Abstract
Background:
MicroRNAs are involved in hepatocellular carcinoma metastasis, a principal cause of hepatocellular carcinoma–related death in patients worldwide. MiR-212 is a microRNA that has been identified in several types of cancers and is postulated to influence cell signaling and subsequent malignant pathogenesis. Despite emerging reports suggesting that miR-212 plays a significant role in the onset, progression, and migration of these types of malignant tumors, its involvement in the development of hepatocellular carcinoma has not been fully elucidated.
Materials and Methods:
Quantitative reverse transcription polymerase chain reaction, wound healing, transwell migration and invasion assays, Western blotting, and xenograft tumor growth models were performed to test the expression levels and functions of miR-212 in hepatocellular carcinoma. Luciferase reporter assay, quantitative reverse transcription polymerase chain reaction, Western blotting, and immunohistochemistry were used to identify and verify the target of miR-212.
Results:
In this study, we identify significant repression of miR-212 in hepatocellular carcinoma and demonstrate that overexpression of miR-212 inhibits the migration of hepatocellular carcinoma cells in vitro and in vivo. Furthermore, we identify forkhead box M1, whose expression is inversely related to that of miR-212, as a direct target of miR-212. Additionally, reexpression of forkhead box M1 rescues the miR-212-mediated inhibition of cell migration. We observed that inhibition of miR-212 activates forkhead box M1 but inhibits the Wnt/β-catenin pathway by suppressing Wnt, LEF-1, c-Myc, and nuclear β-catenin. Finally, in vivo studies confirmed the inhibitory effect of miR-212 on hepatocellular carcinoma growth.
Conclusion:
Our present findings indicate that miR-212 is a potential prognostic biomarker of hepatocellular carcinoma and that the miR-212/forkhead box M1 regulatory axis may represent a new therapeutic objective for hepatocellular carcinoma treatment.
Keywords: hepatocellular carcinoma, miR-212, forkhead box M1, migration, β-catenin
Introduction
Hepatocellular carcinoma (HCC) is malignancy of the liver parenchyma. Frequently, HCC is diagnosed during middle age (40 years or older).1 Hepatocellular carcinoma ranks fourth among the most common malignancies and is the third leading cause of cancer-associated death worldwide.2 This low survival rate is linked to delayed diagnosis, metastasis, resistance to therapy, and recurrence.2 Recent reports have demonstrated the involvement of microRNAs (miRNAs) in cancer development and elucidated their function both as cancer inducers (oncogenes) and tumor suppressor genes, depending on the role of their cellular target.3 Because of their role in gene expression and cellular signaling, studies on their mode of action might aid in the identification of biomarkers that can serve as diagnostic and therapeutic targets for cancer treatment. Elucidation of the molecular mechanisms involved in hepatocellular carcinogenesis is critical for the identification of innovative prognostic strategies for therapy; the majority of patients with HCC are ineligible for curative surgery, largely because of distant metastasis present upon initial diagnosis and the high recurrence rate of HCC.4
MicroRNAs belong to a class of small noncoding RNAs that block messenger RNA (mRNA) translation or the degradation of downstream target genes by binding to the 3′ untranslated region (3′-UTR).5,6 Accumulating evidence implicates miRNA dysfunction in the cellular proliferation, apoptosis, chemoradioresistance, and metastasis of tumors.7,8 Recent reports have shed light on miRNA involvement in HCC tumorigenesis, including roles for miR-221/222,9 miR-124,10 miR-449,11 miR-26,12 miR-34a,13 and miR-148a.14 Previously, we reported that miR-539 upregulation inhibits growth and induces apoptosis in HCC cells by targeting fascin homolog 1. However, the functions and mechanisms of miRNAs involved in HCC metastasis are unclear, as recent gene profiling studies have yielded unpredictable results.
MiR-212 is a new cancer-associated miRNA and has been found to participate in the progression in different types of human cancers. Evidences have revealed that miR-212 was declined in renal cell carcinoma,15 ovarian cancer,16 gastric cancer,17 and lung cancer as tumor suppressive role.18,19 On the contrary, miR-212 was increased in pancreatic cancer and promotes tumor development.20,21 The expression level of miR-212 is decreased in renal cell carcinoma and negatively correlated with renal cell carcinoma prognosis. X-linked inhibitor of apoptosis protein was identified to be the direct downstream target of miR-212.15 Functional study showed that miR-212 suppresses the growth of HCC by inhibiting FOXA1.22 In gastric cancer, miR-212 inhibited the proliferation of cancer cells via targeting retinoblastoma binding protein 2.23 Therefore, it is important to study the direct target genes of miR-212, which helps us understand their role in pathological mechanism of HCC.
In this study, we determined that miR-212 is significantly downregulated in HCC. We observed that overexpression of miR-212 suppressed the migration of HCC cells in vitro and in vivo. Additionally, we demonstrated that forkhead box M1 (FOXM1) is a direct target of miR-212 and that FOXM1 expression exhibits an inverse relationship with the expression of miR-212. Reexpression of FOXM1 rescued the miR-212-mediated inhibition of cell migration. Furthermore, we observed that inhibition and overexpression of miR-212 activated and decreased Wnt/β-catenin activity, respectively, by altering the levels of LEF-1, c-Myc, and nuclear β-catenin. In vivo studies confirmed the repression of HCC growth by miR-212. Hence, our results suggest that miR-212 is a putative biomarker of HCC and that the miR-212/FOXM1 regulatory axis may represent a novel therapeutic marker for HCC treatment. These findings reveal a potential mechanism underlying the tumor-suppressing role of miR-212 and suggest that miR-212 is a potentially useful marker and therapeutic target in HCC.24,25
Materials and methods
Cell Culture and Reagents
The human liver cell line, HL7702, and human liver cancerous cell lines, SK-HEP1, HCCLM3, HepG2, and SMMC7721, were purchased from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (HyClone; Thermo-Fisher Scientific, Waltham, Massachusetts). The medium was supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, California). Penicillin/streptomycin was added to the medium at a concentration of 100 IU/mL and applied to the cells, followed by incubation in a humidified incubator set at 37°C with 5% CO2.
MicroRNA Real-Time Reverse Transcription Polymerase Chain Reaction
Cell lines and mice tissues were used as sources for the isolation of total RNA using TRIzol reagent (Invitrogen, Carlsbad, California). A miR-212 TaqMan MicroRNA Assay Kit (Applied Biosystems, Foster City, California) was used for miRNA real-time reverse transcription polymerase chain reaction (PCR) assays, according to the manufacturer’s protocol. Briefly, complementary DNA was synthesized from total RNA (10 ng) by reverse transcription. Real-time PCR was subsequently performed using specific primers for miR-212. A U6 endogenous control was used to normalize miR-212 levels. All experiments were repeated 3 times. The miRNA expression level was determined using the 2−ΔΔCT method for relative quantitative gene expression.
Transwell Migration Assays
A test to assess HCC cell migration was performed using a MilliCell chamber (12-mm diameter with 8-μm pores; Millipore, Bedford, Massachusetts). Approximately 1 × 105 serum-starved HCC cells were added to the upper chamber in media supplemented with 0.1% FBS, and the chambers were placed into 24-well plates with medium supplemented with 10% FBS. After 24 hours of incubation, a cotton swab was used to scrape the top surface of the transwell chamber. Migrated cells were fixed with paraformaldehyde (4%) and stained with Giemsa solution. The cells were photographed and counted in 5 randomly selected regions under an optical microscope. Labeled cells were counted from images using ImageJ software (https://imagej.nih.gov/ij/).
Wound Healing Assay
A wound healing assay was utilized to evaluate tumor cell migration as previously described.26 Briefly, cells were grown to the exponential phase and harvested, and the cell density was adjusted to 1 × 106 cells/mL. Subsequently, 1 mL of the cell suspension was added to each well of a 6-well plate. Cells were grown overnight and transfected with miR-212 or anti-miR-212, with mock-transfected respective controls. When the cells reached close to 90% confluence, the cell monolayer was scraped in the center with a sterile plastic tip to generate a gap in the cell monolayer and then washed gently twice with media. Pictures of the plates were taken, and the cells were recultured with a serum-reduced medium containing 1% FBS for 24 hours. At various time points, cell migration to the gaps in the center of each plate was monitored by taking photographic images of the plates, which were taken under an inverted microscope, and the data were analyzed at each time point based on sextuplet assays.
Preparation of Cell Extracts and Analysis by Western Blot
Protein lysates preparation was performed using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl [pH 8.0], 1% NP40, 150 mM NaCl, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) on ice for 30 minutes. Subsequently, freshly prepared 0.1 mg/mL phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, and 1 mg/mL aprotinin were added. After centrifugation at 12 000g at 4°C for 20 minutes, the supernatants were collected. The Bio-Rad protein assay kit (Bio-Rad Laboratories, Richmond, California) was used to determine the protein concentration of each sample. Cell extract aliquots, containing 20 to 50 μg of total protein, were resolved on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to a 0.22-μm polyvinylidene fluoride membrane (Millipore, Temecula, California). Blotto A (5% nonfat milk powder in TBS-T: 10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.05% Tween-20) was used to block the membranes for 1 hour at room temperature. The membranes were subsequently incubated with antibody solution containing Blotto A with a 1:1000 dilution of mouse anti-FOXM1, anti-β-catenin, anti-c-Myc, anti-LEF-1, or anti-GAPDH monoclonal antibodies for 1 hour at room temperature. Membranes were washed in TBS-T buffer [10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.05% Tween-20] (5 minutes, room temperature) and incubated in Blotto A containing peroxidase-conjugated anti-mouse secondary antibody at a 1:10 00 dilution (Amersham, Arlington Heights, Illinois) for 1 hour at room temperature. After washing in TBS-T, an enhanced chemiluminescence luminol reagent (Pierce, Rockford, Illinois) was used to detect the protein signals, according to the manufacturer’s recommendation. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) protein expression was used in the assay to normalize protein loading.
Luciferase Reporter Assay
Wild-type (WT) and mutant FOXM1 3′-UTRs were amplified by PCR and cloned into a pMIR-REPORT (Ambion, Austin, Texas) vector with firefly luciferase. A total of 5 × 104 cells treated with control, miR-212 mimics, or miR-212 inhibitors (anti-miR-212) were transfected with WT or mutant FOXM1 3′-UTR luciferase reporters together with Renilla plasmid. Approximately 48 hours posttransfection, the cell culture medium was collected; luciferase activities were determined by Dual Luminescence Assay Kits (Promega, Madison, Wisconsin) according to the manufacturer’s protocols. The firefly luciferase activities were normalized to Renilla luciferase activities. Three independent experiments were performed in duplicate to gather data for analysis.
Lentivirus-Based miR-212 Overexpression
To elucidate the role of miR-212 in vivo, we constructed a recombinant lentivirus termed LV-miR-212 or the control lentivirus LV-miR-con to generate stable gain-of-function miR-212 or miR-con in HCC cells. The recombinant lentivirus LV-miR-212 and its control, LV-miR-con, were purchased from GeneChem (Shanghai, China).
Tumor Studies
For in vivo studies, a suspension of 5 × 106 HCCLM3 cells was stably transfected with LV-miR-212 or control LV-miR-con to generate tumor cells. These infected cells were injected into mouse to create mice tumor models. Each athymic nude BALB/C mouse (7 in each group, female BALB/c-nu/nu at 5-6 weeks old) was injected subcutaneously in the hip region with 0.1 mL of the tumor cells mixture using a microsyringe under anesthesia. Tumor growth was monitored by perpendicular measurements of the tumor diameters using an electronic digital caliper over a 24-day period. Calculation of tumor volume was obtained using the tumor size formula ab2/2, with a and b representing the larger and smaller of the 2 dimensions, respectively. Mice were killed after 24 days, and the tumors were harvested, weighed, fixed with phosphate-buffered neutral formalin, and prepared for standard hematoxylin and eosin (H&E) histological examination. The Animal Ethics Committee of Xi’an Jiaotong University approved our protocols and experimental procedures (No.XJTULAC2016-675). We followed the guidelines for all study protocols in congruence with the Animal Care and Use Committee established by the Xi’an Jiaotong University Health Science Center.
Statistical Analysis
Values are presented as the means (standard deviation) and were analyzed using the Statistical Package for Social Scientists (version 18.0; IBM, Armonk, New York). The statistical significance between 2 selected groups was analyzed using 2-tailed Student t test with the assumption of a normal distribution of the data and equal sample variance. The statistical significance among multiple groups was examined by analysis of variance. P < .05 was considered statistically significant.
Results
Downregulation of MiR-212 in HCC Cell Lines and Tissues
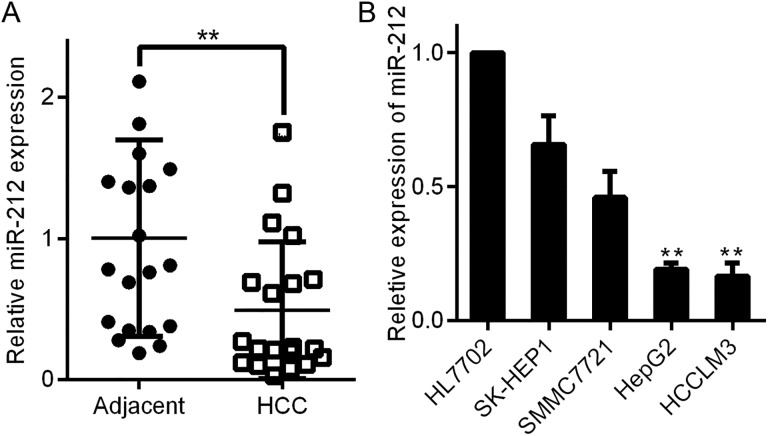
We first measured the expression of miR-212 in HCC by initially comparing the levels of miR-212 in 20 pairs of HCC tissues and parallel nontumor adjacent tissues. The expression of miR-212 in HCC tissues was considerably lower than that in corresponding nontumor surrounding tissues (**P < .01, Figure 1A). Second, we evaluated the relative expression of miR-212 in a normal liver cell line (HL7702) and a panel of human HCC cell lines (SK-HEP1, SMMC7721, HCCLM3, and HepG2). We observed reduced expression of miR-212 in all 4 HCC cell lines compared with the HL7702 cell line (**P < .01, Figure 1B). Notably, miR-212 expression in SK-HEP1 and SMMC7721 cells was higher than that in HCCLM3 and HepG2 cells (P < .01, Figure 1B). Our data indicate that reduced levels of miR-212 may be correlated with the development of HCC.
Figure 1.
MiR-212 expression in HCC tissues and cell lines. A, The expression of miR-212 in HCC tissues was significantly lower than that in matched tumor-adjacent tissues. **P < .01 by t test. B, Comparison of the differences in miR-212 expression between the HL7702 normal liver cell line and HCC cell lines. n = 3 independent experiments, ** P < .01 by analysis of variance. HCC indicates hepatocellular carcinoma.
MiR-212 Inhibits HCC Cell Migration In Vitro
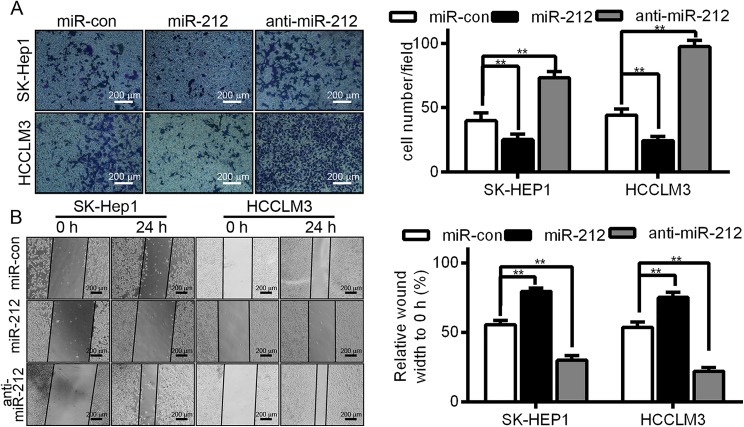
To explore the biological function of miR-212 in HCC, miR-212 mimic or miR-212 inhibitor (anti-miR-212) was used for transient transfection to investigate the function of miR-212 compared with the cells transfected with a negative control mimic (miR-con), which has no specific human gene product target. Our transwell and wound healing assays demonstrated that forced expression of miR-212 in SK-HEP1 and HCCLM3 cells produced a significant decrease in cell migration (**P < .01, as shown in Figure 2A and B, respectively). Furthermore, this downregulation observed with miR-212 in SK-HEP1 and HCCLM3 cells led to a remarkable increase in cell migration compared with control cells (**P < .01, also shown in Figure 2A and B).
Figure 2.
The effect of miR-212 on HCC cell migration. A, Representative images of a transwell migration assay of SK-HEP1 and HCCLM3 cells with miR-con, miR-212 mimic, or anti-miR-212 treatment (**P < .01, compared with miR-con). B, Representative images of a wound healing migration assay in SK-HEP1 and HCCLM3 cells with miR-con, mock miR-212, or anti-miR-212 treatment (**P < .01, compared with miR-con). HCC indicates hepatocellular carcinoma.
MiR-212 Directly Targets the 3′-UTR of FOXM1 and Inhibits the Wnt/β-Catenin Signaling Pathway in HCC Cell Lines
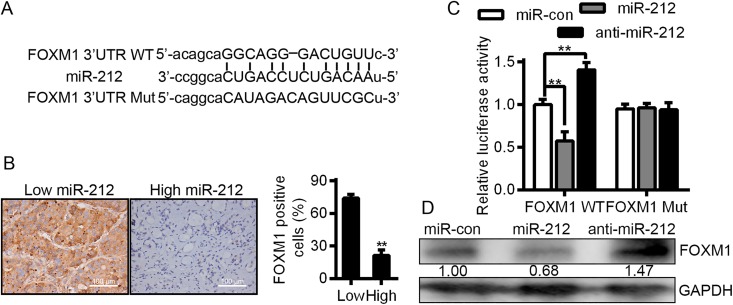
In order to determine the fundamental molecular mechanisms of miR-212 in HCC cells, we performed a literature search for representative target genes of miR-212 through bioinformatic algorithms. The public databases including miRanda (http://microrna.org and miRbase) and TargetScan (http://www.targetscan.org/) were used. As a result, FOXM1 has been showed to a be a potential target site of miR-212, as shown in Figure 3A. Moreover, the 3′-UTR of FOXM1 mRNA containing the complementary sequence of miR-212 was found (Figure 3A). To affirm this prediction, we first detected the expression of miR-212 and FOXM1 in HCC cases. Figure 3B shows that the expression of FOXM1 in miR-212 high-expressing HCC tissues was obviously lower than that in miR-212 low-expressing cases (P < .01). Then a dual-luciferase reporter gene assay was used to evaluate whether miR-212 could directly target the 3′-UTR of FOXM1 mRNA to modulate its expression. As predicted, miR-212 notably suppressed the luciferase activity of FOXM1 containing a WT 3′-UTR but did not inhibit the activity of FOXM1 with a mutant (Mut) 3′-UTR (P < .01, Figure 3C). In addition, through anti-miR-212 transfection, a significant increase was observed in the luciferase activity of FOXM1 3′-UTR WT; however, there was no significant alteration in FOXM1 containing FOXM1 3′-UTR Mut (P < .01, Figure 3C). Furthermore, overexpression of miR-212 in HCCLM3 cells dramatically reduced the expression of FOXM1 and downregulation of miR-212 resulted in elevated expression of FOXM1 in HCCLM3 cells (P < .01, Figure 3D). After all, our results strongly suggest that FOXM1 is a direct downstream target of miR-212 in HCC cells.
Figure 3.
Forkhead box M1 (FOXM1) acts as a direct target downstream of miR-212 in HCC. A, Schematic diagram of the presumed binding site of miR-212 and on the 3′-UTR of FOXM1 (WT, wild-type; Mut, mutant). B, The expression of miR-212 and FOXM1 in HCC specimens. Bar = 100 μm. n = 20, **P < .01. C, The results of luciferase reporter assay. n = 3, ** P < .01. D, Representative images of Western blot. HCC indicates hepatocellular carcinoma.
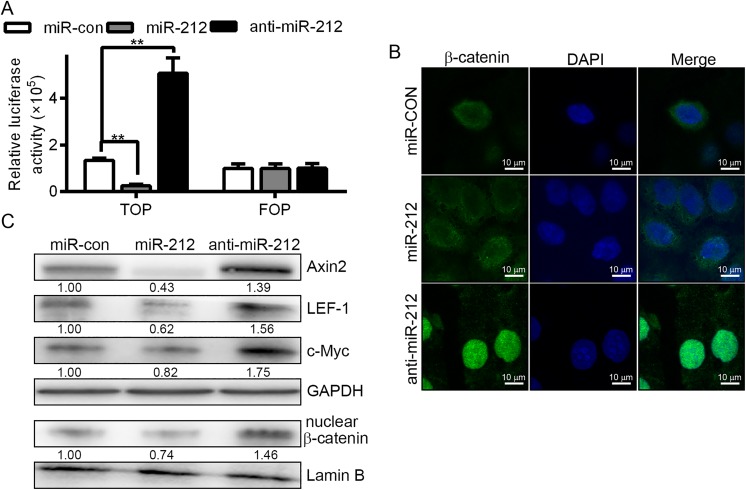
Wnt signaling is involved in embryonic development, adult tissue self-renewal, tissue repair, and cancer development and progression.27 Furthermore, several miRNAs have been shown to influence tumorigenesis by affecting the activity of the Wnt pathway.28 We assessed the relationship between miR-212 expression and the function of the Wnt/β-catenin pathway. First, we used TOPflash and FOPflash reporter molecules, which are widely used to evaluate β-catenin-dependent signaling activity, to determine the effects of miR-212 and FOXM1 on Wnt/β-catenin signal transduction in SK-HEP1 and HCCLM3 cells. As speculated, the luciferase activity of the cells changed, shown in Figure 4A. When the level of miR-212 was downregulated, inhibition of Wnt/β-catenin signaling was observed. Fluorescence microscopy of β-catenin (Figure 4B) revealed that β-catenin translocated from the nucleus to the cytoplasm upon increased expression of miR-212. Additionally, the levels of nuclear β-catenin decreased according to Western blot analysis (Figure 4C). The protein expression of Axin2, LEF-1, and c-Myc was affected by miR-212 and anti-miR-212 transfection (Figure 4C). Together, these data indicate that miR-212 binds to the FOXM1 3′-UTR and that miR-212 expression correlates with Wnt/β-catenin signaling activity.
Figure 4.
Effect of miR-212 on Wnt/β-catenin signaling pathway activity. A, Luciferase reporter gene evaluating β-catenin Transcription factor/Lymphoid enhancer-binding factor (TCF/LEF) promoter activity using TOPflash and FOPflash vectors. The binding site of 3-TCF were localized in the TOPflash reporter vector, and the mutated binding sites of TCF were localized in TOPflash. As indicated, HCCLM3 cells were cotransfected with different expression vectors. MiR-212 treatment reduced β-catenin TCF/LEF promoter activity, whereas anti-miR-212 treatment increased β-catenin TCF/LEF promoter activity. B, Immunofluorescence assay for β-catenin indicated that β-catenin translocated from the nucleus to the cytoplasm upon increased expression of miR-212. When cells were transfected with anti-miR-212, the localization of β-catenin in the cells shifted from the cytoplasm to the nucleus, compared with miR-con. *P < .05. C, Western blot of Axin2, LEF-1, and c-Myc in miR-212-transduced HCCLM3 cells or anti-miR-212-transduced HCCLM3 cells. Evaluation of the expression levels of β-catenin in the cell nuclei of miR-212- and anti-miR-212-transduced HCCLM3 cells. GAPDH, control for cytoplasmic protein; Lamin B, control for nuclear protein. HCC indicates hepatocellular carcinoma.
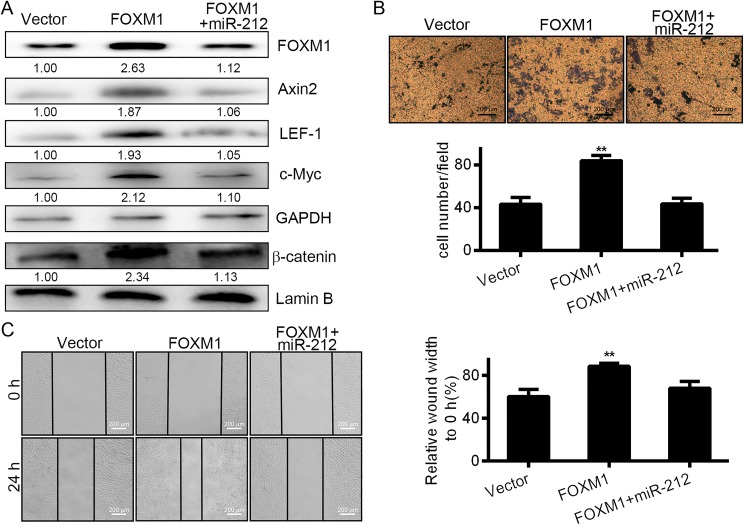
Rescue of FOXM1 in MiR-212-Expressing Cells Partially Restores Cell Migration
To confirm that miR-212 suppresses the growth of HCCLM3 cells directly via downregulation of FOXM1, “rescue” experiments were performed using a FOXM1 expression vector (pcDNA3-FOXM1) containing the FOXM1 gene. When we cotransfected HCCLM3 cells with miR-212 and pcDNA3-FOXM1, the inhibition of FOXM1 expression, wound healing, and cell migration were rescued, as shown in Figure 5A, B, and C. These results indicate that FOXM1 is a direct target of the miR-212-mediated suppression of cellular proliferation in HCCLM3 cells.
Figure 5.
Rescue of FOXM1 in miR-212-expressing cells partially restored cell migration. A, Western blot analysis of FOXM1 expression and related proteins in HCCLM3 cells transfected with empty vector as control, pCDNA3-FOXM1 or pCDNA3-FOXM1, and miR-212. Lower panel: Densitometric analysis of immunodetected bands. Data were normalized using GAPDH expression. B, Transwell analysis for FOXM3 and miR-212 in HCCLM3 cells transfected with empty vector, pCDNA3-FOXM1 or pCDNA3-FOXM1 and miR-212. The histograms represent the migrated cells ± SD from 3 independent experiments after normalization. C, Wound healing analysis for FOXM3 and miR-212 in HCCLM3 cells transfected with empty vector pCDNA3-FOXM1 or pCDNA3-FOXM1 and miR-212. The histograms represent the migration distance ± SD from 3 independent experiments after normalization. FOXM1 forkhead box M1; HCC, hepatocellular carcinoma.
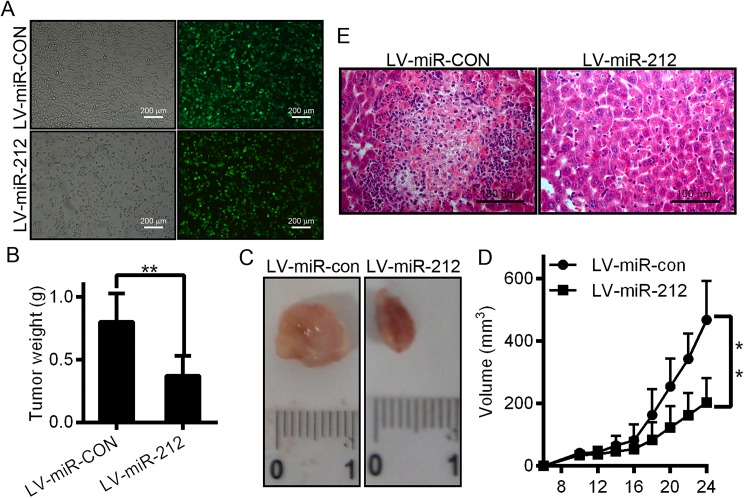
MiR-212 Inhibits the Tumor Growth of Hepatoma Xenografts in Nude Mice
After the consideration of the important roles of miR-212 in HCC, we applied in vivo model of HCC xenograft to further confirm the above findings. Due to significant downregulation of miR-212 in HCC, we successfully constructed a recombinant lentiviral vector (LV-miR-212) to transduce HCCLM3 cells (Figure 6A) and increase the expression of miR-212. Meanwhile, lentiviral vector LV-miR-con was used as a control. Subsequently, LV-miR-212 or LV-miR-con-infected HCCLM3 cells were administered by injection into the hip area of athymic nude mice for HCC xenograft establishment. Twenty-four days of postinjection, tumors were excised and gauged. The results showed that the size of tumor in the LV-miR-212 group were strongly smaller than in the LV-miR-con group (Figure 6B, C, and D; tumor incidence for LV-miR-212 vs LV-miR-con groups). Additionally, mice were killed 24 days after inoculation, and the resected liver tissues from mice with subcutaneous xenograft tumors were analyzed using H&E staining (Figure 6E). Shown in Figure 6E is the LV-miR-212 group, which exhibited a reduced number of tumor cells in the liver tissues. Consistently, the LV-miR-212 group exhibited proliferation inhibition compared with the LV-miR-con group (Figure 6E). These data provide strong evidence that miR-212 can inhibit HCC proliferation in vivo.
Figure 6.
miR-212 impairs the growth of hepatocellular carcinoma xenografts in vivo. A, Efficient infection of HCCLM3 cells with lentivirus shown by fluorescence microscopy. The black and white pictures show the cells in the same field under normal white light. B, The tumor weights were measured and calculated. C, Mice were killed and photographed 24 days after injection of HCCLM3 cells. D, HCCLM3 cells infected with lentivirus LV-miR-212 or LV-miR-con were injected subcutaneously into nude mice. Tumor size measurements began on the sixth day and occurred every other day for 24 consecutive days. The recorded data were used to generate tumor growth curves, as shown. Data are presented as the mean (SD) tumor volume. E, Mice were injected with tumor cells and killed after 24 days. Livers were dissected and fixed in formalin, embedded in paraffin, and sectioned for hematoxylin and eosin staining. *P < .05. HCC indicates hepatocellular carcinoma.
Discussion
Previous reports indicate that human cancers exhibit deregulated miRNA expression relative to the surrounding noncancer tissue, and the role of this aberrant regulation during oncogenesis has been highlighted.29 Several reports have shed light on the roles of miRNAs in treatments and as therapeutic targets in HCC.30–32 For example, robust expression of miR-182 is related to intrahepatic metastasis and poor prognosis,33 whereas low levels of miR-124 has a significant association with a more aggressive phenotype and poor prognosis.34 MiR-212 directly regulates the expression of retinoblastoma binding protein 2 (RBP2) and inhibits cell growth in gastric cancer in vitro and in vivo.23 Histone demethylase RBP2 is overexpressed in HCC and negatively regulated by miR-212, which is important in the pathogenesis of HCC.35
In the present study, we demonstrated that miR-212 is downregulated in HCC tissues compared with nontumorous liver tissues and that the level of miR-212 positively correlates with cancer progression. Our prospective study indicates that miR-212 may have therapeutic value in the treatment of HCC in clinical practice. To date, several reports have demonstrated the important roles of miR-212 in a variety of human tumors. In chronic lymphocytic leukemia, there is an increase in tumor cell proliferation upon IgM stimulation sustained by the enhancement of miR-212 expression.36 MiR-212 may improve the current prognostic risk stratification of acute myeloid leukemia with cytogenetic and molecular abnormalities.37 MiR-212 is frequently downregulated in different types of cancer. For example, this phenomenon has been confirmed in gastric, lung, and colorectal cancers as well as in head and neck squamous cell carcinomas.17,38 In our present study, we have demonstrated that ectopic miR-212 expression markedly repressed HCC migration in vitro and in vivo. Therefore, our results suggested that miR-212 is a novel potential therapeutic target for HCC treatment.
Previous studies have demonstrated an association between miR-25 and Dickkopf-related protein 3 (DKK3) expression with HCC migration.39 However, how miR-212 modulates HCC is not well understood. To increase the specificity of our studies, we first used 2 computational prediction tools: miRanda and TargetScan. The results predicted that FOXM1 is a potential functional target of miR-212 in HCC. MiR-370 may also represent a direct regulator of FOXM1 in acute myeloid leukemia, as previously reported.40 MiR-370 may function as a tumor suppressor by targeting FOXM1; thus, the epigenetic downregulation of miR-370 would lead to derepression of FOXM1 expression and consequently contribute to acute myeloid leukemia development and progression. However, FOXM1 can be regulated by many factors, including miRNAs.40 We determined that miR-212 regulates FOXM1 expression by overexpressing FOXM1 in HCCLM3 cells and examining the changes in cellular proliferation and migration. The introduction of miR-212 significantly inhibited the proliferation and migration of FOXM1-overexpressing cells compared to controls. Additionally, we observed that overexpression of miR-212 significantly downregulated FOXM1 protein levels. In contrast, knockdown of miR-212 in FOXM1 cells resulted in significant increases in cell proliferation and migration. Similarly, we observed noticeable upregulation of FOXM1 protein levels. Reports indicate that the multiple downstream target genes of the Wnt/β-catenin pathway increase in various malignancies, correlating with tumor progression and prognosis.41 Our study suggests that miR-212 inhibits HCC cell proliferation and tumorigenesis through regulation of LEF-1, c-Myc, and β-catenin, which have been demonstrated to act as positive regulators of the Wnt signaling pathway. Furthermore, we demonstrated that FOXM1 is a direct functional target of miR-212. Consequently, we conducted dual-luciferase reporter assays to demonstrate that FOXM1 is a functional downstream target of miR-212. This finding suggests that miR-212 suppresses FOXM1 expression by interacting with the 3′-UTR of FOXM1 mRNA. Furthermore, ectopic expression of FOXM1 significantly increased the proliferation and migration of HCCLM3 cells, and the addition of miR-212 induced effects that were opposite to those stimulated by FOXM1. These results confirm that FOXM1 is a functional target gene of miR-212 in HCC. Thus, the reported targets for miR-212 are consistent with our data and indicate that miR-212 regulates Wnt signaling pathways and that the loss of miR-212 results in HCC tumor progression. These findings reveal a potential mechanism underlying the tumor-suppressive role of miR-212 and suggest that miR-212 is a potentially useful marker and therapeutic target for HCC.
Conclusions
Several reports have revealed the critical roles of miRNA in the development and progression of cancer. miR-212 is one of the several genetic variants identified to have an association with the development of HCC. We analyzed the cancer-specific miR-212 and its downstream targets, FOXM1 and the Wnt/β-catenin pathway. Mapping these pathways may provide insight into the identification of new biomarkers and therapeutic targets for the rescue of tumor suppression in HCC. Collectively, our present study suggests that overexpression of miR-212 inhibits HCC cell proliferation by downregulating the FOXM1 gene. In contrast, inhibition of miR-212 expression positively correlated with Wnt/β-catenin expression and downstream targets. Our findings underline the potential clinical value of miR-212 in HCC treatment and support the use of effective therapeutic strategies that directly targets miR-212 or its associated factors such as FOXM1, Wnt, or β-catenin.
Abbreviations
- FBS
fetal bovine serum
- FOXM1
forkhead box M1
- HCC
hepatocellular carcinoma
- mRNA
messenger RNA
- miRNA
microRNA
- MUT
mutant
- PCR
polymerase chain reaction
- RBP2
retinoblastoma binding protein 2
- 3′-UTR
3′ untranslated region
- WT
wild-type.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Natural Science Foundation of China (No. 81572734) and the Scientific and Technological Development Research Project Foundation by Shaanxi Province (2016SF-121).
ORCID iD: Xuqi Li  http://orcid.org/0000-0002-2497-020X
http://orcid.org/0000-0002-2497-020X
References
- 1. Papatheodoridis GV, Lampertico P, Manolakopoulos S, Lok A. Incidence of hepatocellular carcinoma in chronic hepatitis B patients receiving nucleos(t)ide therapy: a systematic review. J Hepatol. 2010;53(2):348–356. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. [DOI] [PubMed] [Google Scholar]
- 3. Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song T. Recent advances in surgical treatment of hepatocellular carcinoma. Drug Discov Ther. 2015;9(5):319–330. [DOI] [PubMed] [Google Scholar]
- 5. Mori M, Triboulet R, Mohseni M, et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156(5):893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Djuranovic S, Nahvi A, Green R. MiRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meng F, Glaser SS, Francis H, et al. Functional analysis of microRNAs in human hepatocellular cancer stem cells. J Cell Mol Med. 2012;16(1):160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He M, Zhou W, Li C, Guo M. MicroRNAs, DNA damage response, and cancer treatment. Int J Mol Sci. 2016;17(12):E2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. MiR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12(1):27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai QQ, Dong YW, Wang R, et al. MiR-124 inhibits the migration and invasion of human hepatocellular carcinoma cells by suppressing integrin αV expression. Sci Rep. 2017;7:40733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sandbothe M, Buurman R, Reich N, et al. The microRNA-449 family inhibits TGF-β-mediated liver cancer cell migration by targeting SOX4. J Hepatol. 2017;66(5):1012–1021. [DOI] [PubMed] [Google Scholar]
- 12. Jin F, Wang Y, Li M, et al. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8(1):e2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gougelet A, Sartor C, Bachelot L, et al. Antitumour activity of an inhibitor of miR-34a in liver cancer with β-catenin-mutations. Gut. 2016;65(6):1024–1034. [DOI] [PubMed] [Google Scholar]
- 14. Jung KH, Zhang J, Zhou C, et al. Differentiation therapy for hepatocellular carcinoma: multifaceted effects of miR-148a on tumor growth and phenotype and liver fibrosis. Hepatology. 2016;63(3):864–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gu C, Wang Z, Jin Z, et al. MicroRNA-212 inhibits the proliferation, migration and invasion of renal cell carcinoma by targeting X-linked inhibitor of apoptosis protein (XIAP). Oncotarget. 2017;8(54):92119–92133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei LQ, Liang HT, Qin DC, Jin HF, Zhao Y, She MC. MiR-212 exerts suppressive effect on SKOV3 ovarian cancer cells through targeting HBEGF. Tumour Biol. 2014;35(12):12427–12434. [DOI] [PubMed] [Google Scholar]
- 17. Wada R, Akiyama Y, Hashimoto Y, Fukamachi H, Yuasa Y. MiR-212 is downregulated and suppresses methyl-CpG-binding protein MeCP2 in human gastric cancer. Int J Cancer. 2010;127(5):1106–1114. [DOI] [PubMed] [Google Scholar]
- 18. Incoronato M, Urso L, Portela A, et al. Epigenetic regulation of miR-212 expression in lung cancer. PLoS One. 2011;6(11):e27722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Incoronato M, Garofalo M, Urso L, et al. MiR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 2010;70(9):3638–3646. [DOI] [PubMed] [Google Scholar]
- 20. Park JK, Henry JC, Jiang J, et al. MiR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011;406(4):518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma C, Nong K, Wu B, et al. MiR-212 promotes pancreatic cancer cell growth and invasion by targeting the hedgehog signaling pathway receptor patched-1. J Exp Clin Cancer Res. 2014;33:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dou C, Wang Y, Li C, et al. MicroRNA-212 suppresses tumor growth of human hepatocellular carcinoma by targeting FOXA1. Oncotarget. 2015;6(15):13216–13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiping Z, Ming F, Lixiang W, et al. MicroRNA-212 inhibits proliferation of gastric cancer by directly repressing retinoblastoma binding protein 2. J Cell Biochem. 2013;114(12):2666–2672. [DOI] [PubMed] [Google Scholar]
- 24. Yao M, Wang L, Qiu L, Qian Q, Yao D. Encouraging microRNA-based therapeutic strategies for hepatocellular carcinoma. Anticancer Agents Med Chem. 2015;15(4):453–460. [DOI] [PubMed] [Google Scholar]
- 25. Yang N, Ekanem NR, Sakyi CA, Ray SD. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev. 2015;81:62–74. [DOI] [PubMed] [Google Scholar]
- 26. Duan W, Chang Y, Li R, et al. Curcumin inhibits hypoxia inducible factor-1α-induced epithelial-mesenchymal transition in HepG2 hepatocellular carcinoma cells. Mol Med Rep. 2014;10(5):2505–2510. [DOI] [PubMed] [Google Scholar]
- 27. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. [DOI] [PubMed] [Google Scholar]
- 28. Zhao JJ, Lin J, Zhu D, et al. MiR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res. 2014;74(6):1801–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 30. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. [DOI] [PubMed] [Google Scholar]
- 31. Zhou SL, Hu ZQ, Zhou ZJ, et al. MiR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63(5):1560–1575. [DOI] [PubMed] [Google Scholar]
- 32. Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. MiR-122—a key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448–457. [DOI] [PubMed] [Google Scholar]
- 33. Wang J, Li J, Shen J, Wang C, Yang L, Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer. 2012;12:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng F, Liao YJ, Cai MY, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61(2):278–289. [DOI] [PubMed] [Google Scholar]
- 35. Liang X, Zeng J, Wang L, et al. Histone demethylase retinoblastoma binding protein 2 is overexpressed in hepatocellular carcinoma and negatively regulated by hsa-miR-212. PLoS One. 2013;8(7):e69784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tavolaro S, Colombo T, Chiaretti S, et al. Increased chronic lymphocytic leukemia proliferation upon IgM stimulation is sustained by the upregulation of miR-132 and miR-212. Genes Chromosomes Cancer. 2015;54(4):222–234. [DOI] [PubMed] [Google Scholar]
- 37. Sun SM, Rockova V, Bullinger L, et al. The prognostic relevance of miR-212 expression with survival in cytogenetically and molecularly heterogeneous AML. Leukemia. 2013;27(1):100–106. [DOI] [PubMed] [Google Scholar]
- 38. Hatakeyama H, Cheng H, Wirth P, et al. Regulation of heparin-binding EGF-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PLoS One. 2010;5(9):e12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tu H, Wei G, Cai Q, et al. MicroRNA-212 inhibits hepatocellular carcinoma cell proliferation and induces apoptosis by targeting FOXA1. Onco Targets Ther. 2015;8:2227–2235. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40. Zhang X, Zeng J, Zhou M, et al. The tumor suppressive role of miRNA-370 by targeting FoxM1 in acute myeloid leukemia. Mol Cancer. 2012;11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Teufel A, Marquardt JU, Galle PR. Next generation sequencing of HCC from European and Asian HCC cohorts. Back to p53 and Wnt/β-catenin. J Hepatol. 2013;58(3):622–624. [DOI] [PubMed] [Google Scholar]