A novel activation state of a plant legumain reconciles previous data on its protease and ligase stabilities and activities, which allow it to cyclize or fuse peptides with unique functions.
Abstract
The vacuolar cysteine protease legumain can cleave and selectively rebuild peptide bonds, thereby vastly expanding the sequential repertoire of biomolecules. In this context, plant legumains have recently attracted particular interest. Furthermore, legumains have important roles in many physiological processes, including programmed cell death. Their efficient peptide bond ligase activity has gained tremendous interest in the design of cyclic peptides for drug design. However, the mechanistic understanding of these dual activities is incomplete and partly conflicting. Here, we present the crystal structure of a plant legumain, Arabidopsis thaliana isoform-γ (AtLEGγ). Employing a conserved legumain fold, the plant legumain AtLEGγ revealed unique mechanisms of autoactivation, including a plant-specific two-chain activation state, which remains conformationally stable at neutral pH, which is a prerequisite for full ligase activity and survival in different cell compartments. The charge distribution around the α6-helix mediates the pH-dependent dimerization and serves as a gatekeeper for the active site, thus regulating its protease and ligase activity.
INTRODUCTION
In contrast to mammals with only one isoform, plants encode at least four functional isoforms of the cysteine protease legumain (C13 family, EC 3.4.22.34). Legumains were found to be glycosylated in planta (Kembhavi et al., 1993) and can be grouped into seed type and vegetative type isoforms, reflecting their isoform-specific localizations and functions (as reviewed in Yamada et al., 2005; Müntz and Shutov, 2002). The seed-type legumains, as exemplified by isoform β from Arabidopsis thaliana (AtLEGβ), are important for the processing and maturation of seed storage proteins within protein storage vacuoles. Therefore, plant legumains are referred to as vacuolar processing enzymes (Hara-Nishimura et al., 1993) as well as asparaginyl endopeptidases (AEPs), the latter being correlated to their strict specificity for Asn/Asp in the so-called P1 position (Abe et al., 1993; Becker et al., 1995), i.e., the strict preference to cleave the peptide bond immediately after Asn or Asp. According to this nomenclature by Schechter and Berger, the substrate residues are numbered from the cleavage site, whereby the nonprimed residues (P1, P2, …) are counted in N-terminal and the primed residues (P1’, P2’, …) in C-terminal direction (Schechter and Berger, 1967).
The vegetative type legumains, like Arabidopsis isoform γ, have critical roles in plant programmed cell death (reviewed in Hatsugai et al., 2015) and thus partially substitute the orthologous caspases, which are absent in plants (Bonneau et al., 2008). In fact, AtLEGγ exhibits caspase-1 like activity and its expression in vegetative tissues correlates with hypersensitive or wound response and senescence (Yamada et al., 2004; Rojo et al., 2004; Hatsugai et al., 2004; Kinoshita et al., 1999b). Zymogenic AtLEGγ precursors (or proAtLEGγ), which are inactive “single-chain” precursors, accumulate within endoplasmic reticulum (ER)-derived proteinase storing bodies (ER bodies) in epidermal cells of healthy seedlings (Hayashi et al., 2001; Kinoshita et al., 1999a; Chrispeels and Herman, 2000). Upon a pathogen response, ER bodies start to fuse with acidic lytic vacuoles, thereby triggering the pH-dependent autoactivation of proAtLEGγ and promoting cell death (Hayashi et al., 2001).
Besides proteolysis, certain plant legumains are known to be efficient peptide ligases and cyclases (Conlan et al., 2010; Craik and Malik, 2013; Nguyen et al., 2014, 2016; Bernath-Levin et al., 2015a; Harris et al., 2015; Yang et al., 2017; Saska et al., 2007). Ligase activity has recently also been reported in mammalian legumain (Dall et al., 2015; Zhao et al., 2014). However, in vitro proof for ligase activity of Arabidopsis legumain isoforms is lacking.
Plant legumains convert functionally cryptic linear polypeptides into active cyclic compounds with diverse biological activities, as exemplified by the sunflower trypsin inhibitor 1 and the insecticidal cyclotide kalata B1, among many others (Nguyen et al., 2014, 2015; Mylne et al., 2011; Bernath-Levin et al., 2015a). Cyclic peptides, like the cyclotides, combine fold robustness with a large sequence variability, making them unique scaffolds for diverse biotechnological and medical applications, including multivalent drugs against HIV or cancer (Gould et al., 2011; Lesner et al., 2011; Ireland et al., 2008; Gerlach et al., 2010).
Such applications have fueled enormous interest in the ligation mechanism of plant legumain isoforms, which vary in their ligase/cyclase efficiency. For example, legumain isoforms from Clitoria ternatea (butelase 1) or Helianthus annuus (sunflower) have been reported to be particularly efficient peptide cyclases of the cyclotides kalata B1 and sunflower trypsin inhibitor 1, respectively (Nguyen et al., 2014; Mylne et al., 2011). Comparative studies must be taken with some caution and may lead to controversial results because while the ligation efficiency increases with pH (Dall et al., 2015; Ozawa and Laskowski, 1966; Nguyen et al., 2014), this effect will be modulated and counteracted by the pH-dependent conformational destabilization of the catalytic domain only (AEP) at the ligation pH regime and depends on proper posttranslational modifications/glycosylations (Bernath-Levin et al., 2015b; Dall and Brandstetter, 2013; Harris et al., 2015).
Biochemical experiments suggested that the enzymatic latency of plant legumain zymogens is conferred by the C-terminal pro-domain, which was reported to become (auto-)proteolytically cleaved at acidic pH (Kuroyanagi et al., 2002; Hiraiwa et al., 1999, 1997), in agreement with the crystal structures of mammalian legumains (Dall and Brandstetter, 2013; Zhao et al., 2014; Dall and Brandstetter, 2016; Li, 2014). It was reported that the catalytic domain of legumain is instable at neutral pH (Dall and Brandstetter, 2013). The different subcellular localizations of fully activated mammalian legumain can be explained by integrin interactions, which stabilize it at neutral pH (Dall and Brandstetter, 2013).
How plants, which lack integrins, can develop legumain activities in the absence of stabilizing integrins in different subcellular localizations over a broad pH range remained enigmatic so far and is addressed in this work. We present the first recombinant expression of a glycosylated plant legumain in Leishmania tarentolae and its purification to homogeneity. The crystal structure and biochemical analysis of a plant legumain, AtLEGγ, described here enabled us to discover and understand its unexpected novel autoactivation mechanism and ligase activity (Bernath-Levin et al., 2015a). We identified a plant-specific two-chain activation intermediate, which was stable at neutral pH, where ligase activity is favored. The two-chain intermediate results from cleaving the single-chain zymogen into the catalytic domain and the C-terminal pro-domain, which remain noncovalently bound. After proteolytic cleavage, the C-terminal pro-domain modulates rather than inhibits legumain’s activity and confers stability particularly at neutral pH. Hence, we refer to the C-terminal pro-domain as LSAM (legumain stabilization and activity modulation) domain. We discuss the relevance of our findings in the context of cell biology and peptide ligation. This study not only broadens our understanding of plant (patho)physiology and crop design, but also enables the design of efficient peptide ligases and will bring such a technology closer to broad application.
RESULTS
Overall Structure
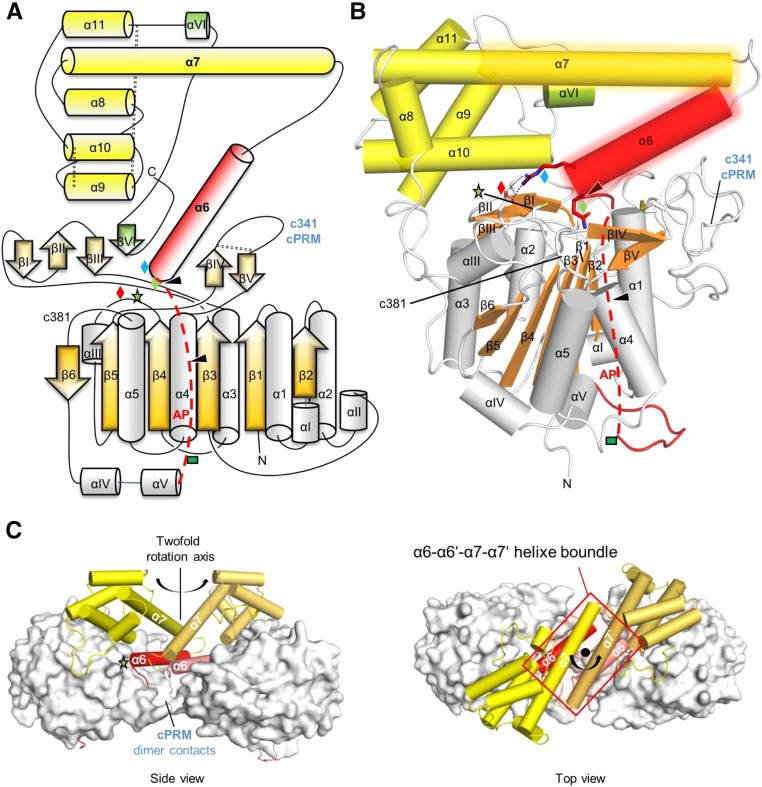
The sequence numbering in this work refers to AtLEGγ, with the corresponding human residues being indicated as superscript, e.g., the catalytic cysteine C219hC189. Comparison of legumain sequence conservation suggested independently evolved roles for the activation peptide and LSAM domain in mammals and plants (Supplemental Figure 1). For a detailed mechanistic understanding of plant legumain’s function, we determined the crystal structure of AtLEGγ. The x-ray structure at 2.75-Å resolution revealed a caspase-like catalytic domain that consists of a central six-stranded β-sheet (β1-β6) surrounded by five major (α1-α5) and five minor α-helices (αI-αV) (Figures 1A and 1B; Supplemental Figure 1; for data statistics, see Table 1). Furthermore, the catalytic domain is covered by two short antiparallel β-sheets (βI-βIII and βIV-βV), serving as primed and nonprimed substrate recognition sites in the mature AEP form and as an anchoring interface to the C-terminal LSAM domain in the zymogen. The latter exhibits an overall death domain-like fold with its typical all α Greek key motif, similar to the fold found in the homologous caspases. Caspases undergo dimerization to gain enzymatic activity (Fuentes-Prior and Salvesen, 2004). Importantly, the crystal structure revealed a dimeric quaternary structure of AtLEGγ, which is mediated by an antiparallel four-helix bundle comprising helices α6-α7 and the equivalent helices α6’-α7’, shaded in Figures 1B and 1C. The asymmetric unit consists out of two such dimers. The dimer was further stabilized by a plant-specific disulphide-linked (Cys252-Cys266) proline-rich insertion (Figure 1C) that extends the c341 specificity loop and resembles a typical cyclic protein recognition motif (cPRM) (Zarrinpar et al., 2003). This cPRM-mediated interaction increases the dimer interface and renders it partly redox sensitive. Mass spectrometric analysis (Supplemental Figure 2) revealed a sugar moiety consisting of an N-glycan core, very likely on its only N-glycosylation site at N336. Due to its location at the relatively disordered activation peptide (AP), only an undefined electron density blob was found, which did not allow a clear modeling of the sugar.
Figure 1.
Topology and Domain Organization of Two-Chain AtLEGγ.
(A) Topology diagram of AtLEGγ. The catalytic domain is colored in gray and orange, the activation peptide (AP) including the α6-helix in red, the LSAM in yellow, and the putative VSS in green. The red dashed line indicates part of the AP, which would be intact in the precursor form. The blue, red, and green diamonds and the green star represent Arg-355, Glu-220, Gln-354, and the catalytic cysteine (Cys-219), respectively. The green rectangle represents an N-glycosylation site. Autocleavage sites are indicated as black triangles and the three disulphide bridges by black dashed lines.
(B) The X-ray structure of AtLEGγ monomer molecule in cartoon representation with identical color code as in (A). Dimer interface regions are shaded areas around the α6- and α7-helices.
(C) Dimeric structure of two-chain AtLEGγ. Catalytic domains are displayed as surface, the AP, including the α6-helix, and the LSAM as red and yellow cartoons, respectively. The cPRM contacts within the dimer are highlighted. Left: Front view, perpendicular to the 2-fold axis. Down: Top view along the 2-fold axis.
Table 1. Data Collection and Refinement Statistics for Two-Chain AtLEGγ.
| Data Collection | Statistical Values |
|---|---|
| Space group | P41 |
| Cell dimensions | |
| a, b, c (Å) | 147.8, 147.8, 101.6 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 72.9–2.75 |
| Rmerge | 0.09 (0.77) |
| I/σI | 8.6 (1.6) |
| Completeness (%) | 99.9 (99.8) |
| Redundancy | 3.4 (3.5) |
| Refinement | |
| Resolution (Å) | 66.1–2.75 |
| No. reflections | 56952 |
| Rwork/Rfree | 0.21/0.25 |
| No. molecules/asu | 4 |
| No. atoms | 13474 |
| Protein | 13329 |
| Ligand/ion | 66 |
| Water | 79 |
| B-factors | |
| Protein | 60.5 |
| Ligand/ion | 62.5 |
| Water | 49.5 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.7 |
Data were collected from a single crystal. Values in parentheses correspond to the highest resolution shell. R.m.s., root mean square; asu, asymmetric unit.
Active-Site and Substrate Recognition Elements
To understand the mechanistic relevance of individual amino acid substitutions, we superimposed the active sites of AtLEGγ with that of human prolegumain and of activated human legumain in complex with the covalent chloromethylketone-based substrate analog Ac-YVAD-CMK (Supplemental Figure 3). The catalytic residues, including the oxyanion hole, are arranged similar to those found for human legumain. Asp176hD147 was found to be converted to a succinimide, with a potential role in peptide bond ligation (Dall et al., 2015). Two prominent loops, c341 and c381 (caspase 1 numbering) located between strands β5 and β6 of the central sheet, define the nonprimed substrate recognition sites (Fuentes-Prior and Salvesen, 2004; Dall and Brandstetter, 2016). Both loops feature plant-specific differences such as insertions of seven and six amino acids in length relative to the human protein, which are conserved in plants and contribute to the formation of the nonprimed substrate recognition sites (Supplemental Figure 1). The c341 specificity loop is segmented into the antiparallel strands βIV and βV and a plant-specific disulphide-linked cPRM, with the disulphide bridge defining the S3 recognition site.
The Dimer Represents the Biological Storage Form of AtLEGγ
The structural analysis suggested that dimerization stabilizes the α6-helix via the four-helix bundle (Figure 1C) and thus contributes to keeping AtLEGγ enzymatically silent. Specifically, access to the primary S1 pocket of the active site was blocked by Gln354hS307, which is located at the beginning of the α6-helix. Binding of the Gln354 carboxamide group mimics that of a canonical Asn-P1 substrate, while escaping peptide bond cleavage by its longer side chain (Supplemental Figure 3A). The zymogenicity is further stabilized by two plant-specific salt bridges that stabilize the α6-helix of the AP in the active site, Asp358hP311 with Arg74hR44 and His177hH148 and Arg355hP308 with Glu220hE190. These three plant-specific interactions stabilize the AP/α6-helix and hint at a characteristic difference in the activation mechanism of human and plant legumain.
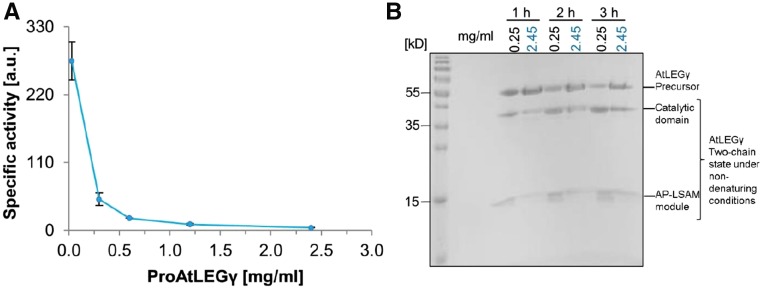
To directly confirm the proposed critical role of dimerization and the α6-helix in conferring enzymatic latency, we tested the enzymatic activity of AtLEGγ in dependence of its protein concentration which, by the law of mass action, favors dimerization at higher concentrations. At first sight, counterintuitively, since autoactivation happens in trans (Dall and Brandstetter, 2013), the specific activity of AtLEGγ decreased almost exponentially with protein concentration, in excellent agreement with our crystal structure analysis (Figures 1 and 2). This finding is of critical relevance to store plant legumains in dense ER bodies, as discussed below.
Figure 2.
AtLEGγ Can Be Stored Efficiently at High Concentrations.
(A) The specific activity of zymogenic AtLEGγ (proAtLEGγ) at pH 4.0 is inversely correlated to its concentration, indicating a strongly autoinhibitory effect of dimerization. Error bars represent standard deviations of triplicate measurements.
(B) SDS-PAGE shows the concentration-dependent autoactivation of proAtLEGγ at a low (0.25 mg/mL; black) and high concentration (2.45 mg/mL; blue).
pH-Dependent Dimerization and Autoactivation, Resulting in a pH-Stable Two-Chain Form of AtLEGγ
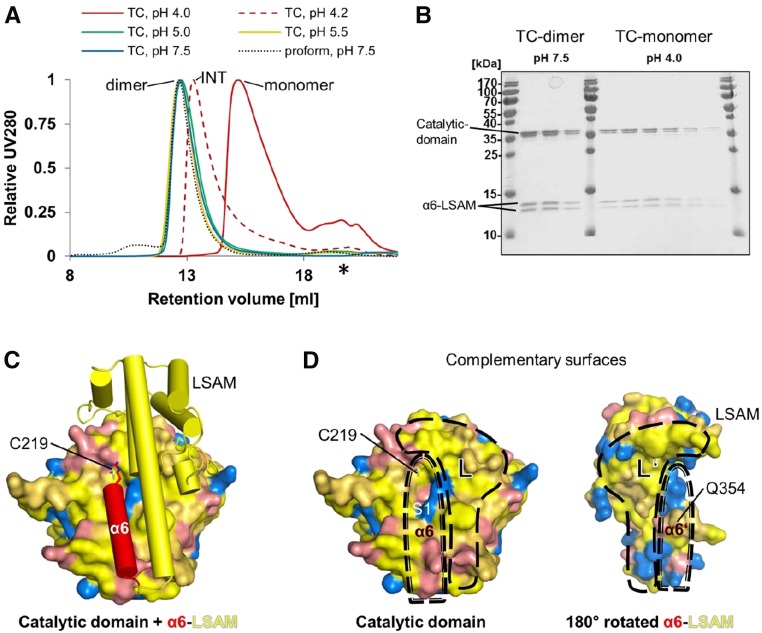
Given our observation that dimerization contributed to zymogenicity and the previous report that, in monomeric human legumain, activation was triggered by acidic pH, resulting in AP cleavage and the release and subsequent degradation of the LSAM domain (Dall and Brandstetter, 2012, 2013), we tested whether AtLEGγ dimerization depends on pH. We found that two-chain AtLEGγ migrated on gel filtration chromatography as a monomer only at pH 4, and as a dimer between pH 5.0 and 7.5 (Figure 3A). At pH 4.2, we observed intermediate retention volumes, indicating a fast monomer-dimer equilibrium at this pH.
Figure 3.
pH-Dependent Oligomerization and the pH-Independent Protease:LSAM Interface.
(A) Gel filtration chromatograms of proAtLEGγ and two-chain form under pH values. proAtLEGγ, zymogen form of AtLEGγ; AtLEGγ-TCγ, two-chain form of AtLEGγ. Intermediate retention volume (INT) was observed at pH 4.2 (around the pKa of carboxyl groups), indicating a fast equilibrium between monomer and dimer. The asterisk indicates a subpeak arising from buffer components <3000 D.
(B) The peptidase and the AP-LSAM domains of two-chain AtLEGγ comigrate on size-exclusion chromatography at neutral and pH 4.0, as evidenced by the corresponding SDS-PAGE fractions of the corresponding main peaks. At pH 4.0, Asp and Glu side chains are protonated. Upper band of α6-LSAM begins with residue S344 and lower band with Q354.
(C) Orientation of the α6-helix and LSAM domain relative to the catalytic domain.
(D) Display of the protease:α6-LSAM interface. Left, Catalytic domain interface with the α6 and LSAM (L) binding region indicated; right, the complementary surface of α6-LSAM (rotated by 180°). Color code of the surface: yellow, hydrophobic residues, i.e., Trp, Phe, Tyr, Leu, Ile, Val, Gly, Ala, Met, and Pro; orange, polar residues, i.e., Ser, Cys, Thr; Asn, and Gln; red, negatively charged residues, i.e., Asp and Glu; blue, positively charged residues, i.e., Arg, His, and Lys.
Moreover, we could induce proteolytic activation by incubating proAtLEGγ at pH 4.0 (without reducing agents), resulting in AP cleavage. Unexpectedly, however, and in contrast to the situation in human legumain, this pH-induced activation resulted in a stable two-chain form of AtLEGγ rather than the catalytic peptidase domain (AEP) only. The two cleavage sites (Asn343-Ser344 and Asn353-Gln354; Supplemental Figure 4) are located in the flexible part of the AP and immediately preceding the ordered α6-helix, respectively. No further degradation or release of the LSAM domain was detected for at least 16 h at pH 4.0 and 30°C. Instead, the LSAM and the peptidase domains of the two-chain form comigrated on gel filtration chromatography at pH values of between 4.0 and pH 7.5 (Figures 3A and 3B). The peptidase-only (AEP) form of AtLEGγ can be generated at acidic pH by the addition of reducing agents, which destabilize disulphide bridges in structural elements important for dimerization, like the cPRM and LSAM, and consequently the dimer interface.
The Protease-LSAM Interface Is Dominated by pH-Independent Hydrophobic Interactions
In mammalian legumain, the electrostatically anchored LSAM domain is released from the protease domain upon protonation of the peptidase-harbored Asp and Glu side chains (Dall and Brandstetter, 2013; Zhao et al., 2014). By contrast, we found a predominantly hydrophobic protease-LSAM interface (yellow in Figures 3C and 3D; Supplemental Figure 5). To further judge the functional relevance of this hydrophobic and thereby pH-independent interface, as already shown by the comigration of AEP and AP-LSAM over the pH range from 4.0 to 7.5 (Figures 3A and 3B), we analyzed the effect of the nonionic detergent N-lauroylsarcosine (LS) on autoactivation. Already 400 µM of lauroylsarcosine (critical micelle concentration = 13.7 mM) led to a complete activation of the zymogen and significant degradation of the LSAM under conditions where only 50% zymogen activation to the two-chain form was observed in the absence of lauroylsarcosine (Supplemental Figure 6A). To confirm that this effect is due to the release of the hydrophobic protease:LSAM interface rather than to a stimulation of the protease domain, we measured the proteolytic activities for the two-chain and the protease only in the presence and absence of 400 µM lauroylsarcosine. Indeed, the stimulatory effect was most prominent for the two-chain state (Supplemental Figure 6B).
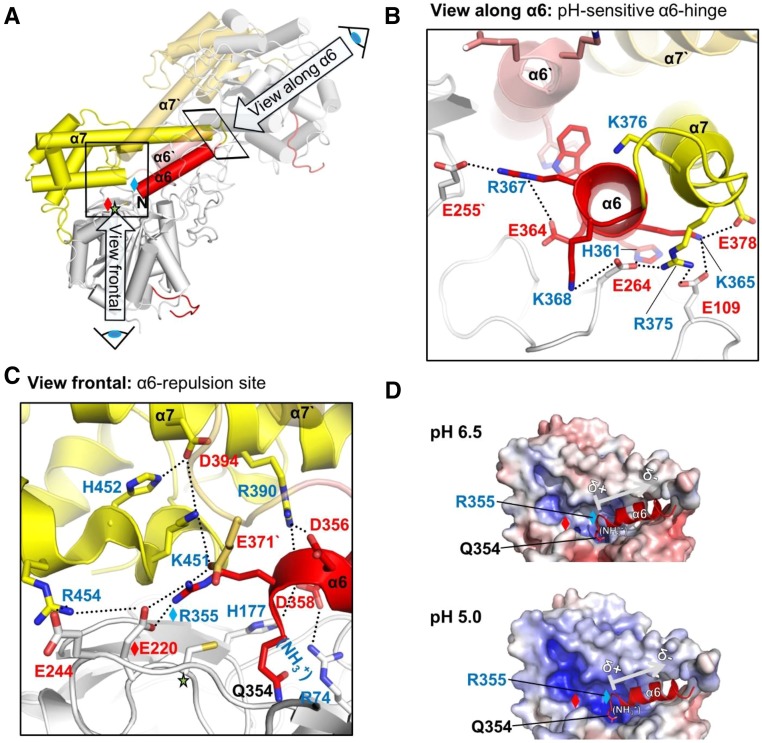
Charge Distributions at α6-Helix Explains the pH Dependency of AtLEGγ Dimerization
The structure analysis of the symmetric AtLEGγ dimer interface (buried surface area of around 2900 Å2) revealed important clues to its marked pH dependency. As illustrated in Figures 4A to 4C, the autoinhibitory α6-helix is engaged in the α6, α7, α6′, and α7′ four-helix bundle (Supplemental Figure 7B) by both intra- and intermolecular ionic interactions. We could identify two prominent positively charged clusters (Arg, Lys, and His) at either side of the α6-helix, which are neutralized by negatively charged glutamate and aspartate patches (Figures 4A to 4C). The latter become protonated at pH 4.0, thereby breaking the charge balance. This overshooting positive charge at the termini of helices α6 and α6′ will loosen their intra- and intermolecular anchorage and, consequently, loosen the four-helix bundle, explaining the monomerization of two-chain AtLEGγ at pH 4.0 (Figure 3A). Additionally, at the α6-repulsion site (Figure 4C), the positively charged (R355 and to a lesser extent the helix dipole; Figure 4D) will unlock the α6-helix at acidic pH, although its secondary structure integrity should remain unaffected. Indeed, in case of the two-chain form with the cleavage at N353-Q354, the N terminus of the α6-helix is even more positively charged due to the free N terminus starting with Q354. This pH-dependent electrostatic repulsion of the α6 helix can be qualitatively visualized by means of the electrostatic surface potentials (Figure 4D), which highlight the abrupt change from electrostatic repulsion to a balanced attraction in a narrow pH range from 5.0 to 6.5. These computational analyses are approximate in their nature and need to be taken with caution. However, consistent results were obtained using electrostatic isosurfaces (Supplemental Figure 7A), thus corroborating the qualitative finding of a rather abrupt decrease in positive charge at the α6-repulsion site from pH 5.5 to pH 6.0, in agreement with enzymatic data (Figure 5A).
Figure 4.
The α6-Helix Is Highly pH-Dependently Regulated.
(A) Overall view of the two-chain AtLEGγ dimer, indicating the two pH-sensitive areas of the four-helix bundle α6, α7, α6′, α7′. The catalytic domain, LSAM, and α6-helix are colored in gray, yellow, and red, respectively.
(B) Close-up view onto the pH-sensitive α6 hinge region, highlighting the intra- and intermolecular electrostatic clamping of the α6-helix, which is released at acidic pH. This view along the α6-helix is rotated around 90° compared with (A).
(C) Close up into the α6-repulsion site (view as in [A]). Protonation of histidine and acidic residues leads to an electrostatic repulsion of the clustered positively charged residues, which subsequently destabilizes the α6-helix.
(D) pH-Dependent electrostatics of the α6-repulsion site. Red, negative potential; blue, positive potential. Contouring was set from −10 to +10. Red or blue diamonds represent R355 or E220, respectively. The arrow indicates the helix dipole moment, which will have presumably only a minor contribution to the electrostatic regulation.
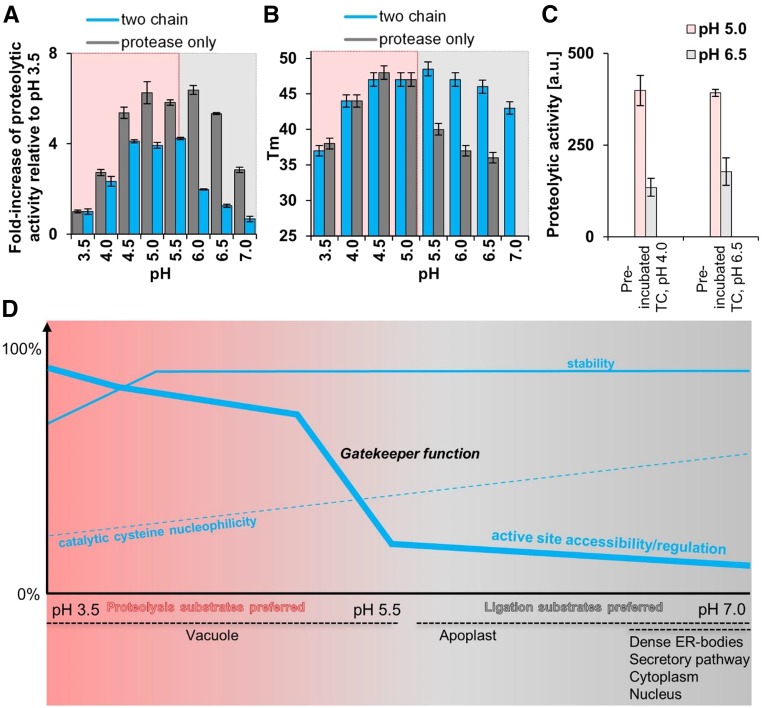
Figure 5.
The Two-Chain State Is Highly Regulated and Is Able to Survive at More Neutral pH.
(A) pH-dependent proteolytic activities (Z-VAN-AMC) of the two-chain (blue) and protease-only (gray) form. Error bars represent standard deviations of duplicate measurements (sample size = 3).
(B) pH-dependent melting temperatures derived by dynamic scanning fluorimetry for the two-chain (blue) and protease-only (gray) form. Data processing was performed according to Niesen (2010) (sample size = 2). Error bars represent a 2% deviation range.
(C) The two-chain form was preincubated at pH 4.0 or pH 6.5. After preincubation, the samples were measured at pH 5.0 (dark blue) and 6.5 (blue). Error bars are standard deviations of triplicate measurements.
(D) Red- and blue-shaded areas are pH regimes with preferential turnover of proteolysis and ligation substrates, respectively. The schematic lines indicate mechanistic details of autoactivation and activity regulation of the two-chain state. Dashed, thick, and thin blue lines represent the nucleophilicity of the catalytic cysteine, accessibility, and stability of the two-chain state, respectively. Note that the two-chain state remains stable at the ligation-relevant pH range and the more restricted accessibility of the active site (“gatekeeper” function of α6-helix).
Functional Relevance of the Electrostatic Anchoring of the α6-Helix: Peptidase Activity Control
Canonical autoactivation of AtLEGγ at acidic pH resulted in a stable two-chain form (Figure 3B; Supplemental Figure 4), whereas only the addition of reducing agents and/or lipophilic compounds generated the AEP form (Supplemental Figure 6A).
Our structure analysis suggests (1) an opening of the active site of the two-chain form at acidic pH, resembling that of the AEP (protease only) form; and (2) a blockage of the active site by the α6-helix in the two-chain form at more neutral pH. To test this model, we determined the pH-dependent proteolytic activities of the two-chain and protease-only forms (Figure 5A). For better comparison, we normalized the proteolytic pH dependencies for the two-chain as well as the protease-only forms at a pH of 3.5 each, because both forms are monomeric, with most active sites accessible and, thus, most similar at this pH. As depicted in Figure 5A, the protease-only form showed a typical bell-shaped pH dependency, which can be seamlessly explained by an initial increase in nucleophilicity with pH, which is then overruled by conformational destabilization at a more neutral pH of ∼6.5 (Dall and Brandstetter, 2013) and also at pH ≤4.0 (Figure 5D). By comparison, the activity of the two-chain form was dampened already at a lower pH of ∼5.5, despite being significantly more stable than the protease-only form, as evidenced by their melting temperatures (Figure 5B). Furthermore, this pH-dependent inhibition in the two-chain form was reversible, as it could be induced after preincubation at low or high pH (Figure 5C). In summary, the two enzyme forms are comparable with respect to activity and stability in the acidic range (red area, Figures 5A, 5B, and 5D), but differ drastically in the near neutral pH range (blue area, Figures 5A, 5B, and 5D).
AtLEGγ Has Dual Protease/Ligase Activity at a More Neutral pH
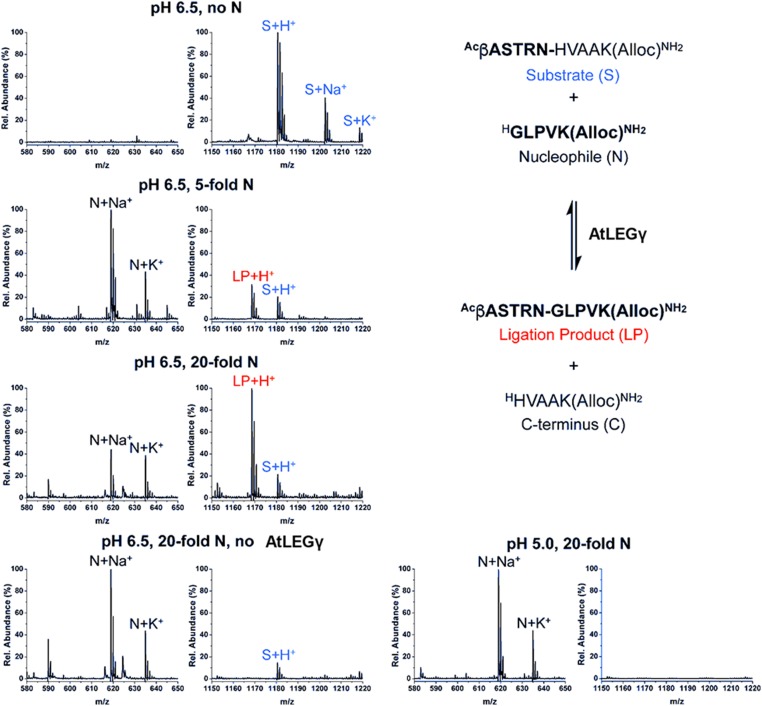
After studying the pH-dependent accessibility of the AtLEGγ active site, we further investigated by mass spectrometry whether the catalytic domain of legumain from Arabidopsis (AtLEGγ) would develop peptide ligase activity. We tested modified peptides based on the sequence of the prototypic cyclotide kalata B1 because this cyclotide was reported to be cyclized by a C. ternatea legumain isoform (Saska et al., 2007; Nguyen et al., 2014). The primed side residues of the peptides were modified accordingly to an optimized sequence (HV), as reported for butelase 1 (C. ternatea legumain) (Nguyen et al., 2014). Specifically, AtLEGγ cleaved and religated the two linear peptides AASTRN-HVAAK and GLPVK to ASTRN-GLPVK. This transpeptidation reaction was remarkably efficient, as a 5-fold excess of the nucleophile substrate GLPVK was sufficient to shift the equilibrium from AASTRN-HVAAK to ASTRN-GLPVK, and at a 20-fold excess of GLPVK the equilibrium was completely shifted toward the ASTRN-GLPVK ligation product (Figure 6). Transpeptidation was observed at pH 6.5, but not at pH 5.0. No ligation product was detected in the absence of the enzyme, AtLEGγ, or, alternatively, in the absence of the nucleophile GLPVK (Figure 6).
Figure 6.
AtLEGγ Is a Peptide Ligase at More Neutral pH.
The upper panel clarifies the experiment design. The substrate S (blue) was incubated with or without AtLEGγ with 5-, 10-, or 20-fold excess of the nucleophile N peptide at pH 5.0 or 6.5. The ligation product (red) and the C terminus were detected by MALDI-TOF-MS. The MS spectra of the ligation mixtures and controls are shown. Acetyl-βASTRNHVAAK(Alloc)-amide (C49H82N18O16) calculated 1179.31, found (M+H+) 1180.02. GLPVK(Alloc)-amide (C28H49N7O7) calculated 595.38, found (M+Na+) 618.65. Acetyl-βASTRNGLPVK(Alloc)-amide (C50H86N16O16) calculated 1667.34, found (M+H+) 1168.07. HVAAK(Alloc)-amide (C27H45N9O7) calculated 607.35, found (M+H+) 608.44.
DISCUSSION
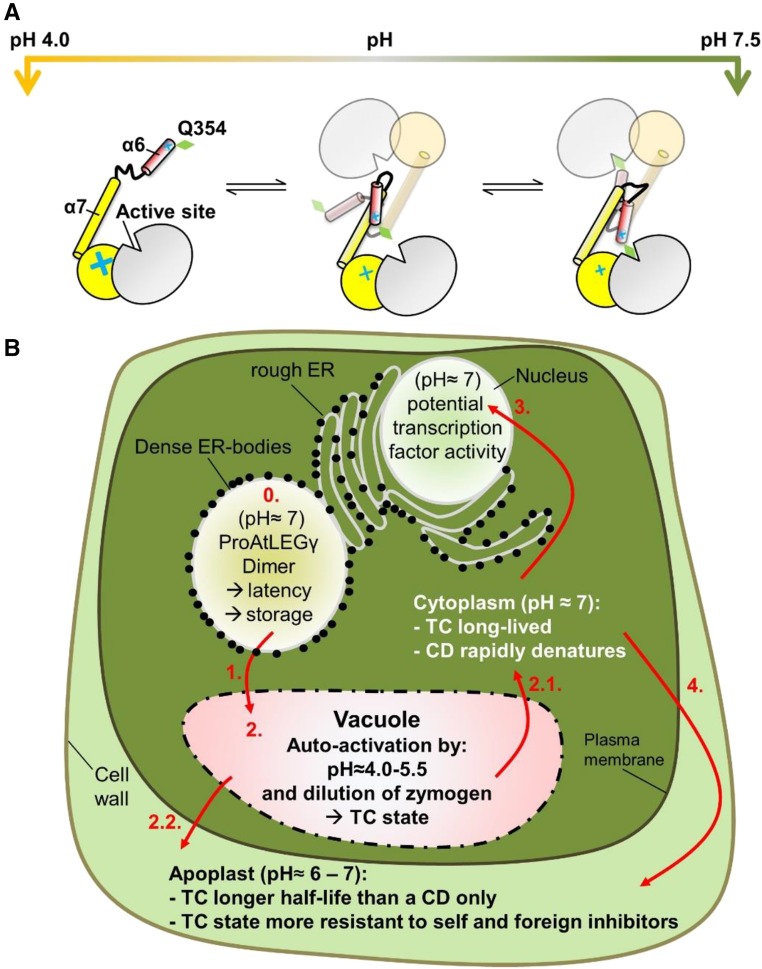
In this study, we recombinantly expressed and purified the Arabidopsis isoform γ legumain (AtLEGγ) in a fully N-glycosylated form (Supplemental Figure 2) and present the crystal structure of an Arabidopsis legumain in its two-chain form. We identified an independently evolved and mechanistically drastically different proteolytic autoactivation compared with mammalian legumain, with a stable two-chain intermediate (Figure 3; Supplemental Figure 4); the latter can be further activated to the classical asparaginyl endopeptidase (AEP) only under reducing conditions. The activated two-chain state of AtLEGγ comprises the catalytic domain and the LSAM domain including the α6 helix, i.e., the α6-LSAM module (Figure 1B; Supplemental Figure 4). This unique, plant-specific α6-LSAM module accounts for the increased pH stability at neutral pH and the significantly stronger interaction in AtLEGγ rather than in the mammalian legumains, although an intermediate comigration of human and mouse peptidase domain with LSAM at pH >4.5 was reported (Zhao et al., 2014; Dall and Brandstetter, 2013). For a direct (α6-)LSAM:catalytic-domain interface comparison between mammalian and AtLEGγ, see Supplemental Figure 5. The mammalian interface is significantly based on electrostatics (Dall and Brandstetter, 2013), contrasting the AtLEGγ interface (Figure 3D; Supplemental Figure 5). Additionally, mammalian legumain has specific autocleavage sites, which lead to a disruption of the dimer interface, in contrast to AtLEGγ. Figure 7A schematically illustrates the deduced model of autoactivation and the pH-dependent regulation of the two-chain state. At pH 4.0, the carboxylate groups of the glutamates in the pH-sensitive regions (Figure 4) are protonated, losing their stabilizing properties in the charge arrays at helix α6. As a result, the N-terminally positively charged (free N terminus, R355, and helix dipole moment) α6-helix is repelled from the active site, rendering it more accessible to substrates. The α6-helix repulsion at pH 4.0 will distort the four-helix bundle α6-α7-α6′-α7′, leading to a dissociation and monomerization at this pH (Figure 3A). Conversely, at more neutral pH, the α6-helix is stabilized, blocking access to the active site, consistent with an apparent inhibition with higher pH that is independent of protein stability (Figure 5). The pH-dependent activity profile of two-chain AtLEGγ (Figure 5A) was associated with reported average pKa values of glutamates (4.2 ± 0.9) and histidines (6.6 ± 1.0) in folded proteins (Grimsley et al., 2009), although specific pKa assignments to individual amino acids remain challenging. In summary, the α6-helix serves as a plant-specific pH-dependent switch, controlling access to the active site.
Figure 7.
Schematic Model for the pH-Dependent (Auto-)Activity Regulation of AtLEGγ and Its (Patho-)Physiological Consequence within a Plant Cell.
(A) At pH below 4.5, the carboxylates of Asp and Glu are protonated, resulting in excess positive charges at both pH-sensitive α6-anchoring sites. As a result, the positioning of the α6-helices is destabilized, and the four-helix bundle is also destabilized, consequently. Such destabilization and repulsion ultimately lead to monomerization of AtLEGγ. At pH 4.5 to 5.5, the positive charges at the pH-sensitive α6 areas start to get compensated, with a concomitant stabilization of the α6-helix, partly rendering the S1 pocket less accessible. This now dimeric form is moderately active. At pH values >6.0, the positive charges at the pH-sensitive α6-clamp and repulsion regions are compensated for by counter charges, stabilizing the α6-helix within the four-helix bundle and blocking the S1 pocket with Gln-354 (green diamond). This stabilized α6-helix translates functionally into a state of reduced activity of the two-chain state. The transitions between these intermediate states are reversible.
(B) (Patho-)physiological relevance: 0. Efficient storage of proAtLEGγ is achieved by a high concentration within ER-dense bodies and neutral pH, which induces inactivation by dimerization. 1. Stress induces the fusion of ER dense bodies with acidic vacuoles. 2. Dilution and acidic pH triggers autoactivation. Now, limited proteolysis can occur. A pH of 6.5 will not denature the two-chain state, contrasting the situation of the CD. Ligation may occur, if a suitable substrate is present. 2.1. Programmed cell death starts with the disassembly of the vacuole, which releases the two-chain state into the neutral cytoplasm. The two-chain state is stable and can still perform proteolytic or ligating functions. A CD would be denatured rapidly. 2.2. The fusion of the vacuole with the plasma membrane would release AtLEGγ out of a cell. ProAtLEGγ and the two-chain form will have a higher half-life and additionally will be more resistant to self and foreign protein inhibitors like phytocystatins or fungi derived macrocypins. 3. Because a TC state is still fully functional within the cytoplasm, nuclear transport is more likely than for a CD (e.g., for tomato LEG, transcription factor activity was reported; Rosin et al., 2005). 4. Transport to the apoplast from the cytoplasm is more likely. These effects broaden the possible roles of legumains and functions within a plant cell significantly.
It is especially interesting to see how the structural and biochemical findings described here can be directly translated into plant cell biology. It has been reported that AtLEGγ is stored in ER bodies, which fuse to the vacuole, if the cell senses stress (Hayashi et al., 2001). We hypothesize that the inhibitory capacity of the dimer state (Figure 2) enables an efficient storage of concentrated AtLEGγ precursors within dense ER bodies (Figure 7B). Importantly, the broad pH stability (Figure 5B) of the two-chain state explains how legumain activity can be found in different subcellular localizations with different functions within a plant cell (Figure 7). For example, during the initial phase of PCD, the vacuole disintegrates (Hatsugai et al., 2006). The vacuole contents, including activated AtLEGγ, is exposed to more neutral pH. However, the vacuole can also fuse with the plasma membrane to release its content to more neutral apoplast micromilieu (Hara-Nishimura and Hatsugai, 2011). For tomato (Solanum lycopersicum) legumain (LeCp), a transcription factor activity was reported (Rosin et al., 2005). However, how different activation states of LeCp are translocated to the nucleus remains unresolved, despite the presence of a nuclear localization sequence (Supplemental Figure 1). A two-chain state would be a perfect candidate, since it can survive the neutral pH compartments (Reguera et al., 2015; Martinière et al., 2013; Otegui and Spitzer, 2008) like cytoplasm, the late endosome/prevacuolar compartment or the nucleus (Figure 7). This is especially interesting, since plants lack integrins and thereby cannot stabilize the protease-only domain (AEP) at neutral pH, as observed for human legumain (Dall and Brandstetter, 2013). Furthermore, inhibition of legumain activity by phytocystatins or fungi-derived macrocypins will be prevented by the LSAM domain because these inhibitors and the LSAM domain compete for the same binding region. Therefore, the two-chain state will be presumably more resistant to endogenous and exogenous inhibitors. In contrast to the protease-only form, the two-chain state will be long lived and may fulfill specialized functions like efficient peptide ligation (Figure 7).
Interestingly, the peptide ligase activity of AtLEGγ is confined to near neutral pH (Figure 6) (Nguyen et al., 2015; Dall and Brandstetter, 2016; Dall et al., 2015). While this pH dependence is consistent with the ligase activity found here and in C. ternatea (butelase-1) or human legumain (Nguyen et al., 2015; Dall and Brandstetter, 2016; Dall et al., 2015), it is also the pH-regime where the plant-specific two-chain activation state described herein is stabilized and the access to the active site is tightly controlled by the α6-helix, which may be a paradox at first sight. However, an efficient peptide ligase needs to suppress premature hydrolysis of ligation intermediates, otherwise initiated ligation reactions would be futile. Given the correlation in pH dependency, it seems that the α6-helix serves as a selective gatekeeper enhancing ligation rates by productive steering of the involved substrates and retarding unproductive hydrolysis of ligation reaction intermediates such as the enzyme-acyl complex. This selection might be accomplished by electrostatic exclusion or dislocation of the catalytic water from the active site, as on a different structural basis described for the nonhomologs bacterial macrocyclaze PatG (Koehnke et al., 2012). Presumably AtLEGγ utilizes a combination of electrostatic and steric exclusion, e.g., the positively charged nucleophilic substrate might serve as a key to electrostatically unlock the α6-helix gate and subsequently allow access for the nonprimed cosubstrate for ligation. Finally, we wish to emphasize the critical requirement of proper glycosylation for AtLEGγ and probably many plant legumains, particularly for their ligase activity. Ligase activity requires near neutral pH, where many activated legumain enzymes rapidly degrade in the deglycoslyated form (Dall and Brandstetter, 2012). Consequently, reports on the (lack of) ligase activities of Escherichia coli produced plant legumain must be taken with some caution, e.g., among four bacterially produced legumains such as AtLEGβ, castor bean (Ricinus communis) legumain (RcAEP1), sunflower legumain HeAEP1, and jack bean (Canavalia ensiformis) legumain, only the latter was reported to display significant ligase activity (Bernath-Levin et al., 2015a).
Recently, an E. coli-produced zymogen structure of OaAEP1 from Oldenlandia affinis was published and displayed a similar mode of dimerization as AtLEGγ (Yang et al., 2017). Yang and colleagues performed the activation of OaAEP1 under reducing conditions, resulting in the peptidase-only form, consistent with our findings. Although the authors did not report on the functional relevance of dimerization or on the existence of the two-chain state of OaAEP1, structural comparison suggests that the here derived activity and stability regulation principles similarly apply to OaAEP1, including a two-chain form of OaAEP1. In fact, the conformational stability of the two-chain form at neutral pH and the role of the α6-helix as a pH-dependent master switch, will cast light on the enigmatic dual protease and ligase activities of legumain isoforms (Nguyen et al., 2015), not only restricted to Arabidopsis.
The structural and mechanistic insights presented here on the autoactivation, stabilization, and activity regulation of plant legumains will expand our molecular understanding of plant (patho-)physiology and provides a rational framework to engineer autoactivating legumain variants as tools for manifold biotechnological and medical applications, including peptide ligation and cyclization.
METHODS
Materials
Arabidopsis thaliana AEP (legumain) isoform γ (AtLEGγ) full-length clone U10153 (locus: AT4G32940) was obtained from the TAIR database. Restriction enzymes and T4 ligase were obtained from Fermentas and Pfu Ultra II Fusion HS DNA polymerase was obtained from Stratagene. Custom-made primers were obtained from Eurofins Genomics, and sequence analyses were performed at Eurofins MWG Operon. Escherichia coli strain XL1 Blue (Stratagene) was used for subcloning expression constructs. To produce fully glycosylated protein, the leishmania tarentolae expression system (LEXSY; Jena Bioscience) was used (Breitling et al., 2002). LS was obtained from AppliChem. All reagents used were of the highest standard available from Sigma-Aldrich or AppliChem. Acetonitrile (ACN; LC-MS grade) was purchased from VWR. Formic acid (98.0–100%), trifluoroacetic acid (TFA; ≥99.5%), ammonium bicarbonate (LC-MS grade), and Tris(2-carboxyethyl) phosphine hydrochloride (≥98.0%) were purchased from Sigma-Aldrich Chemie. The N-glycosidase PNGase F was obtained from New England Biolabs.
Cloning
An N-terminal truncated mutant (S56-A494) of Arabidopsis proLEG isoform γ (referred in this work as proAtLEGγ) was amplified by polymerase chain reaction (Eppendorf Mastercycler ep gradient thermal cycler) to exclude the N-terminal ER signal peptide and vacuolar sorting signal (Christoff et al., 2014). Arabidopsis legumain isoform γ full-length clone U10153 was used as a template. An appropriate forward primer containing an XbaI restriction site, hexahistidine tag, and a TEV-cleavage site, AGCTCTCGAGTCTAGAGCACCACCATCACCACCACGAAAACCTGTATTTTCAGTCCGGTACTAGGTGGGCTGTTCTAGTCGCCG, and a reverse primer containing a NotI restriction site, AGCTGCTCAGCGCGGCCGCCTATGCACTGAATCCACGGTTAAGCGAGCTCCAAGGAC, were used. Subsequently, the PCR product was cloned into the pLEXSY-sat2 vector utilizing the XbaI and NotI restriction sites. The expression constructs carried an N-terminal signal sequence for secretory expression in the LEXSY supernatant. Correctness of all constructs was confirmed by DNA sequencing.
Cell Culture, Protein Expression, and Purification
Expression constructs were stably transfected into the LEXSY P10 host strain and grown at 26°C in Brain Heart Infusion medium (Jena Bioscience) supplemented with 5 µg/mL haemin, 50 units/mL penicillin, and 50 mg/mL streptomycin (Carl Roth). Positive clones were selected by addition of nourseothricin (Jena Bioscience). Protein expression was performed as described elsewhere (Dall and Brandstetter, 2013). Recombinant protein was removed from the LEXSY supernatant via Ni2+ purification using Ni-NTA Superflow resin (Qiagen). The wash buffer contained 20 mM HEPES, pH 7.2, 300 mM NaCl, and 10% glycerol. The elution buffer was composed of 20 mM HEPES, pH 7.2, 300 mM NaCl, 10% glycerol, 250 mM imidazole, and 0.3 mM MMTS (S-methyl methanethiosulfonate). The elution fractions were concentrated using Amicon Ultra centrifugal filter units (3-kD molecular mass cutoff; Millipore) and desalted using PD-10 columns (GE Healthcare) to the final buffer: 20 mM HEPES, pH 7.2, and 50 mM NaCl.
Preparative Autoactivation to Yield Two-Chain State and Protease Only
proAtLEGγ (2–3 mg/mL) was incubated in autoactivation buffer A (100 mM Tris, 100 mM Bis-Tris, 100 mM citrate, pH 4.0, and 100 mM NaCl) for 16 h at 30°C to generate two-chain AtLEGγ. To prepare the protease only, 2 to 3 mg/mL proAtLEGγ was incubated at 30°C in autoactivation buffer B (100 mM Tris, 100 mM Bis-Tris, 100 mM citrate, pH 4.0, 100 mM NaCl, and 2 mM DTT) for 2 h. All samples were checked for the presence or absence of the α6-LSAM domain by SDS-PAGE. After autoactivation, two-chain or protease only samples were subjected to gel filtration chromatography utilizing an Äkta-FPLC system (SEC 200 10/300 GL column, buffer: 20 mM citrate, pH 4.2, and 100 mM NaCl) to remove degradation products and DTT. Afterwards, the respective fractions were either used directly for enzymatic assays or aliquoted and frozen at −20°C. Purities of all activation states can be seen in Supplemental Figure 4.
Identification of N-Glycosylation and Cleavage Sites
For the determination of the N-linked glycosylation of proAtLEGγ (1.0 mg/mL), the sample was diluted to a concentration of 0.10 mg/mL in 50 mM ammonium bicarbonate (pH 7.83) and denatured for 30 min at 60°C and 900 rpm on an Eppendorf Thermoshaker. Subsequently 2 μL PNGase F was added to each sample and the reaction took place for 3 h at 37°C. For cleavage site determination, a two-chain sample (Supplemental Figure 4) was inhibited by 0.8 mM MMTS and diluted in a ratio of 1:3 in 0.10% (v/v) aqueous formic acid to a final concentration of 0.10 mg/mL. The analysis was performed on a high-performance liquid chromatography system (Ultimate 3000; Thermo Fisher Scientific) at a flow rate of 200 μL min−1. A Discovery BIO wide pore C18 column (150 × 2.1-mm i.d., 3.0-μm particle size, 300-Å pore size; Supelco) was operated at a column temperature of 50°C. Five microliters of sample was injected in in-line split-loop mode. Separation was performed with an initial equilibration at 5.0% (v/v) ACN in 0.050% (v/v) TFA for 5 min, followed by a linear gradient of 5.0 to 50.0% ACN + 0.050% TFA in 20 min, column regeneration at 99.95% ACN in 0.050% TFA for 10 min and reequilibration at 5.0% in 0.050% TFA for 15 min. UV detection was performed with a 2.5 μL flow cell at 214 nm. Mass spectrometry was conducted on a quadrupole-Orbitrap instrument (QExactive) equipped with an Ion Max source with a heated electrospray ionization probe from Thermo Fisher Scientific. Mass calibration of the instrument was conducted with Pierce LTQ Velos ESI Positive Ion Calibration Solution from Life Technologies. The instrument settings were as follows: source heater temperature of 200°C, spray voltage of 4.0 kV, sheath gas flow of 15 arbitrary units, auxiliary gas flow of 5 arbitrary units, capillary temperature of 300°C, S-lens RF level of 60.0, in-source CID of 20.0 eV, AGC target of 1e6, and a maximum injection time of 200 ms. The measurements were performed in full scan mode with a range of m/z 1000 to 2500 at a resolution at m/z 200 of 17,500 and 140,000, respectively. Deconvolution of the ion spectra measured at a resolution of 17,500 was performed with the algorithm ReSpect integrated into BioPharma Finder version 1.0.76.10 (Thermo Fisher Scientific). The ion spectra measured at a resolution of 140,000 were deconvoluted using the Xtract algorithm in the software Xcalibur 3.0.63 (Thermo Fisher Scientific).
pH-Dependent Proteolytic Activity Measurements
The proteolytic activities of selected activation intermediates were measured using 100 µM (if not stated otherwise) of the fluorogenic substrate Z-VAN-MCA (AMC) in activity buffer A adjusted to the desired pH value (100 mM Tris, 100 mM Bis-Tris, 100 mM citrate, and 100 mM NaCl) at 293K. For each measured pH value, the reaction was started by adding 1 μL of a respective two-chain or protease only sample to the premixed 49 μL mixture. The concentration of the enzyme in the assay was <1.5 µM. The substrate turnover was measured at an excitation and emission wavelength of 370 and 450 nm, respectively, in an Infinite M200 Plate Reader (Tecan). Each experiment was determined at least in duplicate and each individual pH series was done with 10 and 100 µM of substrate, both showing the same basic overall curve progression. Proteolytic activity was determined by calculating the initial slopes of the time-dependent substrate turnover. For better comparison, activities were each normalized to pH 3.5 because at such pH the two-chain state resembles the protease-only the most.
Reversibility Test for the pH-Dependent Proteolytic Activities of Two-Chain AtLEGγ
A two-chain sample was set to pH 6.5 or 4.0 by adding the desired volume of 10-fold buffer A at the desired pH for at least the time window that was normally used to determine the proteolytic activity in a direct measurement (2–4 min) at room temperature. Afterwards, the pH 6.5 or 4.0 pretreated samples were measured for proteolytic activity at pH 5.0 or 6.5 (see proteolytic activity measurements). The measurements were done in triplicates.
Effect of LS on Autoactivation
proAtLEGγ (0.6 mg/mL) was incubated in buffer A with increasing concentrations of LS in the micromolar range far below the critical micelle concentration (CMC = 14.57 mM) at pH 4.5 and 37°C for 45 min. Then, the samples were quenched by the addition of 0.7 mM MMTS and consequently analyzed by SDS-PAGE using a nonreducing SDS-PAGE loading buffer. Additionally, a two-chain sample was analyzed for LS-induced stimulation of proteolytic activity by testing turnover of 10 µM of Z-VAN-AMC substrate in the absence or presence of 400 µM LS. A protease only sample was used as control. The measurements were done in triplicates.
Size-Exclusion Chromatography
An Äkta-FPLC system equipped with an S200 10/300 GL column was used to evaluate the pH-dependent oligomerization state of two-chain and proAtLEGγ. The latter was used after Ni2+-affinity chromatography (3 mg/mL) and two-chain samples (1.5 mg/mL) were injected after autoactivation. Completeness of turnover was analyzed using SDS-PAGE. The SEC running buffer was composed of 20 mM HEPES, pH 7.5, and 100 mM NaCl for proAtLEGγ. For the two-chain form, we used 20 mM citric acid and 100 mM NaCl or 20 mM HEPES and 100 mM NaCl at pH 4.0, 4.2, 5.0, 5.5, or 7.5, respectively. After each run, corresponding fractions were analyzed by SDS-PAGE, confirming the comigration of the protease and the α6-LSAM domains.
Concentration-Dependent Autoactivation Assay
Proteolytic activity measurements were done using different concentrations of zymogenic AtLEGγ (proAtLEGγ) with 50 µM Z-VAN-MCA (AMC). The respective gains in relative fluorescence were divided by the particular used concentration to get the specific activities and plotted against the used proAtLEGγ concentration. Autoactivation was also analyzed by SDS-PAGE. proAtLEGγ (2.45 and 0.25 mg/mL) was incubated in 100 mM buffer A at pH 4.0 at 20°C. After 1, 2, or 3 h, a sample was taken. Two micrograms for each time point was analyzed by SDS-PAGE.
Differential Scanning Fluorimetry
A differential scanning fluorimetry assay (thermal shift assay) was performed as described (Ericsson et al., 2006). SYPRO Orange (Invitrogen) was used as unfolding sensitive fluorophore. Melting curves were determined using 2× SYPRO Orange (Invitrogen) and 0.05 to 0.1 mg/mL protein. The pH of the screen solution was varied from 3.5 to 7.0 in steps of 0.5 using buffer A (100 mM Tris, 100 mM Bis-Tris, 100 mM citrate, and 100 mM NaCl). Thermal denaturation was measured in a 7500 Real Time PCR System (Applied Biosystems) from 20 to 95°C. Data processing was performed according to (Niesen, 2010). Each measurement was done in duplicate.
Protein Crystallization
AtLEGγ was purified as described above and crystallization screening was performed using the sitting-drop vapor-diffusion method utilizing a Hydra II Plus One (Matrix) liquid-handling system. Crystals grew within 5 to 10 d in a condition consisting of 0.1 M MES, pH 6.5, and 1.6 M MgSO4. To avoid further autodegradation, we modified the catalytic cysteine during crystallization with MMTS.
Data Collection and Processing
An x-ray diffraction data set was collected on beamline ID30A-3 at the ESRF. The beamline was equipped with a Pilatus3 2M detector. Data collection was performed using a crystal-to-detector distance of 237.613 mm and a wavelength of 0.9677 Å. The exposure time was 0.02 s at 10.2% transmission. Three hundred and sixty images were collected with a 0.5° oscillation range at 100K. Data processing was performed using iMOSFLM (Battye et al., 2011) and Aimless from the CCP4 program suite (Winn et al., 2011). Packing density was calculated according to Matthews (1968). An initial model could be generated by molecular replacement with the human prolegumain (PDB ID entry: 4FGU, around 46.8% sequence identity in total, whereas the C-terminal AP and LSAM domain only has a sequence identity of 14.4%). The structure was fine-tuned manually and was refined using Refmac 5 (Murshudov et al., 1997) and phenix.refine (Afonine et al., 2012) . The structure was deposited into the protein data bank under the PDB-ID: 5NIJ.
Peptide Ligation
Interpeptide ligation was accomplished upon incubation of 400 µM of Acβ-ASTRN-HVAAK(Alloc)-amide and 5-, 10-, or 20-fold molar excess of HGLPVK(Alloc)-amide in the presence of 1 µM AtLEGγ, overnight and at 37°C. The reaction buffer consisted of 100 mM NaCl, 100 mM of Tris, Bis-Tris, and citric acid at a pH of 5.0 or 6.5. The ligation mixture was desalted using the ZipTip Pipette Tip C-18 (Merck Millipore) and analyzed by MALDI-TOF-MS (Autoflex; Bruker Daltonics; matrix: α-cyano-4-hydroxycinnamic acid). Control samples consisted of two mixtures without AtLEGγ or HGLPVK(Alloc)-amide.
Electrostatic and RMSD Calculations
Electrostatics were calculated using the PDB2PQR server version 2.0.0 (Dolinsky et al., 2004). Calculations were performed using the proAtLEGγ residues 56 to 337 and 370 to 494. The protonation state at the desired pH values was assigned using Propka, and the corresponding protons were positioned using the PARSE force field. The electrostatic surface potential and positive as well as negative isosurfaces were displayed using Pymol 1.7.2.0 and the APBS tool 2.1 plug-in. The software Superpose from the CCP4 suite was used to superimpose two-chain states and to get a list of Cα root mean square deviation values.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Uniprot accession number Q39119 and AGI code AT4G32940 for Arabidopsis vacuolar-processing enzyme γ-isozyme; andUniprot accession number Q4GWU5 for sunflower trypsin inhibitor SFTI (Helianthus annuus).
Supplemental Data
Supplemental Figure 1. Multiple sequence alignment of legumains from mammals and plants.
Supplement Figure 2. Peptide:N-Glycosidase F (PNGase F) treatment in combination with mass spectrometry measurements reveal an N-glycosylation specific for an N-glycan core.
Supplement Figure 3. Structural alignment of the latent active site and wall-eye stereoview of the electron density around the active site.
Supplement Figure 4. Purities of the activation states and determination of autocleavage sites at the AP-LSAM domain leading to the two chain state.
Supplement Figure 5. Comparison of the LSAM binding sites between plant and mammalian legumains.
Supplement Figure 6. The nonionic detergent lauroylsarcosine induces autoactivation by interfering with the hydrophobic catalytic domain:LSAM interface.
Supplement Figure 7. pH-dependent isosurfaces of the α6-repulsion area and the helix loop helix bundles of AtLEGγ, and mouse and human legumain.
Acknowledgments
We thank Guy Salvesen for invaluable discussions, Stefan Senn for assisting with scripting and mass spectrometry data analysis, and Peter Goettig for fruitful discussions in the structure solving process. This work was supported by the Austrian Science Fund (FWF, project W_01213) and the Austrian Federal Ministry of Science, Research, and Economy and the National Foundation for Research, Technology, and Development. We thank the anonymous reviewers for their extraordinarily constructive comments to improve the manuscript.
AUTHOR CONTRIBUTIONS
F.B.Z. designed and performed most experiments. F.B.Z., E.D., and H.B. discussed and interpreted all experiments. C.R. and C.G.H. analyzed the N-glycosylation and autocleavage sites by mass spectrometry. L.G. and C.C. synthesized the peptides for ligation, and assayed and interpreted the ligation by mass spectrometry. F.B.Z. and H.B. wrote the manuscript, and all authors proofread and agreed with the article.
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Abe Y., Shirane K., Yokosawa H., Matsushita H., Mitta M., Kato I., Ishii S. (1993). Asparaginyl endopeptidase of jack bean seeds. Purification, characterization, and high utility in protein sequence analysis. J. Biol. Chem. 268: 3525–3529. [PubMed] [Google Scholar]
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68: 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye T.G.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.W. (2011). iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C., Shutov A.D., Nong V.H., Senyuk V.I., Jung R., Horstmann C., Fischer J., Nielsen N.C., Müntz K. (1995). Purification, cDNA cloning and characterization of proteinase B, an asparagine-specific endopeptidase from germinating vetch (Vicia sativa L.) seeds. Eur. J. Biochem. 228: 456–462. [PubMed] [Google Scholar]
- Bernath-Levin K., Nelson C., Elliott A.G., Jayasena A.S., Millar A.H., Craik D.J., Mylne J.S. (2015a). Peptide macrocyclization by a bifunctional endoprotease. Chem. Biol. 22: 571–582. [DOI] [PubMed] [Google Scholar]
- Bernath-Levin K., Nelson C., Elliott A.G., Jayasena A.S., Millar A.H., Craik D.J., Mylne J.S. (2015b). Peptide macrocyclization by a bifunctional endoprotease. Chem. Biol. 22: 571–582. [DOI] [PubMed] [Google Scholar]
- Bonneau L., Ge Y., Drury G.E., Gallois P. (2008). What happened to plant caspases? J. Exp. Bot. 59: 491–499. [DOI] [PubMed] [Google Scholar]
- Breitling R., Klingner S., Callewaert N., Pietrucha R., Geyer A., Ehrlich G., Hartung R., Müller A., Contreras R., Beverley S.M., Alexandrov K. (2002). Non-pathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr. Purif. 25: 209–218. [DOI] [PubMed] [Google Scholar]
- Chrispeels M.J., Herman E.M. (2000). Endoplasmic reticulum-derived compartments function in storage and as mediators of vacuolar remodeling via a new type of organelle, precursor protease vesicles. Plant Physiol. 123: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoff A.P., Turchetto-Zolet A.C., Margis R. (2014). Uncovering legumain genes in rice. Plant Sci. 215–216: 100–109. [DOI] [PubMed]
- Conlan B.F., Gillon A.D., Craik D.J., Anderson M.A. (2010). Circular proteins and mechanisms of cyclization. Biopolymers 94: 573–583. [DOI] [PubMed] [Google Scholar]
- Craik D.J., Malik U. (2013). Cyclotide biosynthesis. Curr. Opin. Chem. Biol. 17: 546–554. [DOI] [PubMed] [Google Scholar]
- Dall E., Brandstetter H. (2012). Activation of legumain involves proteolytic and conformational events, resulting in a context- and substrate-dependent activity profile. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68: 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall E., Brandstetter H. (2013). Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation. Proc. Natl. Acad. Sci. USA 110: 10940–10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall E., Brandstetter H. (2016). Structure and function of legumain in health and disease. Biochimie 122: 126–150. [DOI] [PubMed] [Google Scholar]
- Dall E., Fegg J.C., Briza P., Brandstetter H. (2015). Structure and mechanism of an aspartimide-dependent peptide ligase in human legumain. Angew. Chem. Int. Ed. Engl. 54: 2917–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A. (2004). PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32: W665–W667. [DOI] [PMC free article] [PubMed]
- Ericsson U.B., Hallberg B.M., Detitta G.T., Dekker N., Nordlund P. (2006). Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357: 289–298. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P., Salvesen G.S. (2004). The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 384: 201–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach S.L., Rathinakumar R., Chakravarty G., Göransson U., Wimley W.C., Darwin S.P., Mondal D. (2010). Anticancer and chemosensitizing abilities of cycloviolacin 02 from Viola odorata and psyle cyclotides from Psychotria leptothyrsa. Biopolymers 94: 617–625. [DOI] [PubMed] [Google Scholar]
- Gould A., Ji Y., Aboye T.L., Camarero J.A. (2011). Cyclotides, a novel ultrastable polypeptide scaffold for drug discovery. Curr. Pharm. Des. 17: 4294–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley G.R., Scholtz J.M., Pace C.N. (2009). A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 18: 247–251. [DOI] [PMC free article] [PubMed]
- Hara-Nishimura I., Hatsugai N. (2011). The role of vacuole in plant cell death. Cell Death Differ. 18: 1298–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I., Takeuchi Y., Nishimura M. (1993). Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni. Plant Cell 5: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K.S., Durek T., Kaas Q., Poth A.G., Gilding E.K., Conlan B.F., Saska I., Daly N.L., van der Weerden N.L., Craik D.J., Anderson M.A. (2015). Efficient backbone cyclization of linear peptides by a recombinant asparaginyl endopeptidase. Nat. Commun. 6: 10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N., Kuroyanagi M., Nishimura M., Hara-Nishimura I. (2006). A cellular suicide strategy of plants: vacuole-mediated cell death. Apoptosis 11: 905–911. [DOI] [PubMed] [Google Scholar]
- Hatsugai N., Kuroyanagi M., Yamada K., Meshi T., Tsuda S., Kondo M., Nishimura M., Hara-Nishimura I. (2004). A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858. [DOI] [PubMed] [Google Scholar]
- Hatsugai N., Yamada K., Goto-Yamada S., Hara-Nishimura I. (2015). Vacuolar processing enzyme in plant programmed cell death. Front. Plant Sci. 6: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Yamada K., Shimada T., Matsushima R., Nishizawa N.K., Nishimura M., Hara-Nishimura I. (2001). A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42: 894–899. [DOI] [PubMed] [Google Scholar]
- Hiraiwa N., Nishimura M., Hara-Nishimura I. (1997). Expression and activation of the vacuolar processing enzyme in Saccharomyces cerevisiae. Plant J. 12: 819–829. [DOI] [PubMed] [Google Scholar]
- Hiraiwa N., Nishimura M., Hara-Nishimura I. (1999). Vacuolar processing enzyme is self-catalytically activated by sequential removal of the C-terminal and N-terminal propeptides. FEBS Lett. 447: 213–216. [DOI] [PubMed] [Google Scholar]
- Ireland D.C., Wang C.K.L., Wilson J.A., Gustafson K.R., Craik D.J. (2008). Cyclotides as natural anti-HIV agents. Biopolymers 90: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembhavi A.A., Buttle D.J., Knight C.G., Barrett A.J. (1993). The two cysteine endopeptidases of legume seeds: purification and characterization by use of specific fluorometric assays. Arch. Biochem. Biophys. 303: 208–213. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Yamada K., Hiraiwa N., Kondo M., Nishimura M., Hara-Nishimura I. (1999a). Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J. 19: 43–53. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Yamada K., Hiraiwa N., Kondo M., Nishimura M., Hara-Nishimura I. (1999b). Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J. 19: 43–53. [DOI] [PubMed] [Google Scholar]
- Koehnke J., et al. (2012). The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Nat. Struct. Mol. Biol. 19: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi M., Nishimura M., Hara-Nishimura I. (2002). Activation of Arabidopsis vacuolar processing enzyme by self-catalytic removal of an auto-inhibitory domain of the C-terminal propeptide. Plant Cell Physiol. 43: 143–151. [DOI] [PubMed] [Google Scholar]
- Lesner A., Łęgowska A., Wysocka M., Rolka K. (2011). Sunflower trypsin inhibitor 1 as a molecular scaffold for drug discovery. Curr. Pharm. Des. 17: 4308–4317. [DOI] [PubMed] [Google Scholar]
- Li W. (2014). Structural Analysis of Bacterial and Viral Infections and a Cysteine Proteinase Legumain. PhD dissertation (Braunschweig, Germany: Technical University of Braunschweig). [Google Scholar]
- Martinière A., Bassil E., Jublanc E., Alcon C., Reguera M., Sentenac H., Blumwald E., Paris N. (2013). In vivo intracellular pH measurements in tobacco and Arabidopsis reveal an unexpected pH gradient in the endomembrane system. Plant Cell 25: 4028–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B.W. (1968). Solvent content of protein crystals. J. Mol. Biol. 33: 491–497. [DOI] [PubMed] [Google Scholar]
- Müntz K., Shutov A.D. (2002). Legumains and their functions in plants. Trends Plant Sci. 7: 340–344. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Mylne J.S., Colgrave M.L., Daly N.L., Chanson A.H., Elliott A.G., McCallum E.J., Jones A., Craik D.J. (2011). Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat. Chem. Biol. 7: 257–259. [DOI] [PubMed] [Google Scholar]
- Nguyen G.K.T., Kam A., Loo S., Jansson A.E., Pan L.X., Tam J.P. (2015). Butelase 1: a versatile ligase for peptide and protein macrocyclization. J. Am. Chem. Soc. 137: 15398–15401. [DOI] [PubMed] [Google Scholar]
- Nguyen G.K.T., Qiu Y., Cao Y., Hemu X., Liu C.-F., Tam J.P. (2016). Butelase-mediated cyclization and ligation of peptides and proteins. Nat. Protoc. 11: 1977–1988. [DOI] [PubMed] [Google Scholar]
- Nguyen G.K.T., Wang S., Qiu Y., Hemu X., Lian Y., Tam J.P. (2014). Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat. Chem. Biol. 10: 732–738. [DOI] [PubMed] [Google Scholar]
- Nguyen G.K., Wang S., Qiu Y., Hemu X., Lian Y., Tam J.P. (2014). Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat. Chem. Biol. 10: 732–738. [DOI] [PubMed] [Google Scholar]
- Niesen F. (2010). Excel Script for the Analysis of Protein Unfolding Data Acquired by Differential Scanning Fluorimetry. www.beta-sheet.org/resources/T24-DSF-Analysis-Manual-v3.0.pdf. [Google Scholar]
- Otegui M.S., Spitzer C. (2008). Endosomal functions in plants. Traffic 9: 1589–1598. [DOI] [PubMed] [Google Scholar]
- Ozawa K., Laskowski M. Jr (1966). The reactive site of trypsin inhibitors. J. Biol. Chem. 241: 3955–3961. [PubMed] [Google Scholar]
- Reguera M., Bassil E., Tajima H., Wimmer M., Chanoca A., Otegui M.S., Paris N., Blumwald E. (2015). pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 27: 1200–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Martín R., Carter C., Zouhar J., Pan S., Plotnikova J., Jin H., Paneque M., Sánchez-Serrano J.J., Baker B., Ausubel F.M., Raikhel N.V. (2004). VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr. Biol. 14: 1897–1906. [DOI] [PubMed] [Google Scholar]
- Rosin F.M., Watanabe N., Lam E. (2005). Moonlighting vacuolar protease: multiple jobs for a busy protein. Trends Plant Sci. 10: 516–518. [DOI] [PubMed] [Google Scholar]
- Saska I., Gillon A.D., Hatsugai N., Dietzgen R.G., Hara-Nishimura I., Anderson M.A., Craik D.J. (2007). An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J. Biol. Chem. 282: 29721–29728. [DOI] [PubMed] [Google Scholar]
- Schechter I., Berger A. (1967). On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27: 157–162. [DOI] [PubMed] [Google Scholar]
- Winn M.D., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Nishimura M., Hara-Nishimura I. (2004). The slow wound-response of gammaVPE is regulated by endogenous salicylic acid in Arabidopsis. Planta 218: 599–605. [DOI] [PubMed] [Google Scholar]
- Yamada K., Shimada T., Nishimura M., Hara-Nishimura I. (2005). A VPE family supporting various vacuolar functions in plants. Physiol. Plant. 123: 369–375. [Google Scholar]
- Yang R., Wong Y.H., Nguyen G.K.T., Tam J.P., Lescar J., Wu B. (2017). Engineering a catalytically efficient recombinant protein ligase. J. Am. Chem. Soc. 139: 5351–5358. [DOI] [PubMed] [Google Scholar]
- Zarrinpar A., Bhattacharyya R.P., Lim W.A. (2003). The structure and function of proline recognition domains. Sci. STKE 2003: RE8. [DOI] [PubMed] [Google Scholar]
- Zhao L., et al. (2014). Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage. Cell Res. 24: 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]