Summary
New diagnostics are needed to facilitate appropriate use of antimicrobial agents. The Master Protocol for Evaluating Multiple Infection Diagnostics (MASTERMIND) concept for advancement of infectious diseases diagnostics is described, wherein a single subject’s sample(s) evaluates multiple diagnostic tests.
Keywords: molecular diagnostic techniques, communicable diseases, reference standards.
Abstract
New diagnostics are urgently needed to address emerging antimicrobial resistance. The Antibacterial Resistance Leadership Group proposes a strategy called MASTERMIND (Master Protocol for Evaluating Multiple Infection Diagnostics) for advancement of infectious diseases diagnostics. The goal of this strategy is to generate the data necessary to support US Food and Drug Administration clearance of new diagnostic tests by promoting research that might not have otherwise been feasible with conventional trial designs. MASTERMIND uses a single subject’s sample(s) to evaluate multiple diagnostic tests at the same time, providing efficiencies of specimen collection and characterization. MASTERMIND also offers central trial organization, standardization of methods and definitions, and common comparators.
The lack of available diagnostics to rapidly assess for the presence or absence of bacterial infection and the consequent challenges in defining appropriate treatment contribute to the overuse of antimicrobial agents in clinical practice [1, 2]. Although conventional approaches, such as bacterial culture and routine antimicrobial susceptibility testing, can provide a diagnosis for some infectious syndromes, the turnaround time from specimen collection to final result is often too long to provide actionable results. Traditionally, the issue of diagnostic uncertainty has been addressed by using empiric broad-spectrum antibiotic therapy in an effort to treat the most likely bacterial pathogen(s). This strategy is proving to be increasingly ill-advised in the current era of antimicrobial resistance. New diagnostics that are affordable, rapid, and accurate could potentially mitigate overuse of antimicrobial agents. However, validating and obtaining US Food and Drug Administration (FDA) clearance for clinically needed diagnostics is often beyond the reach of diagnostics companies, especially when the return on investment is uncertain and the needed clinical trials are complex. Another challenge is access to good-quality, well-characterized clinical specimens that provide the highest level of validation data, especially when companies need to find exactly the right kind of patients for their evaluations. Moreover, when multiple companies are working on similar assays, they may be competing with one another for limited clinical trial sites. MASTERMIND is a novel approach that overcomes many of the monetary and logistical hurdles associated with clinical trials of new diagnostics.
MASTERMIND: The Master Protocol for Multiple Infection Diagnostics
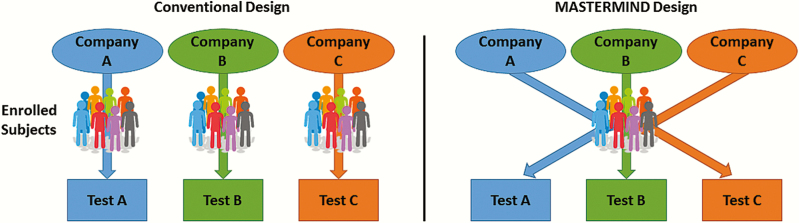
Given the clinical need and availability of interested partners with suitable test platforms, the Antibacterial Resistance Leadership Group (ARLG) has developed a scheme labeled MASTERMIND (Master Protocol for Evaluating Multiple Infection Diagnostics) to advance infectious diseases diagnostics. The scheme is based on novel study designs, such as basket and umbrella designs that were developed in the oncology trial community and which promote clinical research that may have been untenable using conventional study designs [3]. The MASTERMIND concept uses a single subject’s sample(s) to evaluate multiple tests, providing efficiencies of scale for multiple simultaneous or successive investigations (Figure 1). To develop MASTERMIND studies for specific diagnostic tests, the ARLG is bringing together infectious diseases physicians, clinical microbiologists, statisticians, and potentially interested companies.
Figure 1.
Conventional vs MASTERMIND diagnostic study. When multiple similar diagnostics are being evaluated independently, the necessary resources (subjects, costs, effort) are multiplied. In a MASTERMIND design, many of these resources are shared, while maintaining independence of study outcomes.
MASTERMIND Study Possibilities
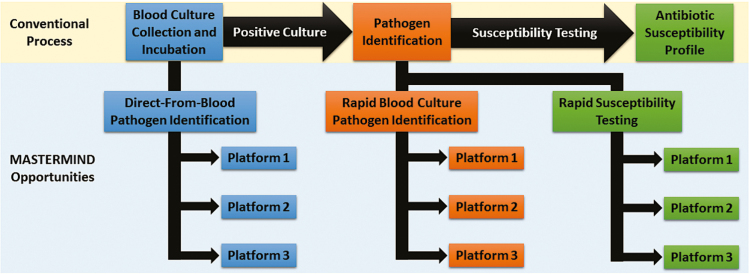
A number of MASTERMIND studies can be conceived by identifying areas of need within existing microbiological workflows. One example is diagnostics for detection and characterization of bacteremia. One need only consider the existing bloodstream infection evaluation paradigm to identify areas for diagnostic development (Figure 2). Beginning with blood culture collection, there is growing interest and success in direct-from-blood pathogen detection assays. Some progress has already been made in this domain, including SeptiFast [4], IRIDICA [5], and T2 Biosystems [6], although only the last is FDA cleared, and only for detection of Candida species. A MASTERMIND protocol for direct-from-blood pathogen detection could theoretically collect enough blood from each enrolled subject to submit for multiple test evaluations (Figure 3), though required volumes may sometimes be limiting and may not enable a large number of diagnostics to be run on a single specimen.
Figure 2.
Opportunities for a MASTERMIND design pertaining to bloodstream infections. There are 3 active areas of development: direct-from-blood pathogen detection, rapid pathogen identification from positive blood culture bottles, and rapid antibacterial susceptibility testing. A MASTERMIND study could evaluate diagnostics at any step in the culture processing pathway.
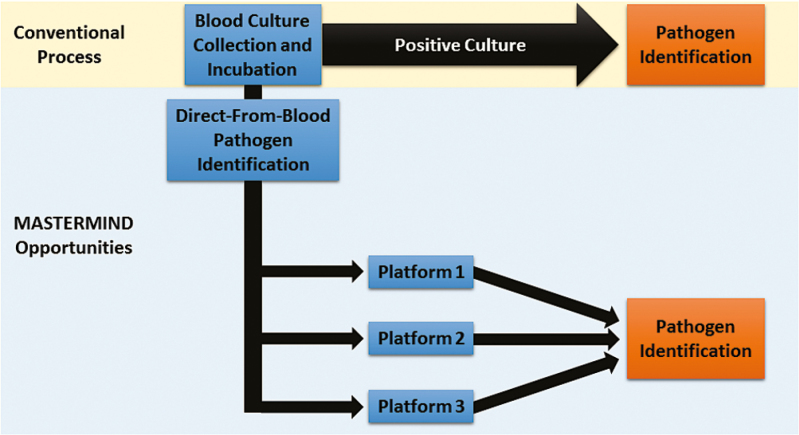
Figure 3.
MASTERMIND design for one diagnostic focus area. In this conceptualized scheme, a single patient would support the evaluation of multiple tests that focus on the same diagnostic task. In this scenario, each test can be compared to a composite comparator comprised of the routine clinical test and the other investigational platforms.
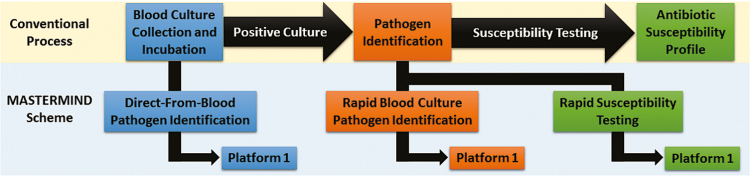
Upon detection of microbial growth in a blood culture bottle, a number of steps typically ensue, including organism identification and antimicrobial susceptibility testing. For both activities, efforts are in place or under way to improve time to result [7]. Unlike the case with direct-from-blood pathogen detection assays, specimen volume is not limiting and therefore this approach is especially conducive to a MASTERMIND design. In another variation of such a design, the diagnostics being evaluated could focus on different tasks. As depicted in Figure 4, blood collected from a single subject could be used to evaluate a direct-from-blood pathogen detection assay. If the subject’s standard blood culture is positive, a sample could be used to evaluate a rapid pathogen identification assay. Moreover, that positive blood culture could be used to evaluate a new phenotypic or genotypic antimicrobial susceptibility platform. In this way, a single subject is still being used to evaluate multiple diagnostics, although the diagnostics focus on different clinical and microbiological questions. Although Figure 4 suggests that only one platform would be used at each stage, the sample volume in a positive blood culture bottle is adequate to allow evaluation of multiple investigational identification or susceptibility platforms, depending on their availability.
Figure 4.
MASTERMIND design for multiple diagnostic focus areas. In this conceptualized scheme, a single patient’s sample would support the evaluation of multiple tests that focus on different diagnostic tasks. Only 1 investigational platform is depicted for each step, although >1 could be evaluated, provided additional platforms are available and sample is not limiting. When there is only 1 test in each domain, the standard clinical assay could serve as the comparator assay.
A number of rapid urine diagnostics are emerging to assess for the presence of infection and, if infection is present, characterize the organism and its antimicrobial susceptibility pattern. Considering the relative ease in obtaining a large volume of urine, urine diagnostics are also considered suitable for a MASTERMIND trial design. Direct pathogen detection and characterization of antimicrobial susceptibility in respiratory specimens is yet another possibility, among many others that can be imagined. One other scenario is to nest the evaluation of one or more diagnostic platforms within an anti-infective clinical trial. Many anti-infective trials rely on supplemental microbiology testing to help define the microbiological intention-to-treat population. Integrating a novel diagnostic strategy could potentially support that diagnostic test, while also supporting the overall clinical trial outcome. Finally, MASTERMIND studies could conceivably allow for collection of data for exploratory purposes, such as setting critical values.
Challenges in Setting a Comparator Method
Although not unique to MASTERMIND, one of the challenges in evaluating new diagnostics is defining appropriate comparator methods. When a nearly perfect comparator method is readily available, it should be used. However, if a broadly acknowledged reference standard does not exist, which is often the case, an imperfect comparator may be considered, especially if a consensus can be reached among imperfect standards. With an imperfect comparator method alone, however, the study cannot be designed to measure sensitivity and specificity (ie, relationship to the “truth”). Instead, the study needs to be designed to estimate the positive percent agreement (PPA) and negative percent agreement (NPA) with the imperfect comparator method. Ideally, a comparator method that utilizes a different strategy than the tests being evaluated should be considered, because tests that use similar approaches may have correlated errors, meaning there could be test agreement with incorrect results [8]. The comparator method could also be multifaceted, involving clinical and laboratory components, clinical adjudication, or an algorithmic approach. In the absence of a perfect comparator method, one idea that is being considered for some MASTERMIND studies is to use a compilation of the test results being evaluated.
Several issues should be brought to bear when considering this design strategy (Table 1). In some planned MASTERMIND studies, nucleic acid amplification tests (NAATs) are the modality being investigated. This may be problematic if all of the investigational tests, as well as the comparator method, are NAATs. In this case, there is a risk of correlated errors, particularly when the investigational and comparator assays have similar targets. To reduce the likelihood of correlated errors, they should, if possible, target different types of nucleic acid (eg, ribosomal RNA vs DNA), use primers that target different parts of the microbial genome, and/or use different methods of target amplification and detection. Sequencing of amplified nucleic acids can be considered to resolve uncertainty.
Table 1.
Challenges and Potential Solutions for MASTERMIND Studies
| Challenges | Potential Solutions | |
|---|---|---|
| Comparator method considerations | • There may be no available, good-quality comparator method, particularly for novel diagnostics | • Use a composite of tests, while excluding the one being evaluated |
| • Tests used for the comparator method may themselves have poor performance characteristics | • Utilize only tests with a high level of preclinical validation or include clinical and laboratory components in the comparator, or apply clinical adjudication, or use an algorithmic approach | |
| Industry commitment | • If a company withdraws, there are deleterious consequences to the remaining components of the protocol | • Ensure high-level commitment from participating companies through early and ongoing engagement |
| Statistical considerations | • Indeterminate/equivocal results | • Include alternative tests, clinical data, or short-term follow-up data to clarify the diagnosis |
| • Variations in specimen quality due to repeated collections | • Randomize order of specimen collection | |
| • Operational bias | • Blind users of the investigational test to clinical information and comparator test result(s) and vice versa | |
| • Determination of sample size | • May need to be adjusted during study based on the prevalence of infection |
Abbreviation: MASTERMIND, Master Protocol for Evaluating Multiple Infection Diagnostics.
Study Design and Analytical Considerations
Randomization of the order of specimen collection and testing should be considered to avoid systematic issues with quality and ensure balance of specimen order. This also allows for evaluation of the impact of specimen collection order on test performance.
Because several tests will be performed as part of a single MASTERMIND study, there is a risk of operational bias, whereby knowing results of one test may influence results of another. To avoid operational bias, users of the investigational tests should be blinded to clinical information and results of other tests. Likewise, users of the comparator method should be blinded to clinical information as well as all investigational test results.
During statistical analysis, it is inappropriate to exclude indeterminate or equivocal results as they may be associated with test performance. Equivocal results are valid results that are neither positive nor negative. If a test is designed with an equivocal zone, the equivocal result should be considered a third category of test result. Every effort should be made to prevent and minimize their frequency through creative design and careful conduct. Additional tests or the collection of clinical data (eg, signs/symptoms) or short-term follow-up data should be planned to be utilized if the primary comparator method is indeterminate. Sensitivity analyses for indeterminate comparator methods results are needed.
For each investigational diagnostic test, performance characteristics, including sensitivity (or PPA for an imperfect test), specificity (or NPA for an imperfect test), and positive and negative likelihood ratios should be estimated. Estimates of positive and negative predictive values should be plotted as a function of prevalence with confidence bands.
Newly developed methods can be used to evaluate the expected diagnostic yield (ie, the expected distribution of true positives, true negatives, false positives, and false negatives) associated with the diagnostics if put into practice. Diagnostics can also be systematically compared using these methods [9]. When designing and publishing clinical trial results, researchers should consult the Standards for the Reporting of Diagnostic Accuracy Studies (STARD) Statement (www.stard-statement.org) [10, 11].
Study Monitoring
MASTERMIND studies are not expected to have toxicity concerns; however, data monitoring will be important given study complexity. Issues to monitor include data completeness and quality, design assumptions used in study sizing, specimen collection methods, and frequency of indeterminate and equivocal test (including comparator) results. To estimate sensitivity with the desired precision, an appropriate number of infected participants is needed. Similarly, an appropriate number of uninfected participants is needed to estimate specificity with the desired precision. If the infection rate is expected to be low, it may be prudent to “enrich” for participants with a high likelihood of infection. However, when enriching for subjects likely to be infected, there is potential bias due to higher disease severity. Spectrum bias may be introduced if healthy normal subjects serve as reference negatives in lieu of enrolling from the intended use population, which may include individuals who have other characteristics that could affect test performance [12].
As with any study evaluating diagnostic assays, infection rates can be monitored on an interim basis to determine whether sample size adjustments are warranted. This can be accomplished using a data monitoring committee. If infection is more prevalent than expected, a smaller sample size may be acceptable, as long as broad generalizability can be maintained. However, if infection is less prevalent than anticipated, increases to sample size should be considered. Note that in this case, sample size is not being adjusted based upon test performance. In fact, knowledge of test performance is not needed. Thus, there are no statistical error control concerns with monitoring disease prevalence. Researchers may choose to monitor test performance if the tests are extremely expensive or invasive but only with appropriate error control strategies. Appropriate planning for batching samples, if required, and laboratory analyses are needed as well.
Ideally, study monitoring should be conducted independently, and particularly if the tests being evaluated also serve as the comparator for other tests. In that case, infection rates or indeterminate/equivocal rates for each test being evaluated may differ and variation among these rates across tests can convey information regarding test performance.
Operational Challenges
The MASTERMIND study design is not intended to be a panacea. Diagnostic platforms being evaluated should be in similar stages of their developmental life cycle. There must be adequate biological matrix available to allow multiple aliquots. And finally, there must be willingness for a certain level of collaboration between industry and academia, as well as between industry partners. Given the involvement of academia and industry, as well as regulatory agencies, it is important to address control over study design, conduct, analysis, reporting, data access, and data ownership at the start. In this study design, it is ultimately each company’s decision as to whether to submit their data to the FDA at the end of the trial. Whereas an ideal outcome is that all tests perform well, it may be problematic if this is not so. There will be different priorities and interests among industry partners, regulatory agencies, practitioners, investigators, and patients. Because the funding agency for ARLG-related research is the National Institutes of Health, the associated data will ultimately be publicly available. Clearly, it is important to have neutral and central party control due to competing interests, as there are opportunities for integrity to be threatened. And, during the course of the study, strong firewalls and blinding of interim test performance are necessary to protect data integrity. A publication policy, such as that established by the ARLG, is needed since MASTERMIND studies are run by academic investigators with government funding. Data monitoring may be complex but rapid access to data and rapid analyses are needed, especially if it is necessary to open/close arms rapidly or otherwise modify the study. Overall, MASTERMIND studies will be complex and need flexibility to adapt to changes. They will take a large effort, require funding for people who focus exclusively on the trial (eg, operations, statistics, data management), and require standardization and robust informatics infrastructure linking elements together for the participating laboratories.
CONCLUSIONS
The MASTERMIND concept, like any new idea, will not be without challenges. However, MASTERMIND has the promise to enable new diagnostics to be brought to the clinic, particularly diagnostics that may not be clinically evaluable without it. Coming to consensus among participants as to protocol design and management may be difficult. However, our experiences so far have been positive and suggest that the many challenges can be overcome.
Notes
Acknowledgments. We thank Fred C. Tenover, PhD, Ellen Jo Baron, PhD, and David H. Persing, MD, PhD, from Cepheid for their thoughtful review.
Financial support. This work was supported by the Antibacterial Resistance Leadership Group of the National Institutes of Health (grant number UM1 AI104681).
Potential conflicts of interest. R. P. reports grants from BioFire, Check-Points, Curetis, 3M, Merck, Hutchison Biofilm Medical Solutions, Accelerate Diagnostics, Allergan, and The Medicines Company; is a consultant to Curetis, Roche, Qvella, and Diaxonhit (monies are paid to Mayo Clinic); has a patent on Bordetella pertussis/parapertussis polymerase chain reaction issued, a patent on a device/method for sonication with royalties paid by Samsung to Mayo Clinic, and a patent on an anti-biofilm substance issued; serves on an Actelion data monitoring board; receives travel reimbursement and an editor’s stipend from the American Society for Microbiology and Infectious Diseases Society of America; and receives honoraria from the National Board of Medical Examiners (NBME), UpToDate, and the Infectious Diseases Board Review Course. E. L. T. has served as a consultant for Immunexpress, CytoVale, and Liquidia; has received research support from Novartis, Defense Advanced Research Projects Agency (DARPA), National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID), DTRA, Bill & Melinda Gates Foundation, and Veterans Health Administration (VHA); has an equity relationship in Host Response, Inc; has filed a patent for methods of identifying infectious disease and assays for identifying infectious disease; and has a patent pending for host gene expression signatures of Staphylococcus aureus and Escherichia coli infections. V. G. F. has received grants from NIH; has received personal fees from Merck, Pfizer, Novartis, Galderma, Novadigm, Durata, Debiopharm, Genentech, Achaogen, Affinium, The Medicines Company, Cerexa, Tetraphase, Trius, MedImmune, Bayer, Theravance, Cubist, Basilea, Affinergy, Jenssen, Contrafect, and xBiotech; has received grants from NIH, MedImmune, Cerexa/Forest/Actavis/Allergan, Pfizer, Advanced Liquid Logics, Theravance, Novartis, Cubist/Merck, Medical Biosurfaces, Locus, Affinergy, Contrafect, Karius, and Centers for Disease Control and Prevention; has received personal fees from Green Cross, Cubist, Cerexa, Durata, Theravance, Debiopharm, and UpToDate; and has a patent pending in sepsis diagnostics. S. E. has received grants from the NIAID/NIH, and Fogarty; has received personal fees from the American Statistical Association, Society for Clinical Trials, Drug Information Association, FDA, NIH, City of Hope, Huntington’s Study Group, Initiative on Methods, Measurement, and Pain Assessment in Clinical Trials (IMMPACT), Preclinical Pain Research Consortium for Investigating Safety and Efficacy (PPRECISE), Muscle Study Group, DeGruyter (Statistical Communications in Infectious Diseases), Takeda, Pfizer, Roche, Novartis, Merck, Achaogen, Auspex, Alcon, Chelsea, Mannkind, QRx Pharma, Genentech, Affymax, FzioMed, Amgen, GlaxoSmithKline, Sunovion, Boehringer-Ingelheim, Cubist, AstraZeneca, Teva, Repros, Dexcom, Zeiss, University of Rhode Island, New Jersey Medical School/Rutgers, University of Vermont, Osaka University, and the National Cerebral and Cardiovascular Center of Japan. E. P. and J. D. K. report no potential conflicts. The authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Chambers HF, Bartlett JG, Bonomo RA, et al. Antibacterial resistance leadership group: open for business. Clin Infect Dis 2014; 58:1571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caliendo AM, Gilbert DN, Ginocchio CC, et al. ; Infectious Diseases Society of America (IDSA) Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 2013; 57:S139–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mandrekar SJ, Dahlberg SE, Simon R. Improving clinical trial efficiency: thinking outside the box. Am Soc Clin Oncol Educ Book 2015; e141–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Westh H, Lisby G, Breysse F, et al. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin Microbiol Infect 2009; 15:544–51. [DOI] [PubMed] [Google Scholar]
- 5. Vincent JL, Brealey D, Libert N, et al. ; Rapid Diagnosis of Infections in the Critically Ill Team Rapid diagnosis of infection in the critically ill, a multicenter study of molecular detection in bloodstream infections, pneumonia, and sterile site infections. Crit Care Med 2015; 43:2283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis 2015; 60:892–9. [DOI] [PubMed] [Google Scholar]
- 7. Banerjee R, Ozenci V, Patel R, Individualized approaches are needed for optimized blood cultures. Clin Infect Dis 2016; 63:1332–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glasziou P, Irwig L, Deeks JJ. When should a new test become the current reference standard? Ann Intern Med 2008; 149:816–22. [DOI] [PubMed] [Google Scholar]
- 9. Evans SR, Pennello G, Pantoja-Galicia N, et al. ; Antibacterial Resistance Leadership Group Benefit-risk evaluation for diagnostics: a framework (BED-FRAME). Clin Infect Dis 2016; 63:812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bossuyt PM, Reitsma JB, Bruns DE, et al. ; STARD Group STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015; 351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bossuyt PM, Reitsma JB, Bruns DE, et al. ; Standards for Reporting of Diagnostic Accuracy The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. Ann Intern Med 2003; 138:W1–12. [DOI] [PubMed] [Google Scholar]
- 12. Lachs MS, Nachamkin I, Edelstein PH, Goldman J, Feinstein AR, Schwartz JS. Spectrum bias in the evaluation of diagnostic tests: lessons from the rapid dipstick test for urinary tract infection. Ann Intern Med 1992; 117:135–40. [DOI] [PubMed] [Google Scholar]