of AtRecQ4A in ΔPpRecQ4, the sensitivity was intermediate between ΔPpRecQ4 and the wild type. Even though AtRecQ4A_ΔPpRecQ4 lines still showed a strong sensitivity after treatment with bleomycin or cisplatin, the high sensitivity of ΔPpRecQ4 was partially rescued by the expression of AtRecQ4A. These experiments indicate an at least partially conserved function of PpRecQ4 and AtRecQ4A in DNA damage repair, with the moss protein being more effective.
Figure 6.
Bleomycin and Cisplatin Treatments Result in Dose-Dependent Growth Impairments.
ΔPpRecQ4 plants are affected most severely while the expression of AtRecQ4A in ΔPpRecQ4 background leads to an intermediate phenotype compared with wild-type and ΔPpRecQ6 plants. Protonema of the wild type, ΔPpRecQ4-1 (ΔRecQ4), ΔPpRecQ6-1 (ΔRecQ6), and AtRecQ4A_ΔPpRecQ4-1#2 (AtRecQ4A_ΔRecQ4) was subjected to treatment with 50 µg/mL (middle) or 200 µg/mL (bottom) bleomycin (1.5–2.0 units/mg) or incubated in Knop medium as a control (top) for 24 h (A) or 0.33 mM (middle), 0.66 mM (bottom) cisplatin, or Knop medium with 0.66% DMSO (corresponding amount of DMSO to the treatment with 0.66 mM cisplatin; top) for 48 h (B). After washing with minimal medium, spot inocula were transferred to solid medium to regenerate for 4 weeks. Bars = 1 mm.
PpRecQ4 and PpRecQ6 Are Involved in Gene Targeting
One distinctive feature of P. patens compared with other model systems is its high rate of HR in somatic cells, which enables precise genome engineering via GT since 1998 (Strepp et al., 1998; Girke et al., 1998; Kamisugi et al., 2006). To quantify GT frequencies in this moss reliably and easily, we developed a fluorescence-based GT reporter. Our strategy was similar to the fluorescent seed marker for Arabidopsis (Shaked et al., 2005), displaying GFP fluorescence only if correctly integrated into the Cruciferin gene. As moss plants are haploid, the fluorescence can already be detected in regenerating filaments in which integration via HR was successful and is stable over the generations without segregation due to homozygous or heterozygous lines. We chose the locus of carbonic anhydrase 2 (Pp3c1_19190V3.1) due to its high expression throughout the life cycle and tissues including freshly isolated and regenerating protoplasts (Hiss et al., 2014). The reporter was designed in a way that citrine fluorescence, driven by the endogenous promoter of the carbonic anhydrase gene, which is not part of the targeting construct, is visible only after its correct integration in frame at the targeted locus (Figure 7A). Additionally, a neomycin phosphotransferase II (nptII), hygromycin (hpt), or blasticidin S (BSD) cassette allows selection of stable transgenic plants. GT frequencies after transformation, regeneration, and selection were determined as proportion of plants with citrine fluorescence from all plants that survived selection (Figure 7B). These reporter plants did not show any morphological or developmental deviations from the wild type under standard growth conditions.
Figure 7.
Citrine-Based Reporter Construct to Assay Gene Targeting Rates.
(A) Schematic representation of the genomic locus of carbonic anhydrase 2 (Pp1s43_118V6.1/ Pp3c1_19190C1.2) (top), the targeting construct (middle), and the genomic region upon successful integration of the construct via homologous integration into the moss genome (bottom). Dark gray boxes and lines represent exons and introns, respectively, for the gene model Pp3c1_19190C1.2, while light-gray boxes represent 5′- and 3′-UTRs, light-gray bars adjacent to intergenic genomic regions. The targeting construct consists of the citrine marker (yellow) including a nos terminator (orange), followed by a selection marker (nptII, hpt, BSD; light green) under the control of the 35S promoter and terminator (dark green) flanked at both sides by ∼1000-bp sequence homologous to the genomic sequence (black) for insertion via HR. The insertion of the transgene via HR takes place directly in front of the stop codon without deletion of any endogenous sequence information. If the construct is inserted via gene targeting into the genomic locus as depicted, the promoterless citrine is expressed under the control of the native promoter of the carbonic anhydrase 2.
(B) Citrine fluorescence can be observed upon gene targeting of this construct in regenerating protoplasts 7 d after the transformation process (left panel, bar = 50 µm) as well as in protonema (middle; bar = 500 µm) and gametophores (right; bar = 500 µm) of plants surviving selection.
Assayed after two cycles of selection on hygromycin, the GT frequency in the wild type was 65.9% (±2.7, three independent transformations). Surprisingly, in ΔPpRecQ6, the GT frequency was drastically reduced to 14.6% (±7.0, ΔPpRecQ6-2; three independent transformations; unpaired t test P ≤ 0.0006), whereas it was significantly increased in ΔPpRecQ4 to 94.0% (±4.7, ΔPpRecQ4-1 and ΔPpRecQ4-2, data pooled, three independent transformations; unpaired t test P ≤ 0.002) (Figure 8A). These experiments reveal a strong and opposite impact of RecQ4 and RecQ6 on GT frequencies in P. patens, suggesting RecQ4 is a repressor and RecQ6 is a potent enhancer of gene targeting.
Figure 8.
Rate of GT and Proportion of GT versus Untargeted Transgene Integration (IR) Is Higher in ΔPpRecQ4 Compared with the Wild Type but Reduced in ΔPpRecQ6 and Lines Expressing AtRecQ4A in ΔPpRecQ4 Background.
Gene targeting in the different transgenic lines is assayed as percentage of colonies with citrine fluorescence from all regenerated colonies after selection using the test construct allowing selection on hygromycin (HR-hpt) for the wild type, ΔPpRecQ4 (ΔRecQ4: ΔPpRecQ4-1 and ΔPpRecQ4-2, data pooled) and ΔPpRecQ6-2 (ΔRecQ6) with three independent rounds of transformation for each line (A) or the version of the test construct allowing selection on BSDS (HR-BSD) for the wild type, ΔPpRecQ4-1 (ΔRecQ4), and AtRecQ4A_ΔPpRecQ4 (AtRecQ4A_ΔRecQ4: AtRecQ4A_ΔPpRecQ4-1#1 and #2, data pooled) with at least four independent rounds of transformations (C). The total number of surviving colonies with citrine fluorescence (light gray) in which the test construct was integrated via GT into the moss genome compared with colonies without fluorescence indicating IR (dark gray) for HR-hpt transformations into the wild type, ΔPpRecQ4 (ΔRecQ4: ΔPpRecQ4-1 and ΔPpRecQ4-2, data pooled) and ΔPpRecQ6-2 (ΔRecQ6) (B) or HR-BSD into the wild type, ΔPpRecQ4-1 (ΔRecQ4), and AtRecQ4A_ΔPpRecQ4 (AtRecQ4A_ΔRecQ4: AtRecQ4A_ΔPpRecQ4-1#1 and #2) (D).
The overall number of surviving transformants was much lower for ΔPpRecQ4 (28 plants surviving selection from 3 independent transformations, average of 9.3 transformants per transformation) compared with the wild type (94 plants from 3 independent transformations, average of 31.3 transformants per transformation) notwithstanding a standardized number of 300,000 protoplasts used for each transformation (Figure 8B). An opposite effect was observed for ΔPpRecQ6: The total number of surviving plants (137 plants surviving selection, average of 45.7 transformants per transformation) was much higher compared with the wild type and particularly ΔPpRecQ4. To analyze whether this relies on different modes of transgene integration (illegitimate integration versus GT) and not on a reduced regeneration capacity of ΔPpRecQ4 protoplasts, we conducted a control experiment. Protoplasts were regenerated directly after isolation, after mock transformation (water instead of DNA), or transformation with another construct for homologous integration. Rate and speed of protoplast regeneration in general and after transformation with the reporter or a mock transformation did not differ between ΔPpRecQ4 and the wild type (Supplemental Figures 19A and 19B), supporting our conclusion of the strong and opposite impact of RecQ4 and RecQ6 on GT in moss.
To elucidate whether the effect of the only partial complementation of DSBs repair in ΔPpRecQ4 by AtRecQ4A was caused by an insufficient expression or a shift in repair pathway specificity, we analyzed the GT frequencies upon expression of AtRecQ4A in ΔPpRecQ4. Therefore, we used the fluorescence-based reporter construct with a BSD cassette, in order to allow selection of all transgenic lines. A dramatic reduction in GT frequency was observed in the two analyzed AtRecQ4A-expressing lines in the ΔPpRecQ4-1 background to an average of 15% (unpaired t test P ≤ 0.0001; 13.2% in AtRecQ4A_ΔPpRecQ4-1#1 and 17.3% in AtRecQ4A_ΔPpRecQ4-1#2) compared with 74.8% in ΔPpRecQ4 and 48.6% in the wild type (Figure 8C). As described above, the mode of integration was shifted toward HR-mediated GT in ΔPpRecQ4, while upon the expression of AtRecQ4A in ΔPpRecQ4, the integration of the construct predominantly occurred in an untargeted way by illegitimate integration (IR) (Figure 8D). The average number of regenerated plants after selection (GT and IR) from each experiment (at least seven independent transformations per line) was much higher in AtRecQ4A_ΔPpRecQ4 (average of 41.8 surviving colonies, 6.3 with integration via GT) compared to the wild type (average of 25.5 plants, 12.4 via GT) and particularly to ΔPpRecQ4 (average of 8.9 plants, 6.6 via GT).
Taken together, these experiments reveal a strong positive influence of PpRecQ6 and a strong negative influence of PpRecQ4 on overall GT frequencies. The different numbers of regenerating plants suggests that PpRecQ6 represses and PpRecQ4 activates IR. In line with this, AtRecQ4A shifts the DNA repair pathway strongly to IR in P. patens.
AtRecQ4B Does Not Rescue ΔPpRecQ4
For the generation of lines expressing AtRecQ4B in ΔPpRecQ4, the AtRecQ4B CDS of 3453 bp was amplified from pPZP221-K-RecQ4B (Schröpfer et al., 2014) and cloned into the two P. patens expression vectors described above with the transgene driven by PpAct5, additionally containing an hpt marker. For the AtRecQ4B expression construct based on the PIG1 vector, one plant was tested positively for expression of the AtRecQ4B cDNA (Supplemental Figure 20A), which was quantified via RT-qPCR (around 7000 transcripts per 50 ng RNA; Supplemental Figure 20B).
Morphology and development of the AtRecQ4B-expressing line (AtRecQ4B_ΔPpRecQ4-1#1) was monitored regarding colony size, gametophore size, and leaves as well as leaf shape and establishment of sporophytes. Furthermore, DNA damage was induced by bleomycin (Supplemental Figure 21A) and cisplatin (Supplemental Figure 21B). For all criteria tested, no differences were detected compared with the parental ΔPpRecQ4, indicating that AtRecQ4B is neither a functional homolog of AtRecQ4A, nor of PpRecQ4.
HsBlm Does Not Rescue ΔPpRecQ4
To test if human Blm, a putative ortholog of AtRecQ4A (Bagherieh-Najjar et al., 2005; Hartung and Puchta, 2006; Hartung et al., 2008), can also complement the ΔPpRecQ4 phenotype, stable lines expressing HsBlm under the control of PpAct5 were created via GT into the PTA2-locus. The HsBlm cDNA of 4268 bp was amplified from human liver cDNA. Expression of HsBlm was verified in six HsBlm_ΔPpRecQ4 lines (Supplemental Figure 22A). The three lines with the strongest expression were selected and expression levels were quantified via RT-qPCR (HsBlm_ΔPpRecQ4-1#1 with ∼10,000 copies per 50 ng RNA, HsBlm_ΔPpRecQ4-1#2 ∼3000 copies per 50 ng RNA and HsBlm_ΔPpRecQ4-1#3 with high HsBlm expression level of ∼18,000 copies per 50 ng RNA; Supplemental Figure 22B). Phenotypic analysis of colonies, average gametophore and leaf length did not indicate complementation irrespective of the expression strength. Also, sensitivity toward bleomycin (Supplemental Figure 23A) or cisplatin (Supplemental Figure 23B) is not altered. These experiments reveal that HsBlm cannot substitute PpRecQ4. This is most probably not due to inefficient translation of human genes in moss, as P. patens is an excellent production host for recombinant biopharmaceuticals, expressing human genes efficiently without any codon adjustments (reviewed in Reski et al., 2015).
DISCUSSION
The RecQ Protein Family in Plants
So far, knowledge about RecQ proteins in plants was predominantly based on studies in Arabidopsis. However, while RecQs are conserved and play crucial roles in maintaining DNA integrity, additional plant-specific variants, e.g., RecQ6 in rice, exist (Hartung and Puchta, 2006). Thus, we were aiming at broadening our conception of plant RecQs by identifying additional orthologs in a wider taxon set. We reconstructed the six RecQ subfamilies and identified members of each in almost all clades of Viridiplantae. Intriguingly, the five human RecQs each aligned basally to individual clusters, indicating that multiple RecQs were already present in the last common ancestor of plants and humans. In combination with the observable, widely conserved diversity of plant RecQs this further underlines their functional importance for eukaryotic life.
Our data set revealed that P. patens and S. fallax encode orthologs of RecQ6 as well as RecQsim, with P. patens harboring two RecQsim paralogs. In addition, we identified RecQ6 and RecQsim in most other clades with the exception of the orders Sapindales, Malvales, and Brassicales that miss RecQ6 (Supplemental Figure 6), as opposed to algae and liverworts that miss RecQsim (Supplemental Figure 7). Interestingly, our approach places the origin of RecQsim on a basal node of the RecQ6 cluster. Taken together, these observations indicate a different, clade-specific evolutionary fate of RecQ6 and RecQsim following a shared origin in the last common ancestor of all plants, including subfunctionalization of both and loss of either RecQ6 or RecQsim.
RecQ4 and RecQ6 Proteins Accumulate in Meristematic Moss Tissues
According to transcriptomic evidence the overall expression level of PpRecQ4 and PpRecQ6 is low. Nevertheless, correlation of mRNA levels and protein abundance is often low (Greenbaum et al., 2003), as shown for the stress response of a P. patens gene involved in protein degradation (Schuessele et al., 2016). Hence, we generated PpRecQ4:GUS and PpRecQ6:GUS reporter lines to study protein distribution. The experimental data on RecQ4 distribution in P. patens (Figures 2A to 2I) are in accordance with those for the orthologs in Arabidopsis, rice, and humans. Expression levels of AtRecQ4A and AtRecQ4B are detected in tissues with higher meristematic activity, similar to that of HsBlm in proliferating cells (Hartung et al., 2000; Bagherieh-Najjar et al., 2003). In young seedlings, OsRecQ4 mRNA and protein were detected in shoot and root apical meristems (Kwon et al., 2013).
PpRecQ6:GUS shows an accumulation in similar tissues as PpRecQ4 (Figures 2J to 2R), and as for PpRecQ4, no effect of genotoxins on protein accumulation regarding localization or amount was observed. These data are comparable to OsRecQ6, which is expressed mainly in proliferating cells such as shoot apical meristems, leaf primordia, and marginal meristems of young leaves (Saotome et al., 2006).
RecQ Functions in Moss Development
The knockout of PpRecQ4 leads to similar phenotypes as described for mutants of AtRecQ4A (Bagherieh-Najjar et al., 2005; Hartung et al., 2007; Higgins et al., 2011) and OsRecQ4 (Kwon et al., 2012): sensitivity to genotoxins, altered frequency of HR, and reduced fertility in the case of Atrecq4a. In addition to the effects described for seed plant mutants, ΔPpRecQ4 but not ΔPpRecQ6 is severely affected in development and morphology (Figure 5; Supplemental Figures 16 and 18). In mosses, the transition from protonema to gametophores is characterized by a switch from two- to three-dimensional growth (Reski, 1998b). Throughout the P. patens life cycle, development of the different tissues is driven by single meristematic cells, with at least eight different types of stem cells specific for each tissue including two different types of protonema (chloronema and caulonema), gametophores, and leaves (Harrison et al., 2009; Kofuji and Hasebe, 2014). Aberrant leaf shapes of x-ray-treated plants (Harrison et al., 2009) resemble the aberrant leaf shapes of ΔPpRecQ4 and are consistent with deletion of stem cells. As all ΔPpRecQ4 lines described here are haploid, we exclude morphological deviations like changes in leaf shape described for diploid lines (Schween et al., 2005).
Besides the morphological deviations of ΔPpRecQ4 in the gametophyte, no sporophytes develop, even though archegonia and antheridia including spermatozoids formed properly. This is reminiscent of the homeotic ΔBELL1 mutant that is specifically affected in very early embryogenesis (Horst et al., 2016). Regarding meiosis, minor defects are reported for S. cerevisiae sgs1 mutants but not Atrecq4a or mouse blm−/− mutants (Watt et al., 1995; Hartung et al., 2007; Luo et al., 2000). In humans, Blm mutations lead to male infertility and embryonic lethality if homozygous (Bernstein et al., 2010). However, Higgins et al. (2011) showed reduced fertility of Atrecq4a due to dissolution of recombination intermediates between non-self telomeres during meiosis. Nevertheless, during the development of moss sporophytes, meiosis takes place in the last step upon formation of the spore tetrads inside the fully developed spore capsule. As we never observed formation of embryos or sporophytes, we propose that an early step of sporophyte formation, such as the division of the zygote, depends on PpRecQ4 function. This is supported by the observations of protein accumulation in our PpRecQ4:GUS lines and expression according to transcriptomic data (Ortiz-Ramírez et al., 2016) in archegonia and developing sporophytes.
In ΔPpRecQ6, morphology and development are unaltered compared with the wild type, also upon frequent vegetative propagation. However, the establishment of the sporophyte is severely affected, but not completely abolished as in ΔPpRecQ4. Once the sporophyte is established it develops wild-type-like including viable spores. Therefore, we conclude that despite its more widespread expression pattern compared with PpRecQ4, PpRecQ6 functions in moss development less specifically and, hence, its deletion can be partially compensated by another RecQ family member.
RecQ4 Functions in DNA Repair
A common result upon deletion of ScSgs1, HsBlm, AtRecQ4A, and OsRecQ4 is increased sensitivity toward induction of DNA damages. Upon UV-B irradiation, which induces pyrimidine dimers (Cadet et al., 1997), the survival rate of ΔPpRecQ4 protoplasts is drastically reduced. The order of magnitude of hypersensitivity is the same as in mutants affected in key proteins of the HR machinery, such as PpRAD51A or PpRAD51B (Schaefer et al., 2010; Charlot et al., 2014). Also upon treatment with genotoxins the deletion of PpRecQ4 yields the same effect, while ΔPpRecQ6 lines exhibit no altered sensitivity. Atrecq4a is sensitive to MMS, causing stalled replication forks, cisplatin, and UV-C light, but not bleomycin (Bagherieh-Najjar et al., 2005; Hartung et al., 2007).
However, for OsRecQ4 mutants, hypersensitivity to bleomycin as well as to aphidicolin and hydroxyurea, inhibiting DNA replication and preventing S-phase progression, is reported (Kwon et al., 2013). Therefore, we conclude that the sensitivity toward bleomycin as shown for ΔPpRecQ4 and OsRecQl4 mutants is connected to the presence of a single RecQ4 ortholog in moss and rice in contrast to Arabidopsis with its duplicated RecQ4. Collectively, our results reveal an essential role of PpRecQ4 in the repair of three different types of DNA damage.
RecQ4 Is a Repressor of Homologous DNA Recombination
For a direct quantification of the GT frequency in P. patens, we established a fast and reliable system where fluorescence can already be detected in regenerating filaments in which integration of the reporter at the targeted locus via HR was successful. In all plants surviving selection but not showing the fluorescence signal, the transgene was integrated randomly into the genome, so that the promoterless reporter is not expressed. Although theoretically fluorescence may occur after IR of the reporter downstream of another promoter, we did not find such an event in ∼100 different lines showing fluorescence.
The increased number of citrine-positive lines for ΔPpRecQ4 suggests an increase in GT frequency which is in line with increased HR frequencies in somatic cells of Atrecq4a (Bagherieh-Najjar et al., 2005; Hartung et al., 2007) and of OsRecQ4 (Kwon et al., 2013). Additionally, the overall number of plants surviving selection, implying integration of the construct into any locus of the genome in a targeted or untargeted way, is reduced by the PpRecQ4 knockout, while the regeneration capacity after transformation is unchanged, indicating a reduction of the overall rate of integration events with a clear shift toward targeted integrations.
HsBlm and ScSgs1 can act at different steps of the DSB repair pathway. In collaboration with the endonuclease Dna2 and in parallel to EXO1, both are involved in the crucial 5′-3′ resection step that generates an extensive tract of single-stranded DNA, which subsequently serves as substrate for HR (Mimitou and Symington, 2011). HsBlm and ScSgs1 act also as antirecombinase factors on late HR intermediates by limiting the formation of DNA crossovers (CO) and promoting the formation of non-CO events in various DNA substrates (reviewed in Sung and Klein, 2006). Therefore, it is conceivable that a fraction of the HR intermediates that are normally resolved in non-CO events in the wild type are resolved in a CO way in ΔPpRecQ4, leading to GT. This is sustained by the finding that in Arabidopsis RecQ4A is involved in the resolution of aberrant DNA structures arising during replication and if deleted the mutant shows an enhanced level of HR in somatic cells (Hartung et al., 2007). Taken together, these two potential functions of PpRecQ4, generation of the single-stranded DNA substrate for HR and antirecombinase action, could explain the concomitant decrease in overall number of GT events and increase in GT efficiency in the mutant. Nevertheless, the decrease in number of GT events observed in the mutant cannot explain entirely the decrease in the overall rate of integration events. Only RecQ4 being an activator of illegitimate integration could explain this general decrease. Further experiments are needed to demonstrate whether or not this function is linked to an antirecombinase effect of RecQ4/Sgs1/Blm.
RecQ6 Is an Enhancer of Homologous DNA Recombination
In contrast, in ΔPpRecQ6, sensitivity toward genotoxins is wild-type-like but the GT frequency is drastically reduced to 14.6% compared with 65.9% in the wild type. The number of surviving plants is increased to 150% (average of 46 plants per transformation surviving selection with hygromycin) compared with the wild type (32 plants from each transformation). The reduced GT frequency reveals a change of the ratio of targeted gene replacement via HR by two-end invasion toward nonhomologous end joining or similar modes of untargeted transgene integration, reported to occur frequently but not predominantly in P. patens (Wendeler et al., 2015). This is reminiscent of observations in the absence of PpRAD51, a key factor for HR, where GT is abolished and random integration is increased (Markmann-Mulisch et al., 2007; Schaefer et al., 2010), and suggests a major role for RecQ6 in HR. Furthermore, these observations correlate with the Atrecq4b phenotype (Hartung et al., 2007), as the deletion of PpRecQ6 or AtRecQ4B results in an opposite effect compared with the deletion of PpRecQ4 and AtRecQ4A. This supports our hypothesis that PpRecQ6 and OsRecQ6, both sharing higher similarity with bacterial RecQs than other plant RecQs (Saotome et al., 2006), have a similar function to that of AtRecQ4B, which developed its antagonistic function to AtRecQ4A as suppressor of HR only after the genome duplication in the Brassicaceae. To gain further insight into PpRecQ6 function, we will test if its overexpression affects GT frequencies.
Distinct Role of RecQ4 in Development, DNA Repair, and Gene Targeting
Several studies describe knockouts of P. patens genes associated with DNA repair and recombination such as PpRAD51A and PpRAD51B (Markmann-Mulisch et al., 2007; Schaefer et al., 2010; Charlot et al., 2014); PpMSH2, a member of the mismatch repair pathway (Trouiller et al., 2006); PpMRE11 and PpRAD50, two members of the MRN complex (Kamisugi et al., 2012); and several DNA helicases, such as PpALC1, PpCHD5, PpZRL, PpTEB, PpRTEL1, PpERCC6, and PpSRS2 as well as PpCtIP, the initiator of end resection during HR (Kamisugi et al., 2016), and their surprisingly little consequences for development, sensitivity to genotoxins and GT compared with other organisms. These findings suggest a high level of redundancy for processes involved in moss genome maintenance.
Regarding PpRecQ6, our study supports this hypothesis, as the knockout does not result in an obvious growth phenotype in the gametophyte nor hypersensitivity to genotoxins, but in a shift from GT toward nontargeted integration. In contrast, the knockout of PpRecQ4 leads to severely impaired growth and development and hypersensitivity to genotoxins, which is reminiscent of knockouts of the PpRAD51 genes (Markmann-Mulisch et al., 2007; Schaefer et al., 2010), the homolog of bacterial RecA commonly regarded as a key protein for HR conserved throughout evolution (Baumann and West, 1998). On the other hand, we found no impact of ΔPpRecQ4 on genome stability, whereas the GT frequency is significantly increased in combination with a reduction of the overall number of plants in which the transgene is integrated into the genome. Therefore, we infer a distinct role of PpRecQ4 in moss development, repair of DNA damage, and GT, which cannot be rescued by redundant proteins or alternative pathways.
Cross-Species but Not Cross-Kingdom Complementation of PpRecQ4, AtRecQ4A, and HsBlm
The protein pair AtRecQ4A/AtRecQ4B is specialized in different antagonistic functions in suppression and promotion of CO and HR events (Hartung et al., 2007). The specific function of both depends on the N termini that facilitate the interaction with other components of the RTR complex (Schröpfer et al., 2014). This complex consists of AtRecQ4A, a type IA topoisomerase (AtTOP3A) and the structural protein RMI (AtRMI1 and AtRMI2), and plays a pivotal role in the processing of DNA recombination intermediates in all eukaryotes (Knoll et al., 2014; Röhrig et al., 2016). No functional characterization exists for the P. patens RTR complex, but because all members are present, we assume that PpRecQ4 comparably functions together with the other partners in the complex. AtRecQ4A has evolved different and partially antagonistic functions in HR depending on whether the reaction is initiated by a DSB and therefore is not completely equivalent to Sgs1, as deletions of the Arabidopsis and S. cerevisiae genes have opposite effects on gene conversion, but more similar to HsBlm (Mannuss et al., 2010). Our complementation experiments of PpRecQ4 in Atrecq4a as well as AtRecQ4A in ΔPpRecQ4 support this hypothesis of a conserved core set of functions common to PpRecQ4 and AtRecQ4A. Additionally, there are specific functions for the RecQ4 proteins of different species, as in moss, embryogenesis or the mode of integration of linearized DNA constructs by GT versus untargeted integration. The specific function of AtRecQ4B promoting CO does not complement any of the ΔPpRecQ4 defects tested in our study. As the expression of HsBlm in ΔPpRecQ4 also does not compensate any of the morphological aberrations or hypersensitivity toward induction of DNA damage, we infer that specific protein functions and/or HsBlm interaction partners do not allow a cross-kingdom complementation as described for HsBlm in yeast, which partially rescues the sgs1 sensitivity to hydroxyurea (Yamagata et al., 1998).
Taken together, it is likely that the reduction of non-HR events during DSB repair is a conserved feature for the orthologs of plant RecQ4 as shown for PpRecQ4 in this study and AtRecQ4A (Bagherieh-Najjar et al., 2005; Hartung et al., 2006, 2007) and for human Blm and yeast Sgs1. Based on our study, we hypothesize a convergent function of AtRecQ4B and PpRecQ6 as activators of HR, potentially favoring CO instead of non-CO, thus explaining the shift toward untargeted transgene integration in ΔPpRecQ6.
Differences between Gene Targeting and DNA Repair
Arabidopsis and P. patens differ in their GT frequencies by orders of magnitude (Reski, 1998a). One distinctive feature between both at the molecular level is their complement of RecQ4 and RecQ6 genes. Here, we describe a strongly enhancing effect of RecQ6 and a strongly inhibiting effect of RecQ4 on GT in moss. Surprisingly, RecQ4, but not RecQ6, has a crucial role in DNA repair and in the control of development.
Cross-species expression of AtRecQ4A in ΔPpRecQ4 demonstrated a similar function of both proteins. The ΔPpRecQ4 phenotype is restored, except the defect in embryogenesis. Upon expression of AtRecQ4A in ΔPpRecQ4, the resolution of HR intermediates is shifted from CO, leading to HR or GT, respectively, to a non-CO way. In moss, the two helicases have opposite roles in HR, with RecQ6 being an activator of HR potentially through stimulation of CO formation and RecQ4 being an inhibitor of CO formation. RecQ4 has an additional role in DSB repair, which involves the resection of DSBs for repair through single-strand annealing or HR, possibly explaining the mutants’ hypersensitivity to genotoxins. Indeed, without RecQ4, a DSB that has been handled by the MRN complex could not be repaired by nonhomologous end joining anymore and could not be handled by HR, or only if resected by EXO1.
The most likely explanation for these unexpected differences between DNA repair and GT in P. patens is the difference of the tissue types in which these events occur: freshly isolated protoplasts for GT experiments and protonema for treatment with genotoxins. Both are haploid gametophytic cells and moss protoplasts regenerate directly into protonema (Bhatla et al., 2002; Hohe and Reski, 2002). However, differences in cell cycle arrest occur during protoplast isolation, protoplast regeneration, and subsequent protonema development (Schween et al., 2003a) and may account via different expression of RecQ4 and RecQ6 for the differences between DNA repair and transgene integration described here.
METHODS
Phylogenetic Analysis
The main source for RecQ sequences was Phytozome version 12 (https://phytozome.jgi.doe.gov/; Goodstein et al., 2012). All full-length sequences from Arabidopsis thaliana were isolated and annotated in GenBank. For other RecQ proteins, Ensembl was used (Ensembl release 90; Yates et al., 2016; EnsemblGenomes release 37; Kersey et al., 2018). For identification of additional RecQ orthologs, the corresponding HMMs of Physcomitrella patens, Arabidopsis, rice (Oryza sativa), and H. sapiens RecQs were obtained from Panther (version 12; Thomas et al., 2003) and used as input for hmmsearch (version 3.1b; Johnson et al., 2010) to search against the proteomes of all species included in this study (Supplemental Data Set 1). Initial hits were checked for PFAM domains (release 31; Finn et al., 2016), and inconsistent or fragmented sequences were removed. Multiple protein alignments were performed with UPP (version 4.3.2; Nguyen et al., 2015) and converted into codon-aware CDS alignments with PAL2NAL (version 14; Suyama et al., 2006). Gapped columns were removed with trimAl (version 1.4rev22; Capella-Gutiérrez et al., 2009) and AliView (version 1.1.8; Larsson, 2014) was used for visualization and manual curation. Phylogenetic trees were calculated with RAxML (version 8.2.10; Stamatakis, 2014), rooted and plotted with R (R Core Team, 2017) and the ggtree package (Yu et al., 2017).
AtRecQ1 (AT3G05740.1) and AtRecQ3 (AT4G35740.1) were used for BLASTP searches of the 1 KP database clade 21-mosses (Altschul et al., 1990; http://db.cngb.org/blast4onekp/blast; Matasci et al., 2014). Reciprocal BLASTP was done with best hit from 1 KP database on TAIR (https://www.arabidopsis.org/Blast/index.jsp). Accessions were considered as orthologs that found the query sequence as best hit in the reciprocal blast search. In several species, the searches identified short transcripts causing low e-values. These sequences were selected for further analysis by Needleman-Wunsch alignments, identifying them as partial sequences coding for true AtRecQ3 orthologs (Needleman and Wunsch, 1970).
Isolation of PpRecQ4 and PpRecQ6
We aligned the respective protein from AtRecQ4 or OsRecQ6 to early genome versions at cossmoss.org (V1.6; Zimmer et al., 2013) using TBLASTN. After identifying the respective best hits, we compared the completeness of those hits, thereby detecting a number of gaps and a missing start position in RecQ4 and RecQ6. To obtain the full-length cDNA sequences, we isolated RNA from protonema as described below and reverse transcribed using an anchored oligo(dT) primer. Subsequently, we used different forward primers in combination with a fixed reverse primer which was located at or shortly after the defined stop codon for each gene. The forward primers were deduced in areas that showed the highest sequence similarity and using primer walking we moved upstream of this area until we found a cDNA product which codes for an appropriate ATG start codon and contained enough 5′-UTR to encode for a stop codon in this area. All primers are listed in Supplemental Table 1. All resulting full-length sequences are included in the genome version V3.3 at cossmoss.org (Lang et al., 2018).
Cloning of the Complementation Constructs for Arabidopsis
The full-length cDNA for PpRecQ4 and PpRecQ6 was cloned into pKD1, a derivative of pPZP221 containing an additional 35S terminator (Hajdukiewicz et al., 1994). The cDNA was cloned either behind the promoter region of AtRecQ4A or -4B, which was defined as the region upstream of the ATG to the end of the 3′-UTR of the previous gene model, each containing the 5′-UTR and TATA-box at position −30. Both constructs were verified by sequencing and transformed by the floral dip method (Clough and Bent, 1998).
Sensitivity and HR Assays in Arabidopsis
Sensitivity to cisplatin and MMS was assessed as described (Hartung et al., 2007). Ten 7-d-old seedlings were transferred into 5 mL GM (4 mL for assays with addition of genotoxins). Subsequently, 1 mL of GM containing the respective concentration of cisplatin (2.5, 5, 7.5, and 10 µM) or MMS (50 and 100 ppm) was added. After 14 d of incubation, the plants were dried on a paper towel to get rid of excess liquid. The fresh weight for each 10 plants per well was determined. The weight was normalized to that of the untreated plants from the same line and expressed in percentage of the internal control weight. The HR assays were performed as described (Hartung et al., 2007). Each sensitivity and HR assay was repeated at least six times.
Moss Material and Growth Conditions
We used P. patens ‘Gransden 2004’, the strain that was used for genome sequencing (Rensing et al., 2008) and is deposited at the IMSC under accession number 40001. The material was axenically cultured in Knop medium (Reski and Abel, 1985) supplemented with microelements (Schween et al., 2003b). For the adjustment of dry weight, 10 mL of culture were removed prior to subculturing and filtered through gauze (Miracloth; Calbiochem). The dry weight was determined after drying the sample for 2 h at 105°C. The transgenic lines described in this study are deposited in the IMSC with the accession numbers 40636 (ΔPpRecQ4-1), 40641 (ΔPpRecQ4-2), 40643 (ΔPpRecQ6-1), 40649 (ΔPpRecQ6-2), 40815 (AtRecQ4A_ΔPpRecQ4-1#1), 40816 (AtRecQ4A_ΔPpRecQ4-1#2), 40817 (AtRecQ4B_ΔPpRecQ4-1#1), 40820 (HsBlm_ΔPpRecQ4-1#1), 40818 (HsBlm_ΔPpRecQ4-1#2), 40819 (HsBlm_ΔPpRecQ4-1#3), 40821 (PpRecQ6:GUS-1), 40822 (PpRecQ6:GUS-2), 40823 (PpRecQ4:GUS-1), 40824 (PpRecQ4:GUS-2), 40825 (PpRecQ4:GUS-3), 40826 (PpRecQ4:GUS-4), and 40827 (PpRecQ4:GUS-5).
Phenotypic Analysis and Treatments of Moss
To measure the length of gametophores, colonies grown from protonema spot inocula on plates for 8 weeks were dissected into single gametophores. To assess the length and width of leaves, the four bottom leaves and the youngest ones on top of the gametophore were discarded. All other leaves were detached from the stem using a needle. To assay vegetative propagation over the period of 1 year, single gametophores picked from colonies on the most recent plate were transferred to new plates every month. Induction of sporophytes was performed according to Hohe et al. (2002). Microscopy of 4′,6-diamidino-2-phenylindole (DAPI)-stained spermatozoids was performed according to Horst and Reski (2017).
The spontaneous mutation frequency was determined according to Trouiller et al. (2006). Non-sense mutations in PpAPT confer resistance to 2-FA. Protoplasts were regenerated for 6 d on PpNH4 supplemented with 8.5% mannitol and 0.5% glucose and then transferred on PpNH4 supplemented with 10 µM 2-FA. After 2 weeks, the number of colonies was counted. The percentage is calculated as the number of 2-FA-resisting colonies on the total number of regenerated plants. Experiments were repeated three times starting from independent cultures followed by isolation and regeneration of protoplasts.
To assay protoplast regeneration, protonema grown in liquid Knop medium pH 4.5 were digested with Driselase according to Hohe et al. (2004). The protoplast density was set to 50,000/mL in regeneration medium. Two hundred microliters of this solution was mixed with 400 μL of 1.2% LMP agarose (Sigma-Aldrich) prepared with regeneration medium. This mixture (550 μL) was transferred to a 3.5-cm Petri dish. Once a liquid film has formed, 300 μL of the mixture was removed and the plates stored for 30 min at 4°C. The solidified film was overlaid with 1 mL of regeneration medium. After 6 d, the medium was changed to Knop medium.
UV-B irradiation was performed according to Kamisugi et al. (2012): Protoplasts were spread (∼50,000/plate) on protoplast agar medium (PpNH4 + 0.5% glucose + 8.5% mannitol). Plates were immediately irradiated with UV-B light (308 nm, 60 J/s) in a Stratalinker. The intensity of the irradiation was controlled using the internal probe of the Stratalinker and one plate of each strain was treated simultaneously. The experiment was repeated four times. Plates were immediately transferred to darkness for 24 h and then to standard growth conditions. Survival was determined after 1 week by microscopy observation.
To test the sensitivity to DNA damaging agents, 0.8 mg of dry weight protonema was incubated in a volume of 500 μL bleomycin sulfate solution (1.5–2.0 units/mg; Calbiochem) in a concentration of 50 or 200 µg/mL for 24 h or with cisplatin (Calbiochem) in a concentration of 0.33 or 0.66 µM for 48 h. According to the manufacturer, the bleomycin activity is 1.5 to 2.0 units/mg and an aqueous stock solution of 2 mg/mL was prepared. For cisplatin, a stock solution of 100 mM was prepared in DMSO. All further dilutions were done in Knop. Knop medium was used as a negative control for the bleomycin treatment (Knop medium with DMSO according to the highest concentration in the treatment for cisplatin). After treatment, the material was sedimented, washed twice, and resuspended in Knop to a density of 2 mg per mL. Five-microliter drops of material were put on solid medium to monitor colony growth.
Images were acquired with a Zeiss ICc1 CCD camera at an Olympus SZX7 binocular microscope or with an Mrc5 CCD camera at an Axioplan2 (Zeiss). AxioVision software version 4.8 (Zeiss) was used for imaging and length measurements.
Flow Cytometry Analysis
The fluorescence intensity of nuclei prepared from 10 to 30 mg freshly chopped protonema stained with a DAPI buffer (Schween et al., 2003a) was measured using a Cyflow Space flow cytometry (Partec) as described (Schuessele et al., 2016).
Cloning of P. patens Targeting Constructs and Generation of Transgenic Moss Lines
We constructed PpRecQ4:GUS and PpRecQ6:GUS fusions by Gibson assembly (Gibson et al., 2009). Prior to assembly, overhangs with at least 25 bp complementary to the adjoining fragments were added via PCR with Phusion polymerase (Thermo Scientific). For PpRecQ4:GUS, the 5′ homologous region consisting of the last 798 bp of the PpRecQ4 gene in front of the stop codon and the 3′ homologous region of 803 bp starting with the native stop codon were amplified from gDNA using RecQ4_5′_fw and RecQ4_5′_rev or RecQ4_3′_fw and RecQ4_3′_rev, respectively. In the same way, the 5′ and 3′ homologous regions were amplified for PpRecQ6 with the primers RecQ6_5′_fw and RecQ6_5′_rev or RecQ6_3′_fw and RecQ6_3′_rev. The CDS for GUS, 5′-prime including a 17-bp linker, was amplified from the ATE:GUS construct (Schuessele et al., 2016) flanked with the appropriate overlaps using RecQ4_GUS_fw and Recq4_GUS_rev or RecQ6_GUS_fw and RecQ6_GUS_rev. These three fragments for each of the constructs were inserted into pJET1.2 linearized with Eco32I. For moss transformation, 60 µg of this plasmid linearized using BglII for PpRecQ4:GUS or XhoI PpRecQ6:GUS was used mixed with 10 µg of a circular plasmid carrying the nptII marker. Protoplast isolation and transformation were performed as described (Hohe et al., 2004), and selection was modified for the cotransformation growing the regenerating plants on media supplemented with 12.5 mg/L G418 (Sigma-Aldrich) for 4 weeks without any release phase (Decker et al., 2015).
To generate PpRecQ4 knockout construct, a 4.3-kb fragment of the gene comprising the exons 13 to 25 was amplified from P. patens gDNA using the primers sc243-470555f and sc243-474840r inserting ScaI sites at both sides. This fragment was subcloned into pCRII (Life Technologies) linearized using BamHI and SalI, subsequently, releasing a 2.6-kb genomic fragment comprising exons 15 to 22 in which the nptII cassette, linearized with the same restriction sites, was inserted. Prior to transformation, the construct was linearized using ScaI. For the PpRecQ6 knockout construct, a 3-kb genomic fragment from the 14th intron to the 3′-UTR was amplified with the primers sc128-302139f and sc128-305142r inserting BglII restriction sites on both ends and subcloned into pJET2.1 (Thermo Scientific). This fragment was linearized using EcoRI excising the region from the beginning of exon 16 to the first bases of intron 18. The same enzyme was used to release the nptII cassette from the vector FT-nptII (Koprivova et al., 2004) and the two fragments were ligated. Prior to transformation, the targeting construct was released from the vector backbone with BglII. To allow purification of the construct, the backbone was additionally cut with BglI. Protoplast isolation, transformation, and regeneration were performed as described (Hohe et al., 2004). Selection was done in two successive cycles of selection on solid media complemented with 25 mg/L G418 (Promega) and release of 2 weeks duration as described (Hohe et al., 2004).
For the overexpression of AtRecQ4A and AtRecQ4B in ΔPpRecQ4, the complete CDS was amplified from pPZP221-K-RecQ4A or pPZP221-K-RecQ4B (Schröpfer et al., 2014), respectively, with Phusion Polymerase. In the first approach, we cloned the construct consisting of PpAct5 (Büttner-Mainik et al., 2011), the cDNA of AtRecQ4A or AtRecQ4B, and a nos terminator into a vector containing homologous regions for targeted integration into the PpPIG1 locus (Okano et al., 2009) and a hpt marker. For the AtRecQ4A construct, the cDNA was amplified using AtRecQ4A_KpnI_fw and AtRecQ4A_SmaI_rev, for the AtRecQ4B construct using AtRecQ4B_KpnI_fw and AtRecQ4B_SmaI_rev and subcloned into pJET2.1. To generate the integration vector, the AtRecQ4A or AtRecQ4B cDNA released by a KpnI and SmaI was inserted between the sequence coding for PpAct5 and nos terminator flanked by the PpPIG1 sequences, which was linearized with KpnI and Ecl136II. The resulting plasmids were named pPIG_hpt_pAct5_AtRecQ4A_nosT and pPIG_hpt_pAct5_AtRecQ4B_nosT and contain an hpt marker. Fifty micrograms of the constructs linearized using PaeI and SgsI (Thermo Scientific) was used for transformation. Selection and regeneration was done with two successive cycles of selection on media containing 25 mg/L Hygromycin B and release.
In the second approach, the construct consisting of PpAct5, the cDNA of AtRecQ4A or AtRecQ4B, and a nos terminator was inserted into a vector containing homologous regions for targeted integration into the PTA2 locus (Kubo et al., 2013; Mueller and Reski, 2015). Therefore, the GFP sequence from pJET1.2_PTA2_Actin5_GFP_nosT was released via digestion with SalI and Ecl136II. Prior to ligation, the SalI sticky ends were filled using Klenow Fragment, exo− (Thermo Scientific). The cDNAs of AtRecQ4A and AtRecQ4B were amplified with the oligos AtRecQ4A_fw and AtRecQ4A_rev or AtRecQ4B_fw and AtRecQ4B_rev, respectively. The PCR fragment was phosphorylated using T4 Polynukleotid Kinase (Thermo Scientific), purified, and inserted between the sequences coding for PpAct5 and nos terminator. For transformation, 45 µg of the plasmid linearized using BspQI (New England Biolabs) for the AtRecQ4A vector or BglII (Thermo Scientific) for the AtRecQ4B vector were used together with 10 µg of a circular vector containing the hpt marker. The selection was adapted to the cotransformation, growing the regenerating plants on media supplemented with 25 mg/L Hygromycin B for 4 weeks without any release phase.
For the overexpression of HsBlm, human cDNA (NM_001287246.1) was used for the amplification with Phusion polymerase and the primers Blm_XhoI_fw and Blm_SacI_rev. The resulting fragment, including XhoI and SacI recognition sites, was ligated with the linearized pJET1.2_PTA2_Actin5_GFP_nosT (Mueller and Reski, 2015) from which the GFP sequence was released with SalI and SacI. For transformation of ΔPpRecQ4-1, 45 µg of the plasmid linearized using LguI (Thermo Scientific) was used together with 10 µg of a circular vector containing the hpt marker. The selection was adapted to the cotransformation, growing the regenerating plants on media supplemented with 25 mg/L Hygromycin B for 4 weeks without any release phase.
Molecular Validation of Transgenic Moss Lines
Plants were screened for 5′- and 3′-integration via direct PCR (Schween et al., 2002). As a positive control for successful extraction of DNA, the primers EF1α_fw and EF1α_rev were used. Correct 5′-integration of PpRecQ4:GUS or PpRecQ6:GUS fusion construct was assayed using the primers RecQ4_GUS_5′_fw or RecQ6_GUS_5′_fw and GUS_5′_rev. Correct 3′-integration was assayed with GUS_3′_fw and RecQ4_GUS_3′_rev or RecQ6_GUS_3′_rev, respectively. For correct 5′-integration of the ΔPpRecQ4 construct, the primers Sc243-470451f and RT1, and for 3′-integration the primers Sc243-474892r and RT4 were used. For ΔPpRecQ6, the primers Sc128f together with RT1 and Sc128r with RT4 were used. For the PpPIG1 locus approach the plants were first screened via direct PCR for correct 5′- and 3′-integration with p1bL_WTB_fw and H3 or p1bL_WTB_rev and H4. As no plants with integration into the PpPIG1 locus were identified, we screened for presence of the respective cDNA in the moss genome with the primers AtRecQ4A_KpnI_fw and AtRecQ4A_723rev or AtRecQ4B_KpnI_fw and AtRecQ4B_749rev, respectively. For correct 5′-integration of the AtRecQ4A or AtRecQ4B construct into the PpPTA2 locus, direct PCR was performed with the primers PTA2_5′_fw and Act5_rev, for 3′-integration with 4A_3′_fw or 4B_3′_fw and PTA2_3′_rev. The presence of the HsBlm cDNA in the genome was detected by direct PCR using Blm_fw and Blm_rev to amplify a fragment of ∼1 kb of the 5′-part of the HsBlm cDNA. Expression of the HsBlm cDNA was assayed via RT-PCR using Blm3265_fw with Blm_SacI_rev, resulting in the amplification of the last 990 bp of the cDNA.
To estimate the number of integrations of the PpRecQ4 and PpRecQ6 constructs into the genome of the respective knockout lines, DNA gel blot analysis was performed as described (Wiedemann et al., 2010) using 500 ng of HindIII-digested genomic DNA and a DIG-labeled probe for the CDS of nptII. Additionally for ΔPpRecQ6 plants, a qPCR-based method (Noy-Malka et al., 2014) was used to quantify copy number using the primers q_RecQ6_5f and q_RecQ6_5r for amplification of the 5′-HR region and q_npt_fw and q_npt_rev for the nptII cassette with pCLF_5915_qf and pCLF_5981_qr or pCLF_7739_qf and pCLF_7804_qr, respectively, as for the single-copy gene PpCLF as internal control. As control lines, we used the wild type and ΔPpRecQ4-2 (single integration of nptII cassette according to DNA gel blot analysis). Genomic DNA for this approach was isolated using the GeneJET plant genomic DNA purification mini kit (Thermo Scientific).
Expression Analysis of Transgenic Moss Lines
After isolation of total RNA using a CTAB method (Beike et al., 2015) or Trizol (Life Technologies) and DNaseI digestion, cDNA synthesis was performed using the RevertAid H Minus M-MuLV reverse transcriptase (Thermo Scientific) or Superscript III (Life Technologies). As a control, wild type- or ΔPpRecQ4-cDNA was always prepared in the same way. RT-PCR was performed using a cDNA amount corresponding to 125 ng total RNA. To check for successful cDNA synthesis, primers for the ribosomal gene L21 were used (L21fwd and L21rev).
The correct expression of the RecQ4:GUS or RecQ6:GUS fusion product was validated using the primers RecQ4_GUS_RT_fw or RecQ6_GUS_RT_fw and GUS_RT_rev. To screen for absence or presence of the PpRecQ4 or PpRecQ6 transcript, the primers RecQ4-RT-f and RecQ4-RT-r or RecQ6-RT-f and RecQ6-RT-r were used. To prove the expression of AtRecQ4A, AtRecQ4B, or HsBlm cDNAs, respectively, RT-PCR was performed using the primers AtRecQ4A_KpnI_fw and AtRecQ4A_749_rev, AtRecQ4B_KpnI_fw and AtRecQ4B_749_rev, or Blm3265_fw and Blm_SacI_rev, resulting in the amplification of the last 990 bp of the HsBlm cDNA, respectively.
Quantification of AtRecQ4A or AtRecQ4B expression in the overexpression lines was performed via RT-qPCR with At4A_1346_fw and At4A_1407_rev or At4B_999_fw and At4B_1064_rev, respectively. Preparation of cDNA and RT-qPCR was performed according to Beike et al. (2015). Absolute quantification was performed with a serial dilution of pPZ221-K-RecQ4A or pPZP221-K-RecQ4B. As reference for normalization of differences in cDNA content the product of EF1α_qf and EF1α_qr was used. Following the same procedure, the level of HsBlm expression was quantified using the primers Blm_qRT1_fw and Blm_qRT1_rev, generating products from the middle or the 3′- part of the cDNA, respectively. For quantification, a serial dilution of pJET1.2_PTA2_Actin5_HsBlm_nosT was used. All RT-qPCR experiments were performed in biological triplicates with material harvested at different time points and independent isolation of RNA and cDNA synthesis.
Histochemical GUS Assay for Moss
According to Bierfreund et al. (2003), moss samples were incubated in X-gluc solution (1 mM 5-bromo-4-chloro-3-indolyl-β-glucoronide and 100 mM sodium phosphate, pH 7.0) at 37°C for 12 to 16 h. Fixation was done in 5% (v/v) formalin for 10 min followed by an incubation for 10 min in 5% (v/v) acetic acid. Pigments in the plant tissues were removed by serial incubations in 30%, 50%, 70%, and finally 96% (v/v) ethanol.
Gene Targeting Assay for P. patens
For quantification of GT frequencies, protoplast transformation was performed with a promoterless citrine-fusion targeting construct for the Carbonic Anhydrase 2 (CA2; Pp3c1_19190V1.1). The citrine tag followed by the marker nptII, hpt, or BSD (Ishikawa et al., 2011) was inserted immediately in front of the stop codon of the genomic sequence. The different parts for the three versions of the construct were amplified with overlapping fragments to the next fragment via PCR with Phusion polymerase and joined by Gibson assembly into a KpnI linearized pUC18. For assembly of the initial version of the construct containing the nptII cassette, the following fragments were amplified: the 5′-homologous region for targeting from genomic DNA using CA5′_KpnI_pUCO_fw and CA5′_CitO_rev, for the 3′ homologous region CA3′_nptO_fw and CA3′_KpnI_pUCO_rev. The sequences for citrine including N- and C-terminal linkers (Tian et al., 2004) and nptII, including a 35S promoter and terminator (Girke et al., 1998), were amplified using the oligos Citrin_CA5′O_fw and Citrin_nptO_rev or nptII_CitO_fw and nptII_CA3′O_rev, respectively. To generate the fragments for replacement of nptII by hpt, we used the oligos CA5′_KpnI_pUCO_fw and 35S-P_hptO_rev or 35S-T_hptO_fw and CA3′_KpnI_pUCOrev, respectively. Thus, by using the GT-reporter construct with nptII as template, one fragment consisting of the 5′-homologous region, the citrine CDS, and the 35S promoter and another consisting of the 35S terminator and the 3′-homologous region were generated. The CDS of hpt was amplified with hpt_35S-PO_fw and hpt_35S-TO_rev. For insertion of the BSD marker, the resulting GT reporter construct with a hpt marker including a 35S promoter and terminator was used as a template to generate the fragments consisting of the 5′-homologous region together with the citrine CDS or the 3′-homologous region with the oligos CA5′_KpnI_pUCO_fw and nosT_BSDSo_r or CA3_BSDSo_f and CA3′_KpnI_pUCOrev, respectively. For the amplification of the BSD cassette including a 35S promoter and terminator from p35S_loxP_BSD (GenBank AB537973.1), BSDS_nosTo_f and BSDS_CA3o_r were used. For protoplast transformation with 65 µg of the KpnI linearized plasmid, corresponding to 40 µg of the targeting construct, was used. For the regenerating plants transformed with the nptII and hpt version of the construct, selection was performed as described (Decker et al., 2015). For selection of stable transformants with the BSD cassette containing a version of the construct with 75 mg/L Blasticidin S HCl (Thermo Scientific) the selection procedure started in the liquid regeneration medium 5 d after transformation when at least half of the protoplasts were divided. Ten days after transformation, the plants were transferred to solid medium containing 75 mg/L BDS. Two successive cycles of selection and release of 3 weeks duration each were performed. To quantify the GT frequencies, all plants surviving the respective selection were screened for citrine fluorescence using a Zeiss Axiovert S100. All plants exhibiting citrine fluorescence were categorized as stable integrations via GT, while, in all, plants surviving selection but not showing citrine fluorescence an integration of the construct must have occurred elsewhere in the genome.
As control for transformation efficiencies at different days and in different lines, a transient transformation was performed with a construct consisting of the CDS of PpCA2 fused to citrine under control of PpAct5 and nos terminator. The PpCA2 CDS was amplified with CA2_BglII_fw and CA2_BglII_rev and cloned into the BglII linearized vector containing the PpAct5 promoter, citrine with linkers (Tian et al., 2004), nos terminator, and an hpt cassette.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT3G05740.1, AT1G31360.1, AT4G35740.1, AT1G10930.1, AT1G60930.1, AT1G27880.1, AT5G27680.1, PNR54208.1, PNR59261.1, PNR44826.1, PNR43304.1, PNR32642.1, PNR29543.1, NP_000048.1, NP_116559.1, NP_004251.3, NP_004250.4, NP_000544.2, KZV09040.1, CAA91177.1, and XP_015645653.1. Accession numbers for genes used in phylogenetic analysis can be found in Supplemental Figures 1 to 7.
Supplemental Data
Supplemental Figure 1. Phylogram of the RecQ1 cluster.
Supplemental Figure 2. Phylogram of the RecQ2 cluster.
Supplemental Figure 3. Phylogram of the RecQ3 cluster.
Supplemental Figure 4. Phylogram of the RecQ4 cluster.
Supplemental Figure 5. Phylogram of the RecQ5 cluster.
Supplemental Figure 6. Phylogram of the RecQ6 cluster.
Supplemental Figure 7. Phylogram of the RecQsim cluster.
Supplemental Figure 8. Gene models for the six PpRecQs according to the P. patens genome annotation V3.3 available on cosmoss.org.
Supplemental Figure 9. In silico expression analysis of PpRecQ4, PpRecQ6, PpRecQ2, PpRecQ5, PpRecQsim1, and PpRecQsim2 based on microarray experiments.
Supplemental Figure 10. Targeting strategy and molecular validation of PpRecQ4:GUS and PpRecQ6:GUS reporter lines.
Supplemental Figure 11. Bisulfite sequencing the partial 5′-UTR of AtRecQ4A, potential versus sequenced methylation sites.
Supplemental Figure 12. Targeting strategy and molecular validation of ΔPpRecQ4.
Supplemental Figure 13. Molecular validation of AtRecQ4A expressing lines in ΔPpRecQ4.
Supplemental Figure 14. Targeting strategy and molecular validation of ΔPpRecQ6.
Supplemental Figure 15. Biomass gain and development of ΔPpRecQ4 is altered in comparison to WT and ΔPpRecQ6.
Supplemental Figure 16. Test for mutator phenotype indicates no genetic instability in ΔRecQ4 or ΔRecQ6.
Supplemental Figure 17. ΔPpRecQ6 produces spore capsules and viable spores with a delayed growth upon germination.
Supplemental Figure 18. Hypersensitivity of ΔRecQ4 to UV treatment.
Supplemental Figure 19. Regeneration of WT and ΔPpRecQ4 protoplasts after transformation is similar to WT.
Supplemental Figure 20. Molecular validation of AtRecQ4B expressing lines in the ΔPpRecQ4 background.
Supplemental Figure 21. Expression of AtRecQ4B in ΔPpRecQ4 background does not change the ΔPpRecQ4 sensitive phenotype upon treatment with bleomycin or cisplatin.
Supplemental Figure 22. Molecular validation of HsBlm-expressing lines in the ΔPpRecQ4 background.
Supplemental Figure 23. Expression of HsBlm in the ΔPpRecQ4 background does not change the ΔPpRecQ4 sensitive phenotype upon treatment with bleomycin or cisplatin.
Supplemental Table 1. Primers used for PCR.
Supplemental Data Set 1. Overview of proteome and CDS data sets.
Supplemental Data Set 2. Identification of AtRecQ1 and AtRecQ3 orthologs in the clade of mosses in the 1000 plants (1KP) database.
Supplemental File 1. Text file of alignment used for phylogenetic analysis in Figure 1.
Supplemental File 2. Text file of alignment used for phylogenetic analysis in Supplemental Figure 1.
Supplemental File 3. Text file of alignment used for phylogenetic analysis in Supplemental Figure 2.
Supplemental File 4. Text file of alignment used for phylogenetic analysis in Supplemental Figure 3.
Supplemental File 5. Text file of alignment used for phylogenetic analysis in Supplemental Figure 4.
Supplemental File 6. Text file of alignment used for phylogenetic analysis in Supplemental Figure 5.
Supplemental File 7. Text file of alignment used for phylogenetic analysis in Supplemental Figure 6.
Supplemental File 8. Text file of alignment used for phylogenetic analysis in Supplemental Figure 7.
Acknowledgments
This work was supported by the Excellence Initiative of the German Federal and State Governments (EXC 294 to R.R.), the Federal Ministry of Education and Research (GABI-PRECISE 0315057D to R.R.), and Deutsche Forschungsgemeinschaft (TRR 141, project B02 to R.R.). It was cofunded by the European Union (European Regional Development Fund) in the framework of the program INTERREG IV Upper Rhine (Project A17 TIP-ITP to R.R.). The IJPB benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS). We thank Holger Puchta for the excellent opportunity to start the work on Arabidopsis RecQ4A complementation and many fruitful discussions of the results. We thank Agnes Novakovic for excellent technical assistance, Lei Zhu for her contribution to the generation of ΔRecQ6 lines and the molecular validation of ΔRecQ4 and ΔRecQ6 lines, and Anne Katrin Prowse for language editing.
AUTHOR CONTRIBUTIONS
G.W., E.L.D., F.N., F.H., and R.R. designed the research. G.W., N.v.G., F.K., L.O., K.S., L.M., and F.H. performed research and analyzed data. G.W., F.N., E.L.D., F.H., and R.R. wrote the manuscript. All authors discussed data and approved the final version of the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bagherieh-Najjar M.B., de Vries O.M.H., Kroon J.T.M., Wright E.L., Elborough K.M., Hille J., Dijkwel P.P. (2003). Arabidopsis RecQsim, a plant-specific member of the RecQ helicase family, can suppress the MMS hypersensitivity of the yeast sgs1 mutant. Plant Mol. Biol. 52: 273–284. [DOI] [PubMed] [Google Scholar]
- Bagherieh-Najjar M.B., de Vries O.M.H., Hille J., Dijkwel P.P. (2005). Arabidopsis RecQI4A suppresses homologous recombination and modulates DNA damage responses. Plant J. 43: 789–798. [DOI] [PubMed] [Google Scholar]
- Baumann P., West S.C. (1998). Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 23: 247–251. [DOI] [PubMed] [Google Scholar]
- Beike A.K., von Stackelberg M., Schallenberg-Rüdinger M., Hanke S.T., Follo M., Quandt D., McDaniel S.F., Reski R., Tan B.C., Rensing S.A. (2014). Molecular evidence for convergent evolution and allopolyploid speciation within the Physcomitrium-Physcomitrella species complex. BMC Evol. Biol. 14: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beike A.K., Lang D., Zimmer A.D., Wüst F., Trautmann D., Wiedemann G., Beyer P., Decker E.L., Reski R. (2015). Insights from the cold transcriptome of Physcomitrella patens: global specialization pattern of conserved transcriptional regulators and identification of orphan genes involved in cold acclimation. New Phytol. 205: 869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein K.A., Gangloff S., Rothstein R. (2010). The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 44: 393–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatla S.C., Kiessling J., Reski R. (2002). Observation of polarity induction by cytochemical localization of phenylalkylamine-binding sites in regenerating protoplasts of the moss Physcomitrella patens. Protoplasma 219: 99–105. [DOI] [PubMed] [Google Scholar]
- Bierfreund N.M., Reski R., Decker E.L. (2003). Use of an inducible reporter gene system for the analysis of auxin distribution in the moss Physcomitrella patens. Plant Cell Rep. 21: 1143–1152. [DOI] [PubMed] [Google Scholar]
- Büttner-Mainik A., Parsons J., Jérôme H., Hartmann A., Lamer S., Schaaf A., Schlosser A., Zipfel P.F., Reski R., Decker E.L. (2011). Production of biologically active recombinant human factor H in Physcomitrella. Plant Biotechnol. J. 9: 373–383. [DOI] [PubMed] [Google Scholar]
- Cadet J., Berger M., Douki T., Morin B., Raoul S., Ravanat J.L., Spinelli S. (1997). Effects of UV and visible radiation on DNA-final base damage. Biol. Chem. 378: 1275–1286. [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlot F., Chelysheva L., Kamisugi Y., Vrielynck N., Guyon A., Epert A., Le Guin S., Schaefer D.G., Cuming A.C., Grelon M., Nogué F. (2014). RAD51B plays an essential role during somatic and meiotic recombination in Physcomitrella. Nucleic Acids Res. 42: 11965–11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G. (1994). Cellular responses to cisplatin. The roles of DNA-binding proteins and DNA repair. J. Biol. Chem. 269: 787–790. [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Croteau D.L., Popuri V., Opresko P.L., Bohr V.A. (2014). Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83: 519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker E.L., Wiedemann G., Reski R. (2015). Gene targeting for precision glyco-engineering: production of biopharmaceuticals devoid of plant-typical glycosylation in moss bioreactors. Methods Mol. Biol. 1321: 213–224. [DOI] [PubMed] [Google Scholar]
- Favaudon V. (1982). On the mechanism of reductive activation in the mode of action of some anticancer drugs. Biochimie 64: 457–475. [DOI] [PubMed] [Google Scholar]
- Finn R.D., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44: D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. III, Smith H.O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Girke T., Schmidt H., Zähringer U., Reski R., Heinz E. (1998). Identification of a novel delta 6-acyl-group desaturase by targeted gene disruption in Physcomitrella patens. Plant J. 15: 39–48. [DOI] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D., Colangelo C., Williams K., Gerstein M. (2003). Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Harrison C.J., Roeder A.H.K., Meyerowitz E.M., Langdale J.A. (2009). Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr. Biol. 19: 461–471. [DOI] [PubMed] [Google Scholar]
- Hartung F., Puchta H. (2006). The RecQ gene family in plants. J. Plant Physiol. 163: 287–296. [DOI] [PubMed] [Google Scholar]
- Hartung F., Plchová H., Puchta H. (2000). Molecular characterisation of RecQ homologues in Arabidopsis thaliana. Nucleic Acids Res. 28: 4275–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Bergmann T., Puchta H. (2006). The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 34: 4438–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Puchta H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Knoll A., Wurz-Wildersinn R., Puchta H. (2008). Topoisomerase 3α and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 4: e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Ferdous M., Osman K., Franklin F.C.H. (2011). The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant J. 65: 492–502. [DOI] [PubMed] [Google Scholar]
- Hiss M., et al. (2014). Large-scale gene expression profiling data for the model moss Physcomitrella patens aid understanding of developmental progression, culture and stress conditions. Plant J. 79: 530–539. [DOI] [PubMed] [Google Scholar]
- Hohe A., Reski R. (2002). Optimisation of a bioreactor culture of the moss Physcomitrella patens for mass production of protoplasts. Plant Sci. 163: 69–74. [Google Scholar]
- Hohe A., Rensing S.A., Mildner M., Lang D., Reski R. (2002). Day length and temperature strongly influence sexual reproduction and expression of a novel MADS-box gene in the moss Physcomitrella patens. Plant Biol. (Stuttg.) 4: 595–602. [Google Scholar]
- Hohe A., Egener T., Lucht J.M., Holtorf H., Reinhard C., Schween G., Reski R. (2004). An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene-knockouts in a moss, Physcomitrella patens. Curr. Genet. 44: 339–347. [DOI] [PubMed] [Google Scholar]
- Horst N.A., Reski R. (2017). Microscopy of Physcomitrella patens sperm cells. Plant Methods 13: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst N.A., Katz A., Pereman I., Decker E.L., Ohad N., Reski R. (2016). A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat. Plants 2: 15209. [DOI] [PubMed] [Google Scholar]
- Ishikawa M., et al. (2011). Physcomitrella cyclin-dependent kinase A links cell cycle reactivation to other cellular changes during reprogramming of leaf cells. Plant Cell 23: 2924–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.S., Eddy S.R., Portugaly E. (2010). Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y., Schlink K., Rensing S.A., Schween G., von Stackelberg M., Cuming A.C., Reski R., Cove D.J. (2006). The mechanism of gene targeting in Physcomitrella patens: homologous recombination, concatenation and multiple integration. Nucleic Acids Res. 34: 6205–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y., Schaefer D.G., Kozak J., Charlot F., Vrielynck N., Holá M., Angelis K.J., Cuming A.C., Nogué F. (2012). MRE11 and RAD50, but not NBS1, are essential for gene targeting in the moss Physcomitrella patens. Nucleic Acids Res. 40: 3496–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y., Whitaker J.W., Cuming A.C. (2016). The transcriptional response to DNA-double-strand breaks in Physcomitrella patens. PLoS One 11: e0161204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P.J., et al. (2018). Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 46: D802–D808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A., Schröpfer S., Puchta H. (2014). The RTR complex as caretaker of genome stability and its unique meiotic function in plants. Front. Plant Sci. 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji R., Hasebe M. (2014). Eight types of stem cells in the life cycle of the moss Physcomitrella patens. Curr. Opin. Plant Biol. 17: 13–21. [DOI] [PubMed] [Google Scholar]
- Koprivova A., Stemmer C., Altmann F., Hoffmann A., Kopriva S., Gorr G., Reski R., Decker E.L. (2004). Targeted knockouts of Physcomitrella lacking plant-specific immunogenic N-glycans. Plant Biotechnol. J. 2: 517–523. [DOI] [PubMed] [Google Scholar]
- Kubo M., Imai A., Nishiyama T., Ishikawa M., Sato Y., Kurata T., Hiwatashi Y., Reski R., Hasebe M. (2013). System for stable β-estradiol-inducible gene expression in the moss Physcomitrella patens. PLoS One 8: e77356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.-I., Abe K., Osakabe K., Endo M., Nishizawa-Yokoi A., Saika H., Shimada H., Toki S. (2012). Overexpression of OsRecQl4 and/or OsExo1 enhances DSB-induced homologous recombination in rice. Plant Cell Physiol. 53: 2142–2152. [DOI] [PubMed] [Google Scholar]
- Kwon Y.-I., Abe K., Endo M., Osakabe K., Ohtsuki N., Nishizawa-Yokoi A., Tagiri A., Saika H., Toki S. (2013). DNA replication arrest leads to enhanced homologous recombination and cell death in meristems of rice OsRecQl4 mutants. BMC Plant Biol. 13: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D., Zimmer A.D., Rensing S.A., Reski R. (2008). Exploring plant biodiversity: the Physcomitrella genome and beyond. Trends Plant Sci. 13: 542–549. [DOI] [PubMed] [Google Scholar]
- Lang D., et al. (2018). The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 93: 515–533. [DOI] [PubMed] [Google Scholar]
- Larsson A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30: 3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Santoro I.M., McDaniel L.D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., Bradley A. (2000). Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26: 424–429. [DOI] [PubMed] [Google Scholar]
- Mannuss A., Dukowic-Schulze S., Suer S., Hartung F., Pacher M., Puchta H. (2010). RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 22: 3318–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmann-Mulisch U., Wendeler E., Zobell O., Schween G., Steinbiss H.-H., Reiss B. (2007). Differential requirements for RAD51 in Physcomitrella patens and Arabidopsis thaliana development and DNA damage repair. Plant Cell 19: 3080–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N., et al. (2014). Data access for the 1,000 Plants (1KP) project. Gigascience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. (2011). DNA end resection: Unraveling the tail. DNA Repair 10: 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ries G., Bonhoeffer S., Hohn B. (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.J., Reski R. (2015). Mitochondrial dynamics and the ER: the plant perspective. Front. Cell Dev. Biol. 3: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S.B., Wunsch C.D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48: 443–453. [DOI] [PubMed] [Google Scholar]
- Noy-Malka C., Yaari R., Itzhaki R., Mosquna A., Auerbach Gershovitz N., Katz A., Ohad N. (2014). A single CMT methyltransferase homolog is involved in CHG DNA methylation and development of Physcomitrella patens. Plant Mol. Biol. 84: 719–735. [DOI] [PubMed] [Google Scholar]
- Nguyen N.P., Mirarab S., Kumar K., Warnow T. (2015). Ultra-large alignments using phylogeny-aware profiles. Genome Biol. 16: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano Y., Aono N., Hiwatashi Y., Murata T., Nishiyama T., Ishikawa T., Kubo M., Hasebe M. (2009). A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc. Natl. Acad. Sci. USA 106: 16321–16326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ramírez C., Hernandez-Coronado M., Thamm A., Catarino B., Wang M., Dolan L., Feijó J.A., Becker J.D. (2016). A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol. Plant 9: 205–220. [DOI] [PubMed] [Google Scholar]
- Proost S., Pattyn P., Gerats T., Van de Peer Y. (2011). Journey through the past: 150 million years of plant genome evolution. Plant J. 66: 58–65. [DOI] [PubMed] [Google Scholar]
- R Core Team (2017). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing).
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Reski R. (1998a). Physcomitrella and Arabidopsis: the David and Goliath of reverse genetics. Trends Plant Sci. 3: 209–210. [Google Scholar]
- Reski R. (1998b). Development, genetics and molecular biology of mosses. Bot. Acta 111: 1–15. [Google Scholar]
- Reski R., Abel W.O. (1985). Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta 165: 354–358. [DOI] [PubMed] [Google Scholar]
- Reski R., Parsons J., Decker E.L. (2015). Moss-made pharmaceuticals: from bench to bedside. Plant Biotechnol. J. 13: 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig S., Schröpfer S., Knoll A., Puchta H. (2016). The RTR complex partner RMI2 and the DNA helicase RTEL1 are both independently involved in preserving the stability of 45S rDNA repeats in Arabidopsis thaliana. PLoS Genet. 12: e1006394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome A., Kimura S., Mori Y., Uchiyama Y., Morohashi K., Sakaguchi K. (2006). Characterization of four RecQ homologues from rice (Oryza sativa L. cv. Nipponbare). Biochem. Biophys. Res. Commun. 345: 1283–1291. [DOI] [PubMed] [Google Scholar]
- Schaefer D.G., Delacote F., Charlot F., Vrielynck N., Guyon-Debast A., Le Guin S., Neuhaus J.M., Doutriaux M.P., Nogué F. (2010). RAD51 loss of function abolishes gene targeting and de-represses illegitimate integration in the moss Physcomitrella patens. DNA Repair (Amst.) 9: 526–533. [DOI] [PubMed] [Google Scholar]
- Schröpfer S., Kobbe D., Hartung F., Knoll A., Puchta H. (2014). Defining the roles of the N-terminal region and the helicase activity of RECQ4A in DNA repair and homologous recombination in Arabidopsis. Nucleic Acids Res. 42: 1684–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuessele C., Hoernstein S.N.W., Mueller S.J., Rodriguez-Franco M., Lorenz T., Lang D., Igloi G.L., Reski R. (2016). Spatio-temporal patterning of arginyl-tRNA protein transferase (ATE) contributes to gametophytic development in a moss. New Phytol. 209: 1014–1027. [DOI] [PubMed] [Google Scholar]
- Schween G., Fleig S., Reski R. (2002). High-throughput-PCR screen of 15,000 transgenic Physcomitrella plants. Plant Mol. Biol. Report. 20: 43–47. [Google Scholar]
- Schween G., Gorr G., Hohe A., Reski R. (2003a). Unique tissue-specific cell cycle in Physcomitrella. Plant Biol. (Stuttg.) 5: 50–58. [Google Scholar]
- Schween G., Hohe A., Koprivova A., Reski R. (2003b). Effects of nutrients, cell density and culture techniques on protoplast regeneration and early protonema development in a moss, Physcomitrella patens. J. Plant Physiol. 160: 209–212. [DOI] [PubMed] [Google Scholar]
- Schween G., Hohe A., Schulte J., Reski R. (2005). Effect of ploidy level on growth, differentiation and phenotype in Physcomitrella patens. Plant Cell Rep. 23: 513–521.15558285 [Google Scholar]
- Shaked H., Melamed-Bessudo C., Levy A.A. (2005). High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc. Natl. Acad. Sci. USA 102: 12265–12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp R., Scholz S., Kruse S., Speth V., Reski R. (1998). Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 95: 4368–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Klein H. (2006). Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7: 739–750. [DOI] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. (2003). PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouiller B., Schaefer D.G., Charlot F., Nogué F. (2006). MSH2 is essential for the preservation of genome integrity and prevents homeologous recombination in the moss Physcomitrella patens. Nucleic Acids Res. 34: 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt P.M., Louis E.J., Borts R.H., Hickson I.D. (1995). Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81: 253–260. [DOI] [PubMed] [Google Scholar]
- Wendeler E., Zobell O., Chrost B., Reiss B. (2015). Recombination products suggest the frequent occurrence of aberrant gene replacement in the moss Physcomitrella patens. Plant J. 81: 548–558. [DOI] [PubMed] [Google Scholar]
- Wiedemann G., Hermsen C., Melzer M., Büttner-Mainik A., Rennenberg H., Reski R., Kopriva S. (2010). Targeted knock-out of a gene encoding sulfite reductase in the moss Physcomitrella patens affects gametophytic and sporophytic development. FEBS Lett. 584: 2271–2278. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Kato J., Shimamoto A., Goto M., Furuichi Y., Ikeda H. (1998). Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA 95: 8733–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A., et al. (2016). Ensembl 2016. Nucleic Acids Res. 44 (D1): D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T.-Y. (2017). ggtree : an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8: 28–36. [Google Scholar]
- Zimmer A.D., Lang D., Buchta K., Rombauts S., Nishiyama T., Hasebe M., Van de Peer Y., Rensing S.A., Reski R. (2013). Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics 14: 498. [DOI] [PMC free article] [PubMed] [Google Scholar]