The small peptide PSK acts as a damage-associated molecular pattern and when perceived by the tomato receptor PSKR1 results in increased cytosolic Ca2+ that initiates auxin-mediated plant immunity to the pathogen B. cinerea.
Abstract
Phytosulfokine (PSK) is a disulfated pentapeptide that is an important signaling molecule. Although it has recently been implicated in plant defenses to pathogen infection, the mechanisms involved remain poorly understood. Using surface plasmon resonance and gene silencing approaches, we showed that the tomato (Solanum lycopersicum) PSK receptor PSKR1, rather than PSKR2, functioned as the major PSK receptor in immune responses. Silencing of PSK signaling genes rendered tomato more susceptible to infection by the economically important necrotrophic pathogen Botrytis cinerea. Analysis of tomato mutants defective in either defense hormone biosynthesis or signaling demonstrated that PSK-induced immunity required auxin biosynthesis and associated defense pathways. Here, using aequorin-expressing tomato plants, we provide evidence that PSK perception by tomato PSKR1 elevated cytosolic [Ca2+], leading to auxin-dependent immune responses via enhanced binding activity between calmodulins and the auxin biosynthetic YUCs. Thus, our data demonstrate that PSK acts as a damage-associated molecular pattern and is perceived mainly by PSKR1, which increases cytosolic [Ca2+] and activates auxin-mediated pathways that enhance immunity of tomato plants to B. cinerea.
INTRODUCTION
In natural environments, plants are exposed to attack by a wide variety of herbivores and microbial pathogens. These biotic threats to crops pose a significant risk to agriculture and often result in tremendous economic losses to the farmer. During evolution, plants have acquired a sophisticated innate immune system that serves to mitigate the adverse effects of pathogen attack. Therefore, accurate signal perception is a prerequisite for the elicitation of appropriate defense responses in the host. Plants employ an array of pattern recognition receptors on the host cell surface that detect various pathogen-derived elicitors in the apoplast between cells. Particularly important are the receptor-like kinases and receptor-like proteins that detect these elicitors. Pattern recognition receptors such as FLAGELLIN-SENSING2 (FLS2), FLS3, EF-Tu receptor, and chitin elicitor receptor kinase 1 directly sense pathogen/microbe-associated molecular patterns (Chinchilla et al., 2006; Zipfel et al., 2006; Miya et al., 2007; Hind et al., 2016). Upon recognition, these receptors trigger both local and systemic pathogen-defense signaling cascades leading to basal immunity and non-host resistance (Böhm et al., 2014). In addition, plant cell surface receptors are also involved in the perception of endogenous plant compounds. These are referred to as damage-associated molecular patterns (DAMPs) and they also trigger immune responses (Boutrot and Zipfel, 2017). In particular, phytosulfokine [PSK; Tyr(SO3H)-Ile-Tyr(SO3H)-Thr-Gln], a disulfated pentapeptide secreted by plants, is thought to be a DAMP acting in the immune response.
PSK is generated by the processing of precursors of ∼80 amino acids that are encoded by the PSK gene family (Yang et al., 2001; Matsubayashi et al., 2006), which appears to be ubiquitous in higher plants. PSK precursors undergo tyrosine sulfation by a tyrosylprotein sulfotransferase (TPST) in the cis-Golgi followed by proteolytic cleavage in the apoplast (Srivastava et al., 2008; Komori et al., 2009). Mature PSK peptides are recognized at the cell surface by membrane-bound PSK receptors (PSKRs), which belong to the leucine-rich repeat receptor-like kinase (LRR-RLK) class of receptors (Matsubayashi et al., 2002). However, the detailed signaling mechanisms and pathways that PSK activates are unknown.
Arabidopsis thaliana PSKR1 and PSKR2 were shown to be PSK receptors that contain extracellular LRRs, an island domain, a transmembrane domain, and a cytoplasmic kinase domain (Matsubayashi et al., 2006; Amano et al., 2007). The extracellular domains consist of 21 LRRs with an island domain and are required for PSK perception (Matsubayashi et al., 2002, 2006; Amano et al., 2007). Recognition between the PSK-PSKR pair functions as a master switch for a complex, but poorly defined, intracellular signaling pathway. Moreover, the plant PSKR family and their ligand binding properties have not been characterized other than in Arabidopsis and carrot (Daucus carota), and little is known about the molecular steps that link PSK to downstream signaling events.
The cytoplasmic kinase domain of Arabidopsis PSKR1 overlaps a canonical guanylate cyclase (GC) core. Overexpression of At-PSKR1 in Arabidopsis leaf protoplasts resulted in an increase in the endogenous levels of guanosine 3′,5′-cyclic phosphate (cGMP), suggesting that At-PSKR1 has GC activity (Kwezi et al., 2011). Similar to At-PSKR1, the cytosolic domains of other LRR-RLKs such as the Arabidopsis Pep receptor (At-PepR1) and the brassinosteroid receptor (At-BRI1) have a GC core with GC activity. Moreover, the generation of cGMP from GTP has been demonstrated by analysis in vitro of the recombinant At-PepR1 and At-BRI1 proteins (Kwezi et al., 2007; Qi et al., 2010). cGMP is a potential activating ligand for cyclic nucleotide-gated channels in plants. These cation channels are thought to facilitate Ca2+ and other cation fluxes (Ladwig et al., 2015). Intriguingly, Pep-At-PepR1 signaling leads to the expression of pathogen defense genes, and this is suggested to be mediated by the cGMP-activated Ca2+-conducting channel CNGC2 as well as the elevation of cytosolic Ca2+ levels (Qi et al., 2010; Ma et al., 2012). Similarly, brassinosteroid perception by the receptor At-BRI1 leads to immediate cGMP-dependent cytosolic Ca2+ release in vivo, a process that was abolished when the CNGC2 gene was mutated in Arabidopsis (Zhao et al., 2013a). Interestingly, PSK not only promotes cell expansion via a cGMP-dependent pathway but also activates a cation channel protein encoded by CNGC17 in Arabidopsis (Ladwig et al., 2015). It is thus reasonable to hypothesize that the PSK receptors may induce an intracellular Ca2+ burst at the cell surface, leading to an immune response in cells where this pathway is triggered.
PSK signaling pathways frequently act together with phytohormones to fine-tune plant responses to external and metabolic stimuli via interactions that can be either synergistic or antagonistic (Mosher et al., 2013; Rodiuc et al., 2016). For example, PSKR1 decreases the resistance of Arabidopsis plants to the hemibiotrophic bacterial pathogen Pseudomonas syringae but enhances defenses against the necrotrophic fungal pathogen Alternaria brassicicola. Such observations were suggested to be related to PSK suppression of salicylic acid (SA)-mediated defense responses (Mosher et al., 2013). PSK has also been demonstrated to suppress ethylene (ET) production; thus, it is involved in regulating Arabidopsis copper homeostasis (Wu et al., 2015). In addition, PSK requires auxin to stimulate nonembryogenic proliferation in carrot cell cultures (Eun et al., 2003).
Necrotrophic fungal pathogens such as the gray mold fungus Botrytis cinerea are a major threat to crop security worldwide. B. cinerea has a remarkable host range, encompassing over 200 plant species. This pathogen alone causes annual losses of several hundreds of millions of U.S. dollars worldwide, mainly because of its adverse effects on tomato production (Dean et al., 2012). To date, accessions with complete resistance to this pathogen have not yet been identified in tomato (Solanum lycopersicum). Therefore, understanding the molecular basis of PSK-triggered immunity to B. cinerea not only has intrinsic scientific value with significant translational opportunities in tomato, but it also has the potential to provide new knowledge that can help to enhance disease resistance in a wide range of crops. Here, we present compelling evidence for a tomato PSK-PSKR1 signaling pathway that involves intracellular Ca2+ release and leads to downstream auxin-dependent signaling cascades that trigger appropriate immune responses against B. cinerea in tomato.
RESULTS
PSK Signaling Conferred Immunity against B. cinerea in Tomato
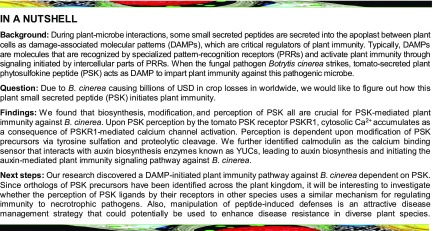
To investigate the function of PSK in the tomato immune response to B. cinerea, exogenous PSK-induced defense responses were induced by applying 0.5 to 20 µM PSK. The application of PSK decreased fungal growth, as measured by B. cinerea actin mRNA accumulation, in a concentration-dependent manner, and the effect was maximal at the 10 and 20 µM concentrations (Supplemental Figure 1A). Based on preliminary dose-dependent trial experiments, the 10 µM concentration was selected as the optimal level required to induce the required response in successive experiments. Leaves were sprayed with either PSK, the inactive desulfated PSK peptide (dPSK) (Mosher et al., 2013; Wang et al., 2015), or water (control) 12 h before the pathogen inoculation. Chlorophyll a fluorescence imaging was used to determine the damage response to B. cinerea infection by measuring the changes in the quantum yield of photosystem II (ΦPSII). Whereas ΦPSII was significantly decreased 2 d postinoculation (dpi) with B. cinerea, this parameter remained higher in PSK-treated plants (Figures 1A and 1C). These observations were consistent with a lower level of pathogen infection in PSK-treated plants compared with dPSK- or non-pretreated controls, as determined by both the extent of cell death assessed by trypan blue staining and B. cinerea-specific actin mRNA accumulation (Figures 1B and 1D).
Figure 1.
PSK Signaling Confers Tomato Plants Immunity against B. cinerea.
(A) to (D) Tomato defense to B. cinerea is promoted by exogenous PSK. Four-week-old tomato plants were treated with 10 µM PSK, 10 µM dPSK, or water 12 h before B. cinerea inoculation.
(A) Representative chlorophyll fluorescence imaging of ΦPSII at 2 dpi. Bar = 1 cm.
(B) Representative images of trypan blue staining for cell death in tomato leaves at 2 dpi. Bar = 250 µm.
(C) Quantification of ΦPSII at 2 dpi.
(D) Relative B. cinerea actin transcript abundance in infected leaves at 1 dpi.
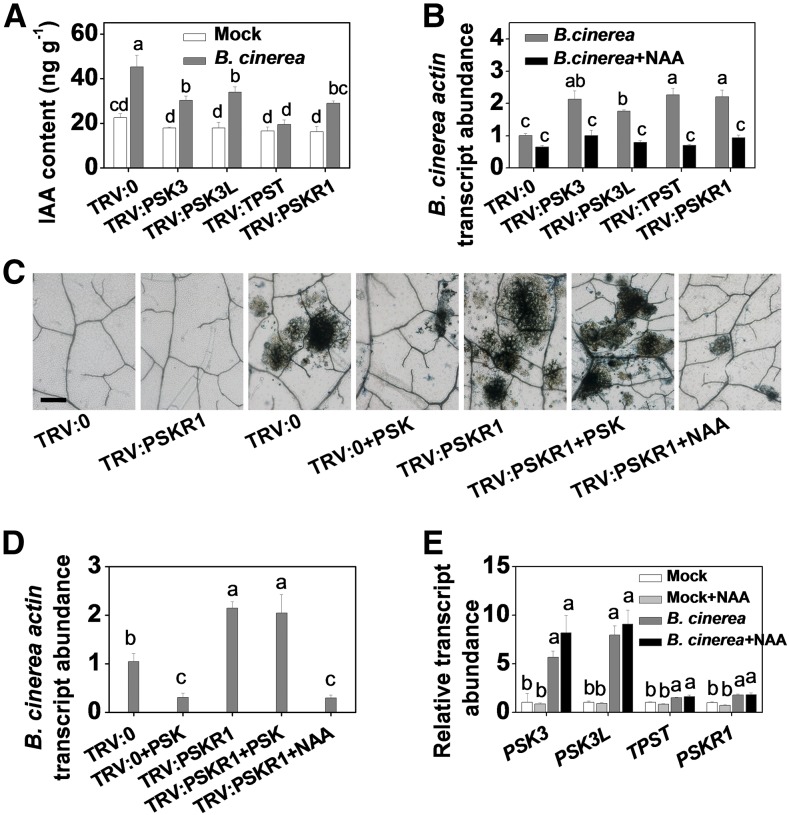
(E) Effects of B. cinerea inoculation on the transcript abundance of PSK precursor genes and tyrosine sulfation processing gene TPST in tomato leaves at 0.5 dpi. The transcript abundance of each gene under mock-inoculated condition was defined as 1. An asterisk indicates a significant effect of B. cinerea inoculation of tomato plants relative to mock-inoculated plants. nd, not detected.
(F) to (I) Effects of silencing of PSK-biosynthesis related genes on tomato innate immunity against B. cinerea.
(F) Representative chlorophyll fluorescence imaging of ΦPSII at 2 dpi. Bar = 1 cm.
(G) Trypan blue staining for cell death in tomato leaves at 2 dpi. Bar = 250 µm.
(H) Quantification of ΦPSII at 2 dpi.
(I) Relative B. cinerea actin transcript abundance in infected leaves at 1 dpi.
The results in (C) to (E), (H), and (I) are presented as mean values ± sd; n = 3 leaves from different plants in (C) and (H); n = 3 independent pooled samples with each sample being from two plants in (D), (E), and (I). Different letters indicate significant differences between treatments (P < 0.05, Tukey’s test). The above experiments were repeated three times with similar results.
Eight PSK precursor genes were identified in the tomato genome based on homology to the Arabidopsis PSK genes (Supplemental Figure 1B and Supplemental File 1.) At the transcriptional level, the expression of four of these tomato precursor genes (PSK3, PSK3L, PSK4, and PSK7) as well as the single-copy tyrosine sulfation processing gene TPST was significantly affected by B. cinerea infection (Figure 1E). Transcript abundance of these genes was reduced using virus-induced gene silencing (VIGS) to examine their roles in innate immunity. This approach decreased the levels of target gene transcripts by up to 75% compared with the empty bipartite tobacco rattle virus (TRV) vector negative control (TRV:0) plants (Supplemental Figure 1C). Following B. cinerea inoculation, gene silencing generally had little or no effect on the expression of nontarget homologous genes (Supplemental Figure 1D and Supplemental Figure 2). However, B. cinerea-induced PSK3L transcription was partially attenuated in the TRV:PSK3 plants (Supplemental Figure 1D). This may be due to the high degree of similarity between the two PSK precursors, resulting in cosilencing of the PSK3 and PSK3L genes in the TRV:PSK3 plants. Upon B. cinerea inoculation, even though gene silencing in the TRV:PSK4 and TRV:PSK7 plants did not influence responses to B. cinerea inoculation, the susceptibility of TRV:PSK3 and TRV:PSK3L, as well as the TRV:TPST plants were significantly enhanced, as shown by the significant decrease in ΦPSII and by the presence of more dead cells in the leaves and increases in leaf B. cinerea actin mRNA accumulation (Figures 1F to 1I). These results indicated that PSK3L, TPST, and possibly PSK3 played a positive role in the immunity against B. cinerea. Taken together, these findings suggest that PSK signaling is vital for the induction of tomato immunity against B. cinerea.
Identification of a PSK Receptor and Its Biological Functions in Tomato Immunity
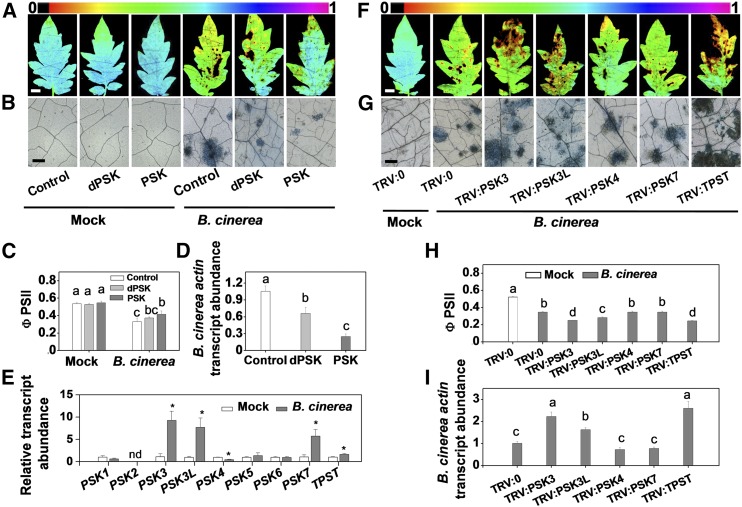
Based on overall amino acid similarity to Arabidopsis At-PSKR1/R2 and carrot Dc-PSKR, tomato Solyc01g008140.3 (Sl-PSKR1) and Solyc07g063000.3 (Sl-PSKR2) were determined as putative PSKR paralogs. The homologous PSKRs belong to the large LRR-RLK family, each of which contains conserved extracellular tandem copies of LRRs, an island domain that is required for PSK perception, a transmembrane domain, and a cytoplasmic kinase domain (Supplemental Figures 3A, 3B, and 4 and Supplemental File 2). To determine the subcellular localization of the tomato PSKR paralogs, the binary vectors 35S:PSKRs fused to GFP and 35S:FLS2-mCherry as a plasma membrane localization marker, were transiently coexpressed in Nicotiana benthamiana using Agrobacterium tumefaciens-mediated transformation (Robatzek et al., 2006). Tomato PSKR1-GFP and PSKR2-GFP colocalized with FLS2-mCherry at the plasma membrane (Figure 2A).
Figure 2.
Characterization of Tomato PSK Receptors.
(A) Subcellular localization of PSKR1 and PSKR2. Tomato PSKRs-GFP and FLS2-mCherry (a marker for plasma membrane localization) plasmids were transiently coexpressed in N. benthamiana leaves. The GFP and mCherry signals were visualized using confocal microscopy 48 h after infiltration. Bar = 50 µm.
(B) SPR analysis of the binding of PSK to potential tomato PSK receptors, PSKR1 and PSKR2. The curves represent the concentrations of the injected PSK and dPSK. From bottom to top: 0.39, 0.78, 3.125, 6.25, 12.5, and 25 µM were used for PSK. An additional 50 µM concentration was also used for dPSK. The recombinant extracellular portion of PSKR1 and PSKR2 was immobilized onto the sensor chip. The obtained kinetic constants for specific binding are shown in each panel. RU, resonance units.
(C) to (F) Effects of PSKR1 and/or PSKR2 silencing on tomato innate immunity against B. cinerea.
(C) Representative chlorophyll fluorescence imaging of ΦPSII at 2 dpi with B. cinerea. Bar = 1 cm.
(D) Representative images of trypan blue staining for cell death in tomato leaves at 2 dpi.
(E) Quantification of ΦPSII at 2 dpi. Bar = 250 µm.
(F) Relative B. cinerea actin transcript abundance in infected leaves at 1 dpi.
The results in (E) and (F) are presented as mean values ± sd; n = 3 leaves from different plants. Different letters indicate significant differences between treatments (P < 0.05, Tukey’s test). The above experiments were repeated two times with similar results.
The binding affinities of PSK to Sl-PSKR1 and Sl-PSKR2 were determined using surface plasmon resonance (SPR) analysis. The recombinant-expressed extracellular portions of these two proteins were immobilized onto the surface of a CM5 sensor chip via amine coupling. Concentration-dependent binding was recorded following the application of PSK or dPSK. Both Sl-PSKR1 and Sl-PSKR2 interacted with PSK. The binding affinity constant (Kd) values were in the similar micromolar range to that described for PSK and the recombinant Dc-PSKR protein (Wang et al., 2015). PSK exhibited a slightly stronger binding affinity to Sl-PSKR1 than to Sl-PSKR2. The association rate constant (Ka) for the Sl-PSKR1-PSK interaction was higher than that of the Sl-PSKR2-PSK interaction (Figure 2B). SPR assays confirmed the important role of the sulfate group in mediating PSK recognition. The naturally occurring form of PSK displayed stronger binding affinity and higher Ka values with regard to both Sl-PSKR1 and Sl-PSKR2 compared with dPSK. This result is in agreement with the observation that dPSK elicits defense responses against B. cinerea in tomato but to a lower extent than PSK (Figures 1A to 1D). Similarly, the application of relatively high concentrations of dPSK also triggered a weak PSK response in Arabidopsis (Kutschmar et al., 2009). Notably, the silencing of Sl-PSKR1 impaired leaf immunity, whereas silencing Sl-PSKR2 had no significant effects. The cosilencing of the Sl-PSKR1/R2 genes did not lead to any further changes compared with the silencing of Sl-PSKR1 alone, confirming the limited role for Sl-PSKR2 in defenses against B. cinerea (Figures 2C to 2F; Supplemental Figures 3C and 5). Furthermore, B. cinerea infection induced a slight but significant increase in Sl-PSKR1 transcript abundance, but again such changes were not observed in the expression of the Sl-PSKR2 gene (Supplemental Figure 3C). Taken together, these observations suggest that Sl-PSKR1, rather than Sl-PSKR2, plays a crucial role in PSK perception for plant immunity to B. cinerea.
PSK-Induced Immunity Required Downstream Auxin Biosynthesis and Associated Defense Pathways
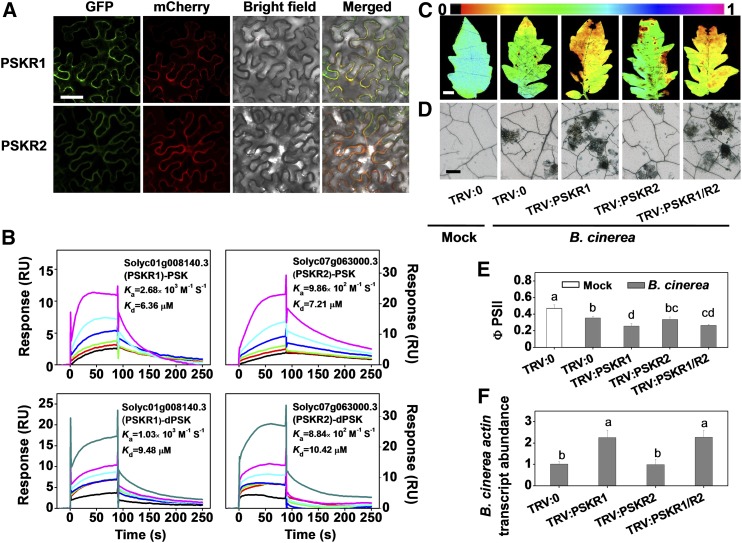
Considerable crosstalk between PSK signaling pathways and hormone pathways has been suggested to occur in the regulation of plant growth and stress responses (Eun et al., 2003; Mosher et al., 2013; Wu et al., 2015; Rodiuc et al., 2016). Therefore, we analyzed the relative levels of pathogen defense-related hormones (Glazebrook, 2005; Kazan and Manners, 2009) under mock- and B. cinerea-inoculated conditions. Significant increases in the levels of phytohormones, including SA, jasmonic acid (JA), ET, and indole-3-acetic acid (IAA), were observed in response to B. cinerea (Figure 3A). However, only IAA content was increased in response to the PSK treatment in both mock- and B. cinerea-inoculated plants. In contrast, the levels of other hormones were either constant or decreased in response to PSK application (Figure 3A). The changes in the transcript abundance of marker genes for the signaling-related pathways of these hormones are consistent with the observed changes in hormone contents (Figure 3B).
Figure 3.
PSK Promotes Endogenous IAA Accumulation, and PSK-Induced Tomato Immunity against B. cinerea Is Blocked in an Auxin Signaling Mutant.
(A) Effects of PSK application on endogenous leaf hormone content. Four-week-old tomato plants were treated with 10 μM PSK, dPSK, or water 12 h before B. cinerea inoculation, and leaf samples were collected at 0.5 dpi.
(B) Effects of PSK application on the transcript abundance of hormone signaling-related marker genes in tomato leaves at 0.5 dpi. The elicitor application was as described in (A). PR1b, SA-related gene; COI1, JA-related gene; ERF1, ET-related gene; ARF5, auxin-related gene.
(C) Effects of PSK application on trypan blue staining for cell death in hormone signaling-defective and control plants at 2 dpi. Tomato wild type, mutants, or transgenic lines were treated with 10 μM PSK or a water control 12 h before B. cinerea inoculation. The following tomato lines were used: SA accumulation-defective transgenic NahG and its wild-type line cv MM, the JA-signaling mutant jai1-1 and its wild-type line cv CM, the ET-signaling mutant Nr and its wild-type line cv Pearson, and the auxin signaling-insensitive mutant dgt and its wild-type line VFN8. Bar = 250 μm.
(D) Relative B. cinerea actin transcript abundance in infected hormone-related mutants in the presence or absence of PSK at 1 dpi. Elicitor application is as described in (C).
The results in (A), (B), and (D) are presented as mean values ± sd; n = 3 independent pooled samples with each sample being from two plants. Different letters indicate significant differences between treatments (P < 0.05, Tukey’s test). The above experiments were repeated three times with similar results.
The responses of mutants that are defective in hormone accumulation or signaling also provide evidence for the roles of phytohormones in PSK-induced immunity. For example, tomato NahG plants do not accumulate SA because of salicylate hydroxylase overexpression (Brading et al., 2000). The jai1-1 mutants of tomato are defective in CORONATINE-INSENSITIVE1 (COI1) and show impaired JA signaling (Li et al., 2004). Never ripe (Nr), which is mutated at the dominant tomato ethylene receptor NEVER RIPE, shows greatly reduced sensitivity to ethylene (Tieman et al., 2000). The diageotropica (dgt) mutants are defective in a type A cyclophilin protein, leading to suppression of the TIR1/AFB auxin receptor-induced signaling cascade (Lavy et al., 2012). The jai1-1, Nr, and dgt mutants exhibited increased susceptibility to B. cinerea (Figures 3C and 3D), whereas no changes in susceptibility were observed in the NahG plants. Strikingly, the application of PSK promoted plant defenses to the same extent in the wild-type and the NahG plants, as well as in the jai1-1 and Nr mutant lines. However, the PSK treatment had no effect on the susceptibility of the dgt plants to B. cinerea (Figures 3C and 3D).
We next verified the involvement of auxin pathways in PSK-induced immunity using the gene-silenced tomato plants described above. Impairing the expression of the PSK synthesis and signaling component genes, PSK3, PSK3L, TPST, and PSKR1, led to lower IAA content and to a higher level of disease susceptibility (Figures 4A and 4B; Supplemental Figure 6A). This increased susceptibility could be largely reversed by the exogenous application of 1-naphthaleneacetic acid (NAA) (Figure 4B). PSKR1 silencing compromised tomato immunity, an effect that could be complemented by the application of NAA but not PSK (Figures 4C and 4D; Supplemental Figure 6B). Interestingly, minor changes in the transcript abundance of genes encoding PSK signaling components were observed following NAA application under both mock- and B. cinerea-inoculated conditions (Figure 4E). These results strongly suggest that auxin functions downstream of PSK-PSKR1 signaling in tomato immunity against B. cinerea.
Figure 4.
Auxin Functions Downstream of PSK-PSKR1 Signaling in Tomato Immunity against B. cinerea.
(A) Changes in IAA content in PSK signaling component gene-silenced tomato plants. Leaf samples were collected at 0.5 dpi with B. cinerea.
(B) Effects of NAA application on leaf B. cinerea actin transcript abundance in target gene-silenced tomato plants at 1 dpi. PSK signaling component gene-silenced and TRV:0 control plants were treated with 50 nM NAA or water 12 h before B. cinerea inoculation.
(C) and (D) Immunity compromised by PSKR1 silencing was complemented by NAA but not by PSK.
(C) Trypan blue staining for cell death, as affected by PSKR1 silencing and application of PSK and NAA. Tomato PSKR1-silenced plants were treated with 10 µM PSK or 50 nM NAA 12 h before B. cinerea inoculation, and leaf samples were collected at 2 dpi. Bar = 250 µm.
(D) Relative B. cinerea actin transcript abundance in infected leaves at 1 dpi.
(E) The effects of 50 nM NAA application on the transcript abundance of PSK signaling component genes in tomato leaves under both mock- and B. cinerea-inoculated conditions. Samples were taken at 0.5 dpi.
The results in (A), (B), (D), and (E) are presented as mean values ± sd; n = 3 independent pooled samples with each sample being from two plants. Different letters indicate significant differences between treatments (P < 0.05, Tukey’s test). The experiments in (A), (B), and (E) were repeated three times, and others were repeated two times with similar results.
PSK Interaction with PSKR1 Triggered Cytosolic Ca2+ Signaling in the Immune Response
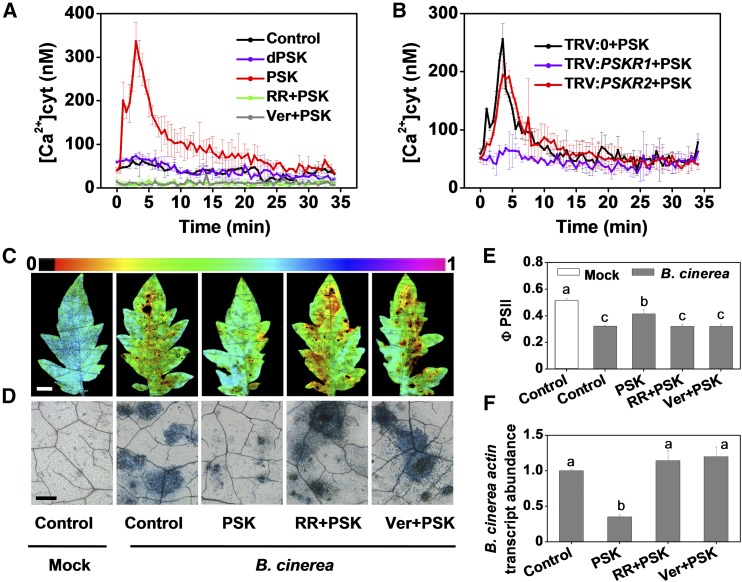
PSK-induced Ca2+ signaling was determined by monitoring the effect of PSK on cytosolic Ca2+ levels in tomato leaf discs that express the Ca2+ reporter protein, aequorin. Cytosolic Ca2+ concentrations [Ca2+] rapidly increased upon PSK application compared with the dPSK or control treatments (Figure 5A; Supplemental Figure 7A), suggesting that Ca2+ channels are activated by PSK in planta. In confirmation of this hypothesis, PSK did not cause cytosolic Ca2+ release in the presence of the Ca2+ channel inhibitors, ruthenium red (RR) or verapamil (Ver) (Figure 5A). PSK-induced immunity to B. cinerea was also compromised by these inhibitors (Figures 5C to 5F). Furthermore, PSK-mediated increase in cytosolic [Ca2+] was impaired in TRV:PSKR1 but not TRV:PSKR2 tomato plants (Figure 5B; Supplemental Figure 7B). Taken together, these observations strongly suggest that cytosolic Ca2+ signaling functions in PSK-PSKR1 binding-induced events. These findings provide compelling evidence of a signaling cascade elicited in tomato defense responses against B. cinerea.
Figure 5.
Cytosolic Ca2+ Elevation Is Induced and Is Required for PSK-Induced Tomato Immunity against B. cinerea.
(A) and (B) PSK-induced cytosolic Ca2+ elevation in leaves of aequorin-expressing tomato plants as affected by Ca2+ channel inhibitors (A) or PSKR1 or PSKR2 gene silencing (B). Tomato leaf discs were preincubated for 30 min with RR or Ver at 20 μM, and 10 μM PSK ligand was then added at time 0. The signals shown at 0.5-min intervals are mean values ± sd (n = 10 to 12 leaf discs). In (A), 50 total leaf discs obtained from at least five plants were used for experiment and each treatment had 10 leaf discs. In (B), 12 leaf discs obtained from five independent plants served as one treatment.
(C) to (F) Effects of Ca2+ channel inhibitors on innate immunity against B. cinerea in tomato. Four-week-old tomato plants were treated with 10 µM PSK, 20 μM each Ca2+ channel inhibitor, or water 12 h before B. cinerea inoculation.
(C) Representative chlorophyll fluorescence imaging of ΦPSII at 2 dpi. Bar = 1 cm.
(D) Representative images of trypan blue staining for cell death in tomato leaves at 2 dpi. Bar = 250 µm.
(E) Quantification of ΦPSII at 2 dpi.
(F) Relative B. cinerea actin transcript abundance in infected leaves at 1 dpi. The results in (E) and (F) are presented as mean values ± sd; n = 3 leaves from different plants. Different letters indicate significant differences between treatments (P < 0.05, Tukey’s test).
The above experiments were repeated three times with similar results.
Calmodulin Binding to the Auxin Biosynthetic Protein YUCs Mediated PSK-Induced Immunity against B. cinerea
Having established that both auxin and cytosolic Ca2+ signaling are required for PSK-induced immunity against B. cinerea, we explored the connection between these two signaling pathways further. Calmodulins (CaMs) are Ca2+ binding proteins whose affinities to target proteins, and hence activities, are modified upon Ca2+ binding. This process translates changes in local [Ca2+] into specific physiological responses (McCormack et al., 2005; Hartmann et al., 2014). In tomato, there are six homologous CaM genes encoding a total of four CaM protein isoforms. CaM3, CaM4, and CaM5 are identical in amino acid sequence. CaM1, CaM2, and CaM6 are also highly conserved and share 98%, 99%, and 91% amino acid identity with CaM3/4/5, respectively (Supplemental Figure 8A). Of these, CaM2 exhibits the highest transcript levels in tomato leaves and other organs (Zhao et al., 2013b). Plant YUCCA (YUC) genes encode flavin-containing monooxygenases that are the rate-limiting enzymes in the typical two-step pathway of auxin biosynthesis, which might be required for auxin-modulated pathogen defense responses (Dai et al., 2013; Hentrich et al., 2013). Based on the Arabidopsis YUCs amino acid sequences, nine YUC homologs were identified in tomato (Supplemental Figure 8B and Supplemental File 3). Three of these, i.e., YUC2, YUC8, and YUC9, are barely detectable at the transcriptional level in leaves (Supplemental Figure 8C). All the other six tomato YUC isoforms are predicted to contain putative CaM binding motifs. The four tomato CaM genes (CaM1, CaM2, CaM3, and CaM6) and the six tomato YUC isoforms (YUC1, YUC3, YUC4, YUC5, YUC6, and YUC7) were cloned and expressed in N. benthamiana and used in bimolecular fluorescence complementation (BiFC) assays (Figure 6A; Supplemental Figure 9A).
Figure 6.
Tomato CaMs Bind to Auxin Biosynthetic Protein YUCs.
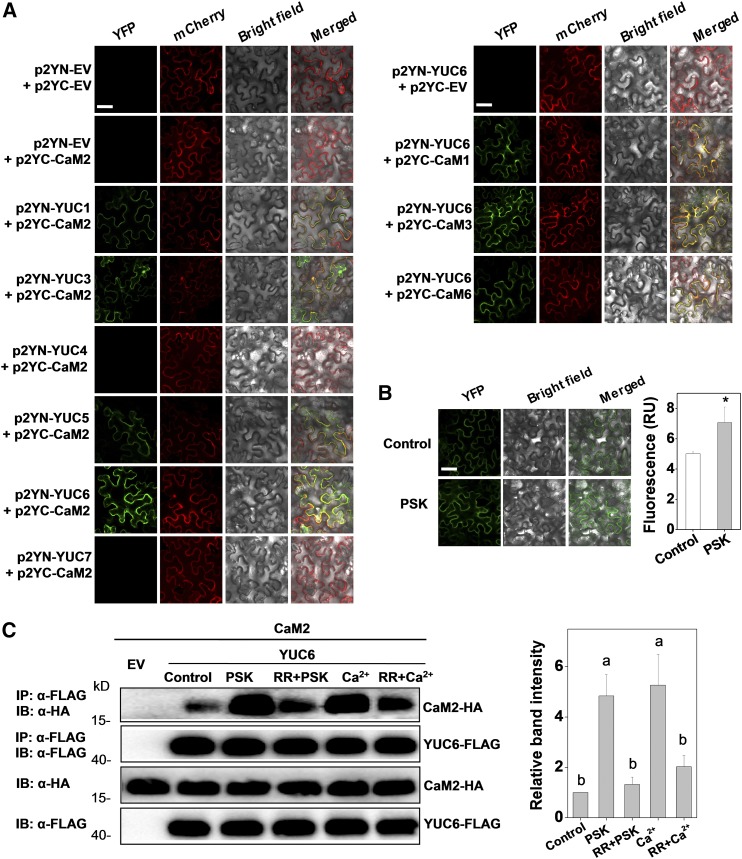
(A) BiFC analyses of the binding between CaM2 and YUCs (left set of panels) and between CaMs and YUC6 (right set of panels). Both spliced YFP constructs and FLS2-mCherry (marker for plasma membrane localization) plasmids were transiently coexpressed in N. benthamiana leaves. The YFP and mCherry signals were visualized under confocal microscopy 48 h after infiltration. Bar = 50 µm.
(B) Changes in the BiFC fluorescence signal between p2YC-CaM2 and p2YN-YUC6 with or without application of PSK (10 µM) for 2 h. Bar = 50 µm. The fluorescence signal intensity from three independent repeats was quantified and the data are shown as means ± sd (n = 3 leaves from different plants). Asterisks indicate a significant effect of PSK application (P < 0.05, Tukey’s test).
(C) Co-IP analysis of the association between HA-tagged CaM2 and FLAG-tagged YUC6 with or without application of 10 µM PSK, 20 µM CaCl2, and 20 μM Ca2+ channel inhibitor RR for 2 h. Total proteins were extracted from leaves transiently expressing the CaM2-HA, YUC6-FLAG construct alone or their combinations after 48 h of infiltration. The extracted proteins were immunoprecipitated with an anti-FLAG antibody and the presence of CaM2-HA and YUC6-FLAG in the immune complex was determined by immunoblot (IB) with the indicated antibody. The co-IP band intensity (top) from three independent repeats was quantified by Image J software. The data are shown as mean ± sd (n = 3 leaves from different plants). Different letters indicate significant differences between treatments (P < 0.05, Tukey’s test).
The experiments in (A) and (B) were repeated three times, and experiments in (C) were repeated two times with similar results.
Taking the most abundant CaM2 isoform (Zhao et al., 2013b) as the target protein, BiFC analysis demonstrated that the YUC1, YUC3, YUC5, and YUC6 proteins, but not the YUC4 or YUC7 proteins, show fluorescence signals when coexpressed with CaM2 (Figure 6A). It should be noted that the expression of YUC7 is relatively low at both transcriptional and protein levels (Supplemental Figures 8C and 9A). Thus, it is still unknown whether YUC7 could interact with CaM2, and further studies are required to elucidate this issue. Furthermore, among these YUC genes, YUC6 transcript abundance was significantly induced upon B. cinerea infection (Supplemental Figure 8C). Therefore, this gene product was used as a target protein to further examine interactions with the other three tomato CaM protein isoforms (CaM1, CaM3, and CaM6). As shown in Figure 6A, CaM1, CaM3, and CaM6 were also able to bind to YUC6, even though the signal was a little weaker for CaM6. The CaM- and YUC-dependent fluorescence signals largely colocalized with those of FLS2-mCherry in the plasma membrane, where YUC6 was specifically localized (Figure 6A; Supplemental Figure 9B). Strikingly, the fluorescence signals were strengthened under PSK-treated conditions in the BiFC assays (Figure 6B). The interactions between the CaM2 and YUC6 proteins were further verified using coimmunoprecipitation (co-IP) following expression of these two proteins with different tags in N. benthamiana (Figure 6C). Intriguingly, CaM2-YUC6 binding was increased by the PSK and CaCl2 treatments. In contrast, binding was weakened in the presence of the Ca2+ channel inhibitor RR. These findings suggest that PSK modulates CaM2 in such a way as to enhance the binding coefficient toward YUC6.
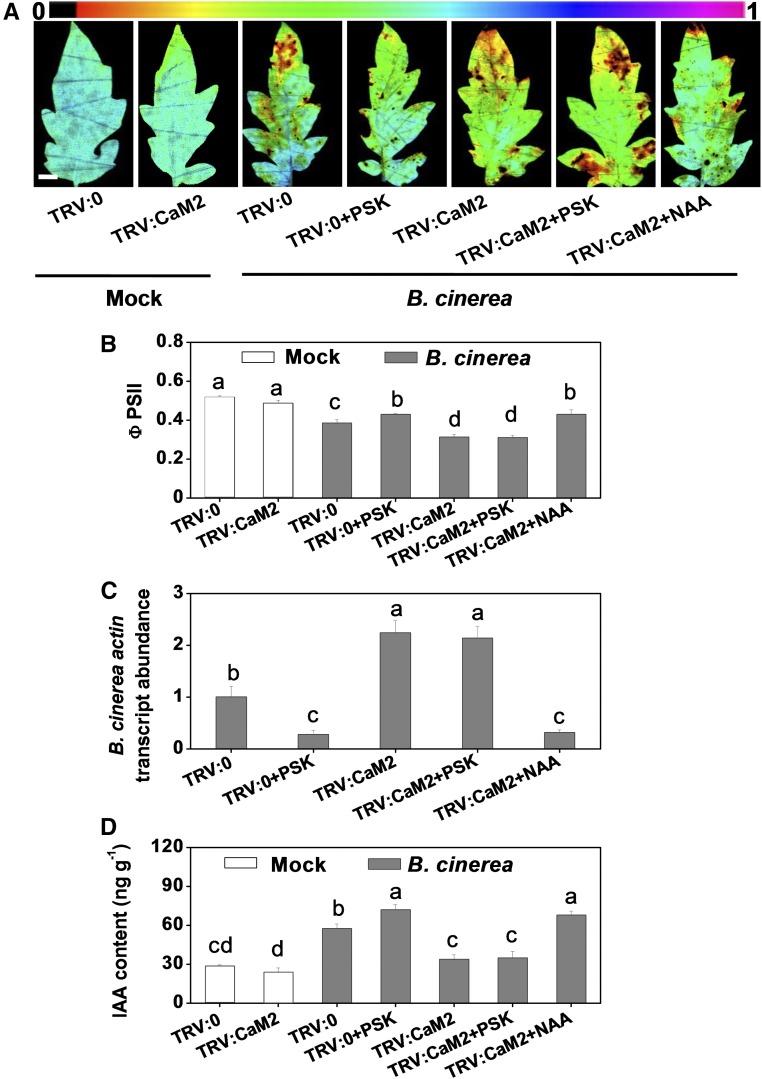
We further investigated the contribution of the CaMs-YUCs interaction to PSK-induced immunity. We constructed a TRV:CaM2 vector based on the targets of the CaM homologs (Supplemental Figure 10). This vector efficiently silenced the expression of most of the CaM genes (Supplemental Figure 11). The TRV:CaM2 plants showed weaker defenses toward B. cinerea infection than the TRV:0 controls. The lower level of immunity observed in these plants was accompanied by lower IAA contents (Figures 7A to 7D). Furthermore, CaM2 silencing compromised PSK-induced immunity and IAA accumulation, an effect that could be complemented with NAA but not PSK (Figures 7A to 7D). Thus, PSK-induced IAA accumulation and immunity were largely dependent on the functions of the Ca2+ sensor CaMs.
Figure 7.
CaM2 Silencing Compromises the PSK-Induced Immunity and IAA Accumulation.
(A) Representative chlorophyll fluorescence imaging of ΦPSII as affected by CaM2 silencing and application of PSK and NAA. Tomato CaM2-silenced plants were treated with 10 µM PSK or 50 nM NAA 12 h before B. cinerea inoculation, and leaf samples were collected at 2 dpi. Bar = 1 cm.
(B) Quantification of ΦPSII at 2 dpi.
(C) Relative B. cinerea actin transcript abundance in infected leaves at 1 dpi.
(D) Changes in IAA content in CaM2-silenced tomato plants at 0.5 dpi, as affected by exogenous PSK and NAA application.
The results in (B) to (D) are presented as mean values ± sd; n = 3 leaves from different plants in (B); n = 3 independent pooled samples with each from two plants in (C) and (D). Different letters indicate significant differences between treatments (P < 0.05, Tukey’s test). The above experiments were repeated three times with similar results.
DISCUSSION
Peptide signaling pathways have been well studied in animals and shown to fulfill many important functions. In contrast, peptide signaling underlying plant responses to environmental stimuli or developmental triggers has received much less attention and remains poorly characterized. Here, we presented several lines of evidence demonstrating that the tomato PSK peptide acts as an immunity-regulating signal to counteract the necrotrophic pathogen B. cinerea. The data not only extended our understanding of the PSK receptor family in tomato but also showed that perception of PSK by PSKR1 initiated a cytosolic Ca2+ signaling cascade leading to the binding of CaMs to YUCs and that this cascade was associated with auxin-dependent immunity against B. cinerea. We presented a model in which the Ca2+ signal triggered by the PSK signaling peptide modulates auxin biosynthesis, leading to innate immune responses to this major plant pathogen (Figure 8). We present several lines of evidence below in support of this conclusion.
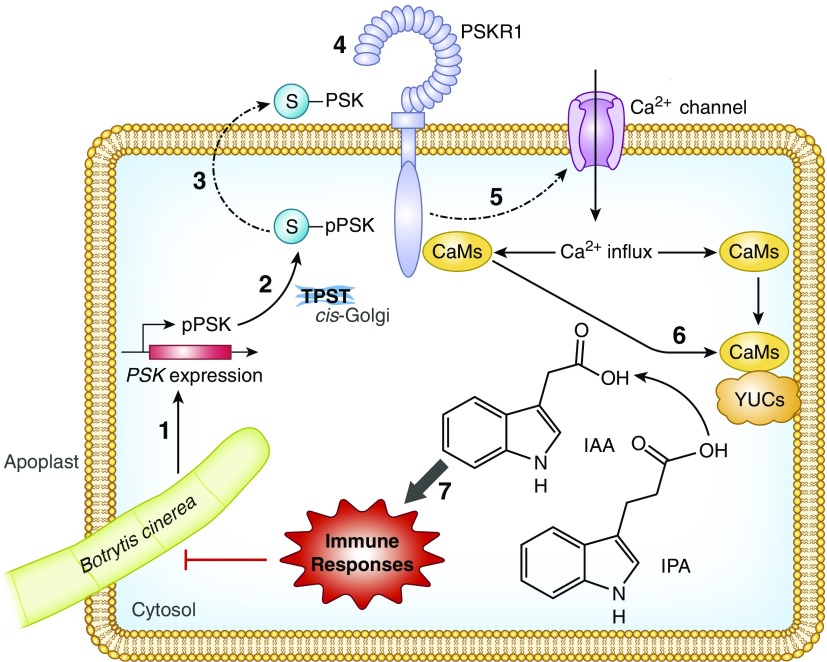
Figure 8.
A Working Model of PSK-Induced Immunity against B. cinerea in Tomato Plants.
PSK is a signaling peptide that acts as a DAMP. Production of PSK precursors (pPSKs) and protein processing are activated upon B. cinerea inoculation (1). pPSKs undergo tyrosine sulfation (blue S) by a TPST in the cis-Golgi (2) followed by proteolytic cleavage in the apoplast (3). At the apoplast, the mature PSK peptide is mainly perceived by its receptor, PSKR1 (4), which transduces the signal into the cytoplasm by initiating cytosolic Ca2+ influx (5). This transient cytosolic Ca2+ signal is further transduced to CaMs which bind to YUCs (6), promoting auxin biosynthesis and associated signaling in order to combat B. cinerea infection (7). pPSK, phytosulfokine precursor; IPA, indole-3-propionic acid; PSKR1, PSK receptor.
First, the expression of several tomato PSK precursor genes was induced in response to B. cinerea inoculation (Figure 1E). The application of the synthetic PSK peptide resulted in enhanced defenses against B. cinerea relative to controls and dPSK treatments (Figures 1A to 1D). Moreover, silencing of the peptide precursor genes (PSK3 and PSK3L), the tyrosine sulfation gene (TPST), and the receptor gene (PSKR1) significantly impaired leaf defense responses, leading to enhanced disease symptoms (Figures 1F to 1I and 2C to 2F). These results supported the conclusion that the PSK signal peptide was essential for immunity to the necrotrophic pathogen B. cinerea in tomato. The positive contribution of PSK signaling to resistance against B. cinerea has not been reported previously, although PSK has been implicated in defenses against another necrotrophic fungal pathogen A. brassicicola in Arabidopsis (Mosher et al., 2013).
Second, the characteristics of the PSK receptor in tomato were described for the first time. LRR-RLK Sl-PSKR1 and Sl-PSKR2 share a high sequence homologies and structural identities to At-PSKR1/R2 (Matsubayashi et al., 2006; Amano et al., 2007) and to Dc-PSKR (Matsubayashi et al., 2002) (Supplemental Figures 3A, 3B, and 4). SPR analysis demonstrated that PSK has a stronger Kd and higher Ka with respect to the recombinant extra cellular region of the Sl-SPKR1 than to Sl-SPKR2 (Figure 2B). A recent study demonstrated that PSK stabilizes the Dc-PSKR island domain, which in turn recruits a somatic embryogenesis receptor-like kinase (SERK) to form a stable PSKR-SERK complex, leading to allosteric activation of Dc-PSKR (Wang et al., 2015). The affinity values obtained in this study were in the similar micromolar range to those reported for the PSK-Dc-PSKR interaction that was measured by microscale thermophoresis method (Wang et al., 2015). However, they were weaker than the [3H]PSK-PSKR interactions measured using ligand-based affinity chromatography of microsomal fractions from either carrot (Matsubayashi et al., 2002) or Arabidopsis (Matsubayashi et al., 2006). The discrepancies in reported affinities likely indicate that the cellular environment provides a more favorable medium for interactions between PSK and its receptor proteins compared with that used in in vitro assays for studies on recombinant proteins. Even though Sl-PSKR2 interacted with PSK and was shown to be involved in PSK perception (Figure 2B), the gene silencing approaches used in this study confirmed that only Sl-PSKR1 was the major transducer of the PSK signal in the regulation of B. cinerea defenses, as silencing of Sl-PSKR2 did not lead to any significant effects on plant responses to B. cinerea (Figures 2C to 2F). Similarly, Arabidopsis loss-of-function and gain-of-function studies demonstrate that PSK perception requires At-PSKR1 for the regulation of root growth, while At-PSKR2 has a more marginal role (Amano et al., 2007). In accordance with this study, mutants lacking At-PSKR2 show similar responses to wild-type Arabidopsis with regard to pathogens such as A. brassicicola and P. syringae (Igarashi et al., 2012; Mosher et al., 2013). Therefore, Sl-PSKR2 functions may be largely inactive or below the level of detection in relation to plant immunity.
Third, the data presented here demonstrated that auxin biosynthesis and associated signaling were required for PSK-induced immunity against B. cinerea. Within the context of phytohormone-mediated immunity, SA-dependent resistance is effective largely against biotrophs and hemibiotrophs, whereas JA- and ET-mediated responses are predominantly effective against necrotrophs (Glazebrook, 2005). While auxin is a classical growth-regulating hormone in plants, it has been shown to positively modulate plant immunity in response to necrotrophic pathogens (Llorente et al., 2008; Kazan and Manners, 2009; Qi et al., 2012). SA has been suggested to be linked to PSK signaling, attenuating responses to the hemibiotrophic P. syringae and pathogen-associated molecular patterns (Igarashi et al., 2012; Mosher et al., 2013; Rodiuc et al., 2016). However, few studies have investigated PSK-induced phytohormone modulation in plant-necrotrophic pathogen interactions. The observed susceptibility phenotype of the Arabidopsis pskr1 mutant to the necrotrophic pathogen A. brassicicola lead us to speculate that SA, JA, or ET pathways may be involved in defense signaling (Mosher et al., 2013). The data presented here demonstrated the absence of significant changes in SA or JA levels or in the transcript accumulation of marker genes related to the signaling pathways of these phytohormones following PSK application (Figures 3A and 3B). A previous transcriptome study using Arabidopsis gain-of-function overexpressing lines and loss-of-function mutants also report that signaling through PSKR1 does not significantly affect SA- or JA-related gene expression (Rodiuc et al., 2016). In addition, ET signaling is important in plant immunity against B. cinerea. Pretreatments with irreversible ethylene perception inhibitors result in significant increases in pathogen susceptibility in tomato (Díaz et al., 2002). However, ET generation was suppressed by PSK application in this study (Figure 3A), a finding that agrees well with a previous study in Arabidopsis (Wu et al., 2015). Moreover, PSK-induced immunity was not changed in the NahG, jai1-1, or Nr mutants (Figures 3C and 3D). Taken together, these findings suggested that SA, JA, and ET signaling were not required for PSK-induced immunity against B. cinerea in tomato.
Based on the observations reported here, we proposed that auxin signaling functions downstream of PSK signaling, leading to immunity against B. cinerea in tomato. This conclusion is based on several lines of evidence: (1) The application of PSK increased auxin levels and the abundance of transcripts associated with auxin signaling under both mock- and B. cinerea-inoculated conditions (Figures 3A and 3B). (2) PSK-induced immunity against B. cinerea was compromised in the auxin signaling mutant dgt compared with the wild type (Figures 3C and 3D). (3) Silencing of the tomato PSK3, PSK3L, TPST, and PSKR1 genes substantially decreased B. cinerea-induced IAA accumulation (Figure 4A). Silencing of the PSK-related genes (PSK3, PSK3L, TPST, and PSKR1) induced susceptibility to B. cinerea in a manner that was effectively complemented by the application of NAA without any effect on the PSK transcript abundance (Figures 4B and 4E). (4) PSKR1 silencing-compromised immunity was complemented by the application of NAA but not by PSK (Figures 4C and 4D). Auxin is involved in most aspects of plant growth and development (Benjamins and Scheres, 2008) and is also required for PSK-induced cell proliferation in asparagus (Asparagus officinalis) (Matsubayashi et al., 1999) and carrot (Eun et al., 2003). In agreement with this study, auxin signaling mutants axr1, axr2, and axr6 in Arabidopsis that are impaired in the auxin-stimulated SCF (Skp1-Cullin-F-box) ubiquitination pathway, exhibited increased susceptibility to B. cinerea and some other necrotrophic fungi (Llorente et al., 2008). The asa1-1 and cyp79b2/b3 mutants in Arabidopsis, which are defective in auxin biosynthesis, were more susceptible to infection by the necrotrophic pathogen A. brassicicola than were wild-type plants (Qi et al., 2012). Therefore, the activation of downstream auxin defense pathways may underlie the effects of PSK on growth, enhancing growth while promoting immunity against necrotrophic pathogens, including B. cinerea.
Further proof in support of this molecular signaling pathway triggered by PSK is the demonstration that cytosolic Ca2+ and the binding of sensor CaMs to auxin biosynthetic proteins were crucial components of the Ca2+-activated auxin signaling pathway that combats B. cinerea infection. The generation of PSK- and PSKR1-dependent Ca2+ accumulation contributed to the innate response to B. cinerea (Figure 5). CaMs are prototypical calcium sensors that translate local changes in [Ca2+] into physiological responses (McCormack et al., 2005). Until now, the role of cytosolic Ca2+ signaling in regulating auxin biosynthesis has been unclear; however, this study has shown that tomato CaMs could regulate auxin biosynthesis by binding to the rate-limiting YUCs in the auxin biosynthesis pathway (Figure 6), providing a potential mechanism by which cytosolic Ca2+ signaling modulates auxin synthesis. Most importantly, the interaction between CaM2 and YUCs was shown to be responsive to PSK (Figures 6B and 6C), a finding that was also essential to PSK-induced IAA generation and immunity against B. cinerea (Figure 7). In accordance with this observation, previous studies have revealed that CaM2 overexpression enhanced B. cinerea defenses in tomato fruit (Peng et al., 2014).
In addition, the PSK-induced cytosolic Ca2+ wave and the elicitation of immunity were completely inhibited by the addition of calcium channels blockers (Figure 5). This suggests that the observed increases in cytosolic [Ca2+] may be generated through the PSKR1 GC activity-induced opening of the cyclic nucleotide-gated channel (Ladwig et al., 2015). It should be noted that tomato CaMs not only act as Ca2+ sensors that bind YUCs, but they can also bind directly to the PSKR1 receptor at the kinase subdomain (Hartmann et al., 2014; Fischer et al., 2017). Binding of At-PSKR1 to CaMs within the kinase subdomain is required for At-PSKR1 functioning in the regulation of growth responses of Arabidopsis roots (Hartmann et al., 2014). The data presented here do not allow any conclusions to be drawn with regard to whether Sl-PSKR1 is directly involved in Ca2+ signal generation or whether this receptor acts as an indirect sensor. Interestingly, prior work with Arabidopsis demonstrated that supplying physiologically relevant concentrations of calcium inhibited PSKR1 kinase activity, while enhancing its GC activity in vitro. These findings suggest that calcium acts as a PSKR1 bimodal switch between the overlapping kinase and GC activities (Muleya et al., 2014). A similar protein structure with dual kinase/GC activities has previously been reported for At-BRI1 (Kwezi et al., 2007) and for Arabidopsis wall-associated kinase-like 10 (Meier et al., 2010), with similar predictions for other kinases. Based on these observations, we concluded that cytosolic Ca2+ levels and CaMs, triggered by PSK signaling, initiated the finely tuned regulation of PSKR activities that were able to respond to different stimuli.
In conclusion, the data presented here show that the signal transduction cascade by which PSK triggers immunity to the necrotrophic pathogen B. cinerea requires tomato PSKR1. This in turn triggers cytosolic Ca2+ signaling and leads to increases in auxin synthesis and associated auxin-dependent immunity in tomato (Figure 8). Since orthologs of PSK precursors have been identified across the plant kingdom, it will be interesting to investigate whether the perception of PSK ligands by their receptors in other species uses a similar mechanism for regulating immunity to necrotrophic pathogens, as well as for the control of plant growth. Manipulation of peptide-induced defenses is an attractive disease management strategy that could potentially be used to enhance disease resistance in many diverse plant species.
METHODS
Plant Materials and VIGS
The tomato (Solanum lycopersicum) lines used in most of the studies were mainly in the Moneymaker (MM) wild-type background. However, some studies were also conducted using several wild-type tomato lines that were used as controls, as appropriate, depending on the mutant background. Specifically, tomato seeds of the JA-signaling mutant jai1-1 and its wild-type progenitor Castlemart (CM) were kindly provided by C. Li (Chinese Academy of Sciences, Beijing, China). Homozygous jai1-1 seedlings were selected from F2 populations as described previously (Li et al., 2004). The ET-signaling mutant Nr and its wild-type line cv Pearson as well as the auxin signaling insensitive mutant dgt and its isogenic wild-type line cv VFN8 were obtained from the Tomato Genetics Resource Center (University of California, Davis, CA). Seeds of the NahG transgenic line (in which overexpression of salicylate hydroxylase abolishes SA accumulation) and its wild-type control line MM were from the laboratory of J.D.G. Jones (Sainsbury Laboratory, Norwich, UK). The MM line expressing aequorin to monitor the effects of PSK treatment on cytosolic Ca2+ concentrations was obtained from Gerald A. Berkowitz (University of Connecticut). Tomato seeds were sown in sterilized soil in 72-well trays and germinated at 25°C. After a 2-week germination period, the seedlings were transplanted into plastic pots (diameter, 10.5 cm; depth, 17.5 cm; one plant per pot) containing soil and perlite in controlled-environment growth chambers (Conviron). The PPFD was 500 μmol m−2 s−1, the photoperiod was 14/10 h (day/night), the day/night air temperature was 25°/20°C, and the relative humidity was 88%.
VIGS was performed by infiltration of fully expanded cotyledons of 10-d-old tomato (MM) seedlings with TRV vectors using a mix of pTRV1 and pTRV2. PCR-amplified cDNA fragments of the target genes were cloned into pTRV2. In the case of cosilencing of tomato PSKR1 and PSKR2, each fragment was cloned into pTRV2 using a different multiple cloning site. The resulting plasmid was transformed into Agrobacterium tumefaciens GV3101. The empty pTRV2 vector was used as a negative control. The success of the VIGS protocol was evaluated according to the method of Liu et al. (2002) using the expression of the phytoene desaturase (PDS) gene, which causes photobleaching, as a marker for silencing in tomato. The infiltrated plants were grown under a 14-h photoperiod at 22°C. Three to four weeks later, transcript abundance of target genes was analyzed by qRT-PCR in each plant. Samples from the uppermost one or two fully expanded leaves were collected by punching out leaf discs. Only plants showing significant silencing were used for experiments. The primers used for VIGS cloning and the qRT-PCR assay are listed in Supplemental Table 1. The nucleotide sequence alignment of homologous genes based on a tomato database (available at http://solgenomics.net/) and an online VIGS tool (available at http://vigs.solgenomics.net/) revealed no significant identical sequence between VIGS targets and nontarget homologous genes (see Supplemental Figures 2, 5, and 10).
Approximately 5 to 10 4-week-old tomato plants at about the five-leaf stage were used for each treatment in all the VIGS or non-VIGS experiments. Except where noted otherwise, samples were randomly collected from lateral leaflets from the uppermost one to two fully expanded leaves. For the VIGS experiments, the transcript levels of target and nontarget homologous genes were determined using the same samples as those used for the Botrytis cinerea actin transcript assays (see Supplemental Figures 1C, 3C, 6A, 6B, and 11). As for the intact leaflets used for the disease symptom assays by an Imaging-PAM chlorophyll fluorometer, samples were taken from the corresponding opposite leaflets for the assays of silencing efficiency and B. cinerea actin transcript.
Pathogens, Elicitor Treatment, and Disease Symptom Assays
Tomato leaves were inoculated with B. cinerea (BO5-10 strain) suspensions at a density of 2 × 105 spores mL−1, and mock inoculations were performed using media buffer (Zhang et al., 2015). The inoculation was performed by spraying the inoculum suspension on the whole leaf portion. For the elicitor treatment, unless otherwise noted, plant leaves were sprayed with water or fresh solutions of 10 μM PSK (Iris Biotech), 10 μM dPSK (ChinaPeptides), 50 nM NAA (Sigma-Aldrich), 20 μM RR (Sigma-Aldrich), or 20 μM Ver (Sigma-Aldrich), individually or in combination. For experiments combining elicitor treatment and B. cinerea inoculation, the leaves were inoculated with B. cinerea 12 h after elicitor pretreatment.
After pathogen inoculation, disease symptoms were assessed by quantifying B. cinerea actin mRNA accumulation by qRT-PCR (Zhang et al., 2015), trypan blue staining (Bai et al., 2012), or by analysis of chlorophyll fluorescence with an Imaging-PAM chlorophyll fluorometer (IMAG-MAXI; Heinz Walz). The quantum efficiency of light-adapted leaves (ΦPSII) was calculated as Fm′–F/Fm′ (Genty et al., 1989).
RNA Isolation, Transcript Analysis, and qRT-PCR
RNA was extracted using an RNA extraction kit (Axygen) followed by DNase digestion (Promega) and reverse transcribed using a ReverTra Ace quantitative (qPCR) RT kit (Toyobo), according to the manufacturer’s instructions. qRT-PCR was performed using the LightCycler 480 real-time PCR system (Roche Diagnostics). Each reaction (20 μL) consisted of 10 μL of SYBR Green PCR Master Mix, 8.2 μL of water, 1 μL of cDNA, and 0.4 μL each of forward and reverse primers. PCR was performed using 35 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C. The specific primers employed for target genes and internal control actin gene are described in Supplemental Table 1.
PSK Receptor Recombinant Protein Expression, Purification, and Analysis of PSK Binding Activity
Protein expression and purification were performed according to previous work (Manohar et al., 2015). Briefly, the extracellular regions of the potential tomato PSKR protein-encoding genes, PSKR1 and PSKR2, were cloned and inserted into the pET28a vector to enable the expression of recombinant proteins with an N-terminal His6 tag. The error-free clones were confirmed by sequencing and then transformed into Escherichia coli strain BL-21 (DE3). Expression of the proteins was induced by IPTG, and the proteins were purified by affinity chromatography on a Ni-NTA His binding resin (Novagen).
SPR analysis of the PSK binding activity was performed with a Biacore T200 instrument (GE Healthcare) with a CM5 sensor chip (GE Healthcare). Activation, deactivation, and preparation of the coupled flow cell as well as the ligand binding assay were performed essentially as described previously (Song et al., 2014). Briefly, the recombinant PSKR1- and PSKR2-encoding proteins were immobilized in parallel-flow channels of the CM5 sensor chip using an amine coupling kit (GE Healthcare). To test PSK binding to potential receptors, serial concentrations of PSK or dPSK (diluted in 0.01 M PBS, pH 7.4) were injected into the flow system (Song et al., 2014). Experiments were conducted with PBS (pH 7.4) as the running buffer, and the analyte was injected at a flow rate of 30 μL/min. The association time was 90 s, the dissociation time was 180 s, and the chip was regenerated for 30 s with 50 mM NaOH. Equilibration of the chip with the running buffer for another 60 s was performed before the next injection. The kinetic constants of binding were obtained using a 1:1 Langmuir binding model in BIA evaluation software.
Measurement of Phytohormone Content
The phytohormones SA, JA, and IAA were extracted from tomato leaves as described previously with minor modifications (Durgbanshi et al., 2005; Wu et al., 2007). Briefly, frozen tomato leaves (100 mg) were homogenized with 1 mL of ethyl acetate spiked with D5-JA, D5-IAA, and D4-SA (OlChemIm) as internal standards to a final concentration of 100 ng mL−1. After shaking for 12 h in the dark at 4°C, the homogenate was centrifuged at 18,000g for 10 min at 4°C. The supernatant was collected, and the pellet was extracted again with 1 mL of ethyl acetate, shaken for 2 h, and centrifuged at 18,000g for 10 min at 4°C. The supernatants from the two centrifugation steps were combined and evaporated to dryness under N2 gas. The residue was resuspended in 0.5 mL of 70% (v/v) methanol and centrifuged at 18,000g for 2 min at 4°C. The final supernatants were pipetted into glass vials and then analyzed by HPLC-MS/MS (Agilent 6460; Agilent Technologies) using the same method described previously (Wang et al., 2016).
Ethylene production was measured as described previously (Yin et al., 2012). Briefly, tomato seedlings were sealed in 500-mL rubber-topped flasks for 1 h at 20°C, and then 1 mL of head-space gas was removed and injected into a gas chromatograph (Agilent 6890N; Agilent Technologies) fitted with a Proapack-Q column. The temperatures of the injector, detector, and oven were 140, 230, and 100°C, respectively.
Measurement of Cytosolic Ca2+ Concentration
Cytosolic Ca2+ levels were evaluated as described previously (Tanaka et al., 2010; Zhao et al., 2013a) with some modifications using aequorin-expressing MM tomato lines. Briefly, leaf discs (0.3 cm diameter) collected from different plants were transferred individually to a 96-well microplate and incubated overnight in 50 μL of reconstitution buffer in the dark to allow binding between coelenterazine-h and aequorin for Ca2+-dependent chemiluminescent assays. The reconstitution buffer contained 12.5 μM coelenterazine-h (Sigma-Aldrich), 1 mM KCl, 1 mM CaCl2, and 10 mM MgCl2, adjusted to pH 5.7 with Tris base. After overnight incubation, 50 μL of PSK solution or water control was added to the wells. RR and Ver were added to the leaves 30 min before adding PSK. Luminescence was measured using a Microplate Luminometer (Titertek Berthold) with a 0.5-min interval reading time over a period of 35 min. At the end of each experiment, the remaining aequorin was discharged by the addition of an equal volume of solution containing 2 M CaCl2 in 30% (v/v) ethanol. Luminescence values were calibrated as calcium concentrations according to previous study (Knight et al., 1996).
Transient Protein Expression and Protein-Protein Interaction Assays
Subcellular localization-associated genes were cloned under control of the 35S CaMV promoter using pCAMBIA2300 (CAMBIA) vectors with a GFP tag at the C-terminus. BiFC assay was performed as previously described (Yang et al., 2007). BiFC vectors p2YC and p2YN were generously provided by C. Mao (Zhejiang University, China), and p2YC-CAMs and p2YN-YUCs were constructed to be fused with C-terminal hemagglutinin (HA) tag upstream of the YFP sequences. Meanwhile, pCAMBIA2300-35S:FLS2-mCherry was coexpressed as membrane location marker. Agrobacterium-mediated transient expression in Nicotiana benthamiana leaves was performed as described (Liao et al., 2015). An Agrobacterium suspension carrying a given construct was infiltrated into young, fully expanded N. benthamiana leaves using a needleless syringe. At 48 h after infiltration, subcellular localization of GFP, YFP, or mCherry-tagged proteins in leaves was determined with a Zeiss LSM 780 confocal microscope, excitation/emission wavelengths were 488 nm/500 to 530 nm for GFP, 514 nm/520 to 560 nm for YFP, and 561 nm/580 to 620 nm for mCherry.
Co-IP was performed as in previous studies with minor modifications (Li et al., 2014). Binary vector pCAMBIA2300-35S:CaM2-HA and pCAMBIA2300-35S:YUC6-FLAG were expressed in N. benthamiana leaves for ∼48 h by Agrobacterium-mediated transformation with empty vector as a negative control and then infiltrated with 10 μM PSK or other chemicals for 2 h. Each set of FLAG-tagged soluble protein immunoprecipitation was operated in 1 mL co-IP buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% Triton, 1× protease inhibitor [Sigma-Aldrich], 2.5 μL 0.4 M DTT, 2 μL 1 M NaF, and 2 μL 1 M Na3VO3 added before using) with 10 μL of α-FLAG agarose beads (Sigma-Aldrich). After 3 h gently shaking at 4°C, the agarose beads were washed four times with co-IP washing buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, and 0.1% Triton) and once with 50 mM Tris-HCl (pH 7.5). The collected agarose beads were used for analyzing the immunoprecipitated proteins by immunoblot with α-FLAG (Sigma-Aldrich; catalog no. F1804) or α-HA (Roche; catalog no. 11867423001) antibodies at a 1:10,000 dilution. In addition, 50-μL samples in co-IP buffer were prepared as a protein loading control before adding agarose beads. Primers used for cloning into binary vectors are listed in Supplemental Table 1.
Statistical Analysis
At least three independent biological replicates were sampled for each determination. Unless otherwise stated, each biological replicate consisted of an independent sample that was pooled of two leaves, each taken from a different plant. The experiments were independently performed two or three times. The data obtained were subjected to analysis of variance using SAS software, version 8 (SAS Institute), and means were compared using Tukey’s test at the 5% level.
Phylogenetic Analysis
Amino acid sequence alignment and phylogenetic tree construction were performed with MEGA version 5.05. A consensus neighbor-joining tree was obtained from 1000 bootstrap replicates of aligned sequences. The percentage at branch points represents the posterior probabilities of amino acid sequences.
Accession Numbers
Sequence data from this article can be found in the TAIR/GenBank database or the Sol genomics network (http://solgenomics.net/) database under the following accession numbers: Sl-PSK1 (Solyc09g009130), Sl-PSK2 (Solyc11g066880), Sl-PSK3 (Solyc02g092110), Sl-PSK3L (Solyc02g092120), Sl-PSK4 (Solyc01g106830), Sl-PSK5 (Solyc10g083580), Sl-PSK6 (Solyc06g074540), Sl-PSK7 (Solyc04g077580), Sl-TPST (Solyc11g069520), Sl-PSKR1 (Solyc01g008140), Sl-PSKR2 (Solyc07g063000), Sl-PR1b (Solyc00g174340), Sl-COI1 (Solyc05g052620), Sl-ERF1 (Solyc05g051200), Sl-ARF5 (Solyc04g081240), Sl-ACTIN (Solyc03g078400), Sl-CaM1 (Solyc01g008950), Sl-CaM2 (Solyc10g081170), Sl-CaM3 (Solyc10g077010), Sl-CaM4 (Solyc11g072240), Sl-CaM5 (Solyc12g099990), Sl-CaM6 (Solyc03g098050), Sl-YUC1 (Solyc06g008050), Sl-YUC2 (Solyc06g065630), Sl-YUC3 (Solyc06g083700), Sl-YUC4 (Solyc08g068160), Sl-YUC5 (Solyc09g064160), Sl-YUC6 (Solyc09g074430), Sl-YUC7 (Solyc09g091090), Sl-YUC8 (Solyc09g091720), Sl-YUC9 (Solyc09g091870), At-PSK1 (AT1G13590), At-PSK2 (AT2G22860), At-PSK3 (AT3G44735), At-PSK4 (AT3G49780), At-PSK5 (AT5G65870), At-PSK6 (AT4G37720), At-PSKR1 (AT2G02220), At-PSKR2 (AT5G53890), At-YUC1 (AT4G32540), At-YUC2 (AT4G13260), At-YUC3 (AT1G04610), At-YUC4 (AT5G11320), At-YUC5 (AT5G43890), At-YUC6 (AT5G25620), At-YUC7 (AT2G33230), At-YUC8 (AT4G28720), At-YUC9 (AT1G04180), At-YUC10 (AT1G48910), Dc-PSKR (AB060167), and B. cinerea actin (XM_001553318).
Supplemental Data
Supplemental Figure 1. The Effect of PSK Concentration on Immunity and the Transcript Abundance of PSK Biosynthesis-Related Genes in VIGS Tomato Plants.
Supplemental Figure 2. Nucleotide Sequence Alignments Based on the mRNA Region of Tomato PSK Homologs.
Supplemental Figure 3. Analysis of PSK Receptor Sequences and the Transcript Abundance of PSKRs in VIGS Tomato Plants.
Supplemental Figure 4. Amino Acid Sequence Alignments of PSKR Homologs from Tomato, Arabidopsis, and Carrot.
Supplemental Figure 5. Nucleotide Sequence Alignments Based on the mRNA Region of Tomato PSKR Homologs.
Supplemental Figure 6. The Transcript Abundance of PSK Signaling-Related Genes in VIGS Tomato Plants.
Supplemental Figure 7. The Effect of PSK Concentration on Cytosolic Ca2+ Levels and the Transcript Abundance of PSKRs in VIGS Tomato Plants.
Supplemental Figure 8. Analysis of Tomato CaM and YUC Homologs.
Supplemental Figure 9. Coexpression of CaMs and YUCs in N. benthamiana Leaves.
Supplemental Figure 10. Nucleotide Sequence Alignments Based on the mRNA Region of Tomato CaM Homologs.
Supplemental Figure 11. Effects of CaM2 Silencing on the Transcript Abundance of CaM Homologs in Tomato Plants.
Supplemental Table 1. Primers Used in This Study.
Supplemental File 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 1B.
Supplemental File 2. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 3A.
Supplemental File 3. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 8B.
Acknowledgments
We thank Gregory B. Martin (Boyce Thompson Institute for Plant Research) for critical reading of the manuscript. We also thank Jianrong Xu (Shanghai Jiao Tong University School of Medicine, China) for assistance with SPR analysis. This work was supported by the National Natural Science Foundation of China (31772355), the National Key Research and Development Program of China (2017YFD0200600), the Modern Agro-industry Technology Research System of China (CARS-25-02A), a grant from the China Scholarship Council (201706325011), and the Fundamental Research Funds for the Central Universities (2016XZZX001-07).
AUTHOR CONTRIBUTIONS
K.S. conceived and designed the experiments. H.Z., Z.H., C.L., C.Z., W.J., S.S., X.L., and K.S. performed the experiments. H.Z., Z.H., and K.S. analyzed the data. X.X., X.C., J.Z., Y.Z., and J.Y. provided technical and intellectual support. J.Y. provided suggestions for the manuscript preparation. K.S. drafted the manuscript. C.F. and K.S. revised the article with contributions from other authors.
Footnotes
Articles can be viewed without a subscription.
References
- Amano Y., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S., Liu J., Chang C., Zhang L., Maekawa T., Wang Q., Xiao W., Liu Y., Chai J., Takken F.L., Schulze-Lefert P., Shen Q.H. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Böhm H., Albert I., Fan L., Reinhard A., Nürnberger T. (2014). Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 20: 47–54. [DOI] [PubMed] [Google Scholar]
- Boutrot F., Zipfel C. (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55: 257–286. [DOI] [PubMed] [Google Scholar]
- Brading P.A., Hammond-Kosack K.E., Parr A., Jones J.D.G. (2000). Salicylic acid is not required for Cf-2- and Cf-9-dependent resistance of tomato to Cladosporium fulvum. Plant J. 23: 305–318. [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Mashiguchi K., Chen Q., Kasahara H., Kamiya Y., Ojha S., DuBois J., Ballou D., Zhao Y. (2013). The biochemical mechanism of auxin biosynthesis by an arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 288: 1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R., Van Kan J.A.L., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J., Foster G.D. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz J., ten Have A., van Kan J.A.L. (2002). The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol. 129: 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgbanshi A., Arbona V., Pozo O., Miersch O., Sancho J.V., Gómez-Cadenas A. (2005). Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography-electrospray tandem mass spectrometry. J. Agric. Food Chem. 53: 8437–8442. [DOI] [PubMed] [Google Scholar]
- Eun C.H., Ko S.M., Matsubayashi Y., Sakagami Y., Kamada H. (2003). Phytosulfokine-α requires auxin to stimulate carrot non-embryogenic cell proliferation. Plant Physiol. Biochem. 41: 447–452. [Google Scholar]
- Fischer C., Sauter M., Dietrich P. (2017). BiFC assay to detect calmodulin binding to plant receptor kinases. Methods Mol. Biol. 1621: 141–149. [DOI] [PubMed] [Google Scholar]
- Genty B., Briantais J.M., Baker N.R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92. [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Hartmann J., Fischer C., Dietrich P., Sauter M. (2014). Kinase activity and calmodulin binding are essential for growth signaling by the phytosulfokine receptor PSKR1. Plant J. 78: 192–202. [DOI] [PubMed] [Google Scholar]
- Hentrich M., Böttcher C., Düchting P., Cheng Y., Zhao Y., Berkowitz O., Masle J., Medina J., Pollmann S. (2013). The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 74: 626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind S.R., et al. (2016). Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2: 16128. [DOI] [PubMed] [Google Scholar]
- Igarashi D., Tsuda K., Katagiri F. (2012). The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. Plant J. 71: 194–204. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2009). Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 14: 373–382. [DOI] [PubMed] [Google Scholar]
- Knight H., Trewavas A.J., Knight M.R. (1996). Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori R., Amano Y., Ogawa-Ohnishi M., Matsubayashi Y. (2009). Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 15067–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschmar A., Rzewuski G., Stührwohldt N., Beemster G.T.S., Inzé D., Sauter M. (2009). PSK-α promotes root growth in Arabidopsis. New Phytol. 181: 820–831. [DOI] [PubMed] [Google Scholar]
- Kwezi L., Meier S., Mungur L., Ruzvidzo O., Irving H., Gehring C. (2007). The Arabidopsis thaliana brassinosteroid receptor (AtBRI1) contains a domain that functions as a guanylyl cyclase in vitro. PLoS One 2: e449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwezi L., Ruzvidzo O., Wheeler J.I., Govender K., Iacuone S., Thompson P.E., Gehring C., Irving H.R. (2011). The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. J. Biol. Chem. 286: 22580–22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladwig F., Dahlke R.I., Stührwohldt N., Hartmann J., Harter K., Sauter M. (2015). Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 27: 1718–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., Prigge M.J., Tigyi K., Estelle M. (2012). The cyclophilin DIAGEOTROPICA has a conserved role in auxin signaling. Development 139: 1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., et al. (2014). Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Tian M., Zhang H., Li X., Wang Y., Xia X., Zhou J., Zhou Y., Yu J., Shi K., Klessig D.F. (2015). Salicylic acid binding of mitochondrial alpha-ketoglutarate dehydrogenase E2 affects mitochondrial oxidative phosphorylation and electron transport chain components and plays a role in basal defense against tobacco mosaic virus in tomato. New Phytol. 205: 1296–1307. [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786. [DOI] [PubMed] [Google Scholar]
- Llorente F., Muskett P., Sánchez-Vallet A., López G., Ramos B., Sánchez-Rodríguez C., Jordá L., Parker J., Molina A. (2008). Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol. Plant 1: 496–509. [DOI] [PubMed] [Google Scholar]
- Ma Y., Walker R.K., Zhao Y., Berkowitz G.A. (2012). Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc. Natl. Acad. Sci. USA 109: 19852–19857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar M., et al. (2015). Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 5: 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Morita A., Matsunaga E., Furuya A., Hanai N., Sakagami Y. (1999). Physiological relationships between auxin, cytokinin, and a peptide growth factor, phytosulfokine-alpha, in stimulation of asparagus cell proliferation. Planta 207: 559–565. [Google Scholar]
- Matsubayashi Y., Ogawa M., Kihara H., Niwa M., Sakagami Y. (2006). Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 142: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Ogawa M., Morita A., Sakagami Y. (2002). An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472. [DOI] [PubMed] [Google Scholar]
- McCormack E., Tsai Y.C., Braam J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10: 383–389. [DOI] [PubMed] [Google Scholar]
- Meier S., Ruzvidzo O., Morse M., Donaldson L., Kwezi L., Gehring C. (2010). The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS One 5: e8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher S., Seybold H., Rodriguez P., Stahl M., Davies K.A., Dayaratne S., Morillo S.A., Wierzba M., Favery B., Keller H., Tax F.E., Kemmerling B. (2013). The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 73: 469–482. [DOI] [PubMed] [Google Scholar]
- Muleya V., Wheeler J.I., Ruzvidzo O., Freihat L., Manallack D.T., Gehring C., Irving H.R. (2014). Calcium is the switch in the moonlighting dual function of the ligand-activated receptor kinase phytosulfokine receptor 1. Cell Commun. Signal. 12: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Yang T., Ii W.M. (2014). Calmodulin gene expression in response to mechanical wounding and Botrytis cinerea infection in tomato fruit. Plants (Basel) 3: 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., et al. (2012). Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 195: 872–882. [DOI] [PubMed] [Google Scholar]
- Qi Z., Verma R., Gehring C., Yamaguchi Y., Zhao Y., Ryan C.A., Berkowitz G.A. (2010). Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. USA 107: 21193–21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodiuc N., et al. (2016). Evolutionarily distant pathogens require the Arabidopsis phytosulfokine signalling pathway to establish disease. Plant Cell Environ. 39: 1396–1407. [DOI] [PubMed] [Google Scholar]
- Song Q., et al. (2014). Lipoprotein-based nanoparticles rescue the memory loss of mice with Alzheimer’s disease by accelerating the clearance of amyloid-beta. ACS Nano 8: 2345–2359. [DOI] [PubMed] [Google Scholar]
- Srivastava R., Liu J.-X., Howell S.H. (2008). Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 56: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Swanson S.J., Gilroy S., Stacey G. (2010). Extracellular nucleotides elicit cytosolic free calcium oscillations in Arabidopsis. Plant Physiol. 154: 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D.M., Taylor M.G., Ciardi J.A., Klee H.J. (2000). The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc. Natl. Acad. Sci. USA 97: 5663–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Guo Z., Li H., Wang M., Onac E., Zhou J., Xia X., Shi K., Yu J., Zhou Y. (2016). Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol. 170: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li H., Han Z., Zhang H., Wang T., Lin G., Chang J., Yang W., Chai J. (2015). Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525: 265–268. [DOI] [PubMed] [Google Scholar]
- Wu J., Hettenhausen C., Meldau S., Baldwin I.T. (2007). Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Kamiya T., Yumoto H., Sotta N., Katsushi Y., Shigenobu S., Matsubayashi Y., Fujiwara T. (2015). An Arabidopsis thaliana copper-sensitive mutant suggests a role of phytosulfokine in ethylene production. J. Exp. Bot. 66: 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Baliji S., Buchmann R.C., Wang H., Lindbo J.A., Sunter G., Bisaro D.M. (2007). Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J. Virol. 81: 11972–11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Matsubayashi Y., Nakamura K., Sakagami Y. (2001). Diversity of Arabidopsis genes encoding precursors for phytosulfokine, a peptide growth factor. Plant Physiol. 127: 842–851. [PMC free article] [PubMed] [Google Scholar]
- Yin X.R., Shi Y.N., Min T., Luo Z.R., Yao Y.C., Xu Q., Ferguson I., Chen K.S. (2012). Expression of ethylene response genes during persimmon fruit astringency removal. Planta 235: 895–906. [DOI] [PubMed] [Google Scholar]
- Zhang S., Li X., Sun Z., Shao S., Hu L., Ye M., Zhou Y., Xia X., Yu J., Shi K. (2015). Antagonism between phytohormone signalling underlies the variation in disease susceptibility of tomato plants under elevated CO2. J. Exp. Bot. 66: 1951–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Liu W., Xu Y.P., Cao J.Y., Braam J., Cai X.Z. (2013b). Genome-wide identification and functional analyses of calmodulin genes in Solanaceous species. BMC Plant Biol. 13: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Qi Z., Berkowitz G.A. (2013a). Teaching an old hormone new tricks: cytosolic Ca2+ elevation involvement in plant brassinosteroid signal transduction cascades. Plant Physiol. 163: 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]