Abstract
The risk of spillover of enzootic paramyxoviruses, and the susceptibility of recipient human and domestic animal populations, are defined by a broad collection of ecological and molecular factors that interact in ways that are not yet fully understood. Nipah and Hendra viruses were the first highly-lethal zoonotic paramyxoviruses discovered in modern times, but other paramyxoviruses from multiple genera are present in bats and other reservoirs that have unknown potential to spill over into humans. We outline our current understanding of paramyxovirus reservoir hosts and the ecological factors that may drive spillover, and we explore the molecular barriers to spillover that emergent paramyxoviruses may encounter. By outlining what is known about enzootic paramyxovirus receptor usage, mechanisms of innate immune evasion, and other host-specific interactions, we highlight the breadth of unexplored avenues that may be important in understanding paramyxovirus emergence.
Keywords: Paramyxovirus, emerging viruses, zoonotic, reservoir, ecology, Henipavirus, Rubulavirus, Morbillivirus, tropism, pathogenesis
1. Introduction
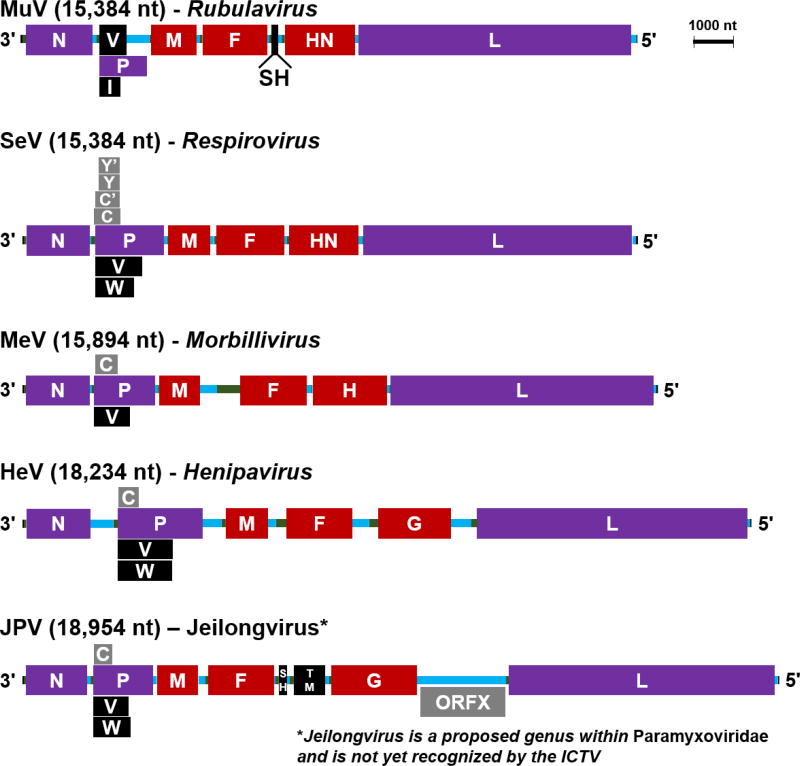
The paramyxovirus family comprises numerous viruses that collectively have a very broad host range, infecting vertebrates from fish to mammals, with transmission primarily via the respiratory route (Lamb & Parks 2006). At the molecular level, paramyxoviruses are enveloped, negative-strand, non-segmented RNA viruses, with six open reading frames encoding the structural genes. N (nucleocapsid), encapsidates genomic and antigenomic viral RNA, while L (polymerase) and P (phosphoprotein) form the RNA-dependent RNA polymerase to transcribe viral messenger RNAs, and coordinate with N to amplify the genome; together these comprise the viral replicase (Figure 1, purple). M (matrix) coordinates virion assembly and budding, while the F (fusion) and G/H/HN (attachment) glycoproteins decorate the surface of the virion and are responsible for entry into the naïve host cell; together with the replicase, these proteins (Figure 1, red) form the virion structure. In most cases, paramyxoviruses are further defined by the use of an alternative start codon and RNA editing to generate two to four additional accessory proteins from the phosphoprotein (P) gene (Figure 1, grey and black; Table 1). Some, but not all, paramyxoviruses have additional open reading frames for non-structural proteins (e.g. Mumps virus – MuV, or Jeilongvirus – JPV, in Figure 1). Collectively, the functions of the various non-structural accessory proteins are incompletely understood, although many have been implicated in manipulating host immune responses (discussed below). An interesting and defining feature of paramyxovirus genome organization is conformity to the “rule of six”: as a consequence of the mode of genome encapsidation by the nucleoprotein, genomes must be a multiple of six nucleotides in length in order for efficient paramyxovirus replication (Kolakofsky et al. 1998; Kolakofsky et al. 2005).
Figure 1. Paramyxovirus genome structures.
Depicted here are scaled genomes of representative virus species in 3' to 5' orientation for each of the mammalian-targeting Paramyxovirus genera. JPV is included to represent the putative Jeilongvirus genus. Protein gene products are the larger lettered blocks: replicase proteins are purple (N, P, L), while the remaining virion proteins are red (M, F, G/H/HN). Non-structural accessory proteins are black (V, W, I, SH, TM) or grey (C, Y, ORFX); grey proteins are produced from alternate start codons of a given mRNA. RNA regions are thinner, unlettered horizontal segments: 3’ leader and 5’ trailer regions are black, while the 5’ UTR of each mRNA is colored green, and the 3’ UTR of each mRNA is colored blue. The scale bar indicates 1000 nucleotides. A color version of this figure is available online.
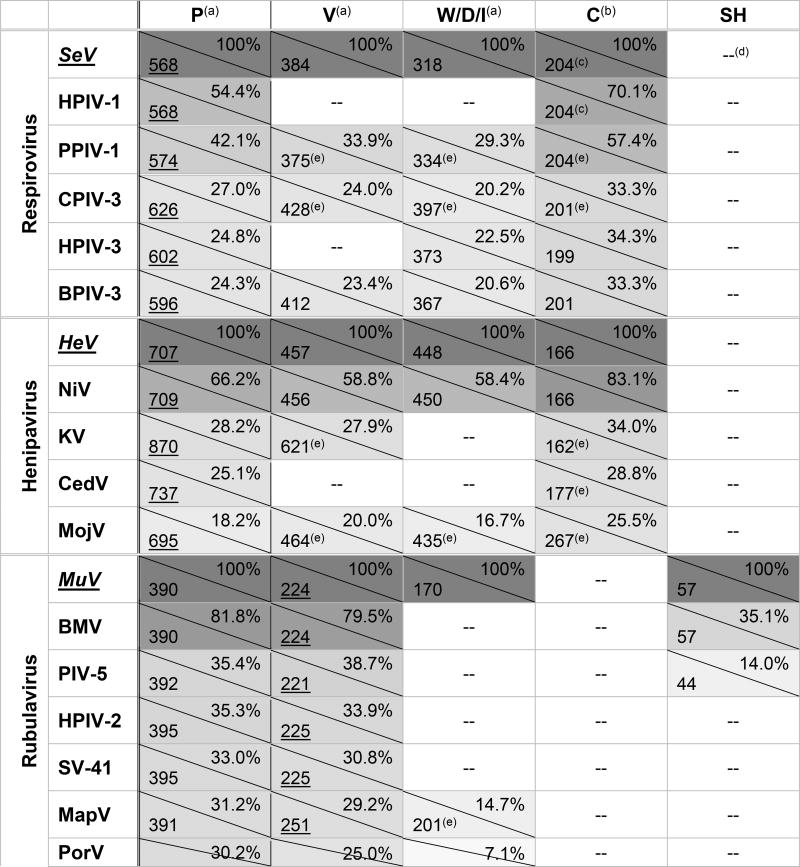
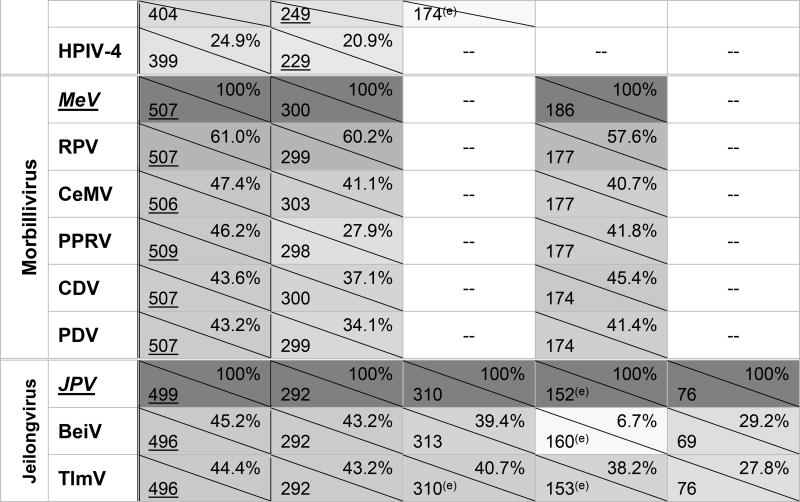
Table 1. Paramyxovirus accessory proteins of interest.
Sizes are given in amino acids (aa; bottom left corner) while percent amino acid identity (progressive pairwise alignment, calculated with default settings of ClustalW in MEGA7.0) to each genus’ type species is given in upper right corner; boxes are shaded to represent percent amino acid identity.
Rubulavirus V proteins (underlined) are the primary transcripts, with P/I proteins being produced through mRNA editing. For all other genera, P proteins (underlined) are the primary transcript, with V/W/D proteins produced through mRNA editing.
C proteins are produced from alternate start codons of the P ORF and are translated in a different open reading frame.
SeV and HPIV-1 produce additional alternate start codon proteins in the same reading frame as C, named C', Y1, and Y2. They share the majority of their sequence with C, but with slightly longer (C') or shorter (Y1, Y2) N termini.
-- = not present or not expressed
putative
Mammalian paramyxoviruses are currently classified into four genera – Morbillivirus, Rubulavirus, Respirovirus, and Henipavirus – each of which contain both human-tropic and animal-tropic viruses (Afonso et al. 2016). However, many recently-identified paramyxoviruses fall outside these existing genera, and may represent additional clusters of closely-related mammalian paramyxoviruses (Figure 2) (Magoffin et al. 2007; Wilkinson et al. 2014). Because there is no evidence that reptilian, piscine, or avian paramyxoviruses pose a major health threat to humans, this review will focus on discovery and understanding of the mammalian paramyxoviruses.
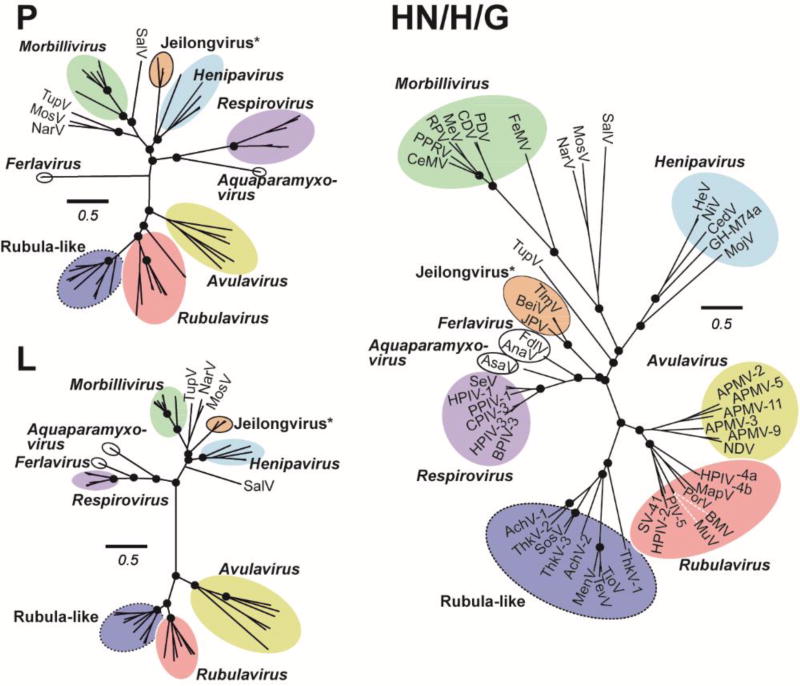
Figure 2. Paramyxovirus attachment proteins demonstrate great diversity within and among genera.
Maximum likelihood phylogenetic trees of P, L and attachment (HN/H/G) protein sequences of Paramyxoviridae. Colored circles encompass phylogenetic groupings best characterized as genera. *Jeilongvirus is a proposed genus that has not yet been recognized by the ICTV, and so are encircled in black. Rubula-like viruses have genetic similarities to recognized members of Rubulavirus, but group separately, and so are encircled in a dotted line. Unclassified-but-fully-sequenced Paramyxoviruses are individually labelled in each tree. Scale bar indicates 0.5 amino acid substitutions per site, and trees are scaled such that the scale bar is the same in all trees. Black circles at nodes represent >75% support from 500 bootstrap replicates. All analyses were performed in MEGA 6 using a complete deletion option and a WAG substitution model. A color version of this figure is available online.
We know from early written accounts of disease that some paramyxoviruses, such as measles virus (MeV), have circulated in human populations and caused substantial morbidity and mortality for well over 1000 years (Furuse et al. 2010). However, in just the last several decades, the repeated zoonotic outbreaks of the highly lethal Hendra virus (HeV) and Nipah virus (NiV) has alerted us to the threat of emergence of as-yet-unknown paramyxoviruses into human populations from wildlife reservoirs. Fully-sequenced, novel enzootic paramyxoviruses are catalogued in Table 2, and their wildlife reservoir(s) are indicated, where they have been identified. In this review, we will discuss our current knowledge of emergent paramyxoviruses, but also look more broadly at whether more divergent paramyxoviruses circulating in wildlife reservoirs also warrant increased attention as having zoonotic potential. In addition, we will discuss what we know, as well as what we cannot yet predict, in terms of the factors anticipated to contribute to the likelihood of emergence of highly pathogenic zoonotic paramyxoviruses.
Table 2. Distribution, classification, and origins of fully-sequenced enzootic and emergent paramyxoviruses.
| Virus | Abbrev. | Proposed Genus |
Geographical distribution | Animal source(s) (Order) |
Spillover hosts |
Evidence | References |
|---|---|---|---|---|---|---|---|
| Achimota virus 1 | AchV-1; AchPV1 | Rubula-like(a) | Accra, Ghana Dar es Salaam, Tanzania São Tomé and Principe Annobón, Equatorial Guinea | Eidolon helvum (Chiroptera) | Humans | Serology | (Baker et al. 2013) |
| Achimota virus 2 | AchV-2; AchPV2 | Rubula-like(a) | Accra, Ghana Dar es Salaam/Muheza, Tanzania São Tomé and Principe | Eidolon helvum (Chiroptera) | (Baker et al. 2013) | ||
| Bat Mumps virus | BMV; BMuV | Rubulavirus | Democratic Republic of Congo | Epomophorus spp. (Chiroptera) | (Drexler et al. 2012) | ||
| Beilong virus | BeiV; BeiPV | Jeilongvirus(b) | Laboratory animal cell line – Beijing, China Hong Kong, China La Réunion, France (Indian Ocean) | Rattus norwegicus, rattus; (Rodentia) | (Li et al. 2006; Wilkinson et al. 2014; Woo et al. 2012) | ||
| Cedar henipavirus | CedV | Henipavirus | Queensland/Victoria, Australia | Pteropus alecto, poliocephalus (Chiroptera) | (Marsh et al. 2012; Burroughs et al. 2016) | ||
| Cetacean morbillivirus | CeMV; DMV; PWMV; | Morbillivirus | Worldwide | Dolphins, porpoises; toothed and baleen whales (Cetacea) | (Van Bressem et al. 2014)(c) | ||
| Caprine parainfluenza virus 3 | CPIV-3 | Respirovirus | Jiangsu/Anhui, China | Capra aegagrus hircus (Artiodactyla) | Goats(d) | Virus, Serology | (Li et al. 2014) |
| Feline morbillivirus | FeMV; FMoV | Morbilli-like(a) | China Japan USA | Felis cattus (Carnivora) | Cats(d) | Virus, Serology | (Furuya et al. 2014; Sharp et al. 2016; Sakaguchi et al. 2014; Woo et al. 2012) |
| Ghanaian bat henipavirus | GH-M74a KV; GhV; | Henipavirus | Ghana Cameroon | Eidolon helvum (Chiroptera) | Humans | Serology | (Drexler et al. 2012; Pernet et al. 2014b) |
| Hendra virus | HeV | Henipavirus | New South Wales/ Northern Territory/ Western Australia/ Queensland, Australia Papua New Guinea | Pteropus alecto, conspicillatus Other fruit bat spp. (Chiroptera) | Horses, humans, dogs | Virus, Serology | (Field 2016) (c) (Field et al. 2013) |
| J paramyxovirus | JPV | Jeilongvirus(b) | Queensland, Australia | Mus musculus (Rodentia) | Humans, pigs, cows | Serology | (Drexler et al. 2012; Jun et al. 1977; Mesina et al. 1974) |
| Mapuera virus | MapV | Rubulavirus | Para, Brazil | Sturnira lilium (Chiroptera) | (Karabatsos 1985) | ||
| Menangle virus | MenV | Rubula-like(a) | New South Wales/ Queensland, Australia Papua New Guinea | Dobsonia magna; Pteropus alecto, conspicillatus, poliocephalus, scapulatus (Chiroptera) | Pigs, humans | Virus, Serology | (Barr et al. 2012; Breed et al. 2010; Chant et al. 1998; Philbey et al. 1998) |
| Mojiang virus | MojV | Henipavirus | Yunnan Province, China | Rattus flavipectus (Rodentia) | Humans | association(e) | (Wu et al. 2014) |
| Mossman virus | MosV | Morbilli-like(a) | Queensland, Australia | Rattus leucopus, fuscipes (Rodentia) | (Campbell et al. 1977) | ||
| Nariva virus | NarV | Morbilli-like(a) | Bush Bush Island, Trinidad | Zygodontomys brevicauda brevicauda (Rodentia) | (Tikasingh et al. 1966) | ||
| Nipah virus | NiV | Henipavirus | Bangladesh Ghana West Bengal, India Malaysia Papua New Guinea Singapore Thailand Philippines | Pteropus spp.; Eidolon helvum, Dobsonia magna (Chiroptera) | Pigs, humans, horses, dogs, goats, cows, cats | Virus, Serology | (Luby and Gurley 2015)(c) (Breed et al. 2010; Ching et al. 2015; Chowdhury et al. 2014; Hayman et al. 2008; Wacharapluesadee et al. 2005) |
| Phocine distemper virus | PDV | Morbillivirus | Worldwide | Seals, sea lions, sea otters, walruses (Carnivora) | (Duignan et al. 2014)(c) | ||
| Porcine rubulavirus | PorV; LPMV-PorPV; PPBED | Rubulavirus | Guanajuato/Jalisco/Michoacan/Estado de Mexico, Mexico | Sus scrofa (Artiodactyla) Rhogeessa parvula major, (Chiroptera) | Pigs, humans | Virus, Serology | (Cuevas-Romero et al. 2015)(c) (Rivera-Benitez et al. 2014; Salas-Rojas et al. 2004) |
| Porcine parainfluenza virus 1 | PPIV-1 | Respirovirus | Hong Kong, China Illinois/Nebraska/Oklahoma, USA | Sus scrofa (Artiodactyla) | Pigs | Virus | (Lau et al. 2013; Palinski et al. 2016) |
| Salem virus | SalV | Morbilli-like(a) | Massachusetts, USA | Equus caballus (Perissodactyla) | Horses(d) | Virus, Serology | (Renshaw et al. 2000) |
| Sosuga virus | SosV | Rubula-like(a) | South Sudan Uganda | Homo sapiens (Primates) Rousettus aegypticus (Chiroptera) | Humans | Virus | (Albariño et al. 2014; Amman et al. 2015) |
| Teviot paramyxovirus | TevV | Rubula-like(a) | Queensland/Victoria, Australia | Pteropus alecto, poliocephalus (Chiroptera) | (Barr et al. 2015; Burroughs et al. 2015) | ||
| Tuhoko virus 1 | ThkV-1 | Rubula-like(a) | Guangdong, China | Rousettus leschenaulti (Chiroptera) | (Lau et al. 2010) | ||
| Tuhoko virus 2 | ThkV-2 | Rubula-like(a) | Guangdong, China | Rousettus leschenaulti (Chiroptera) | (Lau et al. 2010) | ||
| Tuhoko virus 3 | ThkV-3 | Rubula-like(a) | Guangdong, China | Rousettus leschenaulti (Chiroptera) | (Lau et al. 2010) | ||
| Tioman virus | TioV | Rubula-like(a) | Tioman Island, Malaysia Madagascar Papua New Guinea Assam, India | Pteropus spp. (Chiroptera) | Humans, pigs | Virus, Serology | (Breed et al. 2010; Chua et al. 2001; Iehlé et al. 2007; Yadav et al. 2016; Yaiw et al. 2007; Yaiw et al. 2008) |
| Tailam paramyxovirus | TlmV | Jeilongvirus(b) | Hong Kong, China | Rattus andamanensis (Rodentia) | (Woo et al. 2011) | ||
| Tupaia paramyxovirus | TupV; TPMV | Morbilli-like(a) | Bangkok, Thailand | Tupaia belangeri (Scandentia) | (Tidona et al. 1999) |
unclassified, but phylogenetically similar to the indicated genus
proposed genus (Li et al. 2006)
review article
reservoir and spillover host are same species
temporal and geographical association, no molecular or immunological evidence
2. Known highly lethal emergent paramyxoviruses: Nipah virus and Hendra virus
NiV and HeV (both species in the Henipavirus genus) are highly virulent emerging zoonotic paramyxoviruses that circulate in Pteropus species of bats (Halpin et al. 2011), and have caused numerous outbreaks of respiratory and neurological disease in humans and domesticated mammals over the last several decades (reviewed in Eaton et al. 2006). Remarkably high case fatality rates, their repeated re-emergence, and the absence of any vaccines or therapeutics approved for use in humans has led to their classification as pathogens that require biosafety level 4 (BSL-4) containment facilities for handling, and they are designated “Category C” priority pathogens by the National Institute of Allergy and Infectious Diseases (NIAID) Biodefense Research Agenda, reflecting their potentially devastating impact if used as bioterrorism agents (NIAID & NIH 2016). Moreover, in December 2015, NiV was included in the World Health Organization (WHO) list of the seven pathogens it deems most require urgent research and development in order to mitigate potential public health emergencies (WHO 2015).
2.1 Hendra virus
Chronologically, the first zoonotic HeV outbreak occurred in Mackay, Australia in August 1995, resulting in the deaths of two horses and a farm worker (Hooper et al. 1996; Rogers et al. 1996). However, HeV infection was only retrospectively identified as the etiology of these cases, following a larger outbreak almost 1000 km away in the Brisbane suburb Hendra, Australia, approximately one month later. In this second outbreak, 21 horses and two further people presented with severe disease, and a novel zoonotic paramyxovirus was identified as causative (Murray et al. 1995). This highly lethal emerging virus was initially classified as a morbillivirus, but further characterization, as well as the subsequent emergence of NiV (below), a phylogenetically and antigenically related paramyxovirus (Chua et al. 2000), led to reclassification of HeV as the type species in a novel paramyxovirus genus Henipavirus, which also includes NiV (Mayo 2002; Wang et al. 2000; see also Figure 2, “L” bottom left). In the decades following the initial emergence of HeV, it has continued to cause annual spillovers and outbreaks in horses and closely-associated humans (Field 2016).
2.2 Nipah virus
The first documented outbreak of NiV began in September 1998 in Peninsular Malaysia, with respiratory illness in pigs (Mohd Nor et al. 2000), and severe, frequently fatal, febrile illness in humans (Chua et al. 2000). The outbreak continued for over six months, and as a result of the transport of infected pigs, spread to more distant regions of Malaysia, and to Singapore, cumulatively causing 265 human cases of encephalitis and 105 fatalities (Chua et al. 2000; Paton et al. 1999). Following this initial outbreak, NiV re-emerged separately in Bangladesh (Arankalle et al. 2011; Hsu et al. 2004; Rahman & Chakraborty 2012), and in Siliguri, India (Chadha et al. 2006), both during 2001. NiV has subsequently re-emerged almost every year in Bangladesh, with frequent outbreaks also documented in India (Islam et al. 2016; Kulkarni et al. 2013; Luby et al. 2009). These outbreaks differ from the first – Malaysian – NiV outbreak in that transmission appears to occur directly from the bat reservoir to humans, primarily via consumption of date palm sap contaminated with urine or saliva from infected bats (Islam et al. 2016; Luby et al. 2006; Openshaw et al. 2016; Rahman et al. 2012). Furthermore, there is clear evidence of secondary human-to-human transmission during outbreaks (Hegde et al. 2016; Homaira et al. 2010; Sazzad et al. 2013), raising serious concern that NiV has the capacity to cause a devastating pandemic (Luby 2013).
2.3 Broad geographic distribution of potentially zoonotic henipaviruses
Given that documented NiV and HeV outbreaks have so far been relatively restricted in their geographical distribution (South East Asia and Australia, respectively), it is perhaps both surprising and worrisome that in the last several years, survey studies in wildlife and domesticated mammals have revealed evidence of henipa- and henipa-like viruses circulating in animal populations as distantly as West Africa, and perhaps even in the south and central Americas (Drexler et al. 2009; Drexler et al. 2012; Hasebe et al. 2012; Hayman et al. 2008; Hayman et al. 2011; Iehlé et al. 2007; Pernet et al. 2014b; Reynes et al. 2005; Thanapongtharm et al. 2015). These data highlight the role human and ecological factors must play in determining the probability of zoonotic transmission of henipaviruses from animal reservoirs to human populations, which will be discussed in detail below. However, the recent NiV outbreak in the Philippines, a country previously unaffected by NiV, with evidence of a novel route of exposure – consumption of meat from infected horses – serves to highlight that risk factors and drivers of emergence cannot be expected to remain static (Ching et al. 2015).
Given that NiV and HeV are sufficiently different at the molecular level from other paramyxoviruses to have warranted classification of a novel genus (Figure 2), and that they are the only paramyxoviruses confirmed to have fatally infected humans in repeated outbreaks, this poses the question: Are henipaviruses unique in their likelihood to spillover from wildlife reservoirs and cause serious human disease? While the answer to this question is currently unknown, it is clearly of important consequence in terms of how we direct our attention and resources when considering the zoonotic potential of viruses in the paramyxovirus family. We propose that there are several lines of evidence suggesting that viruses from multiple paramyxovirus genera, as well as numerous currently unclassified paramyxoviruses, should be considered alongside henipaviruses as having zoonotic potential.
One aspect of this question is whether henipaviruses represent a genus of viruses that are inherently highly pathogenic to humans, should spillover infection occur. This is particularly pertinent when considering the recent dramatic expansion in our knowledge of both the sequence diversity of henipaviruses present in wildlife, and the geographical areas in which henipaviruses exist (Drexler et al. 2012; Hayman et al. 2008; Iehlé et al. 2007; Peel et al. 2013; Pernet et al. 2014b). While there is currently insufficient evidence to determine whether henipaviruses (as compared to other paramyxoviruses) have a higher probability of being highly pathogenic in humans, the recent characterization of Cedar virus (CedV), a henipavirus closely related to NiV and HeV, suggests that high pathogenicity is at least not a universal feature of the Henipavirus genus. CedV was isolated from Pteropus species bats, displays some antigenic cross-reactivity with both NiV and HeV, and likely uses the same cellular entry receptor (ephrin-B2) (Marsh et al. 2012). However, despite this high degree of relatedness, CedV experimental infection of ferrets and guinea pigs resulted in seroconversion but no overt clinical disease (Marsh et al. 2012), in marked contrast to NiV and HeV, which are highly pathogenic in these model species (Pallister et al. 2009; Pallister et al. 2011). Molecular characterization of CedV demonstrated an absence of production of frameshift accessory proteins from the P gene (Table 1; Marsh et al. 2012), which are produced in almost all other paramyxoviruses, and contribute to antagonism of the human type I interferon (IFN) response and to pathogenesis (reviewed in Chambers & Takimoto 2009), and Lieu et al. (2015) show that relative to NiV and HeV, the CedV P protein is ineffective in counteracting the human IFN signaling pathway. Collectively, these molecular features and limited pathogenesis in non-human mammalian species suggest CedV is relatively unlikely to be capable of causing serious disease in humans. Additional suggestive evidence that some henipaviruses may result in relatively innocuous spillover events comes from a human serosurvey conducted in Cameroon, in which 1–2% of human serum samples were positive for NiV cross-neutralizing antibodies, despite no reports of serious disease in seropositive individuals (Pernet et al. 2014b). Interestingly, NiV seropositivity was found almost exclusively amongst individuals who reported butchering bats for bushmeat (seroprevalence in this high-risk group was ~4%; Pernet et al. 2014b). This relatively high seroprevalence suggests that close contact with body fluids from bats harboring henipaviruses related to NiV might cause spillover infections in the absence of serious clinical disease more frequently than otherwise believed. It is likely that the divergent clades of henipaviruses harbor a similarly diverse virulence spectrum, and that elucidating the determinants of pathogenicity of henipaviruses as a group requires urgent attention.
3. Unknown zoonotic potential of paramyxoviruses
Whether or not we consider additional paramyxoviruses in the henipavirus genus inherently likely to cause serious human disease in the context of spillover, NiV and HeV remain the two major emerging paramyxoviruses that pose a known threat to human health. However, given that three of the four other paramyxovirus genera (Rubulavirus, Morbillivirus, and Respirovirus) include established human pathogens with considerable global health burdens, as well as recently-detected vast sequence diversity in wildlife reservoirs (Table 2; Drexler et al. 2012; Kurth et al. 2012; Wilkinson et al. 2014), it is worth exploring whether unknown zoonotic paramyxoviruses from these genera might also pose a significant threat to human populations.
3.1 Diversity of novel enzootic paramyxoviruses
In a landmark study, Drexler et al. (2012) conducted an (RT)-PCR screen to identify potential paramyxoviruses in bat and rodent samples collected from multiple locations around the world, and identified an estimated 66 novel paramyxoviruses. To put this in context, this exceeds the total number of paramyxoviruses in all genera currently listed by the International Committee on Taxonomy of Viruses (ICTV; Afonso et al. 2016; Drexler et al. 2012). Although this large study sampled over eighty bat species, it is worth noting that this represents less than 10% of known Chiropterans, and represents only certain geographical locations and moments in time. Thus, in terms of our knowledge of paramyxoviruses in wildlife with unknown capacity for spillover infection and disease, the most dramatic shift this study provided is perhaps an increased awareness of the extent of the unknown unknowns. As well as greatly expanding the known paramyxovirus sequence diversity, the identification of viruses closely related to human mumps virus (MuV, Rubulavirus) and canine distemper virus (CDV, Morbillivirus) in bats suggests bats host paramyxoviruses from genera beyond just the henipaviruses that may have the potential to switch hosts and cause disease in other mammalian species (Drexler et al. 2012). This, and subsequent (RT)-PCR- and sero-surveys for paramyxovirus in wildlife (Mélade et al. 2016; Wilkinson et al. 2014) have also identified many paramyxoviruses that fall within the paramyxovirus family, but variously cluster outside of the currently recognized genera, indicating that older isolates of morbilli-like (Nariva virus, NarV; Mossman virus, MosV) and rubula-like (Menangle virus, MenV; Tioman virus, TioV) paramyxoviruses are not isolated outliers.
The mere presence of – or indeed, simply new knowledge of – diverse paramyxoviruses in wildlife is clearly separable from the propensity for zoonotic transmission to humans, and particularly from the likelihood of causing serious disease or secondary human-to-human transmission, and therefore potential to cause an epidemic. However, efforts to model and understand emerging infectious disease in order to try to predict and respond better to future outbreaks have identified a number of trends and risk factors that allow us to begin to consider the relative threat of the emergence of a zoonotic paramyxovirus of public health concern. Firstly, it is worth noting that there has been a statistically significant increase in outbreaks of emerging infectious disease per se since the 1940s, even correcting for biases due to improvements in detection and reporting (Jones et al. 2008; Smith et al. 2014). The majority of these events are due to zoonotic transmission of viruses, primarily from wildlife reservoirs (Jones et al. 2008; Smith et al. 2014), with human disruption of ecosystems due to changes in land usage shown to be a major driver of such events (Daszak et al. 2001; Myers et al. 2013; Patz et al. 2004). Changes in available technology, such as development of pan-paramyxovirus PCR primers (Tong et al. 2008) and next generation sequencing, have revolutionized our ability to detect viral genome diversity in wildlife populations, and while such data in isolation reveals little about the likelihood of serious epidemics, it can facilitate attempts to analyze virological factors associated with elevated risk. Phylogenetic analysis comparing various viral families (Kitchen et al. 2011), as well as looking specifically at paramyxoviruses (Mélade et al. 2016), indicate that the dominant driver of viral macro-evolution is host-switching amongst different, but relatively related, host species. In fact, comparison with RNA viruses of other families suggests that paramyxoviruses may have relatively high rates and probabilities of transfer to new host species (Kitchen et al. 2011). Moreover, such relative flexibility in host species, in addition to molecular traits of paramyxoviruses such as a non-segmented genome and non-vector transmission, has also been identified as a positive correlate of likelihood of human-to-human transmission of emerging zoonotic viruses following a spillover event (Anthony et al. 2015; Geoghegan et al. 2016).
3.2 Human infection by zoonotic rubulaviruses
In addition to the theoretical risks of virulent emerging paramyxoviruses (not limited to the henipavirus genus) discussed above, there are two case reports of human infections with zoonotic rubula-like viruses (Table 2) that were associated with severe clinical disease. Firstly, concurrent with an outbreak of MenV that caused severe birth defects and stillbirths in swine at several affected piggeries in Australia during 1997, two piggery workers with extensive exposure to infected body fluids developed a severe influenza-like illness and rash, with no other identified cause (Chant et al. 1998; Philbey et al. 1998). While it remains possible that this novel bat paramyxovirus (Barr et al. 2012) was not the etiological agent responsible for the reported human disease, these were the only two piggery workers who became seropositive for MenV, making it the most likely cause (Chant et al. 1998). Interestingly, this outbreak somewhat parallels the initial NiV outbreak, which was also transmitted to humans from the bat reservoir via infected domesticated swine, with no evidence of onward human-to-human transmission, in contrast to subsequent outbreaks where direct transmission from bats to humans and secondary transmission are well documented (reviewed in Eaton et al. 2006). The second documented case of likely human disease resulting from zoonotic transmission of a bat rubula-like virus occurred in 2012 when a wildlife biologist suffered severe acute febrile disease following collection and dissection of bat and rodent specimens in South Sudan and Uganda (Albariño et al. 2014). Deep sequencing of patient samples identified the presence of a novel paramyxovirus termed Sosuga virus (SosV), and further investigation confirmed seroconversion in the patient, as well as the presence of SosV in samples collected in the weeks immediately prior to the onset of symptoms in the patient, and more widely in bat samples collected by the researcher at multiple locations across Uganda (Albariño et al. 2014; Amman et al. 2015).
3.3 Zoonotic transmission of morbilliviruses
While the above are currently isolated events, they indicate that emergence of a novel rubula-like virus of global health concern is at least plausible. It is perhaps relevant to also consider the repeated emergence of the relatively highly pathogenic morbillivirus, CDV, in non-human primates (Qiu et al. 2011; Sakai et al. 2013a; Sun et al. 2010; Yoshikawa et al. 1989). As recently reviewed (Nambulli et al. 2016), while CDV was originally thought to be restricted to canine host species, this paramyxovirus actually has a diverse range of potential hosts, and this recent emergence into primate species is perhaps cause for concern. MeV and CDV share significant antigenic similarity, and it has been suggested (discussed below) that CDV has not emerged into human populations already only because pre-existing MeV immunity prevents this jump (Nambulli et al. 2016). However, the identification of hundreds of a paramyxoviruses currently grouped as unclassified morbilli-related viruses in wildlife with genetic diversity that vastly eclipses that of the classified morbilliviruses (Drexler et al. 2012; Mélade et al. 2016; Wilkinson et al. 2014) suggests there is reason to believe there could well be morbilli- or morbilli-related viruses circulating in wildlife with the potential to cause human epidemics unhampered by pre-existing adaptive immune responses.
With these concerns in mind, in the following sections we examine the knowns and unknowns of the determinants of zoonotic emergence of pathogenic paramyxoviruses, considering both ecological and molecular factors.
4. Ecological factors driving zoonotic paramyxovirus emergence
Following initial zoonotic transmission of a pathogenic virus to a human, whether this becomes an isolated spillover event, sparks an outbreak, or ignites an epidemic, is determined by the capacity for human-to-human transmission (Lloyd-Smith et al. 2009; Wolfe et al. 2007). This can be defined by the basic reproductive number (R0) of a virus in the human population, where R0 is the average number of new infections that will occur from a single infected individual, in the absence of pre-existing immunity in the population. Of the two major emergent paramyxoviruses, HeV R0 in human populations is zero, since there is no onward human-to-human transmission, whereas NiV can initiate short chains of secondary human transmission (0<R0<1). The R0 for NiV in human populations in Bangladesh across 23 outbreaks between 2001 and 2007 was calculated to be 0.48, and so the authors conclude that the recurrent emergence of NiV seems unlikely to be capable of igniting an epidemic (Luby et al. 2009), which would require sustained human-to-human transmission, and therefore an R0 at least equal to 1. As noted by Woolhouse et al., (2016) the R0 of any pathogen is dependent on the unique factors of the specific host population in which it is circulating, and as such is far from a constant. Thus, emerging pathogens for which human-to-human transmission is currently self-limiting, but which have an R0 approaching 1, are of particular concern, since re-emergence in a population with even subtle differences in demographics or local ecological factors could be sufficient to provoke epidemic spread (Woolhouse et al. 2016).
From an evolutionary perspective, an important aspect of the emergence of pathogens capable of sustained transmission in human populations (R0≥1), is whether this is driven more by adaptive viral evolution within the new (human) host, or if it instead results primarily from multiple spillovers of diverse viruses, some of which may be pre-adapted for human-to-human transmission (Woolhouse et al. 2016). Woolhouse et al. propose that since virological factors such as tissue tropism and route of transmission, that are likely to determine human-to-human transmission rates, are generally phylogenetically conserved, relatively dramatic adaptive evolution in a short time frame in the new host would be required to induce critical shifts in the R0. Therefore, in most cases, preadaptation seems like the more probable evolutionary mechanism (Woolhouse et al. 2016). In this context, each individual transmission event from an animal reservoir to a human introduces additional viral diversity to the human population, and therefore contributes to the cumulative probability of transmission of a virus capable of initiating an epidemic.
The extent to which human populations are exposed to a potentially zoonotic pathogen is clearly a critical component of the likelihood of spillover infection. However, simply sharing a geographical area with infected animals alone is clearly not sufficient, given that transmission that initiates an outbreak is actually relatively rare, despite many people living at wildlife interfaces, and the vast viral diversity hosted by wildlife species. Beyond basic proximity, the main ecological factors that govern the rates of such primary animal-to-human transmissions can be broadly categorized as either those that affect the prevalence rates and viral diversity in the reservoir host; or those that affect the frequency and nature of reservoir-human contacts (Lloyd-Smith et al. 2009). How these factors theoretically relate to paramyxoviruses with zoonotic potential will be discussed below.
4.1 Viral prevalence and diversity in reservoir hosts
Bat and rodent species have been identified as key wildlife reservoir hosts, collectively harboring a vast diversity of paramyxoviruses with currently unknown zoonotic potential (e.g. Drexler et al. 2012; Mélade et al. 2016; Wilkinson et al. 2014). There are various traits associated with species of these two taxonomic orders (Chiroptera and Rodentia) that have been proposed to drive the observed high prevalence rates of viral infection, and high viral diversity. For example, both rodents and bats have high species diversity (together comprising more than half of all mammalian species); frequently exhibit gregarious behaviors in large colony groups; and often share habitats with numerous other bat and rodent species. All of these are traits thought to contribute to frequent cross-species transmission of viruses (reviewed in Brook & Dobson 2015; Han et al. 2015; Wang et al. 2008). Macro-evolution of viral diversity is thought to be driven predominantly by such host-switching events between relatively related species, where the viral replication fitness landscape is sufficiently different in the donor and recipient hosts as to drive adaptive evolution, but not so extreme as to provide a serious impediment to transmission (Kitchen et al. 2011; Kreuder Johnson et al. 2015; Mélade et al. 2016; Parrish et al. 2008). Thus, the orders Chiroptera and Rodentia have ideal characteristics for amplification of the viral diversity important for virus spillover through optimized intra- and inter-species transmission of viruses.
Paramyxoviruses which spill over into to human and domestic mammal populations in modern times display an obvious association with bat reservoirs (Table 2; HeV, NiV, SosV, MenV, compare reservoir hosts to spillover hosts). Indeed, there are no confirmed instances of recent pathogenic paramyxovirus spillover or emergence from a non-chiropteran reservoir species. The possible exception to this may be Mòjiāng virus (MojV), which was circumstantially associated with three human cases of respiratory illness in mine workers following isolation of MojV from rodents in the same mine (Wu et al. 2014). However, rodents have been identified as contributing to the spread of enzootic paramyxoviruses for which they are not the reservoir host (Wilkinson et al. 2014), so in the case of MojV, it remains to be determined which species constitute the reservoir host(s), as well as whether it was the causative agent of the observed human disease. Efforts in virus isolation, serology, and (RT)-PCR have detected unknown paramyxoviruses in bats from South America, Europe, Africa, Asia, and Australia – see (Baker et al. 2013; Barr et al. 2015; Breed et al. 2010; Epstein et al. 2008; Henderson et al. 1995; Iehlé et al. 2007; Li et al. 2008; Kurth et al. 2012; Pavri et al. 1971; Weiss et al. 2012) for some examples of these diverse surveys. There are paramyxoviruses whose origins are unknown, like SalV, suggesting that researchers have yet to look in all the right hosts to find paramyxovirus reservoirs, but several (RT)-PCR surveys for paramyxoviruses in both bat and rodent wildlife indicate a particular association between paramyxoviruses and bat hosts (Drexler et al. 2012; Muleya et al. 2014; Sasaki et al. 2014; Wilkinson et al. 2014). Drexler et al. (2012) conclude that paramyxovirus host switches occur most frequently from bats to other mammalian species, suggesting that emergence of novel paramyxoviruses will likely continue to occur predominantly from spillover from bat reservoirs. In addition, in a survey of paramyxoviruses in small mammals covering 4 taxonomic orders (Rodentia, Afrosoricida, Soricomorpha, and Chirpotera) in the biodiversity hotspot of the South West Indian Ocean Islands, bat species showed slightly elevated rates of paramyxovirus infection, and importantly, hosted a greater collective paramyxovirus sequence diversity than that observed across any other mammalian order (Wilkinson et al. 2014). Although beyond the scope of this review, bats have received considerable recent attention as possible ‘special’ reservoirs of viruses with zoonotic potential and high risk of virulence, and the possible immunological and ecological factors that may make bats unique in this regard have been recently reviewed by others (Brook & Dobson 2015; Han et al. 2015; Wang et al. 2008).
4.2 Frequency and nature of human-animal contacts
With increasing human population size, and increasing resources demanded by each person, humans are continuously encroaching on previously unsettled land, therefore inevitably perturbing the balance in delicate ecosystems (Patz et al. 2004). For example, ecological change such as deforestation is considered a major driver of virus emergence due to increased mixing of wildlife, particularly bats and rodents, with domestic animals and humans (Jones et al. 2008; Loh et al. 2013). Similarly, urbanization and intensification of farming practices gather large populations of humans and domesticated animals in novel geographical areas, creating vulnerable host populations with sometimes extensive interfaces with wildlife species (Fournié et al. 2015; Luby & Gurley 2015). In addition to these more localized human impacts on the environment, it has been proposed that global climate change will also likely drive changes in human-wildlife contacts, due to expansion or contraction of the ecological niches of reservoir hosts (Daszak et al. 2013). However, while climate change and anthropogenic land use changes have frequently been theorized or modeled as credible major ecological drivers of the documented increase in emergence of zoonotic diseases over the course of the last century (Daszak et al. 2013; Jones et al. 2008; Murray & Daszak 2011; Smith et al. 2014; Wolfe et al. 2005), there largely remains a paucity of evidence in specific cases.
A recent serology study in Cameroon highlighted the importance of the collection of detailed metadata in efforts to identify risk factors for paramyxovirus spillover infections, and provided evidence of likely drivers. Analysis of serum samples from Cameroon found a high prevalence of henipavirus-seropositive bats across all regions tested, as well as a low rate (<2%) but substantial number (7–10 individuals) of human samples positive for NiV and HeV cross-neutralizing antibodies, indicating likely spillover infections with related bat henipaviruses (Pernet et al. 2014b). While neither reported exposure to nor hunting of bats were alone associated with the probability of seropositivity in humans, those who specifically reported butchering bat bushmeat were 29 times more likely to be seropositive than those reporting no contact. Moreover, this study also provided evidence of the role of human land use changes, as individuals living in areas undergoing deforestation were also significantly more likely to be seropositive (Pernet et al. 2014b). While factors are likely to differ between regions, this study points to potential predictive ecological factors for bat henipavirus spillover risk, and suggests that targeted surveillance amongst those who prepare bushmeat in areas undergoing dramatic ecological change such as deforestation could be valuable in early identification of zoonotic emergence, by identifying potentially frequent spillover events that currently go detected unless a noticeable outbreak results.
An improved understanding of the ecological factors that govern the likelihood of zoonotic transmission could reduce the risk of serious epidemics by allowing targeted surveillance efforts, as well as appropriate social and environmental interventions. In order to consider more specific risk factors for emerging paramyxoviruses, we must look to emerging paramyxoviruses that have caused substantial enough numbers of outbreaks to be able to start to draw such conclusions, so in the following sections we will discuss the known risk factors for HeV and NiV outbreaks, as well as the evident gaps in existing knowledge.
4.3 Known risk factors – Hendra virus and Nipah virus
In cases of HeV infection, the clear risk factor for humans is close contact with infected horses, and so far can account for all human cases (Field 2016). Similarly, there is an obvious required enabling factor for the primary outbreak from the bat reservoir to horses, which is the presence of infected bats roosting in fruit trees sufficiently near to horse paddocks (Plowright et al. 2015). However, there are many locations in which these basic enabling conditions are met without spillover, and thus additional, though currently unknown, factors must contribute to the observed clustering and seasonality of outbreaks, as discussed in more detail later. In recent years, development and introduction of an effective equine vaccine means that the vaccination status of horses is now also a key outbreak risk factor. Although vaccine coverage is not complete, and outbreaks still occur, there have so far been no HeV cases reported in vaccinated horses, suggesting that this should be an effective outbreak risk mitigation strategy (Broder et al. 2013; Middleton et al. 2014; Peel et al. 2016). This approach also exemplifies the ‘One Health’ approach to protect humans from emerging zoonotic disease by vaccination of intermediate hosts (Middleton et al. 2014).
The ecological factors that drove the first NiV outbreak have been extensively investigated, and based on the levels of seroprevalence and active infection at the index farm, Pulliam et al. (2012) conclude that NiV circulation amongst pigs occurred at this farm for several years prior to the recorded start of the human outbreak. Moreover, the true human index case was retrospectively identified to have been a worker from the index farm in January 1997, with at least five additional human cases confirmed to have occurred prior to May 1998, months before the outbreak was detected in September of that year. Pulliam et al. (2012) identified intensification of farming practices at the index farm as key factors that facilitated NiV emergence. Firstly, the planting of mango trees (to increase agricultural output by also selling fruit) within contact range of pigsties a few years before the outbreak attracted bats, and dramatically increased the chances of pigs coming into contact with saliva and other body fluids from bats on dropped fruits. There are now restrictions on planting of fruit trees near piggeries throughout Malaysia, and though one cannot conclude from the absence of further outbreaks, it is possible this has helped to prevent a second NiV outbreak in Malaysia. Secondly, to maximize efficiency, pigs were kept in separate pens for breeding, early growth, and finishing, and this turnover and segregation of young naïve piglets therefore maintained a highly susceptible population, whereas adult breeding pigs were more likely to acquire protective immunity (Pulliam et al. 2012). Once the outbreak amongst pigs at the index farm was established, transportation of infected pigs for sale or slaughter dramatically spread the NiV to relatively distant regions of Malaysia, as well as to Singapore, and the outbreak was only contained after almost one million pigs were culled (Mohd Nor et al. 2000).
As noted above, following the initial NiV outbreak in which pigs were an intermediate host, re-emergence has generally occurred directly from the bat reservoir to humans, with the majority of these outbreaks occurring in Bangladesh and India. At the individual level, as for HeV infection, the main risk factors for NiV infection in such outbreaks are very clear. The main risk for initial transmission to humans is consumption of raw date palm sap contaminated with urine or saliva from infected bats (Islam et al. 2016; Luby et al. 2006; Openshaw et al. 2016; Rahman et al. 2012), while close contact with an infected person then enables secondary human-to-human transmission (Hegde et al. 2016; Homaira et al. 2010; Sazzad et al. 2013). However, consumption of date palm sap, as well as hunting bats for bushmeat, are relatively common practices throughout Bangladesh (Openshaw et al. 2016), and yet the majority of outbreaks have occurred in central and northwest Bangladesh, to the extent that these regions are colloquially referred to as the ‘Nipah belt’. This has provoked attempts to correlate ecological factors in these regions with the high outbreak prevalence (Hahn et al. 2014a; Hahn et al. 2014b). One analysis found that Nipah belt villages have significantly higher population densities, and occur in regions where forest land is more fragmented, suggesting these factors could promote NiV outbreaks (Hahn et al. 2014b). A similar analysis sought to identify risk factors by better understanding the features of preferred habitats of P. giganteus, a key NiV reservoir bat species – an approach validated by their finding that outbreak villages were 2.6 times more likely to map to a location their model predicts as a likely preferred habitat (Hahn et al. 2014a). The major contributing factors determining habitat preference included tree species, forest fragmentation, rainfall, temperature gradients and human disturbance levels. Interestingly, this model also identifies a number of villages outside the Nipah belt where outbreaks seem likely, which raises the possibility that outbreaks may go undetected in some regions, due to a lack of local awareness or surveillance bias (Hahn et al. 2014a).
4.4 Observed trends with unknown driving factors
For both NiV and HeV outbreaks, there is a strong seasonality in documented cases, clearly implicating seasonal ecological factors. Excluding the initial NiV outbreak, which began in Malaysia in September 1998, and continued until April 1999, every documented human case of NiV infection has occurred between the months of December and May (Islam et al. 2016; Luby et al. 2009; Wacharapluesadee et al. 2010). HeV spillover events also exhibit strong seasonality, though the timing is shifted relative to NiV outbreaks, occurring almost exclusively between May and October (McFarlane et al. 2011; Plowright et al. 2011). While it is unclear what drives these observed seasonal trends, and whether drivers differ for NiV compared to HeV, current hypotheses regarding the contribution of various factors will be discussed.
Human HeV infection has always been documented to occur via contact with an infected horse, so it is critical to consider what drives the primary host switch from the bat reservoir to cause an outbreak in horses. It is thought that the main route of transmission from bat to horse is a result of horses eating fruit contaminated with saliva and other body fluids from infected bats. One factor proposed to contribute to the seasonality of HeV outbreaks in horses is the relative stability of the virus in the environment, with increased persistence of infectious virions on contaminated fruit during the cooler, dryer season in Queensland and New South Wales when outbreaks occur (Fogarty et al. 2008; McFarlane et al. 2011).
An alternative, but not incompatible, hypothesis posits that there are seasonal changes in the likelihood of bats being infected, or of actively shedding virus, and indeed there is evidence of seasonal increases in levels of HeV seropositivity in bat populations that correlate with the timings of spillover events (Plowright et al. 2008). It has been variously suggested that there could be pulses of infection due to migration; waning population immunity because of seasonal birth pulses and infection of juveniles; or population level stresses such as the metabolic stress of pregnancy and seasonal food shortages (McFarlane et al. 2011; Plowright et al. 2008; Plowright et al. 2011; Plowright et al. 2015). It has been noted that in recent decades, many Australian colonies of Pteropus species bats have dramatically reduced their tendency to migrate during the winter months, perhaps due to increased availability of year-round food sources in urban and peri-urban settings. Plowright et al. (2011) have proposed that this change in migratory behavior might result in increased shedding of HeV during winter months, as a result of nutritional stress due to lower quality food sources in urbanizing areas, as compared to forest environments. Furthermore, they suggest that increasing geographical isolation of colonies could facilitate localized pockets of bats with waning population immunity, and therefore result in more dramatic pulses of infection upon viral re-introduction than when seasonal migration was more prevalent, which could increase risk of viral shedding future outbreaks (Plowright et al. 2011).
The seasonality of outbreaks of NiV in Bangladesh and India have also been correlated with seasonal increases in both the percentages of bats that test positive for NiV antibodies, and levels of shed NiV detected in bat urine (Wacharapluesadee et al. 2010). A longitudinal study of a bat colony in Ghana comprised of wild-caught bats that were then kept in captivity found a corresponding seasonal pattern in seroconversion, and therefore in inferred henipavirus infection. The pattern was specific to adult female bats and coincided with late pregnancy and lactation (Baker et al. 2014). Similar results were obtained in a bat serum survey in Malaysia, in which bats most likely to be positive for henipavirus antibodies were adult females that were either pregnant, lactating, or had a dependent pup (Rahman et al. 2013). These studies collectively suggest that seasonal reproductive stress is a contributory factor causing seasonal increases in NiV infection and shedding, and therefore increased probability of human exposure to contaminated fruit or date palm sap. Seasonal pulses of virus replication and documented human spillover associated with bat reproduction have also been identified for the zoonotic filovirus Marburg virus in Rousettus aegypticus bats (Amman et al. 2012), which can also host paramyxoviruses (Amman et al. 2015; Drexler et al. 2012). However, longitudinal analysis of morbili-related viruses in urine collected from a German maternal Myotis myotis colony over several years saw no temporal changes in shedding, and no evidence that reproduction, parturition, or age affected shedding (Drexler et al. 2012). Thus, these trends remain incompletely understood, could vary by geography, climate, bat host, or virus, and therefore may not be broadly applicable.
A better understanding of what determines spillover risk is clearly critical for mitigating the risks of recurrent NiV and HeV spillover, and for predicting and managing risk associated with emergence of other pathogenic paramyxoviruses. In particular, if birthing pulses or population level stresses increase the likelihood of spillover events, then attempting to curb risk by reducing urban and peri-urban bat colony population size, as has been proposed in Australia, could have the opposite effect to that intended, as recently discussed elsewhere (Plowright et al. 2015; Plowright et al. 2016). In this instance, as suggested by Hahn et al., land management strategies such as forest conservation of known ideal roosting sites for bat reservoir species in areas away from villages could be a useful means of risk management (Hahn et al. 2014a). Similarly, by identifying at-risk communities based on more detailed understanding of ecological drivers of spillover risk, education of the most at-risk human populations could be used to minimize the social and cultural practices that are known to determine infection risk at the individual level (Kamins et al. 2015; Nahar et al. 2010), combined with targeted surveillance to allow early identification of outbreaks.
5. Virus-host molecular interactions affecting paramyxovirus emergence
As discussed above, paramyxoviruses have been observed to switch hosts at a high rate (Kitchen et al. 2011), which has also been associated with higher viral diversity (Brierley et al. 2016; Kreuder Johnson et al. 2015; Mélade et al. 2016) and thus with a potentially increased capacity to overcome the molecular viral and host barriers to spill over into susceptible human and domestic animal populations (Woolhouse et al. 2016). Such barriers in a new host include the need for efficient receptor usage and tissue targeting, interactions with required host factors, and evasion of innate immune signaling; as well as potential pre-existing immunity to antigenically-similar viruses. P (phosphoprotein) and its accessory gene products (Table 1), as well as the viral attachment proteins (G/H/HN), have the most frequent and direct molecular interface with these host processes. P is an essential component of the polymerase, but has immunomodulatory functions for some paramyxoviruses, and P-derived alternative reading-frame proteins (discussed below) are important for controlling the innate immune response in infected cells. Evidence for the evolutionary pressure exerted by the need to interact with different hosts through these proteins can be seen in Figure 3A; note that both P and H/HN/G proteins demonstrate a marked increase in diversity above the other viral proteins in each paramyxovirus genus (see also Figure 2, “P” top left and “HN/H/G” right). What is known, and what is not, about paramyxoviral tools and strategies to overcome these barriers is described below.
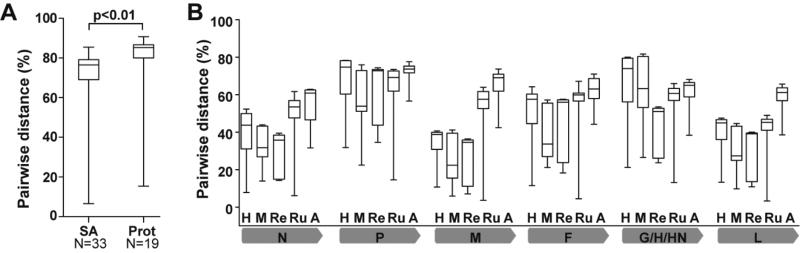
Figure 3. Paramyxoviruses demonstrate greater diversity within each genus at virus proteins with significant host interactions.
(A) Pairwise distances of Sialic acid (SA)-using paramyxoviruses versus protein (Prot)-receptor using viruses, indicating the number of virus species attachment proteins. Genera included in SA: Rubulavirus, Respirovirus, Ferlavirus, Avulavirus, Aquaparamyxovirus, and Unclassified (including Rubula-like viruses). Genera included in Prot: Morbillivirus, Henipavirus, and Unclassified (including Jeilongvirus and Morbilli-like viruses). Pairwise distance was calculated in MEGA 6.0, and T test was done using SPSS software (IBM). (B) Pairwise distances per gene and per genus across Paramyxoviridae. Genera are labelled within the graph; H = Henipavirus, n=5 (HeV, NiV, CedV, GH-M74a and MojV); M=Morbillivirus, n=7 (FeMV, PDV, CDV, MeV, PPRV, CeMV); Re=Respirovirus, n=6 (SeV, HPIV-1, PPIV-1, CPIV-3, HPIV-3, BPIV-3); Ru=Rubulavirus, n= 9 (MuV, BMV, PIV-5, HPIV-2, SV-41, PorV, MapV, HPIV-4a, HPIV4b); and A=Avulavirus, n=6 (APMV-2, APMV-5, APMV-11, APMV-3, APMV-9, NDV). Grey bars indicate the virus gene being evaluated. Whiskers show minimal and maximal variation within the genus, boxes denote the 25% (low) and 75% (high) percentiles, and horizontal lines indicate medians.
5.1 Successful emergence requires host-specific interactions
5.1.1 Effects of receptor specificity on species and tissue tropism
Virus attachment and entry is the first direct interaction between an emergent virus and its new host, and would appear to be one of the clearest barriers that prevent interspecies paramyxovirus transmission. Paramyxovirus entry requires the coordinated action of both the fusion (F) and attachment glycoproteins, the latter variously termed HN, H, or G depending on the nature of the receptor used (Figure 1). While paramyxoviruses are primarily classified based on sequence analysis of L proteins (Figure 2, “L” bottom left), the attachment glycoproteins are much more divergent (Figure 2, compare “L” bottom left to “HN/H/G” right; Figure 3A) and have functional characteristics that provide useful delineations within the virus family as well. HN glycoproteins have sialic acid dependent hemagglutinin and neuraminidase (HN) activities, H glycoproteins have hemagglutinin activity (unrelated to sialic acid binding), and G glycoproteins have neither. H and G designate the attachment glycoproteins of the morbilliviruses and henipaviruses, respectively. The glycoproteins from many of the unclassified paramyxoviruses that have none of the essential sialic acid binding site residues found in HN proteins are presumed to also use protein-based receptors and are thus designated as G (Figure 1; Figure 2, “HN/H/G” right). Morbillivirus H glycoproteins use protein-based receptors, too (see below); the hemagglutination activity in absence of neuraminidase activity that was attributed initially to MeV-H is not sialic acid-dependent, but is rather due to the expression of CD46, an alternative cell culture receptor for the vaccine strain of MeV, on the RBCs from certain non-human primates (Krah 1991; Lee et al. 2011). Although HN-bearing, sialic acid-using viruses comprise a majority of paramyxovirus genera and infect a larger of number host species/orders than protein receptor-using (H or G) paramyxoviruses, the attachment proteins from the latter have significantly greater diversity than the former (Figure 3B). This likely represents the diversity of receptor molecules used by these attachment proteins, since sialic acid-based receptors are relatively structurally constrained compared to the universe of potential protein receptors (discussed below). Despite the divergent nature of the receptors used, paramyxovirus attachment glycoproteins all bear common features such as a receptor-binding globular head domain that adopts a six-bladed beta-propeller fold, which is connected to a flexible stalk region. The stalk region triggers the metastable F protein upon receptor binding, which is what initiates the conformational cascade that eventually results in membrane fusion (reviewed in Bose et al. 2015). Below we will explore whether receptor specificity can be an important contributor to species and tissue tropism at the level of viral entry, and propose that viral use of a highly-conserved receptor molecule may remove at least one barrier to inter-species transmissibility.
5.1.2 Use of sialic acid as a paramyxovirus receptor
Five of the seven recognized genera of paramyxoviruses (Figure 2) use sialic acids (SAs) on cell surface glycoproteins or gangliosides as receptors. These HN-bearing paramyxoviruses infect a wide variety of vertebrate hosts (mammals, birds, reptiles, and fish), but Avulavirus, Ferlavirus, and Aquaparamyxovirus species have not demonstrated consistent infection of mammals, and so are unlikely source genera for zoonotic transmission of emerging paramyxoviruses into humans, so we focus here on respiroviruses and rubulaviruses. Paramyxovirus HN proteins have two important activities: the first is the hemagglutination, or sialic acid-binding activity, that is important for attachment and triggering of fusion during entry, while the second is the neuraminidase, or sialic acid-cleavage activity, that is important to release the virion from the parent cell during budding, and prevent re-infection of the same cell. HN proteins have differences in their binding and cleavage activity for various terminal sialyl linkages. For example, while the HN proteins from most human parainfluenza viruses preferentially bind and cleave α2,3 linked SAs, HPIV-3 HN can also recognize α2,6 linked SAs (Amonsen et al. 2007; Fukushima et al. 2014; Fukushima et al. 2015). For MuV, a single amino acid change can switch the viral HN preference from α2,3 to α2,6 SA linkages (Reyes-Leyva et al. 2007). Importantly, this E335K mutation in the HN protein also results in heightened neurotropism and neurovirulence associated with the differential distribution of these SA linkages on nervous tissues (Santos-López et al. 2009), indicating that receptor usage and tissue distribution can also affect viral pathogenesis.
Along with preference for specific linkages, HN proteins additionally demonstrate preference for the context of terminal SAs: MuV-HN binding to α2,3 sialylated oligosaccharides is significantly weaker when the terminal SAs are present on branched tri- and tetra-antennary complex glycans than on their simple or bi-antennary counterparts. Such preferences could potentially modulate the tissue tropism and transmissibility for HN-bearing paramyxoviruses (Fukushima et al. 2014; Reyes-Leyva et al. 2007; Villar & Barroso 2006) since sialylated glycans with diverse structures and compositions are differentially expressed in different tissues. Although much remains to be determined about the systemic distribution of glycan structures, the classical notion established by lectin immunohistochemistry that α2,6 and α2,3 receptors are concentrated in the human upper and lower respiratory tract, respectively (Wright et al. 2013), are being revised. Glycomic analysis based on lectin binding (Kogure et al. 2006; Nicholls et al. 2007) and mass spectrometry methods (Walther et al. 2013) have revealed that both upper and lower human respiratory tract tissues have a spectrum of α2,6 and α2,3 terminated glycans in varying portions, and at least for influenza viruses, binding to sialyated α2,6 or α2,3 glycans is not necessarily predictive of productive replication in ex vivo human bronchial or lung tissues (Walther et al. 2013). Thus, the paradigm that the preferential α2,3 binding of avian influenza viruses is a determinant of its lack of transmissibility among humans may need to be refined. Overall, detailed study of the systemic distribution of glycan structures – both respiratory, and elsewhere in the body – in different hosts may shed light on the tropism and pathogenesis of HN-bearing paramyxoviruses that disseminate systemically to cause disease, such as MuV and recently-discovered MenV and SosV (Table 2).
In addition to direct interaction with sialic acids on the target cell, respiratory viruses must navigate a respiratory tract that is known to be densely-packed with secreted sialic acid-bearing mucins and free sialylated oligosaccharides (Zanin et al. 2016) that could act as decoys for viral HN (Wasik et al. 2016). The ability to evade such decoys would depend, in part, on the neuraminidase (sialic acid cleavage) activity of the HN, by allowing the virus to penetrate the mucus barrier and reach the susceptible respiratory epithelial cells. Human respiratory secretions differentially restrict avian, porcine, and human-tropic influenza virus entry into the same cells in vitro (Zanin et al. 2016), providing indirect evidence that the lung surfactant environment varies between species, but this has not yet been studied for paramyxoviruses. It is therefore not clear whether evasion of these “decoy” sialic acids is a determinant of HN-bearing paramyxovirus species specificity. Overall, given the heterogeneous expression of α2,6 and α2,3 SAs in the human respiratory tract, it is likely receptor specificity alone for HN-bearing respiroviruses and rubulaviruses does not dictate their potential for cross-species transmission. Furthermore, since single amino acid changes in the HN protein of HPIV-3 (Respirovirus) or MuV (Rubulavirus) can markedly affect α2,3 versus α2,6 receptor binding (Fukushima et al. 2015; Mishin et al. 2010; Reyes-Leyva et al. 2007), it is unlikely that sialic acid linkage receptor specificity is a strong barrier to spillover of HN-bearing paramyxoviruses.
5.1.3 Paramyxoviruses that use protein-based receptors
Two of the currently recognized genera of paramyxoviruses, morbilliviruses and henipaviruses, use protein-based receptors. Structural phylogeny analysis indicates that morbillivirus H and henipavirus G proteins independently evolved from what is thought to be the ancestral HN attachment protein (Bowden et al. 2008; Lee et al. 2011). Interestingly, extant henipavirus G proteins are structurally more related to extant HN proteins than MeV-H (Zeltina et al. 2016; see also Figure 2, “HN/H/G” right), suggesting that morbillivirus H evolved the use of protein-based receptors before henipaviruses (Bowden et al. 2008; Bowden et al. 2010; Lee et al. 2008), assuming that its structural diversity and divergence from other paramyxoviruses is a function of time and not due to other evolutionary constraints. We speculate that morbilliviruses may thus have speciated in their respective hosts for a longer period of time, which may explain in part the species specificity of each morbillivirus in comparison to the species promiscuity of a given henipavirus. These factors may also be responsible for the greater divergence among protein receptor-using viruses demonstrated in Figure 3B. Here, we discuss how for both morbilliviruses and henipaviruses, the use of specific protein receptors is correlated with aspects of transmission, species and tissue tropism, and pathogenesis.
All extant henipaviruses whose receptors have been functionally defined use ephrin-B2, a receptor tyrosine kinase, as an entry receptor (Bonaparte et al. 2005; Lawrence et al. 2014; Lee et al. 2015; Marsh et al. 2012; Negrete et al. 2005; Weis et al. 2014). The presumed mode of transmission and systemic dissemination is consistent with the expression of ephrin-B2 on microvascular endothelial cells, neurons, and some cells in the respiratory epithelium (Bennett et al. 2013; Gale et al. 2001; Pernet et al. 2012); like most other paramyxoviruses, henipaviruses are likely spread via the oropharyngeal route, and often result in fatal neurological and respiratory disease in non-reservoir susceptible hosts (Middleton & Weingartl 2012; de Wit & Munster 2015; Wong & Tan 2012). Ephrin-B2 is highly-conserved across mammalian species from bats to humans (Bossart et al. 2008; Pernet et al. 2012), which explains in part the ability of HeV and NiV to infect a wide range of mammals under natural (bats, horses, pigs, cats, dogs, humans) and laboratory (hamsters, guinea pigs, rodents) conditions (Ching et al. 2015; Mills et al. 2009; Vigant & Lee 2011). Correlation of ephrin-B2 conservation and susceptibility to henipaviruses like Nipah across so many mammalian species supports the proposition that paramyxoviruses using highly-conserved receptors are better-equipped to achieve transmission to new host species.
Morbilliviruses, in contrast, utilize CD150 (SLAM/F1 – signaling lymphocyte activation molecule family member 1) as the major morbillivirus host receptor protein and first point of contact with the new host (Delpeut et al. 2014; Tatsuo et al. 2001). Morbilliviruses are also primarily contracted through respiratory exposure, but spread via initial infection of SLAM/F1-bearing alveolar macrophages, which then disseminate the virus to local lymph nodes where the virus replicates in subsets of B- and T-cells (Delpeut et al. 2014). Incidentally, infection of SLAM/F1 expressing B- and T-cells is likely responsible for both the short- and long-term immune dysfunction that follows infection with MeV (Mina et al. 2015; de Vries et al. 2012). In the respiratory epithelium, morbilliviruses complete their infection cycle by entry at the basolateral surface using nectin-4 (PVRL4), the other major morbillivirus attachment receptor (reviewed in Delpeut et al. 2014), and are then secreted into the respiratory tract lumen for transmission.
Nectin-4 is highly-conserved and does not appear to enforce species-specific limitations in morbilliviruses, since canine distemper virus (CDV) can utilize human Nectin-4 with no adaptation (Bieringer et al. 2013; Sakai et al. 2013b). However, nectin-4 is only required for morbillivirus exit from the host; indeed, recombinant MeV that is selectively blind for nectin-4 can infect and cause primary disease in non-human primates, but is unable to transmit or be shed (Frenzke et al. 2013). Although evidence exists for additional natural MeV attachment receptors (Griffin 2013; Sato et al. 2012), SLAM/F1-specificity is best-characterized and appears to be the main barrier to Morbillivirus entry in hosts of different species. SLAM/F1 from various mammalian species, while functionally conserved, is much more divergent than the henipavirus receptor, ephrin-B2 (Ohishi et al. 2010; Zeltina et al. 2016), and it has been shown experimentally that species specific use of SLAM/F1 is an important determinant of host species restriction of viruses like MeV, CDV, and the recently-eradicated Rinderpest virus (RPV) (reviewed in Nambulli et al. 2016; (Adombi et al. 2011; Baron 2005; Tatsuo et al. 2001; Yanagi et al. 2000)). This provides further support for the concept of receptor conservation contributing to inter-species transmission and restriction. However, this has been overcome in both experimental (Bieringer et al. 2013) and natural settings (Sakai et al. 2013b) with CDV, so other barriers discussed below likely function in concert with receptor specificity to maintain the observed morbillivirus and other paramyxovirus host restriction.
5.1.4 Contribution of host conservation of paramyxovirus receptors to spillover risk
Both henipaviruses and HN-bearing paramyxoviruses like rubulaviruses have demonstrated the ability to attach and enter cells from a broad range of species, eliminating one of the restrictions on interspecies transmission. This receptor promiscuity is likely important for the diversity of these and related viruses in both bat and rodent reservoirs (for example: Baker et al. 2012; Drexler et al. 2012; Wilkinson et al. 2012; Wilkinson et al. 2014), and for the large number of spillovers and infected species associated with these viruses. Thus, paramyxoviruses with the ability to use a receptor molecule that is highly-conserved across animal species – whether carbohydrate or protein – appear better-equipped to overcome the initial molecular restrictions of attachment and entry to new host species, and may therefore have a greater likelihood of spilling over into human and domestic animal populations.
5.1.5 Post-entry essential host factors that are species-specific are not yet known
While human and bovine PIV-3s are extremely similar at both an antigenic and genetic (see Figure 2, and Table 1) level, BPIV-3 is known to be attenuated for human infection (Karron et al. 1995), while HPIV-3 is decidedly not (Pecchini et al. 2015). Interestingly, simply replacing the N (nucleocapsid) protein of HPIV-3 with that of BPIV-3 attenuates the human virus in macaques (Bailly et al. 2000), as does swapping other BPIV-3/HPIV-3 ORFs (Skiadopoulos et al. 2003), although the converse experiment in cattle has not been performed. Infection of macaques and humans with BPIV-3 results in 1-to-3-log lower viral titers recovered, but viral replication does occur and antibody responses are induced (Bailly et al. 2000; Schmidt et al. 2000; Skiadopoulos et al. 2003), which indicates that the species restriction for BPIV-3/HPIV-3 is not absolute. Since we know that BPIV-3 is competent to enter primate cells (Schmidt et al. 2000; Skiadopoulos et al. 2003), and is able to successfully antagonize the human innate immune response, blocking interferon induction with both C and V accessory proteins (Komatsu et al. 2007; Yamaguchi et al. 2014), the species-specific attenuation of BPIV-3 in humans may instead be explained by an incompatibility with a required host factor during RNA replication or virion production. Although examples of species-specific virus-host interactions outside of entry and immune evasion are limited and not well-characterized in paramyxoviruses (Tao & Ryan 1996), a biologically-related example of such can be found in H5N1 avian influenza. A mutation in the PB2 polymerase subunit, E-to-K at amino acid 627, has long been associated with adaptation to replication, transmission, and pathogenicity in mammalian cells (Gabriel et al. 2013). This mutation has recently been determined to restore interaction the influenza polymerase and the mammalian version of ANP32A, a required host factor with significant differences from the avian protein (Long et al. 2016). Paramyxoviruses are likely to have analogous interactions with host proteins that facilitate important stages of the virus life cycle, but since discovery and characterization of enzootic paramyxoviruses in both their current hosts and potential emergent host systems has received less attention and research energy than pandemic and avian influenza viruses, the significance of species-specific host factor restriction remains to be determined and is a ripe area for exploration. As this area of paramyxovirus biology is examined, we may also find that lack of a required host factor negatively synergizes with other barriers like suboptimal receptor interactions to ultimately prevent successful cross-species infection and/or sustained transmissibility in humans.
5.1.6 Antagonism of innate immune responses is known to be key to successful infection
Species-specific mechanisms of immune modulation carried out by paramyxoviruses that infect humans and our domestic mammals are relevant to our understanding of paramyxovirus-host biology and interactions. General paramyxovirus strategies to modulate the host’s immune environment have been most recently reviewed by Audsley & Moseley (2013) and Parks & Alexander-Miller (2013). Broadly speaking, paramyxoviruses have transcriptional frameshift protein products of the phosphoprotein (Figure 1; catalogued in Table 1) that manipulate interferon induction and signaling in the host cells to varying degrees, with the known exceptions of HPIV-1 (Power et al. 1992) and CedV (Marsh et al. 2012), although both HPIV-1 and CedV still produce alternate-start C proteins from the phosphoprotein transcript (Table 1). The phosphoprotein itself (Ciancanelli et al. 2009; Devaux et al. 2013; Gainey et al. 2008), along with the small hydrophobic (SH) protein found in Rubulaviruses and Jeilongviruses (Figure 1, Table 1), have further been implicated in modulating the host innate response to the virus’ benefit (Li et al. 2011; Wilson et al. 2006). As compared in Table 1 and illustrated for P in Figures 2 and 3A, each of these proteins is highly divergent, even within the same genus, which may represent the fact that these proteins must frequently interact with some of the proteins that are so variable within and between host species, like those of the innate immune response (Kwiatkowski 2005; Du Pasquier 2005).
Overall, efforts to determine the species-specificity of these virus-host interactions have been piecemeal, particularly in enzootic viruses that have not yet been detected to jump species. Generalizations or predictions about the effect of specific viral protein-host factor interactions on species tropism are difficult to make, since significant variability in viral innate immune evasion occurs even within a given viral genus. Both the P and accessory proteins of viruses from a given genus typically demonstrate less than 50% identity at the amino acid level (Table 1; see also Figure 2, “P” upper left), and so prediction of host-specific interactions with innate immune signaling by analogy to other viruses within the genus is impractical. As a functional example, rubulavirus V proteins are known to antagonize STAT1-mediated interferon signaling through a common mechanism of facilitating ubiquitination and proteasomal degradation of STAT1 (Audsley & Moseley 2013 Horvath 2004). Although the mechanism is shared, the species restrictions of these viruses are not: human MuV is not known to infect any other animals in the wild, and only infects certain nonhuman primates in a laboratory setting (Xu et al. 2013), while PIV-5 naturally infects a multitude of wild and domestic mammals – often asymptomatically (Chatziandreou et al. 2004). Despite its species promiscuity, PIV-5 only poorly infects mouse cells, and causes no disease in infected mice, because its V protein cannot disrupt murine STAT1-mediated interferon signaling (Young et al. 2001). The PIV-5 V protein can, however, disrupt human STAT1-mediated interferon signaling, and thus, its ability to infect a host is correlated with its ability to control interferon signaling. Conversely, although the capacity of human MuV to manipulate other mammalian interferon induction and signaling pathways has not been systematically characterized to determine the basis of its species restriction, the inability of MuV to productively infect ferret airway epithelial cultures even at high MOIs suggests a very early block involving incompatible host factors prior to innate immune signaling (Elderfield et al. 2015).
Not only do paramyxoviruses from various hosts have to contend with antagonizing innate immune signaling, they must also contend with innate immune effectors. Even such highly human-adapted viruses as MeV, MuV, or HPIV-2 do not completely abrogate innate antiviral signaling (Young et al. 2003) and are therefore under pressure from interferon-stimulated host effectors (Rabbani et al. 2016; Young et al. 2016). As an example, the IFIT family of proteins use a variety of means to recognize and inhibit translation of non-self transcripts (Daugherty et al. 2016), therefore each requiring different mechanisms of viral evasion or antagonism (Diamond & Farzan 2012). Daugherty et al. demonstrate that within the IFITs, principally encompassing IFIT1, 1B, 2, 3, and 5 and their homologs, there is a large diversity in coding and expression of these proteins across the order Mammalia (2016). For example, primates encode both IFIT1 and IFIT1B, although human IFIT1B does not appear to be functional; Chiroptera and Artiodactyla (goats, cows, pigs) lack IFIT1B entirely and only encode IFIT1; and Myomorpha (a sub-order of rodents that includes mice, rats, and voles) lack IFIT1 and encode only IFIT1B (Daugherty et al. 2016). Thus, viruses that are highly-adapted to rodent infection may lack molecular tools to evade or inhibit IFIT1 effector functions and may therefore experience a higher restriction barrier in human and bat cells, while viruses that are adapted to bats may have IFIT1 evasion mechanisms that can function to some degree in humans (or pigs, or cattle) and may therefore not be restricted by the human version of this effector. IFIT proteins are just one family among many, and protein families – effectors, signal transducers, and sensors – throughout the mammalian innate immune network demonstrate analogous diversity in various mammalian species (e.g. NOD-like receptors, or TLR4: Motta et al. 2014; Vaure & Liu 2014), posing an intricate web of obstacles that viruses must navigate when expanding their host repertoire.
5.2 Unknown connection between emergence and pathogenicity
As evidenced by the serosurveillance study discussed above (Pernet et al. 2014b), and by other indications of human serological responses to enzootic paramyxoviruses (Table 2), zoonotic virus spillover does not always result in pathogenesis in the new host. However, factors that impact paramyxovirus inter-species transmission may also play a separate role in defining the pathogenicity of novel viruses. For human parainfluenza viruses (respiroviruses – HPIV-1 and 3; and rubulaviruses – HPIV-2, 4, and PIV-5), transmissibility is associated with upper respiratory tract (URT) infection, while pathogenesis correlates with lower respiratory tract (LRT) infections (Burke et al. 2011). As such, efforts to understand and limit the transmission potential of parainfluenza viruses and any novel zoonotic relatives should not focus solely on the severity of respiratory symptoms (which is a consequence of LRT infection), but on factors that facilitate virus replication and shedding in the URT, which could result in transmission from asymptomatic hosts (Henrickson 2003). Lower temperature in the URT, differential innate immune responses and cytokine milieu, and the presence of antiviral (e.g. surfactant proteins in the lung) or proviral (e.g. secreted proteases required for F cleavage) factors may all contribute in a host-specific manner to increase the capacity for transmissibility and emergence of a potential pathogen.
In some instances, it is possible that manipulation of host immune signaling merely defines the virus’ pathogenicity in a given species, rather than its ability to infect that species overall. For example, Tioman virus (TioV; Chua et al. 2002) is another rubulavirus which is found in flying foxes (Yadav et al. 2016) and which may have asymptomatically infected humans (Yaiw et al. 2007). TioV does not antagonize IFN-β signaling in human cells, yet it also does not appear to interact with STAT2 from its fruit bat reservoir host (Caignard et al. 2013). Thus, TioV appears capable of infection irrespective of its ability to antagonize interferon signaling. This can also be observed with MeV, where specific abolition of its ability to counter STAT1 signaling severely attenuates virus pathogenesis in the Rhesus macaque model of infection (Devaux et al. 2011). However, a counterexample can be found in NiV and HeV evasion of both bat and human interferon induction (Glennon et al. 2015) – and yet these viruses do not appear to cause disease in the bat host (Middleton et al. 2007), but most definitely do in humans. This may relate to the potential differences in immune responses and homeostasis between bats and humans alluded to earlier (see also Han et al. 2015; Mandl et al. 2015; Brook & Dobson 2015; O’Shea et al. 2014), but clearly, further work in exploring paramyxovirus mechanisms of innate immune modulation comparing enzootic, emergent, and non-permissive hosts is needed to gain a better understanding of molecular factors that drive emergence of zoonotic paramyxoviruses.
5.3 Pre-existing immunity may restrict emergence
In addition to the virus’ ability to antagonize the innate immune response in a new host, another important determinant of the capacity to both initiate productive infection and cause disease is the ability of the host immune system to recognize a novel pathogen. Parrish et al. (2008) have proposed that an emergent virus must spill over from just the right host – a species that is genetically similar to the at-risk species, but not so similar that the at-risk species has already encountered a similar virus and has pre-existing immunity. Unlike many other virus families where single virus species can have multiple serotypes, paramyxoviruses can even share the same serotype with other members of the same genus, such as is the case between human, bovine, and caprine parainfluenza virus 3 (HPIV-3, BPIV-3, CPIV-3; Abinanti & Huebner 1959; Barrett & Chalmers 1975; Chowdhury et al. 2014; Ditchfield et al. 1965; Henrickson 2003; Lehmkuhl & Cutlip 1982; Lyon et al. 1997; Maidana et al. 2012). For instance, cross-protection via this antigenic similarity between different parainfluenza viruses has been proposed to restrict infection of BPIV-3 to cattle, and HPIV-3 to humans, since exposure to HPIV-3 in childhood establishes a sufficient adaptive response to limit subsequent infection by BPIV-3 (Karron et al. 1995).
If amplification of human-adapted viruses like HPIV-1 and 3 can be affected by pre-existing immunity (Bhattacharyya et al. 2015), despite the fact that natural immune responses to these viruses are known to be short-lived and poorly protective (Cooney et al. 1975), then it is logical that cross-reactive immunity could be capable of preventing or limiting infection with a poorly-adapted emergent virus. As mentioned earlier, such is believed to be the case with Morbilliviruses, where pre-existing immunity to MeV in humans is sufficient to prevent successful transmission of CDV (canine distemper virus, a closely-related morbillivirus), despite humanity’s frequent encounters – and close relationships – with CDV-infected animals like the domestic dog (Nambulli et al. 2016). MeV in humans is believed to be the result of a successful zoonosis of a Rinderpest (RPV)-like virus from domesticated cattle (Furuse et al. 2010), at a time when humans presumably did not yet have pre-existing immunity from yet another related virus to prevent such a transmission. Rubulaviruses may provide another example of this: recently, Drexler et al. found a mumps-like virus in African fruit bats (BMV; Drexler et al. 2012) that we and others have shown to be sensitive to human and murine antibodies generated against human MuV (Beaty et al. 2016; Katoh et al. 2016; Krüger et al. 2016). Coupled with phylogenetic analysis that suggests BMV could be another strain within the Mumps virus species (see also Figure 2), this suggests that human MuV may have first arisen in bats (Drexler et al. 2012), and that since it has not since fully speciated from BMV, it may have emerged into humans more recently than MeV did. Since then, immunity to human MuV may have been cross-protecting humans from further spillover events of other bat-borne MuV-like viruses. Given these examples of possible cross-protection from zoonotic viruses mediated by exposure to endemic human viruses, it is further possible that the antigenic relatedness of at least some bat-borne henipaviruses (Chua et al. 2000; Eaton et al. 2006; Marsh et al. 2012; Drexler et al. 2012; Pernet et al. 2014a; Pernet et al. 2014b) may also influence future henipavirus emergence dynamics in certain human or animal populations that have been frequently exposed to zoonotic henipavirus spillover.
6. Summary
Spillover of enzootic paramyxoviruses into susceptible human and domestic animal populations is defined by a broad collection of ecological and molecular factors that interact in ways we don’t yet fully understand. Here, we have summarized the key known determinants of paramyxovirus spillover, while also highlighting some of the unexplored avenues defining paramyxovirus emergence. The continued expansion of human populations and consumption of resources ensure that we will continue to see spillovers and emergence of zoonotic viruses from reservoir hosts like bats into both humans and our domestic animals. It is clear that we need a better understanding of the paramyxoviruses that bats and other reservoirs harbor. We require a sense of paramyxovirus diversity and dynamics within their host populations, and mechanisms of immune evasion that can be compared across different species to evaluate risk; we also need a more comprehensive understanding of the species restrictions – or lack thereof – on these immune evasion mechanisms. To this end, systematic research is needed to characterize paramyxoviruses that have emerged with little detectable pathology. We can benefit from understanding paramyxoviruses like the rubulaviruses PIV-5 and Tioman virus, or henipaviruses like CedV and the unidentified Cameroonian henipavirus(es) detected by serosurveillance, to explore the differences between viruses that have not spilled over into humans, those that have but did not cause disease, and those that give us greatest concern: highly pathogenic and lethal emerging paramyxoviruses. We also need to improve our understanding of spillover events, and our detection of “silent” spillovers, before we can establish predictive parameters or algorithms for future paramyxovirus emergence. Characterization of cross-protective immunity provided by less-pathogenic paramyxoviruses and how it may prevent emergence of high-pathogenicity paramyxoviruses will also be valuable, because it could inform preventative approaches in vulnerable populations. Over the last several decades, we have become aware of humanity’s vulnerability to the threat of emerging pathogens, such that it has seeped into popular culture in the form of books, comics, and movies; but concomitant with this awareness we are developing the techniques and resources to begin to accurately assess the threat and hopefully, one day, predict and contain viral outbreaks before they can cause serious repercussions.
Acknowledgments
PAT is funded by a Canadian Institutes for Health Research Postdoctoral Fellowship, CIHR-IRSC:0041001056. REW is funded by a European Molecular Biology Organization Long-Term Fellowship, EMBO ALTF 628-2015.
Abbreviations: (see also Table 2)
- BPIV-3
Bovine parainfluenza virus 3
- CDV
Canine distemper virus
- (HPIV) -1, -2, -3, and -4
Human parainfluenza virus
- MuV
Mumps virus
- MeV
Measles virus
- PIV-5
Parainfluenza virus 5
- PPRV
Peste-des-petits-ruminants virus
- RPV
Rinderpest virus
- SV-41
Simian virus 41
References
- Abinanti FR, Huebner RJ. The serological relationships of strains of parainfluenza 3 virus isolated from humans and cattle with respiratory disease. Virology. 1959;8(3):391–394. doi: 10.1016/0042-6822(59)90041-8. [DOI] [PubMed] [Google Scholar]
- Adombi CM, et al. Monkey CV1 cell line expressing the sheep–goat SLAM protein: A highly sensitive cell line for the isolation of peste des petits ruminants virus from pathological specimens. Journal of Virological Methods. 2011;173(2):306–313. doi: 10.1016/j.jviromet.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso CL, et al. Taxonomy of the order Mononegavirales: update 2016. Archives of Virology. 2016;161(8):2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albariño CG, Foltzer M, Towner JS, Rowe LA, Campbell S, Jaramillo CM, Bird BH, Reeder DM, et al. Novel paramyxovirus associated with severe acute febrile disease, South Sudan and Uganda, 2012. Emerging infectious diseases. 2014;20(2):211–6. doi: 10.3201/eid2002.131620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amonsen M, Smith DF, Cummings RD, Air GM. Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with alpha2-3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol. 2007;81:8341–5. doi: 10.1128/JVI.00718-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, et al. A Recently Discovered Pathogenic Paramyxovirus, Sosuga Virus, is Present in Rousettus aegyptiacus Fruit Bats at Multiple Locations in Uganda. Journal of Wildlife Diseases. 2015;51(3):774–779. doi: 10.7589/2015-02-044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, et al. Seasonal Pulses of Marburg Virus Circulation in Juvenile Rousettus aegyptiacus Bats Coincide with Periods of Increased Risk of Human Infection. PLoS Pathogens. 2012;8(10) doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony SJ, et al. Non-random patterns in viral diversity. Nature Communications. 2015;6:8147. doi: 10.1038/ncomms9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle VA, et al. Genomic characterization of nipah virus, West Bengal, India. Emerging Infectious Diseases. 2011;17(5):907–909. doi: 10.3201/eid1705.100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audsley MD, Moseley GW. Paramyxovirus evasion of innate immunity: Diverse strategies for common targets. World Journal of Virology. 2013;2(2):57–70. doi: 10.5501/wjv.v2.i2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly JE, et al. A recombinant human parainfluenza virus type 3 (PIV3) in which the nucleocapsid N protein has been replaced by that of bovine PIV3 is attenuated in primates. Journal of virology. 2000;74(7):3188–95. doi: 10.1128/jvi.74.7.3188-3195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KS, et al. Co-circulation of diverse paramyxoviruses in an urban African fruit bat population. Journal of General Virology. 2012;93(Pt_4):850–856. doi: 10.1099/vir.0.039339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KS, et al. Novel, Potentially Zoonotic Paramyxoviruses from the African Straw-Colored Fruit Bat Eidolon helvum. Journal of Virology. 2013;87(3):1348–1358. doi: 10.1128/JVI.01202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KS, et al. Viral antibody dynamics in a chiropteran host. Journal of Animal Ecology. 2014;83(2):415–428. doi: 10.1111/1365-2656.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron MD. Wild-type Rinderpest virus uses SLAM (CD150) as its receptor. Journal of General Virology. 2005;86(6):1753–1757. doi: 10.1099/vir.0.80836-0. [DOI] [PubMed] [Google Scholar]
- Barr J, et al. Isolation of multiple novel paramyxoviruses from pteropid bat urine. Journal of General Virology. 2015;96(Pt_1):24–29. doi: 10.1099/vir.0.068106-0. [DOI] [PubMed] [Google Scholar]
- Barr JA, et al. Evidence of bat origin for Menangle virus, a zoonotic paramyxovirus first isolated from diseased pigs. Journal of General Virology. 2012;93(PART 12):2590–2594. doi: 10.1099/vir.0.045385-0. [DOI] [PubMed] [Google Scholar]
- Barrett MW, Chalmers GA. A serologic survey of pronghorns in Alberta and Saskatchewan, 1970–1972. Journal of wildlife diseases. 11(2):157–63. doi: 10.7589/0090-3558-11.2.157. [DOI] [PubMed] [Google Scholar]
- Beaty SM, et al. Cross-reactive and cross-neutralizing activity of human mumps antibodies against a novel mumps virus from bats. Journal of Infectious Diseases. 2016:jiw53. doi: 10.1093/infdis/jiw534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KM, et al. Ephrin-B2 Reverse Signaling Increases α5β1 Integrin–Mediated Fibronectin Deposition and Reduces Distal Lung Compliance. American Journal of Respiratory Cell and Molecular Biology. 2013;49(4):680–687. doi: 10.1165/rcmb.2013-0002OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, et al. Cross-immunity between strains explains the dynamical pattern of paramyxoviruses. Proceedings of the National Academy of Sciences. 2015;112(43):13396–13400. doi: 10.1073/pnas.1516698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieringer M, et al. Experimental Adaptation of Wild-Type Canine Distemper Virus (CDV) to the Human Entry Receptor CD150. In: Schnell MJ, editor. PLoS ONE. 3. Vol. 8. 2013. p. e57488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaparte MI, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(30):10652–7. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Jardetzky TS, Lamb RA. Timing is everything: Fine-tuned molecular machines orchestrate paramyxovirus entry. Virology. 2015;479:518–531. doi: 10.1016/j.virol.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, et al. Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology. 2008;372(2):357–371. doi: 10.1016/j.virol.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bowden TA, et al. Shared paramyxoviral glycoprotein architecture is adapted for diverse attachment strategies. Biochemical Society Transactions. 2010;38(5) doi: 10.1042/BST0381349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden TA, et al. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nature Structural & Molecular Biology. 2008;15(6):567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- Breed AC, et al. Prevalence of henipavirus and rubulavirus antibodies in pteropid bats, Papua New Guinea. Emerging infectious diseases. 2010;16(12):1997–9. doi: 10.3201/eid1612.100879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bressem M-F, et al. Cetacean Morbillivirus: Current Knowledge and Future Directions. Viruses. 2014;6(12):5145–5181. doi: 10.3390/v6125145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley L, et al. Quantifying Global Drivers of Zoonotic Bat Viruses: A Process-Based Perspective. The American Naturalist. 2016;187(2):E53–E64. doi: 10.1086/684391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder CC, et al. A treatment for and vaccine against the deadly Hendra and Nipah viruses. Antiviral Research. 2013;100(1):8–13. doi: 10.1016/j.antiviral.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook CE, Dobson AP. Bats as “special” reservoirs for emerging zoonotic pathogens. Trends in Microbiology. 2015;23(3):172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CW, et al. Illumination of Parainfluenza Virus Infection and Transmission in Living Animals Reveals a Tissue-Specific Dichotomy. In: Fouchier RAM, editor. PLoS Pathogens. 7. Vol. 7. 2011. p. e1002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AL, et al. Complete Genome Sequence of Teviot Paramyxovirus, a Novel Rubulavirus Isolated from Fruit Bats in Australia. Genome Announcements. 2015;3(2):e00177–15. doi: 10.1128/genomeA.00177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AL, et al. Hendra Virus Infection Dynamics in the Grey-Headed Flying Fox (Pteropus poliocephalus) at the Southern-Most Extent of Its Range: Further Evidence This Species Does Not Readily Transmit the Virus to Horses. In: Schneider BS, editor. PLOS ONE. 6. Vol. 11. 2016. p. e0155252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caignard G, et al. The V Protein of Tioman Virus Is Incapable of Blocking Type I Interferon Signaling in Human Cells. In: Hahm B, editor. PLoS ONE. 1. Vol. 8. 2013. p. e53881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RW, et al. Mossman Virus, a Paramyxovirus of Rodents Isolated in Queensland. Search (Sydney) - Journal of the Australian and New Zealand Association for the Advancement of Science. 1977;8(11–1):435–436. [Google Scholar]
- Chadha MS, et al. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerging Infectious Diseases. 2006;12(2):235–240. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R, Takimoto T. Antagonism of innate immunity by Paramyxovirus accessory proteins. Viruses. 2009;1(3):574–593. doi: 10.3390/v1030574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant K, et al. Probable human infection with a newly described virus in the family Paramyxoviridae. Emerging infectious diseases. 1998;4(2):273–5. doi: 10.3201/eid0402.980215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatziandreou N, et al. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5) Journal of General Virology. 2004;85(10):3007–3016. doi: 10.1099/vir.0.80200-0. [DOI] [PubMed] [Google Scholar]
- Ching PKG, et al. Outbreak of henipavirus infection, Philippines, 2014. Emerging Infectious Diseases. 2015;21(2):328–331. doi: 10.3201/eid2102.141433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, et al. Serological Evidence of Henipavirus Exposure in Cattle, Goats and Pigs in Bangladesh. In: Geisbert T, editor. PLoS Neglected Tropical Diseases. 11. Vol. 8. 2014. p. e3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KB, et al. Full length genome sequence of Tioman virus, a novel paramyxovirus in the genus Rubulavirus isolated from fruit bats in Malaysia. Archives of Virology. 2002;147(7):1323–1348. doi: 10.1007/s00705-002-0815-5. [DOI] [PubMed] [Google Scholar]
- Chua KB, et al. Nipah Virus: A Recently Emergent Deadly Paramyxovirus. Science. 2000;288(5470) doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Chua KB, et al. Tioman Virus, a Novel Paramyxovirus Isolated from Fruit Bats in Malaysia. Virology. 2001;283(2):215–229. doi: 10.1006/viro.2000.0882. [DOI] [PubMed] [Google Scholar]
- Ciancanelli MJ, et al. Nipah virus sequesters inactive STAT1 in the nucleus via a P gene-encoded mechanism. [Accessed October 16, 2016];Journal of virology. 2009 83(16):7828–41. doi: 10.1128/JVI.02610-08. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19515782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney MK, Fox JP, Hall CE. The Seattle Virus Watch. VI. Observations of infections with and illness due to parainfluenza, mumps and respiratory syncytial viruses and Mycoplasma pneumoniae. American journal of epidemiology. 1975;101(6):532–51. doi: 10.1093/oxfordjournals.aje.a112125. [DOI] [PubMed] [Google Scholar]
- Cuevas-Romero JS, Blomström A-L, Berg M. Molecular and epidemiological studies of Porcine rubulavirus infection - an overview. Infection ecology & epidemiology. 2015;5:29602. doi: 10.3402/iee.v5.29602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P, et al. Interdisciplinary approaches to understanding disease emergence: the past, present, and future drivers of Nipah virus emergence. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(Suppl 1):3681–8. doi: 10.1073/pnas.1201243109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Tropica. 2001;78(2):103–116. doi: 10.1016/s0001-706x(00)00179-0. [DOI] [PubMed] [Google Scholar]
- Daugherty MD, et al. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. eLife. 2016;5:60–64. doi: 10.7554/eLife.14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpeut S, Noyce R, Richardson C. The Tumor-Associated Marker, PVRL4 (Nectin-4), is the Epithelial Receptor for Morbilliviruses. Viruses. 2014;6(6):2268–2286. doi: 10.3390/v6062268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux P, et al. A recombinant measles virus unable to antagonize STAT1 function cannot control inflammation and is attenuated in rhesus monkeys. Journal of virology. 2011;85(1):348–56. doi: 10.1128/JVI.00802-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux P, Priniski L, Cattaneo R. The measles virus phosphoprotein interacts with the linker domain of STAT1. Virology. 2013;444(1):250–256. doi: 10.1016/j.virol.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nature Reviews Immunology. 2012;13(1):46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield J, Macpherson LW, Zbitnew A. Upper respiratory disease in thouroughbred horses: studies of its viral etiology in the Toronto area, 1960 TO 1963. Canadian journal of comparative medicine and veterinary science. 1965;29(1):18–22. [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, et al. Bats host major mammalian paramyxoviruses. Nature Communications. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, et al. Henipavirus RNA in African Bats. In: Markotter W, editor. PLoS ONE. 7. Vol. 4. 2009. p. e6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duignan P, et al. Phocine Distemper Virus: Current Knowledge and Future Directions. Viruses. 2014;6(12):5093–5134. doi: 10.3390/v6125093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton BT, et al. Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol. 2006;4(1):23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elderfield RA, et al. Ferret airway epithelial cell cultures support efficient replication of influenza B virus but not mumps virus. Journal of General Virology. 2015;96(8):2092–2098. doi: 10.1099/vir.0.000176. [DOI] [PubMed] [Google Scholar]
- Epstein JH, et al. Henipavirus Infection in Fruit Bats (Pteropus giganteus), India. Emerging Infectious Diseases. 2008;14(8):1309–1311. doi: 10.3201/eid1408.071492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H, et al. Henipaviruses and Fruit Bats, Papua New Guinea. Emerging Infectious Diseases. 2013;19(4):670–671. doi: 10.3201/eid1904.111912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HE. Hendra virus ecology and transmission. Current Opinion in Virology. 2016;16:120–125. doi: 10.1016/j.coviro.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Fogarty R, et al. Henipavirus susceptibility to environmental variables. Virus Research. 2008;132(1–2):140–144. doi: 10.1016/j.virusres.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournié G, et al. Spatiotemporal trends in the discovery of new swine infectious agents. Veterinary Research. 2015;46(1):114. doi: 10.1186/s13567-015-0226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzke M, et al. Nectin-4-dependent measles virus spread to the cynomolgus monkey tracheal epithelium: role of infected immune cells infiltrating the lamina propria. Journal of virology. 2013;87(5):2526–34. doi: 10.1128/JVI.03037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima K, et al. Amino acid substitutions contributing to α2,6-sialic acid linkage binding specificity of human parainfluenza virus type 3 hemagglutinin-neuraminidase. FEBS Letters. 2015;589(11):1278–82. doi: 10.1016/j.febslet.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima K, et al. Terminal sialic acid linkages determine different cell infectivities of human parainfluenza virus type 1 and type 3. Virology. 2014;464:424–431. doi: 10.1016/j.virol.2014.07.033. [DOI] [PubMed] [Google Scholar]
- Furuse Y, et al. Origin of measles virus: divergence from rinderpest virus between the 11th and 12th centuries. Virology Journal. 2010;7(1):52. doi: 10.1186/1743-422X-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T, et al. Existence of feline morbillivirus infection in Japanese cat populations. Archives of Virology. 2014;159(2):371–373. doi: 10.1007/s00705-013-1813-5. [DOI] [PubMed] [Google Scholar]
- Gabriel G, Czudai-Matwich V, Klenk H-D. Adaptive mutations in the H5N1 polymerase complex. Virus Research. 2013;178(1):53–62. doi: 10.1016/j.virusres.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Gainey MD, et al. Paramyxovirus-induced shutoff of host and viral protein synthesis: role of the P and V proteins in limiting PKR activation. Journal of virology. 2008;82(2):828–39. doi: 10.1128/JVI.02023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale NW, et al. Ephrin-B2 Selectively Marks Arterial Vessels and Neovascularization Sites in the Adult, with Expression in Both Endothelial and Smooth-Muscle Cells. Developmental Biology. 2001;230(2):151–160. doi: 10.1006/dbio.2000.0112. [DOI] [PubMed] [Google Scholar]
- Geoghegan JL, et al. Virological factors that increase the transmissibility of emerging human viruses. Proceedings of the National Academy of Sciences. 2016;113(15):4170–4175. doi: 10.1073/pnas.1521582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glennon NB, et al. Transcriptome Profiling of the Virus-Induced Innate Immune Response in Pteropus vampyrus and Its Attenuation by Nipah Virus Interferon Antagonist Functions. Journal of virology. 2015;89(15):7550–66. doi: 10.1128/JVI.00302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DE. Measles virus. In: Knipe D, Howley P, editors. Fields Virology. 6. Wolters Kluwer; 2013. pp. 1042–1069. [Google Scholar]
- Hahn MB, et al. Roosting behaviour and habitat selection of Pteropus giganteus reveals potential links to Nipah virus epidemiology. Journal of Applied Ecology. 2014a;51(2):376–387. doi: 10.1111/1365-2664.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MB, et al. The role of landscape composition and configuration on Pteropus giganteus roosting ecology and Nipah virus spillover risk in Bangladesh. American Journal of Tropical Medicine and Hygiene. 2014b;90(2):247–255. doi: 10.4269/ajtmh.13-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin K, et al. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: A comprehensive experimental study of virus transmission. American Journal of Tropical Medicine and Hygiene. 2011;85(5):946–951. doi: 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HJ, et al. Bats as reservoirs of severe emerging infectious diseases. Virus Research. 2015;205:1–6. doi: 10.1016/j.virusres.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasebe F, et al. Serologic Evidence of Nipah Virus Infection in Bats, Vietnam. Emerging Infectious Diseases: Letters. 2012;18(3):536–537. doi: 10.3201/eid1803.111121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DTS, et al. Antibodies to Henipavirus or Henipa-Like Viruses in Domestic Pigs in Ghana, West Africa. In: Johnson WE, editor. PLoS ONE. 9. Vol. 6. 2011. p. e25256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DTS, et al. Evidence of Henipavirus Infection in West African Fruit Bats. In: Montgomery JM, editor. PLoS ONE. 7. Vol. 3. 2008. p. e2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ST, et al. Investigating Rare Risk Factors for Nipah Virus in Bangladesh: 2001–2012. EcoHealth. 2016 doi: 10.1007/s10393-016-1166-0. 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson GW, et al. Characterization of Mapuera virus: structure, proteins and nucleotide sequence of the gene encoding the nucleocapsid protein. Journal of General Virology. 1995;76(10):2509–2518. doi: 10.1099/0022-1317-76-10-2509. [DOI] [PubMed] [Google Scholar]
- Henrickson KJ. Parainfluenza viruses. Clinical microbiology reviews. 2003;16(2):242–64. doi: 10.1128/CMR.16.2.242-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homaira N, et al. Nipah virus outbreak with person-to-person transmission in a district of Bangladesh, 2007. Epidemiology and infection. 2010;138(11):1630–1636. doi: 10.1017/S0950268810000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper PT, et al. The retrospective diagnosis of a second outbreak of equine morbillivirus infection. Aust Vet J. 1996;74(3):244–245. doi: 10.1111/j.1751-0813.1996.tb15414.x. [DOI] [PubMed] [Google Scholar]
- Horvath CM. Weapons of STAT destruction. Interferon evasion by paramyxovirus V proteins. European Journal of Biochemistry. 2004;271(23–24):4621–4628. doi: 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- Hsu VP, et al. Nipah virus encephalitis reemergence, Bangladesh. Emerging Infectious Diseases. 2004;10(12):2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iehlé C, et al. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerging infectious diseases. 2007;13(1):159–61. doi: 10.3201/eid1301.060791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, et al. Nipah Virus Transmission from Bats to Humans Associated with Drinking Traditional Liquor Made from Date Palm Sap, Bangladesh, 2011–2014. Emerging Infectious Diseases. 2016;22(4):664–670. doi: 10.3201/eid2204.151747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun MH, Karabatsos N, Johnson RH. A new mouse paramyxovirus (J virus) The Australian journal of experimental biology and medical science. 1977;55(6):645–7. doi: 10.1038/icb.1977.63. [DOI] [PubMed] [Google Scholar]
- Kamins AO, et al. Characteristics and Risk Perceptions of Ghanaians Potentially Exposed to Bat-Borne Zoonoses through Bushmeat. EcoHealth. 2015;12(1):104–120. doi: 10.1007/s10393-014-0977-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabatsos N. In: International catalogue of arboviruses : including certain other viruses of vertebrates. 3. Karabatsos N, editor. San Antonio: American Society of Tropical Medicine and Hygiene; 1985. [DOI] [PubMed] [Google Scholar]
- Karron RA, et al. A live attenuated bovine parainfluenza virus type 3 vaccine is safe, infectious, immunogenic, and phenotypically stable in infants and children. The Journal of infectious diseases. 1995;171(5):1107–14. doi: 10.1093/infdis/171.5.1107. [DOI] [PubMed] [Google Scholar]
- Katoh H, et al. Cross-Neutralization between Human and African Bat Mumps Viruses. Emerging Infectious Diseases. 2016;22(4):703–706. doi: 10.3201/eid2204.151116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen A, Shackelton LA, Holmes EC. Family level phylogenies reveal modes of macroevolution in RNA viruses. Proceedings of the National Academy of Sciences. 2011;108(1):238–243. doi: 10.1073/pnas.1011090108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure T, et al. Human trachea primary epithelial cells express both sialyl(alpha2-3)Gal receptor for human parainfluenza virus type 1 and avian influenza viruses, and sialyl(alpha2-6)Gal receptor for human influenza viruses. Glycoconjugate journal. 2006;23(1–2):101–6. doi: 10.1007/s10719-006-5442-z. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D, et al. Paramyxovirus mRNA editing, the "rule of six” and error catastrophe: A hypothesis. Journal of General Virology. 2005;86(7):1869–1877. doi: 10.1099/vir.0.80986-0. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D, et al. Paramyxovirus RNA Synthesis and the Requirement for Hexamer Genome Length: the Rule of Six Revisited. Journal of virology. 1998;72(2):891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T, Takeuchi K, Gotoh B. Bovine parainfluenza virus type 3 accessory proteins that suppress beta interferon production. Microbes and Infection. 2007;9(8):954–962. doi: 10.1016/j.micinf.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Krah DL. Receptors for binding measles virus on host cells and erythrocytes. Microbial Pathogenesis. 1991;11(4):221–228. doi: 10.1016/0882-4010(91)90026-7. [DOI] [PubMed] [Google Scholar]
- Kreuder Johnson C, et al. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Scientific Reports. 2015;5:14830. doi: 10.1038/srep14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger N, et al. Recombinant mumps viruses expressing the batMuV fusion glycoprotein are highly fusion-active and neurovirulent. Journal of General Virology. 2016 doi: 10.1099/jgv.0.000596. [DOI] [PubMed] [Google Scholar]
- Kulkarni DD, et al. Nipah virus infection: Current scenario. Indian Journal of Virology. 2013;24(3):398–408. doi: 10.1007/s13337-013-0171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth A, Kohl C, Brinkmann A, Ebinger A, Harper JA, Wang L-FF, et al. Novel paramyxoviruses in free-ranging European bats. In: Vartanian J-P, editor. PLoS ONE. 6. Vol. 7. 2012. p. e38688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DP. The complexity of genetic variation in a simple immune system. Trends in Genetics. 2005;21(4):197–199. doi: 10.1016/j.tig.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Parks GD. Paramyxoviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott, Williams and Wilkins; 2006. pp. 1449–1496. [Google Scholar]
- Lau SKP, et al. Identification and characterization of a novel paramyxovirus, porcine parainfluenza virus 1, from deceased pigs. Journal of General Virology. 2013;94(Pt_10):2184–2190. doi: 10.1099/vir.0.052985-0. [DOI] [PubMed] [Google Scholar]
- Lau SKP, et al. Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology. 2010;404(1):106–16. doi: 10.1016/j.virol.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P, et al. Surface glycoproteins of the recently identified African Henipavirus promote viral entry and cell fusion in a range of human, simian and bat cell lines. Virus Research. 2014;181:77–80. doi: 10.1016/j.virusres.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Lee B, et al. Modes of paramyxovirus fusion: a Henipavirus perspective. Trends in microbiology. 2011;19(8):389–99. doi: 10.1016/j.tim.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, et al. Molecular recognition of human ephrinB2 cell surface receptor by an emergent African henipavirus. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(17):E2156–65. doi: 10.1073/pnas.1501690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Ataman ZA, Jin L. Evil versus “eph-ective” use of ephrin-B2. Nature Structural & Molecular Biology. 2008;15(6):540–542. doi: 10.1038/nsmb0608-540. [DOI] [PubMed] [Google Scholar]
- Lehmkuhl HD, Cutlip RC. Characterization of parainfluenza type 3 virus isolated from the lung of a lamb with pneumonia. American journal of veterinary research. 1982;43(4):626–8. [PubMed] [Google Scholar]
- Li W, et al. A novel parainfluenza virus type 3 (PIV3) identified from goat herds with respiratory diseases in eastern China. Veterinary Microbiology. 2014;174(1):100–106. doi: 10.1016/j.vetmic.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Antibodies to Nipah or Nipah-like Viruses in Bats, China. Emerging Infectious Diseases. 2008;14(12):1974–1976. doi: 10.3201/eid1412.080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology. 2006;346(1):219–228. doi: 10.1016/j.virol.2005.10.039. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. Function of the small hydrophobic protein of J paramyxovirus. Journal of virology. 2011;85(1):32–42. doi: 10.1128/JVI.01673-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu KG, et al. The non-pathogenic Henipavirus Cedar paramyxovirus phosphoprotein has a compromised ability to target STAT1 and STAT2. Antiviral Research. 2015;124:69–76. doi: 10.1016/j.antiviral.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith JO, et al. Epidemic Dynamics at the Human-Animal Interface. Science. 2009;1362(2009):1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh EH, et al. Ecological Approaches to Studying Zoonoses. Microbiology Spectrum. 2013;1(2) doi: 10.1128/microbiolspec.OH-0009-2012. [DOI] [PubMed] [Google Scholar]
- Long JS, et al. Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature. 2016;529(7584):101–104. doi: 10.1038/nature16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby S, Gurley E. Global Virology I - Identifying and Investigating Viral Diseases. New York, NY: Springer New York: 2015. Epidemiology of Henipaviruses; pp. 55–71. [Google Scholar]
- Luby SP, et al. Foodborne transmission of Nipah virus, Bangladesh. Emerging Infectious Diseases. 2006;12(12):1888–1894. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby SP, et al. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerging Infectious Diseases. 2009;15(8):1229–1235. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby SP. The pandemic potential of Nipah virus. Antiviral Research. 2013;100(1):38–43. doi: 10.1016/j.antiviral.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Lyon M, et al. Presence of a unique parainfluenza virus 3 strain identified by RT-PCR in visna-maedi virus infected sheep. Veterinary Microbiology. 1997;57(2):95–104. doi: 10.1016/s0378-1135(97)00104-1. [DOI] [PubMed] [Google Scholar]
- Magoffin DE, Mackenzie JS, Wang L-F. Genetic analysis of J-virus and Beilong virus using minireplicons. Virology. 2007;364(1):103–11. doi: 10.1016/j.virol.2007.01.045. [DOI] [PubMed] [Google Scholar]
- Maidana SS, et al. Isolation and characterization of bovine parainfluenza virus type 3 from water buffaloes (Bubalus bubalis) in Argentina. BMC Veterinary Research. 2012;8(1):83. doi: 10.1186/1746-6148-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl JN, et al. Reservoir Host Immune Responses to Emerging Zoonotic Viruses. Cell. 2015;160(1):20–35. doi: 10.1016/j.cell.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh GA, et al. Cedar Virus: A Novel Henipavirus Isolated from Australian Bats. PLoS Pathogens. 2012;8(8):e1002836. doi: 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo MA. A summary of taxonomic changes recently approved by ICTV. Arch Virol. 2002;147(1997):1655–1656. doi: 10.1007/s007050200039. [DOI] [PubMed] [Google Scholar]
- McFarlane R, Becker N, Field H. Investigation of the climatic and environmental context of hendra virus spillover events 1994–2010. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0028374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélade J, et al. An eco-epidemiological study of Morbilli-related paramyxovirus infection in Madagascar bats reveals host-switching as the dominant macro-evolutionary mechanism. Scientific reports. 2016 Apr;6:23752. doi: 10.1038/srep23752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesina JE, et al. The pathology of feral rodents in North Queensland. Tropenmedizin und Parasitologie. 1974;25(1):116–27. [PubMed] [Google Scholar]
- Middleton D, et al. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerging Infectious Diseases. 2014;20(3):372–379. doi: 10.3201/eid2003.131159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton DJ, et al. Experimental Nipah Virus Infection in Pteropid Bats (Pteropus poliocephalus) Journal of Comparative Pathology. 2007;136(4):266–272. doi: 10.1016/j.jcpa.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Middleton DJ, Weingartl HM. Henipaviruses in Their Natural Animal Hosts. Henipavirus: Ecology, Molecular Virology, and Pathogenesis. 2012:105–121. doi: 10.1007/82_2012_210. [DOI] [PubMed] [Google Scholar]
- Mills JN, et al. Nipah Virus Infection in Dogs, Malaysia, 1999. Emerging Infectious Diseases. 2009;15(6):950–952. doi: 10.3201/eid1506.080453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina MJ, et al. Long-term measles-induced immunomodulation increases overall childhood infectious disease mortality. Science. 2015;348(6235):694–699. doi: 10.1126/science.aaa3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishin VP, et al. N-linked glycan at residue 523 of human parainfluenza virus type 3 hemagglutinin-neuraminidase masks a second receptor-binding site. Journal of virology. 2010;84(6):3094–100. doi: 10.1128/JVI.02331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Nor M, Gan C, Ong B. Nipah virus infection of pigs in peninsular Malaysia Epidemiological findings. Rev Sci Tech. 2000;19(1):160–165. doi: 10.20506/rst.19.1.1202. [DOI] [PubMed] [Google Scholar]
- Motta V, et al. NOD-Like Receptors: Versatile Cytosolic Sentinels. Physiological Reviews. 2014;95(1) doi: 10.1152/physrev.00009.2014. [DOI] [PubMed] [Google Scholar]
- Muleya W, et al. Molecular Epidemiology of Paramyxoviruses in Frugivorous Eidolon helvum Bats in Zambia. Journal of Veterinary Medical Science. 2014;76(4):611–614. doi: 10.1292/jvms.13-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K, et al. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268(5207):94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- Murray KA, Daszak P. Human ecology in pathogenic landscapes: two hypotheses on how land use change drives viral emergence. Current opinion in virology. 2011;72(2):181–204. doi: 10.1016/j.coviro.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SS, et al. Human health impacts of ecosystem alteration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(47):18753–60. doi: 10.1073/pnas.1218656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar N, et al. Date Palm Sap Collection: Exploring Opportunities to Prevent Nipah Transmission. EcoHealth. 2010;7(2):196–203. doi: 10.1007/s10393-010-0320-3. [DOI] [PubMed] [Google Scholar]
- Nambulli S, et al. Mapping the evolutionary trajectories of morbilliviruses: what, where and whither. Current Opinion in Virology. 2016;16:95–105. doi: 10.1016/j.coviro.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete OA, et al. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436(7049):401. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- NIAID & NIH. NIAID Emerging Infectious Diseases/Pathogens: Priority Pathogens. [Accessed November 1, 2016];NIH/NIAID: Biodefence & Emerging Infectious Diseases. 2016 Available at: https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens.
- Nicholls JM, et al. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respiratory Research. 2007;8(1):73. doi: 10.1186/1465-9921-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea TJ, et al. Bat flight and zoonotic viruses. Emerging Infectious Diseases. 2014;20(5):741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K, et al. Host–virus specificity of morbilliviruses predicted by structural modeling of the marine mammal SLAM, a receptor. Comparative Immunology, Microbiology and Infectious Diseases. 2010;33(3):227–241. doi: 10.1016/j.cimid.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Openshaw JJ, et al. Bat Hunting and Bat-Human Interactions in Bangladeshi Villages: Implications for Zoonotic Disease Transmission and Bat Conservation. Transboundary and Emerging Diseases. 2016:1–7. doi: 10.1111/tbed.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinski RM, et al. Widespread detection and characterization of porcine parainfluenza virus 1 in pigs in the USA. Journal of General Virology. 2016;97(2):281–286. doi: 10.1099/jgv.0.000343. [DOI] [PubMed] [Google Scholar]
- Pallister J, et al. A recombinant Hendra virus G glycoprotein-based subunit vaccine protects ferrets from lethal Hendra virus challenge. Vaccine. 2011;29(34):5623–5630. doi: 10.1016/j.vaccine.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallister J, et al. Chloroquine administration does not prevent Nipah virus infection and disease in ferrets. Journal of virology. 2009;83(22):11979–11982. doi: 10.1128/JVI.01847-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks GD, Alexander-Miller MA. Paramyxovirus Activation and Inhibition of Innate Immune Responses. Journal of Molecular Biology. 2013;425(24):4872–4892. doi: 10.1016/j.jmb.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish CR, et al. Cross-Species Virus Transmission and the Emergence of New Epidemic Diseases. Microbiology and Molecular Biology Reviews. 2008;72(3):457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Pasquier L. Meeting the Demand for Innate and Adaptive Immunities During Evolution. Scandinavian Journal of Immunology. 2005;62(s1):39–48. doi: 10.1111/j.1365-3083.2005.01608.x. [DOI] [PubMed] [Google Scholar]
- Paton NI, et al. Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet. 1999;354(9186):1253–1256. doi: 10.1016/S0140-6736(99)04379-2. [DOI] [PubMed] [Google Scholar]
- Patz JA, et al. Unhealthy landscapes: Policy recommendations on land use change and infectious disease emergence. Environmental health perspectives. 2004;112(10):1092–8. doi: 10.1289/ehp.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri KM, Singh KR, Hollinger FB. Isolation of a new parainfluenza virus from a frugivorous bat, Rousettus leschenaulti, collected at Poona, India. The American journal of tropical medicine and hygiene. 1971;20(1):125–30. doi: 10.4269/ajtmh.1971.20.125. [DOI] [PubMed] [Google Scholar]
- Pecchini R, et al. Parainfluenza virus as a cause of acute respiratory infection in hospitalized children. The Brazilian Journal of Infectious Diseases. 2015;19(4):358–362. doi: 10.1016/j.bjid.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel AJ, et al. Continent-wide panmixia of an African fruit bat facilitates transmission of potentially zoonotic viruses. Nature Communications. 2013;4:2770. doi: 10.1038/ncomms3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel AJ, et al. The equine Hendra virus vaccine remains a highly effective preventative measure against infection in horses and humans: “The imperative to develop a human vaccine for the Hendra virus in Australia”. Infection ecology & epidemiology. 2016;6:31658. doi: 10.3402/iee.v6.31658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet O, Beaty S, Lee B. Functional rectification of the newly described African henipavirus fusion glycoprotein (Gh-M74a) Journal of virology. 2014a;88(9):5171–6. doi: 10.1128/JVI.03655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet O, et al. Evidence for henipavirus spillover into human populations in Africa. Nature Communications. 2014b;5:5342. doi: 10.1038/ncomms6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet O, Wang YE, Lee B. Henipavirus Receptor Usage and Tropism. In: Lee Benhur, Rota Paul A., editors. Henipavirus: Ecology, Molecular Virology, and Pathogenesis. Springer Berlin Heidelberg: 2012. pp. 59–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philbey AW, et al. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerging infectious diseases. 1998;4(2):269–71. doi: 10.3201/eid0402.980214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, et al. Ecological dynamics of emerging bat virus spillover. Proceedings. Biological sciences / The Royal Society. 2015;282(1798):20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, et al. Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus) Proceedings. Biological sciences / The Royal Society. 2008;275(1636):861–9. doi: 10.1098/rspb.2007.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, et al. Transmission or Within-Host Dynamics Driving Pulses of Zoonotic Viruses in Reservoir–Host Populations. PLOS Neglected Tropical Diseases. 2016;10(8):e0004796. doi: 10.1371/journal.pntd.0004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, et al. Urban habituation, ecological connectivity and epidemic dampening: the emergence of Hendra virus from flying foxes (Pteropus spp.) Proceedings. Biological sciences / The Royal Society. 2011;278(1725):3703–12. doi: 10.1098/rspb.2011.0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power UF, Ryan KW, Portner A. The P genes of human parainfluenza virus type 1 clinical isolates are polycistronic and microheterogenous. Virology. 1992;189(1):340–343. doi: 10.1016/0042-6822(92)90712-x. [DOI] [PubMed] [Google Scholar]
- Pulliam JRC, et al. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. Journal of the Royal Society, Interface / the Royal Society. 2012;9(66):89–101. doi: 10.1098/rsif.2011.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, et al. Canine distemper outbreak in rhesus monkeys, China. Emerging Infectious Diseases. 2011;17(8):1541–1543. doi: 10.3201/eid1708.101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani MAG, et al. Identification of IFN-stimulated gene (ISG) proteins that inhibit human parainfluenza virus type 3. Journal of virology. 2016:JVI.01551–16. doi: 10.1128/JVI.01551-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MA, et al. Date Palm Sap Linked to Nipah Virus Outbreak in Bangladesh, 2008. Vector-Borne and Zoonotic Diseases. 2012;12(1):65–72. doi: 10.1089/vbz.2011.0656. [DOI] [PubMed] [Google Scholar]
- Rahman M, Chakraborty A. Nipah virus outbreaks in Bangladesh: a deadly infectious disease. WHO South-East Asia Journal of Public Health. 2012;1(2):208–212. doi: 10.4103/2224-3151.206933. [DOI] [PubMed] [Google Scholar]
- Rahman SA, et al. Risk factors for Nipah virus infection among pteropid bats, Peninsular Malaysia. Emerging Infectious Diseases. 2013;19(1):51–60. doi: 10.3201/eid1901.120221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw RW, et al. Identification and phylogenetic comparison of Salem virus, a novel paramyxovirus of horses. Virology. 2000;270(2):417–29. doi: 10.1006/viro.2000.0305. [DOI] [PubMed] [Google Scholar]
- Reyes-Leyva J, et al. Amino acid change 335 E to K affects the sialic-acid-binding and neuraminidase activities of Urabe AM9 mumps virus hemagglutinin–neuraminidase glycoprotein. Microbes and Infection. 2007;9(2):234–240. doi: 10.1016/j.micinf.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Reynes JM, et al. Nipah Virus in Lyle’s Flying Foxes, Cambodia. Emerging Infectious Diseases. 2005;11(7):1042–1047. doi: 10.3201/eid1107.041350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Benitez JF, et al. Serological Survey of Veterinarians to Assess the Zoonotic Potential of Three Emerging Swine Diseases in Mexico. Zoonoses and Public Health. 2014;61(2):131–137. doi: 10.1111/zph.12055. [DOI] [PubMed] [Google Scholar]
- Rogers RJ, et al. Investigation of a second focus of equine morbillivirus infection in coastal Queensland. Australian veterinary journal. 1996 Jul;74:243–244. doi: 10.1111/j.1751-0813.1996.tb15413.x. 1994. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, et al. Genetic diversity of feline morbilliviruses isolated in Japan. Journal of General Virology. 2014;95(Pt_7):1464–1468. doi: 10.1099/vir.0.065029-0. [DOI] [PubMed] [Google Scholar]
- Sakai K, et al. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. Journal of Virology. 2013a;87(2):1105–1114. doi: 10.1128/JVI.02419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, et al. Canine distemper virus associated with a lethal outbreak in monkeys can readily adapt to use human receptors. Journal of virology. 2013b;87(12):7170–5. doi: 10.1128/JVI.03479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Rojas M, et al. Prevalence of rabies and LPM paramyxovirus antibody in non-hematophagous bats captured in the Central Pacific coast of Mexico. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2004;98(10):577–84. doi: 10.1016/j.trstmh.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Santos-López G, et al. Structure-function analysis of two variants of mumps virus hemagglutinin-neuraminidase protein. Brazilian Journal of Infectious Diseases. 2009;13(1):24–34. doi: 10.1590/s1413-86702009000100007. [DOI] [PubMed] [Google Scholar]
- Sasaki M, et al. Molecular epidemiology of paramyxoviruses in Zambian wild rodents and shrews. Journal of General Virology. 2014;95:325–330. doi: 10.1099/vir.0.058404-0. [DOI] [PubMed] [Google Scholar]
- Sato H, et al. Morbillivirus Receptors and Tropism: Multiple Pathways for Infection. Frontiers in Microbiology. 2012;3:75. doi: 10.3389/fmicb.2012.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazzad HMS, et al. Nipah virus infection outbreak with nosocomial and corpse-to-human transmission, Bangladesh. Emerging Infectious Diseases. 2013;19(2):210–217. doi: 10.3201/eid1902.120971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AC, et al. Bovine parainfluenza virus type 3 (BPIV3) fusion and hemagglutinin-neuraminidase glycoproteins make an important contribution to the restricted replication of BPIV3 in primates. Journal of virology. 2000;74(19):8922–9. doi: 10.1128/jvi.74.19.8922-8929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp CR, et al. Chronic Infection of Domestic Cats with Feline Morbillivirus, United States. Emerging Infectious Diseases. 2016;22(4):760–762. doi: 10.3201/eid2204.151921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiadopoulos MH, et al. Determinants of the Host Range Restriction of Replication of Bovine Parainfluenza Virus Type 3 in Rhesus Monkeys Are Polygenic. Journal of Virology. 2003;77(2):1141–1148. doi: 10.1128/JVI.77.2.1141-1148.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KF, et al. Global rise in human infectious disease outbreaks. Journal of the Royal Society, Interface [electronic resource] / the Royal Society. 2014;11(101):20140950. doi: 10.1098/rsif.2014.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, et al. Natural infection with canine distemper virus in hand-feeding Rhesus monkeys in China. Veterinary Microbiology. 2010;141(3–4):374–378. doi: 10.1016/j.vetmic.2009.09.024. [DOI] [PubMed] [Google Scholar]
- Tao T, Ryan KW. Host Range Restriction of Parainfluenza Virus Growth Occurs at the Level of Virus Genome Replication. Virology. 1996;220(1):69–77. doi: 10.1006/viro.1996.0287. [DOI] [PubMed] [Google Scholar]
- Tatsuo H, Ono N, Yanagi Y. Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. Journal of virology. 2001;75(13):5842–50. doi: 10.1128/JVI.75.13.5842-5850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanapongtharm W, et al. Spatial characterization of colonies of the flying fox bat, a carrier of Nipah Virus in Thailand. BMC Veterinary Research. 2015;11(1):81. doi: 10.1186/s12917-015-0390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidona CA, et al. Isolation and Molecular Characterization of a Novel Cytopathogenic Paramyxovirus from Tree Shrews. Virology. 1999;258(2):425–434. doi: 10.1006/viro.1999.9693. [DOI] [PubMed] [Google Scholar]
- Tikasingh ES, et al. Nariva virus, a hitherto undescribed agent isolated from the Trinidadian rat, Zygodontomys b. brevicauda. The American journal of tropical medicine and hygiene. 1966;15(2):235–8. doi: 10.4269/ajtmh.1966.15.235. [DOI] [PubMed] [Google Scholar]
- Tong S, et al. Sensitive and Broadly Reactive Reverse Transcription-PCR Assays To Detect Novel Paramyxoviruses. Journal of Clinical Microbiology. 2008;46(8):2652–2658. doi: 10.1128/JCM.00192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaure C, Liu Y. A Comparative Review of Toll-Like Receptor 4 Expression and Functionality in Different Animal Species. Frontiers in Immunology. 2014;5:316. doi: 10.3389/fimmu.2014.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigant F, Lee B. Hendra and Nipah infection: pathology, models and potential therapies. Infectious disorders drug targets. 2011;11(3):315–36. doi: 10.2174/187152611795768097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar E, Barroso IM. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: a minireview. Glycoconjugate journal. 2006;23(1–2):5–17. doi: 10.1007/s10719-006-5433-0. [DOI] [PubMed] [Google Scholar]
- de Vries RD, et al. Measles immune suppression: lessons from the macaque model. PLoS pathogens. 2012;8(8):e1002885. doi: 10.1371/journal.ppat.1002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S, et al. A longitudinal study of the prevalence of Nipah virus in Pteropus lylei bats in Thailand: evidence for seasonal preference in disease transmission. Vector Borne Zoonotic Dis. 2010;10(2):183–190. doi: 10.1089/vbz.2008.0105. [DOI] [PubMed] [Google Scholar]
- Wacharapluesadee S, et al. Bat Nipah virus, Thailand. Emerging infectious diseases. 2005;11(12):1949–51. doi: 10.3201/eid1112.050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T, et al. Glycomic Analysis of Human Respiratory Tract Tissues and Correlation with Influenza Virus Infection. In: Pekosz A, editor. PLoS Pathogens. 3. Vol. 9. 2013. p. e1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L-FF, et al. The Exceptionally Large Genome of Hendra Virus: Support for Creation of a New Genus within the Family Paramyxoviridae. Journal of Virology. 2000;74(21):9972–9979. doi: 10.1128/jvi.74.21.9972-9979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Mackenzie JS, Eaton BT. Disease outbreaks caused by emerging paramyxoviruses of bat origin. Emerging Infections in Asia. 2008:193–208. [Google Scholar]
- Wasik BR, Barnard KN, Parrish CR. Effects of Sialic Acid Modifications on Virus Binding and Infection. Trends in Microbiology. 2016 doi: 10.1016/j.tim.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis M, et al. Characterization of African bat henipavirus GH-M74a glycoproteins. Journal of General Virology. 2014;95(Pt_3):539–548. doi: 10.1099/vir.0.060632-0. [DOI] [PubMed] [Google Scholar]
- Weiss S, et al. Henipavirus-related sequences in fruit bat bushmeat, Republic of Congo. Emerging infectious diseases. 2012;18(9):1536–7. doi: 10.3201/eid1809.111607. A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Blueprint for R&D preparedness and response to public health emergencies due to highly infectious pathogens. WHO workshop on prioritization of pathogens report. 2015 Dec;:1–7. [Google Scholar]
- Wilkinson DA, et al. Highly diverse morbillivirus-related paramyxoviruses in wild fauna of the Southwestern Indian Ocean Islands: Evidence of exchange between introduced and endemic small mammals. Journal of Virology. 2014a;88(15):8268–8277. doi: 10.1128/JVI.01211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DA, et al. Identification of novel paramyxoviruses in insectivorous bats of the Southwest Indian Ocean. Virus Research. 2012;170(1):159–163. doi: 10.1016/j.virusres.2012.08.022. [DOI] [PubMed] [Google Scholar]
- Wilson RL, et al. Function of small hydrophobic proteins of paramyxovirus. Journal of virology. 2006;80(4):1700–9. doi: 10.1128/JVI.80.4.1700-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Munster VJ. Animal models of disease shed light on Nipah virus pathogenesis and transmission. The Journal of Pathology. 2015;235(2):196–205. doi: 10.1002/path.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, et al. Bushmeat hunting, deforesation, and prediction of zoonotic disease emergence. Emerging Infectious Diseases. 2005;11(12):1822–1827. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447(7142):279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KT, Tan CT. Clinical and Pathological Manifestations of Human Henipavirus Infection. Henipavirus: Ecology, Molecular Virology, and Pathogenesis. 2012:95–104. doi: 10.1007/82_2012_205. [DOI] [PubMed] [Google Scholar]
- Woo PCY, et al. Complete Genome Sequence of a Novel Paramyxovirus, Tailam Virus, Discovered in Sikkim Rats. Journal of Virology. 2011;85(24):13473–13474. doi: 10.1128/JVI.06356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, et al. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proceedings of the National Academy of Sciences. 2012;109(14):5435–5440. doi: 10.1073/pnas.1119972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, et al. Novel Variant of Beilong Paramyxovirus in Rats, China. Emerging Infectious Disease journal. 2012;18(6):1022. doi: 10.3201/eid1806.111901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M, et al. Assessing the epidemic potential of RNA and DNA viruses. Emerging Infectious Diseases. 2016;22(12) doi: 10.3201/eid2212.160123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PF, Neumann G, Kawaoka Y. Orthomyxoviruses. In: Knipe DM, Howley P, editors. Fields Virology. 6. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1186–1243. [Google Scholar]
- Wu Z, et al. Novel Henipa-like Virus, Mojiang Paramyxovirus, in Rats, China, 2012. Emerging Infectious Diseases. 2014;20(6) doi: 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, et al. Infection of mice, ferrets, and rhesus macaques with a clinical mumps virus isolate. Journal of virology. 2013;87(14):8158–68. doi: 10.1128/JVI.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P, et al. Isolation of Tioman virus from Pteropus giganteus bat in North-East region of India. Infection, Genetics and Evolution. 2016;45:224–229. doi: 10.1016/j.meegid.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaiw KC, et al. Serological Evidence of Possible Human Infection with Tioman virus, a Newly Described Paramyxovirus of Bat Origin. The Journal of Infectious Diseases. 2007;196(6):884–886. doi: 10.1086/520817. [DOI] [PubMed] [Google Scholar]
- Yaiw KC, et al. Tioman Virus, a Paramyxovirus of Bat Origin, Causes Mild Disease in Pigs and Has a Predilection for Lymphoid Tissues. Journal of Virology. 2008;82(1):565–568. doi: 10.1128/JVI.01660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, et al. An anti-interferon activity shared by paramyxovirus C proteins: Inhibition of Toll-like receptor 7/9-dependent alpha interferon induction. FEBS Letters. 2014;588(1):28–34. doi: 10.1016/j.febslet.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Yanagi Y, et al. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406(6798):893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- Yoshikawa Y, et al. Natural Infection with Canine Distemper Virus in a Japanese Monkey (Macaca fuscata) Veterinary Microbiology. 1989;20(3):193–205. doi: 10.1016/0378-1135(89)90043-6. [DOI] [PubMed] [Google Scholar]
- Young DF, et al. Human IFIT1 inhibits mRNA translation of rubulaviruses but not other members of the Paramyxoviridae family. Journal of Virology. 2016:JVI.01056–16. doi: 10.1128/JVI.01056-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DF, et al. Single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block interferon signaling in human and murine cells. Journal of virology. 2001;75(7):3363–70. doi: 10.1128/JVI.75.7.3363-3370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DF, et al. Virus replication in engineered human cells that do not respond to interferons. Journal of virology. 2003;77(3):2174–81. doi: 10.1128/JVI.77.3.2174-2181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin M, et al. The Interaction between Respiratory Pathogens and Mucus. Cell Host & Microbe. 2016;19(2):159–168. doi: 10.1016/j.chom.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeltina A, Bowden TA, Lee B. Emerging Paramyxoviruses: Receptor Tropism and Zoonotic Potential. In: Dutch RE, editor. PLOS Pathogens. 2. Vol. 12. 2016. p. e1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]