Abstract
Cancer stem cells (CSCs) are an attractive therapeutic target due to their predicted role in both metastasis and chemoresistance. One of the most commonly agreed on markers for ovarian CSCs is the cell surface protein CD133. CD133+ ovarian CSCs have increased tumorigenicity, resistance to chemotherapy, and increased metastasis. Therefore, we were interested in defining how CD133 is regulated and whether it has a role in tumor metastasis. Previously we found that overexpression of the transcription factor, ARID3B, increased the expression of PROM1 (CD133 gene) in ovarian cancer cells in vitro and in xenograft tumors. We report that ARID3B directly regulates PROM1 expression. Importantly, in a xenograft mouse model of ovarian cancer, knockdown of PROM1 in cells expressing exogenous ARID3B resulted in increased survival time compared with cells expressing ARID3B and a control short hairpin RNA. This indicated that ARID3B regulation of PROM1 is critical for tumor growth. Moreover, we hypothesized that CD133 may affect metastatic spread. Given that the peritoneal mesothelium is a major site of ovarian cancer metastasis, we explored the role of PROM1 in mesothelial attachment. PROM1 expression increased adhesion to mesothelium in vitro and ex vivo. Collectively, our work demonstrates that ARID3B regulates PROM1 adhesion to the ovarian cancer metastatic niche.
Keywords: Ovarian cancer, cancer stem cells, metastasis, adhesion, transcription factor
Introduction
Ovarian cancer is the most lethal gynecological cancer in women, accounting for about 5% of all female cancer deaths in the United States, with an estimated 14 080 deaths in 2017 alone (American Cancer Society, Facts and Figures 2017). Patients typically undergo debulking surgery followed by treatment with carboplatin and paclitaxel.1 A common feature of advanced ovarian cancer is the presence of ascites fluid, which often contains cancer stem cells (CSCs).2–6 According to the CSC model, a small percentage of slowly dividing CSCs are resistant to chemotherapy and are capable of repopulating tumors and thus are postulated to promote recurrence.5,7,8 Several criteria are used to define CSCs including high aldehyde dehydrogenase activity; expression of cell surface markers such as CD24, CD44, CD117, and CD133; exclusion of Hoechst dye 33342 (denoted as the side population); or expression of stem cell genes.5,9 Others and we previously demonstrated that the transcription factor ARID3B upregulates cancer stemness markers.10,11 In particular, we found that the stem cell gene marker CD133 was regulated by ARID3B. We wanted to know whether ARID3B directly regulates the CD133 gene PROM1, and how PROM1 regulation contributes to tumor growth and metastasis.
Little is known about the contribution of CD133 to metastasis. CD133 is a transmembrane glycoprotein encoded by the gene PROM1.12 CD133+ cancer cells exhibit increased resistance to drugs used to treat ovarian cancer including paclitaxel and/or cisplatin.13–18 CD133 expression is indicative of poor prognosis for patients with ovarian cancer. Higher CD133 correlates with reduced 2-year survival, advanced disease, and decreased survival time.19,20 The presence of CD133+ cells is associated with increased platinum resistance and increased metastasis to the central nervous system.15 However, these studies do not define how CD133 contributes to ovarian cancer metastasis.
CD133 may interact with multiple signaling pathways involved in invasion or metastasis. CD133 influences ERK and NF-κB (nuclear factor κB) activity to facilitate epithelial to mesenchymal transition in pancreatic cancer cells.21,22 In hepatocellular cancer cells, CD133 contributes to metastasis by affecting Wnt signaling and release of CD133-containing vesicles.23,24 Interestingly, CD133+ cells can be found in collagen I niches near blood vessels, suggesting a role in metastatic spread.25,26 Mounting evidence suggests that CD133 is actively involved in metastasis.
We previously demonstrated that ARID3B increases tumor growth and dissemination in a xenograft model of ovarian cancer and ARID3B increases expression of PROM1.11 Therefore, we hypothesized that ARID3B in part acts through PROM1 to increase tumor spread. In this study, we demonstrate for the first time that ARID3B directly associates with the PROM1 regulator region to activate gene expression and therefore PROM1 is a transcriptional target of ARID3B. Finally, we demonstrate that CD133 promotes peritoneal adhesion, providing a role for CD133 in recruitment of cancer cells including CSCs to the ovarian cancer metastatic niche.
Materials and Methods
Cell culture
Cell lines were grown in a 37°C incubator with 5% CO2. OVCA429 cells (from Dr Bast, MD Anderson Cancer Center, Houston, TX, USA) were grown in minimal essential medium (MEM). Skov3IP cells (Dr Mills, MD Anderson Cancer Center) were grown in McCoys Media 5A. Kuramochi cells (from Dr Mitra, Indiana University, South Bend, IN, USA) were grown in RPMI media with nonessential amino acids and MEM vitamin solution. Media was supplemented with 10% fetal bovine serum (FBS; Peak Serum, Ft. Collins, CO, USA), 0.1 mM Glutamax, 1 mM sodium pyruvate, 50 U/mL penicillin, and 50 μg/mL streptomycin. LP9 cells (from Dr Sharon Stack, University of Notre Dame, Notre Dame, IN, USA) were grown in a 1:1 mix of Media 199 and MCDB 105, 15% FBS, 0.1 mM Glutamax, 50 U/mL penicillin, and 50 μg/mL streptomycin. Cells expressing green fluorescent protein (GFP) (LV105; GeneCopoeia, Rockville, MD, USA), red fluorescent protein (RFP) (GenTarget; San Diego, CA, USA) ARID3B (pLenti-CMV; GenTarget), PROM1 (pReceiver-LV122; GeneCopeia), or short hairpin RNA (shRNA) targeted toward PROM1 (pGFP-C-shLenti; OriGene, Rockville, MD, USA) were produced by transduction with lentiviral particles, supplemented by 1 μg/mL polybrene. Successful transduction was verified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and Western blot. All media components except FBS are from Gibco ThermoScientific (Carlsbad, CA, USA). All cell lines were authenticated in February 2017 by Genetica (Burlington, NC, USA).
Flow cytometry
All antibodies used for flow cytometry staining were obtained from BioLegend (San Diego, CA, USA). Cells were collected and blocked for 15 minutes at room temperature with 1× phosphate-buffered saline (PBS) supplemented with 1% bovine serum albumin. Cells were then stained for 30 minutes at room temperature with human CD133-phycoerythrin. Stained cells were subsequently assessed using Beckman Coulter FC500 Flow Cytometer (Beckman Coulter, Inc., Brea, CA, USA) and data were analyzed using FlowJo software (Tree Star, Ashland, OR, USA). Dead cells were removed from analysis using FSC/SSC (forward/side scatter) gating. Basis of gates was determined using fluorescence minus one controls when necessary.
Western blot
Whole-cell protein lysates were obtained by lysing Kuramochi and Skov3IP (parental and expressing GFP or PROM1-GFP) ovarian cancer cells in radioimmunoprecipitation assay buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% EDTA, 0.1% sodium dodecyl sulfate, and 1× Halt Protease & Phosphatase Inhibitor Cocktail [Pierce, Rockford, IL, USA). Protein concentration was measured using a bicinchoninic acid assay according to standard protocol (Pierce). Proteins were detected using the following antibodies: CD133 (AC133; Miltenyi Biotec, Auburn, CA, USA), β-actin (#AM1829b; Abgent, San Diego, CA, USA), followed by a secondary anti-rabbit horseradish peroxidase-conjugated antibody (GE Healthcare, Knox, IN, USA). Imaging was conducted using a Bio-Rad Chemidoc XRS+ System, with Image Lab Software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Fluorescence microscopy
OVCA429, Kuramochi, and Skov3IP cells were transduced with GFP or PROM1-GFP as described. Fluorescence microscopy was used to verify transduction using EVOS FL microscope (Thermo Fisher Scientific, Waltham, MA, USA).
Chromatin immunoprecipitation
The ChIP was conducted following a protocol from Cold Spring Harbor.27 For each cell line, 5 × 106 cells were cross-linked with 1% formaldehyde (Sigma-Aldrich, St. Louis, MO, USA). Nuclear lysates were harvested using the Pierce Agarose ChIP Kit (Pierce). The chromosomal DNA was sheared with an EpiShear Probe Sonicator (Active Motif, Carlsbad, CA, USA), using a series of 10 × 20-second pulses at 25% amplitude. About 100 μg of sheared chromatin was used for each immunoprecipitation (IP). Immunoprecipitation was performed using an anti-ARID3B antibody (A302-564A; Bethyl Laboratories, Montgomery, TX, USA) or IgG (Pierce) and Protein G-Agarose magnetic beads (Active Motif). Input DNA was collected before IP. The cross-links were reversed by heating the sample to 65°C for 4 hours with 20 μg proteinase K and 250 mM NaCl. DNA obtained by ChIP was purified using a standard phenol-chloroform extraction followed by ethanol precipitation. To determine binding, we conducted qPCR with primers designed to flank a predicted ARID3B-binding site. Relative binding was determined by calculating the amplification of the ARID3B-binding site in ChIP DNA against the corresponding input DNA control. A separate amplification of the IGX1A region (primers from Qiagen) was used as a negative control. Primers for the ARID3B-binding site in the PROM1 were obtained from Integrated DNA Technologies (Coralville, IA, USA). Site 1: Primers: F: TGC TGT TTT TCA ACC GCT TC R: CCG CAT TAG ACC CTT CTG TT. Site 2: Primers: F: TCT CGA TCT CCT GAC CTC GT, R: CTG GGC GAC CTT ACA TGA TT.
Reverse transcription-quantitative polymerase chain reaction
RNA from the cells used in our experiments was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was made using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Waltham, MA, USA), using 1 μg of RNA. The RT-qPCR experiments were conducted using iTaq Universal Probes Supermix (Bio-Rad) for gene expression and Sso Fast EvaGreen Supermix (Bio-Rad) for ChIP. All RT-qPCR gene expression experiments included reference genes GAPDH and β-actin primers for normalization and run in duplicate. Primers were purchased from Integrated DNA Technologies, with the exception of the primers used for Actin (Applied Biosystems, Foster, CA, USA). Procedures and quality controls consistent with accepted RT-qPCR guidelines were followed.28 The following primers were used: GAPDH: Hs.PT.391.22214836, Actin: 4310881E, ARID3B: Hs.PT.58.40614873, and Prom1: Hs.PT53a.3493627.
Cell viability assay
Skov3IP, OVCA429, and Kuramochi cells were treated with10 μM paclitaxel. Cells were counted prior to and after treatment. The percentage of surviving cells was calculated after 72 hours of treatment.
Mesothelial cell adhesion assay
LP9 cells were plated at 30 000 cells/well on gelatin-coated wells of a solid black 96-well plate to form a confluent monolayer. OVCA429 and Skov3IP cells were transduced with either a control GFP vector or the control GFP vector in addition to a PROM1-overexpressing vector (pReceiver-LV122; GeneCopoeia). The appropriate OVCA429 and Skov3IP cells were seeded at a density of 2 × 104 cells per well on top of the LP9 cell layer. After 1 hour of incubation, the GFP signal in each well was determined using a SpectraMax Gemini EM (Molecular Devices, Sunnyvale, CA, USA), with excitation wavelength 490 nm, emission 520 nm. Wells were then washed 3 times with PBS to remove nonadherent cells, and GFP signal was measured again. Each condition was run with 3 or more replicates. Negative control wells containing LP9 monolayers but no other cells were used to correct for background signal. Statistical comparisons were made between control GFP-expressing cells and those overexpressing PROM1.
Ovarian xenografts
Under an approved IACUC (Institutional Animal Care and Use Committee) protocol (ND #14-060), 6-week-old female nude mice nu/nu (Charles River, Wilmington, MA, USA) were maintained at the Freimann Life Science Center (University of Notre Dame). For the survival studies and analysis of tumor growth, 1 × 106 Skov3IP-RFP+shControl (10 mice), Skov3IP-ARID3B+shControl (10 mice), and Skov3IP-ARID3B+shPROM1 (10 mice) cells in 200 μL of PBS were each injected intraperitoneally into nude mice. Starting 14 days after injection, mice were imaged weekly at the Notre Dame Integrated Imaging Facility using the Multispectral FX (Carestream, Rochester, NY, USA).
Explant adhesion assay
An ex vivo peritoneal explant assay was performed to examine adhesion to intact peritoneal mesothelium as described.29 Briefly, all wells of a 24-well tissue culture–treated plate were coated with silicone using the Sylgard 184 Silicone Elastomer Kit (Thermo Fisher Scientific). The wells were filled with sterile PBS prior to dissecting the mice. A 1-cm2 piece of the parietal peritoneum from the ventral abdominal wall was removed from the lower left and right abdominal quadrants. The explants were then pinned mesothelial side up to the silicone support using 0.2-mm pins taking care not to pull the tissue taut. The PBS was removed and fluorescently tagged ovarian cancer cells (1 × 105 cells/mL) were added to the explants. The cells were allowed to adhere to the tissue explant for 2 hours at 37°C and 5% CO2. Nonadherent cells were gently washed away using ice-cold PBS. Adherent cells were visualized using an EVOS FL microscope (Thermo Fisher Scientific) and the cells were counted.
Mesothelial clearance assay
We performed the mesothelial clearance assays as described.30 Briefly, Skov3IP (expressing RFP or PROM1 + RFP) spheroids were prepared in hanging drop culture by suspending 15-μL drops of media containing 200 cells each on the lid of a 10-cm plate. Spheroids were allowed 72 hours to form. LP9-GFP cells were plated at 2.5 × 105 cells/well on a 24-well plate that was coated with fibronectin (4 μg/cm2; Millipore, Burlington, MA), collagen I (8 μg/cm2; Stemcell Technologies, Vancouver, BC, USA) or no coating. Spheroids were seeded onto the LP9 mesothelial cell layer and allowed to attach. The progress of the mesothelial clearance was monitored periodically and imaged using an EVOS FL and a confocal microscope (see below).
Confocal fluorescence microscopy
Confocal fluorescence microscopy was performed using a standard bench-top upright Nikon C2+ Laser Scanning Microscope (Nikon Instruments, Melville, NY, USA) equipped with multichannel fluorescent imaging. Laser excitation was provided by a 405 nm (DAPI [4ʹ,6-diamidino-2-phenylindole]), 488 nm (GFP), and 561 laser (RFP). All images were acquired using a ×40/0.95 oil-immersion objective lens. Three-channel, composite color confocal images were acquired. Frames were 1024 × 1024 pixels in size. Step size between each image slice of the Z-stack was 0.4 µm. Each image slice was frame averaged at least 4× times to reduce background noise. The first image of this image stack was acquired from just below the top of the spheroid, whereas the image of the last slice of the Z-stack was stopped at the focal plane of the glass slide. Image analysis was done using Nikon’s proprietary NIS-Elements C Software. A volume projection in either the XZ or YZ plane was used to determine whether RFP-PROM1 cells cleared the basal layer of GFP-expressing mesothelial cells.
Chemoresistance and extracellular matrix PCR array
RNA from cells was isolated using Trizol according to manufacturer’s instructions. The RT2 First Strand Kit (Qiagen) was used to make the cDNA according to the kit instructions. Briefly, 0.5 μg of each total RNA sample for either OVCA429 control cells or OVCA429 PROM1 cells was incubated in a genomic DNA elimination buffer prior to using the reverse transcription kit to make cDNA. The RT-qPCR was performed according to instructions with the RT2 Profiler Array Format D Handbook using RT2 Syber Green PCR mastermix and the provided 96-well arrays. Each 96-well plate contained primer assays for either cancer drug resistance genes or extracellular matrix (ECM) component genes. Each plate also contained 5 housekeeping genes, 3 reverse transcription controls, a genomic DNA control, and 3 positive PCR controls. Analysis of results was performed as instructed in the RT2 Profiler Array Format D Handbook.
Statistics
Statistical significance for ChIP, RT-qPCR, survival assays, spheroid replating data, and cell adhesion experiments was calculated using the Smith-Satterthwaite procedure with unequal population variances. Statistical significance was assigned to comparisons with a P value of .05 or lower. Significance of ex vivo explant experiments was calculated with a 2-way analysis of variance with either Tukey or Sidak posttests. Animal survival was analyzed using the Kaplan-Meier method with Mantel-Cox log-rank significance test and correlation analysis. Overexpression of PROM1 in cell lines was analyzed using the Kruskal-Wallis method with a Dunn posttest.
Results
PROM1 is a transcriptional target of ARID3B
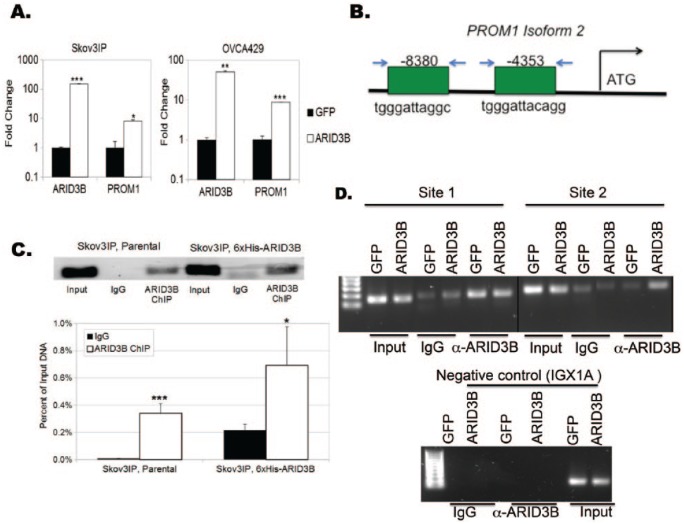
ARID3B nuclear expression correlates with relapse following chemotherapy.11 We were the first to identify direct ARID3B target genes. In doing so, we identified and validated that ARID3B regulates stem- and metastasis-related genes.31 Of the stem cell genes we examined, PROM1 was consistently induced by ARID3B in vitro and in tumor cells recovered from xenograft ascites. Given that PROM1 is a well-studied CSC marker, we sought to understand the importance of its regulation in ovarian cancer. To demonstrate that ARID3B regulates PROM1, we lentivirally transduced 2 different cell lines (Skov3IP and OVCA429) with ARID3B or GFP. We chose these cell lines because they exhibit different morphological features, adhesion properties, genetics, and expression levels of ARID3B. Skov3IP cells have a mesenchymal morphology, form peritoneal tumors (with ascites) in vivo, and express higher ARID3B than OVCA429 cells.11,31,32 OVCA429 cells are morphologically epithelial and do not form tumors readily in animal models. ARID3B expression was confirmed by RT-qPCR (Figure 1A). We then conducted RT-qPCR to quantitate changes in PROM1 expression (Figure 1A). In both cell lines, ARID3B significantly increased PROM1 expression (approximately 10-fold). Using ARID3B consensus site we previously described,31 we identified 2 putative ARID3B-binding sites upstream of the transcription start site of PROM1, isoform 2 (Figure 1B). To ascertain whether this ARID3B-binding site is functional, we performed chromatin immunoprecipitation (ChIP). In Skov3IP cells and OVCA429 cells, ARID3B bound to an ARID3B consensus site 8380 bases upstream of the transcription start site in PROM1 isoform 2 (Figure 1C and D). In the OVCA429 cells, ARID3B bound to the consensus site but was not increased with exogenous expression of ARID3B. Therefore, we used a new set of primers (to a second ARID3B consensus sequence), which revealed increased association of ARID3B with the PROM1 gene. Future analyses will evaluate which binding regions are necessary for induction of PROM1 by ARID3B. These findings are consistent with the conclusion that ARID3B directly regulates PROM1 expression in ovarian cancer cells.
Figure 1.
ARID3B regulates expression of the PROM1. (A) Ovarian cancer cell lines (OVCA429 and Skov3IP) were lentivirally transduced with GFP or ARID3B. Reverse transcription-quantitative polymerase chain reaction was preformed to verify fold change in ARID3B and PROM1 expression. (B) Diagram of ARID3B-binding sites within the PROM1 gene isoform 2. We tested binding of ARID3B to 2 different sites upstream of transcription start for PROM1 isoform 2. Relative locations for amplification are shown. Arrows represent primers (C) Chromatin immunoprecipitation (ChIP) was performed on Skov3IP cells. ARID3B binding to the 5ʹ regulatory region of PROM1 was analyzed by end point polymerase chain reaction (PCR)/electrophoresis and quantitative PCR. All calculations are presented as relative binding compared with input DNA, with significance against the background IgG sample displayed. *P < .05; **P < .005; ***P < .0005. (D) ChIP was performed on OVCA429 cells transduced with either GFP or ARID3B. PCR was conducted using 2 primer sets to detect ARID3B binding to the 5ʹ regulatory region of PROM1. IGXIA (a region of DNA devoid of transcription factor binding sites) was used as a negative control region. GFP indicates green fluorescent protein.
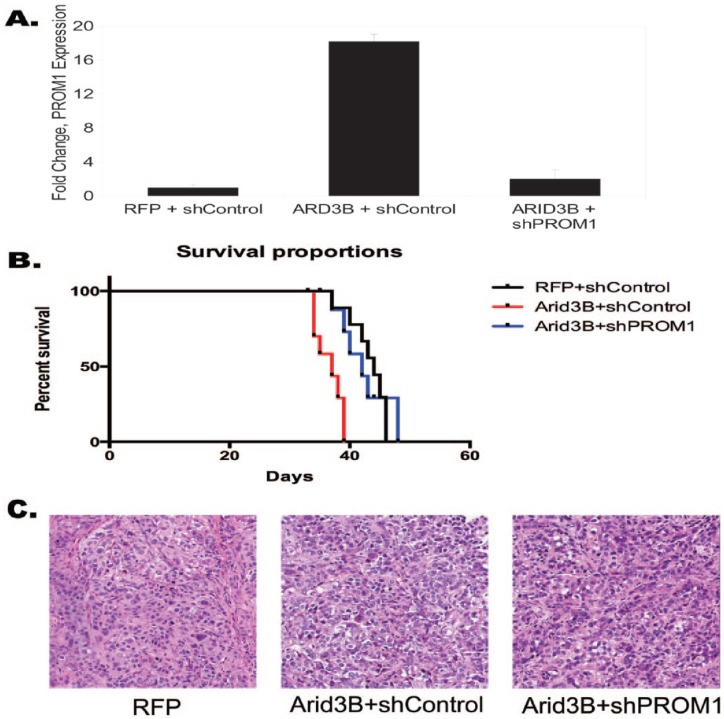
PROM1 is a critical mediator of ARID3B-induced tumor growth
We previously demonstrated that ARID3B increased tumor growth and decreased survival in a xenograft model of ovarian cancer.11 To determine whether PROM1 regulation by ARID3B contributes to tumor growth, we conducted xenograft studies using cells that overexpress ARID3B and express shRNA for PROM1. We generated Skov3IP sublines with the following modifications: (1) RFP and scrambled shRNA (shControl); (2) RFP, ARID3B, and scrambled shRNA; and (3) RFP, ARID3B, and PROM1 shRNA. We verified PROM1 expression for each subset of cells by RT-qPCR on pooled cells just prior to injection (Figure 2A). We injected female 6-week-old mice intraperitoneally with 1 × 106 cells each (10 mice in each group). The mice injected with cells overexpressing ARID3B (ARID3B + shControl) showed decreased survival time compared with mice injected with either RFP + shControl Skov3IP cells or ARID3B + shPROM1 Skov3IP cells (P < .001 using Mantel-Cox log-rank test). There was no difference in survival between mice injected with cells expressing exogenous ARID3B + shPROM1 and mice injected with cells expressing RFP + shControl (Figure 2B). Pathology analysis of the hematoxylin-eosin–stained tumors confirmed that the tumors were poorly differentiated, high grade, and adenocarcinoma (Figure 2C). We detected no difference in histology between treatment groups. Importantly, we found a difference in ascites formation. About 80% of mice injected with cells expressing ARID3B + shControl formed ascites. In the control treatment (RFP + shControl and ARID3B + shPROM1) groups, only 40% of the mice developed ascites. We did not find a significant difference in tumor size between treatment groups because this was an end point/survival study. Our results suggest that ARID3B regulation of PROM1 is important to tumor growth and ascites formation.
Figure 2.
PROM1 knockdown in ARID3B-expressing cells increases median survival in ovarian cancer xenografts. Skov3IP cells were transduced with RFP and a scrambled shRNA control vector (shControl), ARID3B + shControl or ARID3B + shRNA against PROM1 (shPROM1). (A) Reverse transcription-quantitative polymerase chain reaction for expression of PROM1 for each condition (pooled cells that were used in injections). (B) Kaplan-Meier survival curve shows decreased median survival for mice injected with ARID3B + shControl Skov3IP, compared with those injected with Skov3IP with RFP + shControl. Mice injected with ARID3B + shPROM1 Skov3IP cells exhibited median survival similar to RFP + shControl and ARID3B + shPROM1. P < .0001 (Mantel-Cox log-rank test). (C) Hematoxylin-eosin staining in tumor tissue from mice bearing RFP, ARID3B + shControl, and ARID3B + shPROM1–expressing tumors (original magnification ×20). RFP indicates red fluorescent protein; shRNA, short hairpin RNA.
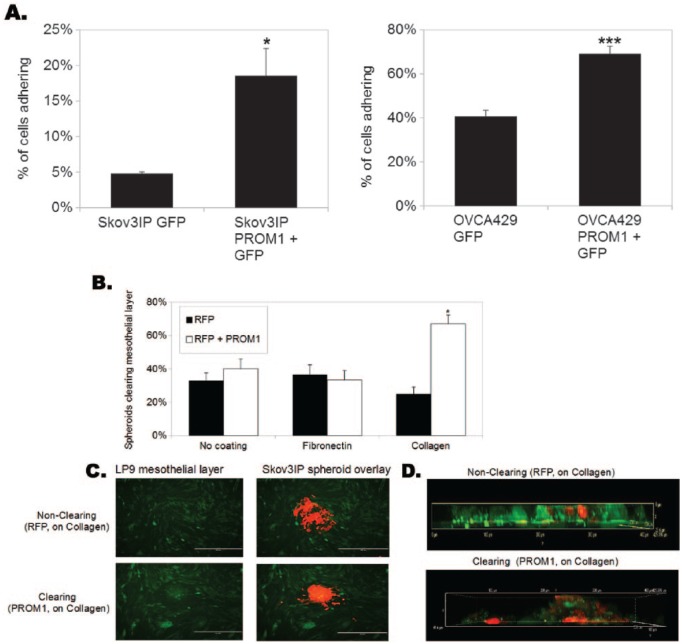
PROM1 expression increases homing to the ovarian cancer metastatic niche
Because the mesothelial lining of the peritoneal cavity is one of the first and most common sites of ovarian cancer metastasis, we examined the impact of CD133 expression on adhesion to mesothelial cells. We began by generating PROM1-expressing cells. Expression is confirmed in Supplemental Figures 1 and 2. We seeded OVCA429 and Skov3IP cells expressing either GFP or PROM1-GFP onto a nonfluorescent monolayer of LP9 mesothelial cells and determined the percentage of ovarian cancer cells adhering by measuring GFP fluorescence. PROM1 expression increased adhesion of OVCA429 cells to mesothelial LP9 cells by 1.7-fold and Skov3IP cells by 3.9-fold (Figure 3A). Iwanicki and colleagues30,33 previously demonstrated that ovarian cancer cells not only bind to mesothelium but they also displace mesothelial cells to contact the underlying basement membrane as part of their metastatic initiation process. To recapitulate this phenomenon in vitro, we generated RFP-expressing Skov3IP or Skov3IP-PROM1 + RFP spheroids using the hanging drop method.30 The GFP-expressing LP9 cells were plated on plastic, fibronectin, or collagen I. After 72 hours, the spheroids were cultured on the GFP-LP9 cells. Invasion of cancer cells was monitored via fluorescence imaging for 24 hours. About 67% of the Skov3IP-PROM1 spheroids cleared the underlying LP9 cells when plated on collagen I, compared with 25% of parental Skov3IP-RFP spheroids (Figure 3B). Fluorescence (Figure 3C) and confocal imaging (Z-stack orthogonal view; Figure 3D) demonstrated the increased invasion and clearing of Skov3IP cells expressing PROM1 compared with control cells. We also conducted mesothelial clearance assays on OVCA429 and Kuramochi cells expressing RFP or PROM1. PROM1 increased clearance of LP9 cells (on collagen I) by the ovarian cancer cells by 2.3-fold and 2.2-fold after 4 hours, respectively (data not shown). Our data demonstrate that PROM1 increases invasion through the mesothelium.
Figure 3.
PROM1 expression increases adhesion to and clearance of mesothelial cells in the presence of collagen. (A) Skov3IP and OVCA429 cells were transduced with either GFP alone or with PROM1 + GFP and then seeded onto confluent layers of mesothelial LP9 cells. The percent adherence of the ovarian cancer cells was determined by reading GFP fluorescence before and after a series of phosphate-buffered saline washes. Significantly increased binding was observed with both cell lines when PROM1 was overexpressed. *P < .05; **P < .005; ***P < .0005. (B) Skov3IP-RFP, Skov3IP-PROM1 + RFP spheroids were seeded onto a confluent layer of fluorescent mesothelial cells (LP9-GFP) on plastic, fibronectin, or collagen 1. After 24 hours, clearance was quantitated. Percentages are provided for the number of RFP+ spheroids that successfully attached and cleared the mesothelial LP9-GFP cells; *P < .05. (C) Representative Skov3IP-RFP and Skov3IP-PROM1 spheroids shown by fluorescence microscopy (original magnification ×10). (D) Representative Skov3IP-RFP and Skov3IP-PROM1 are shown as a Z-stack using confocal fluorescence microscopy (original magnification ×40). GFP indicates green fluorescent protein.
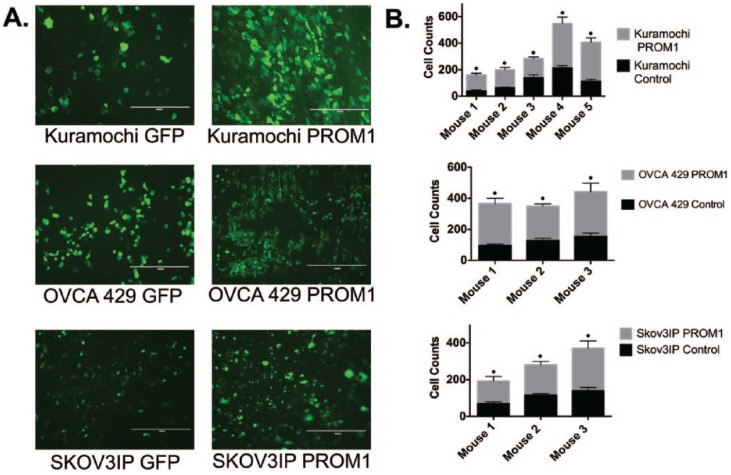
Finally, because PROM1 increased adhesion to a mesothelial cell line, we wanted to establish whether PROM1 induced tumor cell adhesion to mouse mesothelium in a published ex vivo assay.29 For these studies, we also included Kuramochi cells as they are an ovarian cancer cell line with genetics consistent with high-grade serous ovarian cancer34 (PROM1/CD133 expression is shown in Supplemental Figures 1 and 2). Mouse peritoneal tissue was isolated from C57BL/6 mice and adhesion assays were conducted as described.29 We seeded GFP and PROM1-GFP expressing OVCA429, Skov3IP, and Kuramochi cells onto dissected peritoneal tissue, washed the tissue to remove nonadherent cells, and then imaged the tissue using a fluorescent microscope. PROM1 expression significantly increased adhesion of ovarian cancer cells to the peritoneum by 2-fold to 3-fold (Figure 4A and B). In total, our data suggest that PROM1 regulates adhesion to sites of metastasis.
Figure 4.
PROM1 increases adhesion of cells to peritoneal explants. Peritoneal explant adhesion assays were conducted on Kuramochi, OVCA429, and Skov3IP cells expressing GFP or PROM1-GFP. (A) Fluorescent microscopy of Kuramochi, OVCA429, and Skov3IP cells adhering to peritoneal explants (original magnification ×10). (B) The number of fluorescent cells that adhered to the peritoneal explant was quantitated. The effect of CD133 on adhesion for Skov3IP, OVCA429, and Kuramochi cells was considered extremely significant. For experiments with Kuramochi and Skov3IP cells, there was significant mouse to mouse variation with the amount of adhesion for both control GFP and PROM1-GFP Kuramochi cells. There was no significant mouse to mouse variation in the experiments using OVCA429; •P < .0001.
CD133 increases expression of adhesion and stemness-related genes in ovarian cancer cells
To begin to elucidate how CD133 mediates metastatic spread, we conducted gene expression analysis on OVCA429 cells transduced with PROM1 or GFP alone. Focused RT-qPCR arrays from Qiagen (Germantown, MD, USA; chemoresistance and ECM/adhesion) were conducted (Tables 1 and 2). CD133 induces expression of several matrix metalloproteinases (MMPs), which are implicated in ovarian cancer metastasis.35–45 CD133 also induced adhesion molecules such as PECAM1 and ICAM1. Future studies will delineate how CD133 activity leads to activation of adhesion/metastasis-related molecules. Because CD133 is most commonly associated with stemness, we also wanted to determine what stemness-related genes were activated in the presence of exogenous CD133. CD133 induced numerous stem-related genes including FGF2, MYC, HIF1A, EGFR, CCND1, and APC. Although we found that a number of stem-related genes were induced by CD133, PROM1 expression may not be sufficient to activate a full stem cell phenotype. Some classical markers of stemness (Nanog, NESTIN, KIT, and POU5F1) are induced when cells are cultured as spheroids (not shown). These genes were not induced in 2-dimensional cultures expressing exogenous PROM1 (not shown). However, PROM1 increases survival in the presence of paclitaxel (Supplemental Figure 3). Collectively, these data suggest that CD133 may activate a partial stemness phenotype and contribute to ovarian cancer metastasis through activation of MMPs, adhesion molecules, and select stemness genes.
Table 1.
Upregulated extracellular matrix genes in OVCA429 PROM1 cells.
| Fold change | ||
|---|---|---|
| SELE | Selectin E | 4579.49106 |
| MMP-3 | Matrix metallopeptidase 3 | 16.4340217 |
| SELL | Selectin L | 12.43982849 |
| TNC | Tenascin C | 11.34023301 |
| FN1 | Fibronectin 1 | 6.15782159 |
| THBS1 | Thrombospondin 1 | 5.737029383 |
| MMP-14 | Matrix metallopeptidase 14 | 5.18356624 |
| ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 | 5.029293321 |
| CTGF | Connective tissue growth factor | 4.775721739 |
| MMP-8 | Matrix metallopeptidase 8 | 4.589592187 |
| COL8A1 | Collagen type VIII alpha 1 chain | 4.281696999 |
| MMP-9 | Matrix metallopeptidase 9 | 4.188376539 |
| PECAM1 | Platelet and endothelial cell adhesion molecule 1 | 3.847303157 |
| ICAM1 | Intercellular adhesion molecule 1 | 3.597750715 |
Table 2.
Upregulated chemoresistance genes in OVCA429 PROM1 cells.
| Fold change | ||
|---|---|---|
| FGF2 | Fibroblast growth factor 2 | 3.590252709 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A | 3.525707607 |
| ABCC3 | ATP-binding cassette subfamily C member 3 | 3.469434302 |
| ESR2 | Estrogen receptor 2 | 3.334056831 |
| MYC | MYC proto-oncogene, BHLH transcription factor | 2.618260004 |
| HIF1A | Hypoxia-inducible factor 1 alpha subunit | 2.485575885 |
| ATM | ATM serine/threonine kinase | 2.404776967 |
| EGFR | Epidermal growth factor receptor | 2.366241112 |
| XPA | XPA, DNA damage recognition and repair factor | 2.25357438 |
| CCND1 | Cyclin D1 | 2.189750119 |
| APC | Adenomatous polyposis coli | 2.167686844 |
| AHR | Aryl hydrocarbon receptor | 2.158026925 |
| ABCB1 | ATP-binding cassette subfamily B member 1 | 2.119631716 |
| CYP2B6 | Cytochrome P450 family 2 subfamily B member 6 | 2.119631716 |
| CYP2C19 | Cytochrome P450 family 2 subfamily C member 19 | 2.119631716 |
| CYP2C9 | Cytochrome P450 family 2 subfamily C member 9 | 2.119631716 |
| CYP3A4 | Cytochrome P450 family 3 subfamily A member 4 | 2.119631716 |
| ERBB4 | Erb-B2 receptor tyrosine kinase 4 | 2.119631716 |
| BCL2L1 | BCL2 like 1 | 1.913634596 |
| XPC | XPC complex subunit, DNA damage recognition and repair factor | 1.837259659 |
| ARNT | Aryl hydrocarbon receptor nuclear translocator | 1.802262585 |
| RELB | RELB proto-oncogene, NF-κB subunit | 1.727148195 |
| CYP1A2 | Cytochrome P450 family 1 subfamily A member 2 | 1.648582767 |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A | 1.636827721 |
| RB1 | RB transcriptional corepressor 1 | 1.633245096 |
| RARB | Retinoic acid receptor beta | 1.630097904 |
| PPARA | Peroxisome proliferator activated receptor alpha | 1.60966284 |
| TPMT | Thiopurine S-methyltransferase | 1.591838228 |
| CYP2E1 | Cytochrome P450 family 2 subfamily E member 1 | 1.540737499 |
| BAX | BCL2-associated X, apoptosis regulator | 1.531548663 |
| MVP | Major vault protein | 1.461726433 |
| PPARG | Peroxisome proliferator activated receptor gamma | 1.434950731 |
| CYP2C8 | Cytochrome P450 family 2 subfamily C member 8 | 1.431037222 |
| BCL2 | BCL2, apoptosis regulator | 1.381050929 |
| NFKB2 | Nuclear factor κB subunit 2 | 1.378026125 |
| UGCG | UDP-glucose ceramide glucosyltransferase | 1.350627898 |
| IGF2R | Insulinlike growth factor 2 receptor | 1.348659773 |
| MET | MET proto-oncogene, receptor tyrosine kinase | 1.325719435 |
| SULT1E1 | Sulfotransferase family 1E member 1 | 1.318469151 |
| RXRB | Retinoid X receptor beta | 1.309538256 |
| TOP1 | DNA topoisomerase I | 1.290835139 |
| EPHX1 | Epoxide hydrolase 1 | 1.252818997 |
| CLPTM1L | CLPTM1 like | 1.241588366 |
| NFKBIB | NF-κB inhibitor beta | 1.227599665 |
| MSH2 | MutS homolog 2 | 1.213348089 |
| RARA | Retinoic acid receptor alpha | 1.210805565 |
| PPARD | Peroxisome proliferator activated receptor delta | 1.187990737 |
| TOP2B | DNA topoisomerase II beta | 1.167762548 |
| NFKBIE | NF-κB inhibitor epsilon | 1.061546007 |
| NAT2 | N-acetyltransferase 2 | 1.032968675 |
| ABCC1 | ATP-binding cassette subfamily C member 1 | 1.005664789 |
Discussion
Most of the women with ovarian cancer are diagnosed with metastatic disease. Many women respond to chemotherapy but most will relapse. Therefore, curing patients will depend on elimination of metastatic and chemoresistant cells. Cancer stem cells are both putative metastatic units and a therapeutically resistant population of cells.46,47 Thus, elimination of CSCs may benefit patients. We previously found that the transcription factor ARID3B correlates with relapse following therapy in patients with ovarian cancer.11 We also demonstrated that ARID3B increases tumor growth and dissemination.11 ARID3B increases the proportion of CD133+ cancer cells.11 We hypothesized that ARID3B direct regulation of the CD133 gene (PROM1) may contribute to tumor growth. We confirmed that ARID3B directly regulates messenger RNA induction of PROM1. In addition, we found that knockdown of PROM1 in cells that overexpress ARID3B resulted in increased median survival of mice in ovarian xenografts. These studies suggest that ARID3B regulation of PROM1 contributes to tumor growth.
Although CD133 is commonly used as a marker of CSCs, the exact function of CD133 in mediating stemness and metastasis is not well defined. To clarify the role of CD133, we generated cell lines that expressed PROM1. Considering that CD133 is a transmembrane protein and it promotes stemness, we explored the possibility that CD133 affects homing of cells during peritoneal metastasis. PROM1 increased adhesion to and clearance of mesothelial cells in culture, thus implicating CD133 in peritoneal metastasis. Interestingly, mesothelial adhesion and clearing was dependent on ECM. Cells expressing high CD133 preferentially adhered to and cleared LP9 cells on collagen I (Figure 3), suggesting that CD133 may direct homing of tumor cells to collagen I–rich sites of metastasis. As a mimetic of early steps in metastasis, we performed an ex vivo assay to determine whether ovarian cancer cells adhere to mouse peritoneal tissue as they would during tumor progression. PROM1 increased ex vivo peritoneal adhesion. Previous studies implicated CD133 in tumor cell adhesion. In particular, CD133 was demonstrated to promote integrin and FAK signaling.48,49 In gene expression studies, we found that CD133 induced genes related to both metastasis (including MMP-3, MMP-9, and MMP-14) and stemness (including MYC, FGF2, EGFR, and APC). Our data suggest that cells with cell surface CD133 may be more successful at adhering to the metastatic niche and invading the peritoneal tissue during metastasis.
Collectively, we demonstrated that ARID3B regulation of PROM1 gene expression plays an important role in tumor growth. Furthermore, we show for the first time that CD133 mediates metastatic homing to peritoneal tissue in ovarian cancer. Targeting CD133 may lead to improved treatments that reduce the risk of recurrence.
Supplemental Material
Supplemental material, Supplementary_Material for CD133 Promotes Adhesion to the Ovarian Cancer Metastatic Niche by Lynn Roy, Alexander Bobbs, Rachel Sattler, Jeffrey L Kurkewich, Paige B Dausinas, Prakash Nallathamby and Karen D Cowden Dahl in Cancer Growth and Metastasis
Acknowledgments
The Indiana University School of Medicine—South Bend Imaging and Flow Cytometry Core Facility and the Notre Dame Integrated Imaging Facility were used for imaging studies. All mice used in this study were maintained at the Freimann Life Science Center at the University of Notre Dame. The authors would also like to thank Elizabeth Loughran for demonstrating the ex vivo adhesion assay. Austin Boucher assisted JLK in data acquisition.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Department of Defense Ovarian Cancer Research Program (W81XWH-15-1-0071).
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: LR and AB conducted experiments and contributed to the writing. RS, JLK, PBD, and PN assisted in design and execution of experiments. KDCD designed and conducted experiments and contributed to writing and editing.
References
- 1. Coward JI, Middleton K, Murphy F. New perspectives on targeted therapy in ovarian cancer. Int J Womens Health. 2015;7:189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Foster R, Buckanovich RJ, Rueda BR. Ovarian cancer stem cells: working towards the root of stemness. Cancer Lett. 2013;338:147–157. [DOI] [PubMed] [Google Scholar]
- 3. Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177:1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shield K, Ackland ML, Ahmed N, Rice GE. Multicellular spheroids in ovarian cancer metastases: biology and pathology. Gynecol Oncol. 2009;113:143–148. [DOI] [PubMed] [Google Scholar]
- 5. Vathipadiekal V, Saxena D, Mok SC, Hauschka PV, Ozbun L, Birrer MJ. Identification of a potential ovarian cancer stem cell gene expression profile from advanced stage papillary serous ovarian cancer. PLoS ONE. 2012;7:e29079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wintzell M, Hjerpe E, Avall Lundqvist E, Shoshan M. Protein markers of cancer-associated fibroblasts and tumor-initiating cells reveal subpopulations in freshly isolated ovarian cancer ascites. BMC Cancer. 2012;12:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Silva IA, Bai S, McLean K, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yasuda K, Torigoe T, Morita R, et al. Ovarian cancer stem cells are enriched in side population and aldehyde dehydrogenase bright overlapping population. PLoS ONE. 2013;8:e68187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burgos-Ojeda D, Rueda BR, Buckanovich RJ. Ovarian cancer stem cell markers: prognostic and therapeutic implications. Cancer Lett. 2012;322:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liao TT, Hsu WH, Ho CH, et al. let-7 modulates chromatin configuration and target gene repression through regulation of the ARID3B complex. Cell Rep. 2016;14:520–533. [DOI] [PubMed] [Google Scholar]
- 11. Roy L, Samyesudhas SJ, Carrasco M, Li J, Joseph S, Dahl R, Cowden Dahl KD. ARID3B increases ovarian tumor burden and is associated with a cancer stem cell gene signature. Oncotarget. 2014;5(18):8355–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yin AH, Miraglia S, Zanjani ED, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 13. Baba T, Convery PA, Matsumura N, et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 2009;28:209–218. [DOI] [PubMed] [Google Scholar]
- 14. El-Khattouti A, Selimovic D, Haikel Y, Megahed M, Gomez CR, Hassan M. Identification and analysis of CD133(+) melanoma stem-like cells conferring resistance to taxol: an insight into the mechanisms of their resistance and response. Cancer Lett. 2014;343:123–133. [DOI] [PubMed] [Google Scholar]
- 15. Liu BL, Liu SJ, Baskys A, et al. Platinum sensitivity and CD133 expression as risk and prognostic predictors of central nervous system metastases in patients with epithelial ovarian cancer. BMC Cancer. 2014;14:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Steg AD, Bevis KS, Katre AA, et al. Stem cell pathways contribute to clinical chemoresistance in ovarian cancer. Clin Cancer Res. 2012;18:869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Q, Shi S, Yen Y, Brown J, Ta JQ, Le AD. A subpopulation of CD133(+) cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett 2010;289:151–160. [DOI] [PubMed] [Google Scholar]
- 19. Zhang J, Guo X, Chang DY, Rosen DG, Mercado-Uribe I, Liu J. CD133 expression associated with poor prognosis in ovarian cancer. Mod Pathol. 2012;25:456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Q, Chen A, Song H, Tao J, Yang H, Zuo M. Prognostic value of cancer stem cell marker CD133 in ovarian cancer: a meta-analysis. Int J Clin Exp Med. 2015;8:3080–3088. [PMC free article] [PubMed] [Google Scholar]
- 21. Ding Q, Miyazaki Y, Tsukasa K, Matsubara S, Yoshimitsu M, Takao S. CD133 facilitates epithelial-mesenchymal transition through interaction with the ERK pathway in pancreatic cancer metastasis. Mol Cancer. 2014;13:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nomura A, Banerjee S, Chugh R, et al. CD133 initiates tumors, induces epithelial-mesenchymal transition and increases metastasis in pancreatic cancer. Oncotarget. 2015;6:8313–8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rappa G, Mercapide J, Anzanello F, et al. Wnt interaction and extracellular release of prominin-1/CD133 in human malignant melanoma cells. Exp Cell Res. 2013;319:810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rappa G, Mercapide J, Anzanello F, Pope RM, Lorico A. Biochemical and biological characterization of exosomes containing prominin-1/CD133. Mol Cancer. 2013;12:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mak AB, Schnegg C, Lai CY, et al. CD133-targeted niche-dependent therapy in cancer: a multipronged approach. Am J Pathol. 2014;184:1256–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Motegi H, Kamoshima Y, Terasaka S, Kobayashi H, Houkin K. Type 1 collagen as a potential niche component for CD133-positive glioblastoma cells. Neuropathology. 2014;34:378–385. [DOI] [PubMed] [Google Scholar]
- 27. Carey MF, Peterson CL, Smale ST. Chromatin immunoprecipitation (ChIP). Cold Spring Harb Protoc. 2009;2009:pdb.prot5279. [DOI] [PubMed] [Google Scholar]
- 28. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. [DOI] [PubMed] [Google Scholar]
- 29. Bruney L, Conley KC, Moss NM, Liu Y, Stack MS. Membrane-type I matrix metalloproteinase-dependent ectodomain shedding of mucin16/ CA-125 on ovarian cancer cells modulates adhesion and invasion of peritoneal mesothelium. Biol Chem. 2014;395:1221–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davidowitz RA, Iwanicki MP, Brugge JS. In vitro mesothelial clearance assay that models the early steps of ovarian cancer metastasis. J Vis Exp: JoVE. 2012(60):3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bobbs A, Gellerman K, Hallas WM, et al. ARID3B directly regulates ovarian cancer promoting genes. PLoS ONE. 2015;10:e0131961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joseph S, Deneke VE, Cowden Dahl KD. ARID3B induces tumor necrosis factor alpha mediated apoptosis while a novel ARID3B splice form does not induce cell death. PLoS ONE. 2012;7:e42159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iwanicki MP, Davidowitz RA, Ng MR, et al. Ovarian cancer spheroids use myosin-generated force to clear the mesothelium. Cancer Discov. 2011;1:144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mitra AK, Davis DA, Tomar S, et al. In vivo tumor growth of high-grade serous ovarian cancer cell lines. Gynecol Oncol. 2015;138:372–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Afzal S, Lalani EN, Poulsom R, et al. MT1-MMP and MMP-2 mRNA expression in human ovarian tumors: possible implications for the role of desmoplastic fibroblasts. Hum Pathol. 1998;29:155–165. [DOI] [PubMed] [Google Scholar]
- 36. Cowden Dahl KD, Symowicz J, Ning Y, et al. Matrix metalloproteinase 9 is a mediator of epidermal growth factor-dependent e-cadherin loss in ovarian carcinoma cells. Cancer Res. 2008;68:4606–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davidson B, Goldberg I, Gotlieb WH, et al. High levels of MMP-2, MMP-9, MT1-MMP and TIMP-2 mRNA correlate with poor survival in ovarian carcinoma. Clin Exp Metastasis. 1999;17:799–808. [DOI] [PubMed] [Google Scholar]
- 38. Hu X, Li D, Zhang W, Zhou J, Tang B, Li L. Matrix metalloproteinase-9 expression correlates with prognosis and involved in ovarian cancer cell invasion. Arch Gynecol Obstet. 2012;286:1537–1543. [DOI] [PubMed] [Google Scholar]
- 39. Lynch CC, Crawford HC, Matrisian LM, McDonnell S. Epidermal growth factor upregulates matrix metalloproteinase-7 expression through activation of PEA3 transcription factors. Int J Oncol. 2004;24:1565–1572. [PubMed] [Google Scholar]
- 40. Redzic JS, Kendrick AA, Bahmed K, et al. Extracellular vesicles secreted from cancer cell lines stimulate secretion of MMP-9, IL-6, TGF-beta1 and EMMPRIN. PLoS ONE. 2013;8:e71225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stack MS, Ellerbroek SM, Fishman DA. The role of proteolytic enzymes in the pathology of epithelial ovarian carcinoma. Int J Oncol. 1998;12:569–576. [DOI] [PubMed] [Google Scholar]
- 42. Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol. 2000;10:415–433. [DOI] [PubMed] [Google Scholar]
- 43. Van Meter TE, Broaddus WC, Rooprai HK, Pilkington GJ, Fillmore HL. Induction of membrane-type-1 matrix metalloproteinase by epidermal growth factor-mediated signaling in gliomas. Neuro Oncol. 2004;6:188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang BQ, Zhang CM, Gao W, Wang XF, Zhang HL, Yang PC. Cancer-derived matrix metalloproteinase-9 contributes to tumor tolerance. J Cancer Res Clin Oncol. 2011;137:1525–1533. [DOI] [PubMed] [Google Scholar]
- 45. Wu M, Xu G, Xi L, et al. Down-regulation of MT1-MMP expression suppresses tumor cell invasion in metastatic human SW626 ovarian cancer cells. Oncol Rep. 2006;15:501–505. [PubMed] [Google Scholar]
- 46. Deng J, Wang L, Chen H, et al. Targeting epithelial-mesenchymal transition and cancer stem cells for chemoresistant ovarian cancer. Oncotarget. 2016;7:55771–55788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peitzsch C, Tyutyunnykova A, Pantel K, Dubrovska A. Cancer stem cells: the root of tumor recurrence and metastases. Semin Cancer Biol. 2017;44:10–24. [DOI] [PubMed] [Google Scholar]
- 48. Liu C, Li Y, Xing Y, et al. The interaction between cancer stem cell marker CD133 and Src protein promotes focal adhesion kinase (FAK) phosphorylation and cell migration. J Biol Chem. 2016;291:15540–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Su YJ, Lin WH, Chang YW, et al. Polarized cell migration induces cancer type-specific CD133/integrin/Src/Akt/GSK3β/β-catenin signaling required for maintenance of cancer stem cell properties. Oncotarget. 2015;6:38029–38045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_Material for CD133 Promotes Adhesion to the Ovarian Cancer Metastatic Niche by Lynn Roy, Alexander Bobbs, Rachel Sattler, Jeffrey L Kurkewich, Paige B Dausinas, Prakash Nallathamby and Karen D Cowden Dahl in Cancer Growth and Metastasis