Abstract
Diverse plant pathogens export effector proteins to reprogram host cells. One of the most challenging goals in the molecular plant-microbe field is to functionally characterize the complex repertoires of effectors secreted by these pathogens. For bacterial pathogens, the predominant class of effectors is delivered to host cells by Type III secretion. For oomycetes, the predominant class of effectors is defined by a signal peptide that mediates secretion from the oomycete and a conserved RxLR motif. Downy mildew pathogens and Phytophthora species maintain hundreds of candidate RxLR effector genes in their genomes. Although no primary sequence similarity is evident between bacterial Type III effectors (T3Es) and oomycete RXLR effectors, some bacterial and oomycete effectors have convergently evolved to target the same host proteins. Such effectors might have evolved domains that are functionally similar but sequence-unrelated. We reasoned that alignment-free bioinformatics approaches could be useful to identify structural similarities between bacterial and oomycete effectors. To test this approach, we used partial least squares regression, alignment-free bioinformatics methods to identify effector proteins from the genome of the oomycete Hyaloperonospora arabidopsidis that are similar to the well-studied AvrE1 effector from Pseudomonas syringae. This approach identified five RxLR proteins with putative structural similarity to AvrE1. We focused on one, HaRxL23, because it is an experimentally validated effector and it is conserved between distantly related oomycetes. Several experiments indicate that HaRxL23 is functionally similar to AvrE1, including the ability to partially rescue an AvrE1 loss-of-function mutant. This study provides an example of how an alignment-free bioinformatics approach can identify functionally similar effector proteins in the absence of primary sequence similarity. This approach could be useful to identify effectors that have convergently evolved regardless of whether the shared host target is known.
Introduction
Successful plant pathogens must overcome a multilayered system of inducible immune responses that plants have evolved to defend against pathogen attack [1, 2]. The first layer of immunity, termed pattern-triggered immunity (PTI, [3]), is activated when conserved microbial molecules (pathogen-associated molecular patterns or PAMPs) are recognized by pattern recognition receptors (PRRs) in the host [4]. This recognition event triggers a complex immune response that includes the production of reactive oxygen species (ROS), deposition of callose and phenolic compounds in the cell wall, and extensive reprogramming of the transcriptome. Plant pathogenic bacteria, fungi, oomycetes, nematodes, and insects secrete effector proteins that suppress this immunity by directly targeting the PRRs or manipulating regulatory hubs in the PTI signaling network [5].
The second layer of plant immunity is referred to as effector-triggered immunity (ETI, [6]). ETI is based on direct or indirect recognition of pathogen effectors by corresponding plant immune surveillance proteins, known as resistance or “R” proteins [7, 8]. This recognition triggers a robust and effective suite of defense responses that typically includes the hypersensitive response (HR), featuring programmed cell death at the site of infection [9]. Pathogens, in turn, have shuffled their effector repertoire or evolved novel effectors to counteract ETI by either avoiding R protein recognition or suppressing downstream signaling events [10, 11].
In addition to suppressing host defense, bacterial and perhaps other classes of pathogens deploy effectors to control the extracellular plant environment. Availability of suitable nutrients and water are essential to pathogen growth and induction of disease. Numerous T3Es of the Transcriptional Activator-Like (TAL) family control nutrient availability through direct or indirect transcriptional induction of SWEET-family sugar transporters [12–15]. Other T3Es promote the conversion of the plant extracellular space into an aqueous environment [16]. “Water-soaking” is a classic symptom in plants infected with phytopathogenic bacteria that might benefit the bacteria in various ways, including to offset plant defenses associated with limited water availability during the HR [17].
The virulence-promoting activities of effectors are best understood in bacteria, owing to the focus on genetically tractable model strains such as Pseudomonas syringae strain DC3000 [18]. The predominant category of effectors in phytopathogenic bacteria are delivered to the interior of host cells via Type III secretion [19]. Significant progress has been made in elucidating the targets and establishing the functions and targets of bacterial T3Es [5]. In this study, we focused on the highly conserved and widespread AvrE-family of T3Es. AvrE-like proteins are encoded by Pseudomonas, Pectobacterium, Erwinia, Pantoea and Dickeya. AvrE1 from P. syringae belongs to a so-called redundant effector group (REG) comprised of AvrE1, HopM1, and HopR1. AvrE1 resides within the conserved effector locus (CEL) region that also harbors HopM1, HrpW and HopAA1. Conservation of the CEL, and especially an AvrE-family T3E, in diverse genera of bacteria [20] indicates the importance of both the locus and the AvrE-family T3Es.
T3Es of the AvrE-family display each of the general activities discussed above, including elicitation of ETI in nonhost plants and suppression of PTI and elicitation of water-soaking in host plants [21–27]. The AvrE-family effectors are large proteins with low sequence identity. For example, WtsE, the AvrE-family effector protein from the maize pathogen Pantoea stewartii pv. stewartii, shares 27.1% amino acid identity with AvrE1 from P. syringae [21]. AvrE-family effectors share WxxxE motif and a C-terminal endoplasmic reticulum membrane retention/retrieval signal (ER-MRS), which were required for activities of AvrE1 and WtsE in both host and nonhost plants [21]. The ability of both AvrE1 and WtsE to carry out their virulence activities in host plants depends on their interaction at the plasma membrane with specific regulatory subunits of the heterotrimeric protein phosphatase, PP2A [28, 29]. Host proteins whose phosphorylation status is affected by the interaction of the AvrE-family T3Es with PP2A remain to be identified. AvrE1 has recently been linked to water-soaking during infection of Arabidopsis [16], but it is unknown whether this function requires interaction with PP2A subunits.
Oomycetes are filamentous eukaryotic microorganisms, many species of which cause important diseases of crops. Oomycetes secrete effector proteins outside and inside host cells to promote infection and colonization [30–32]. Recently published genome sequences of species from some of the important oomycete genera including Phytophthora [33–35], Pythium [36], Albugo [37, 38] and Hyaloperonospora [39] revealed large repertoires of candidate effectors in these pathogens. A major class of effectors in Phytophthora species and downy mildew pathogens is named “RxLR”. These proteins are defined by an N-terminal signal peptide (SP), followed by the conserved motif, RXLR (where R is Arginine, X is any amino acid and L is leucine). The RXLR motif has been proposed to specify translocation to the interior of host cells [40, 41], but this is controversial [42, 43]. Another family of oomycete effector proteins are named “CRN” for the crinkling and necrosis phenotype that some family members induce when transiently expressed in leaves [44]. These proteins are defined by an N-terminal SP followed by a LXLFLAK motif, and appear to be translocated to the interior of host cells. Virulence-promoting functions have been ascribed to some members of this family [45].
A major area of current interest is to more thoroughly understand how RxLR proteins promote oomycete virulence [46]. This challenge is compounded by two factors: First, RxLR gene families range in size from 130 to over 550 amino acids [32]. Second, almost all of the RxLR effector genes encode novel proteins with no recognizable functional motifs [46]. Thus, new computational approaches for predicting RxLR protein function are needed [47]. A major premise of this study is that it would be useful to leverage information from the study of bacterial type III effectors to frame hypotheses about the function of RxLR effector proteins. Because effectors from bacteria and oomycetes share common, in planta targets [48, 49], convergent evolution may have given rise to effector domains that share no discernable primary sequence similarity, but share similar chemical/structural features that facilitate shared virulence-promoting functions. Such domains could be revealed through the use of recently developed bioinformatic tools that predict structural similarity in unrelated proteins. Partial least squares (PLS) is a regression method that is trained to learn from positives samples (example, AvrE), and negative samples (example, Non-AvrE) for future predictions. We reasoned that this approach could be used to identify oomycete effectors with putative structural similarity to well-studied bacterial effectors such as AvrE. Such effectors would represent potentially promising leads for downstream functional analysis.
As a first step towards testing the validity of this approach, we applied a PLS alignment-free bioinformatics method to identify oomycete RxLR effectors with structural similarity to bacterial AvrE-family type III effectors. We focused on the oomycete Hyaloperonospora arabidopsidis (Hpa), a naturally occurring downy mildew pathogen of Arabidopsis [50]. The genome of a reference Hpa isolate has been sequenced and 134 high-confidence RXLR genes have been annotated [39]. Several Hpa RXLR effectors have been functionally characterized [39, 46]. The PLS alignment-free methods identified five candidate RxLR proteins and four CRN proteins from the H. arabidopsidis genome that are similar to AvrE-family proteins. We predicted the structure of one of the identified proteins (HaRxL23 [51]) and AvrE1 from P. syringae using the iterative threading assembly refinement (I-TASSER [52–54]), and compared their structural similarity using the DaliLite [55] method. Based on this similarity, we hypothesized that AvrE1 and HaRxL23 may have convergently evolved to perturb the same host target(s). As an initial test of this hypothesis, we examined the activity of HaRxL23 and AvrE1 in a variety of biological assays and observed the two proteins to be functionally similar.
Materials and methods
Bioinformatics analysis
Data sets
Twelve AvrE proteins (positives) from the study by Ham and associates [21] and non-AvrE proteins (negatives) were downloaded from National Center for Biotechnology Information (NCBI) websites (http://www.ncbi.nlm.nih.gov/), and were used for training the PLS methods. The H. arabidopsidis RxLR candidate gene sequences used in this study were as described in Baxter et al. [39]. Accession numbers for all sequences are listed in S1–S3 Tables.
Amino acid composition
From each protein sequence, frequencies of 20 amino acids were calculated. In this study, amino acid composition was used as descriptors for a PLS classifier (PLS-AA).
Dipeptide composition
Dipeptide composition represents all 400 frequencies of consecutive amino acid pairs in a protein sequence and corresponds to a 400 (20 X 20) feature vector. It can encapsulate information on composition of amino acids as well as their local order. We used dipeptide composition as descriptors for a PLS classifier (PLS-DIP).
Physicochemical properties of amino acids
In Opiyo and Moriyama [56], we developed five descriptors (the first five principal component scores [PC1- PC5]) using the principal component analysis (PCA) of 12 physicochemical properties of amino acids (mass, volume, surface area, hydrophilicity, hydrophobicity, isoelectric point, transfer of energy solvent to water, refractivity, non-polar surface area, and frequencies of alpha-helix, beta-sheet, and reverse turn). The five descriptors (PC1-PC5) were used in this study.
Auto/Cross covariance transformation
Auto/cross covariance (ACC) transformation method discussed in [56] was used to transform each amino acid sequence using the five descriptors (PC1-PC5). ACC with the maximum lag of 30 residues yielded 775 descriptors for each sequence. The calculation of ACC was performed using the R implementation (version 2.12.0; http://www.R-project.org; 2010).
Partial least squares
As described in detail previously [57] partial least squares (PLS) is a regression used for predictions. PLS is a projection method similar to principal component analysis (PCA) where the independent variables, represented as the matrix X, are projected onto a low dimensional space. PLS uses both independent variables X (sequence descriptors such as amino acid composition) and dependent variables Y (positive or negative label) [58]. PLS using descriptors transformed by ACC (PLS-ACC) was used in [56]. PLS discriminant analysis is performed to separate groups of observations. It consists of a classical PLS where the response variable is a categorical one (replaced by the set of dummy variables describing the categories, e.g., 0 and 1) expressing the class membership of the samples. In this study, each of a training sample, a response variable was assigned 1 for the positive sample (AvrE) and 0 for a negative sample (non-AvrE). The group membership, AvrE or non-AvrE of a new sequence was predicted based on descriptors and y-value. Predicted y-value closer to 1 was considered to be AvrE candidate and closer to 0 considered to be non-AvrE candidate. PLS analysis was performed using an R implementation, the PLS package developed by Wehrens and Mevik (version 1.2.1 [59]).
Performance analysis
Cross-validation analysis (leave-one-out) was performed for all the 24 sequences used for training the methods. One sequence in the training dataset was left out and the learning algorithm was trained on the rest of the sequences. The trained model was used to predict the class (AvrE or non-AvrE) of the proteins left out of the training. For the 24 sequences, the process was repeated 24 times leaving each of the 24 sequences out and creating a model from the remaining 23 sequences.
Predictions were grouped as follows:
True Positives (TP): the number of actual AvrE proteins (AvrEs) that were predicted as AvrEs.
False Positive (FP): the number of actual non-AvrEs that were predicted as AvrEs.
True Negative (TN): the number of actual non-AvrEs that were predicted as non-AvrEs.
False Negative (FN): the number of actual AvrEs that were predicted as non-AvrEs
Minimum error point
The minimum error point [60] was used to determine threshold values of PLS methods. The sequences are ranked based on the values. The threshold value where the minimum number of errors (FN + FP) occurs is the minimum error point (MEP) and the number of false positives and false negatives are assessed at this point. The minimum error point reports the best-case accuracy of a method. The minimum error points for PLS-AA, PLS-DIP, and PLS-ACC were 0.94, 0.96, and 0.94, respectively. The upper cut-off point for all methods was set at 1.00 to further reduce the number of false positives. To be selected as a candidate, a protein has to be identified by all the three methods (PLS-AA, PLS-DIP, and PLS-ACC) as positive.
Goodness of prediction of PLS methods
The goodness of prediction, q2 Eq 1, describes how well the method can predict a data.
| (1) |
where Ya is a sample from a training data, Yp is the omitted sample, Ym is the mean of the samples of the training data, and N is the total number of samples in the training data. The value of q2 > 0.50 is considered good. In this study, the leave-one-out cross-validation procedure was used for the q2 calculation. Detailed results of PLS analyses, and the H. arabidopsidis proteins identified by each method, are given in Tables 1 and 2 and S4 Table for PLS-AA, PLS-DIP, and PLS-ACC, respectively.
Table 1. The number of PLS components and the predictive abilities of PLS-AA, PLS-DIP, and PLS-ACC, respectively from the leave-one-out cross validation procedures.
| Methods | Number of PLS components | q2 |
|---|---|---|
| PLS-AA | 4 | 0.72 |
| PLS-DIP | 3 | 0.67 |
| PLS-ACC | 4 | 0.78 |
Table 2. Nine protein candidates identified from Hyaloperonospora arabidopsidis genome by the three methods.
| Accession number | PLS-AA | PLS-DIP | PLS-ACC |
|---|---|---|---|
| HaRxL23 | 0.98 | 0.99 | 0.95 |
| HaRxL33 | 0.99 | 1.00 | 1.00 |
| HaRxL71 | 1.00 | 0.98 | 0.94 |
| HaRxL94 | 0.94 | 0.96 | 0.94 |
| HaRxL120 | 0.94 | 0.97 | 1.01 |
| HaCRN9 | 0.95 | 0.99 | 1.00 |
| HaCRN10 | 0.97 | 1.00 | 1.00 |
| HaCRN12 | 0.98 | 0.97 | 0.95 |
| HaCRN14 | 0.94 | 0.96 | 0.98 |
Structural prediction of AvrE1 and the nine predicted proteins by I-TASSER, and structural comparison by DaliLite
To predict the structures of AvrE1 and nine predicted protein candidates from H. arabidopsidis, we selected the I-TASSER server because it predicts secondary and tertiary structures, and ligand-binding sites [52–54, 61]. After the predictions, the structural similarity of AvrE1 and the nine protein candidates were compared using DaliLite [62] (S5 Table). The structures of the DspA/E propeller-domain amino acids and the AvrE1 1513–1578 region were predicted using I-TASSER (structural templates are listed in S6 Table). The predicted structures of DspA/E and AvrE1 region were aligned using TM-align [63].
Experimental comparisons of HaRxL23 and AvrE1
Construction of expression plasmids
HaRxL23 was amplified from genomic DNA extracted from Arabidopsis Oy-1 plants, infected with Hpa isolate Emoy2, using proofreading polymerase (Pfu, Invitrogen). Forward and reverse primers were designed to amplify from the signal peptide cleavage site (HaRxL23 NOSP, S7 Table) with or without the stop codon (HaRxL23 S and HaRxL23 NS, respectively, S7 Table) depending on the type of fusion. For cloning into Gateway destination vectors, the sequence CACC was added at the 5’ end of the forward primer and PCR was performed using the genomic DNA as template. PCR products were gel purified (Qiagen) and recombined into pENTR-D-TOPO Gateway entry vector following the manufacture’s protocol (Invitrogen). This step was followed by transformation into Escherichia coli DH5α competent cells. Kanamycin resistant colonies were selected on agarose plates followed by colony PCR with plasmid-specific M13 forward and reverse primers. Colonies having the correct size were selected for plasmid purification and confirmed by sequencing. The pENTR clone generated was then used to create Gateway expression plasmids using LR recombination (Invitrogen).
For Pseudomonas-mediated transient studies, the HaRxL23 gene was shuttled from pENTR into pEDV6 by LR recombination. pEDV6 contains the AvrRPS4 promoter and leader sequence that directs the fusion protein through the Type III secretion system [64]. The EDV constructs with our effectors were transformed into Pseudomonas phaseolicola strains by standard tri-parental mating using E. coli pRK600 as a helper strain. The Pph 3121, Pph AvrE1, ΔavrE1 mutant (CUCPB5374) and ΔavrE1(avrE1) (pCPP5246) strains have been previously described [65, 66]. All gene fusions were confirmed by sequencing.
Plant materials and growth conditions
Arabidopsis and tomato (Lycopersicon esculentum cv. Moneymaker) plants were grown in Sunshine Pro-mix soil mixture number one. For experiments involving inoculation with Pseudomonas spp., Arabidopsis was grown in controlled growth chambers under short day cycles (8h/16h light/dark and 150–200 μE/m2s) at 22°C and 60% relative humidity. For all other experiments, Arabidopsis and tomato were grown under long day cycles (16h/8h light/dark at 90–100 μE/m2s) at 22°C and 60% relative humidity.
Assays for plant cell death, bacterial virulence and callose suppression in Arabidopsis
For assays involving Pseudomonas spp., Arabidopsis Col-0 plants were syringe-infiltrated with 1x105 colony-forming units (cfu)/ml (virulence assays) or 1x108 cfu/ml (cell death and callose suppression assays) bacterial solution in 10mM MgSO4. For cell death assays, a total of 6 plants, 3 leaves each, were infiltrated and visual scoring was performed 16–20 hours later. For bacterial growth assays, leaf discs were scored at zero and three days post infiltration (dpi), surface sterilized with 70% ethanol, and homogenized using a mini-bead beater (Biospec products). Serial dilutions were performed to count colony-forming units. For each sample, three leaf discs were pooled three times per data point. For callose suppression assays, whole leaves were harvested 16 hours post infiltration (hpi), treated with alcoholic lactophenol and stained with 0.01% (w/v) Aniline blue stain in K2HPO4 buffer as described previously [64]. Stained leaves were mounted on glass slides using 50% glycerol and imaged with a Zeiss Axio Imager.M1 using the filter settings for DAPI. Quantification of callose spots was performed using the Autospots software [67]. Statistical analyses for growth curves were performed on the means of log-transformed data using Student’s t-test (*p < 0.01, **p <0.001).
Bacterial lesion assay in tomato Moneymaker plants
For the bacterial lesion assay, 4–5 week old Solanum lycopersicum cv. Moneymaker (tomato) plants were dip-inoculated for 30 seconds with 1x108 cfu/ml bacterial solution in 10mM MgSO4 containing 0.02% Silwet. A total of 3–4 plants were used for each treatment. Disease symptoms on leaves in the form of small, brown, necrotic lesions were monitored for a total of six days. At five days after inoculation, the number of well-developed lesions (≥0.25 mm2) per leaf was quantified.
Results
Mining Hyaloperonospora arabidopsidis candidate RXLR proteins using three PLS methods
In this study, we trained three PLS methods (PLS-AA, PLS-DIP, and PLS-ACC) using AvrE family proteins to identify similar RxLR effector proteins from H. arabidopsidis RXLR proteins. The scores ranged between 0 and 1, with the cut-off point of 0.5 for AvrE candidates, and less than 0.5 for Non-AvrE candidates. The higher the score, the higher the confidence that the protein is an AvrE candidate. A total of 20 HaRxL proteins, five HaCRN proteins, and four non-effector proteins were identified by at least one of the three PLS methods (S4 Table). Of these, five Hpa RxLR protein candidates and four CRN proteins were identified by all three PLS methods with scores > 0.9 (Table 2). Of these nine candidates, only HaRxLR23 has been validated as a bona fide effector gene [51].
Structural predictions of AvrE1 and the nine protein candidates by I-TASSER and comparison with DaliLite
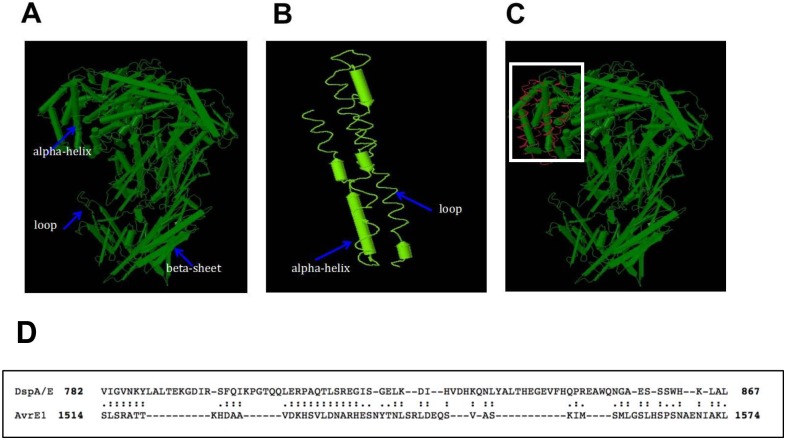
We obtained the DaliLite Z-scores from pairwise comparison of the predicted structures of AvrE1 and the nine protein candidates. Five of the nine (HaRxL23, HaRxL33, HaRxL71, HaRxL94, and HaCRN10) had structural similarity with AvrE1. The predicted structures of AvrE1 and HaRxL23 are presented in Fig 1. AvrE1 is a very large protein of 1795 amino acids and HaRxL23 is a small protein with 142 amino acids. Despite this disparity in size and the fact that the proteins share no significant sequence identity, it can be seen that HaRxL23 overlaps structurally with amino acids 1513 to 1578 of AvrE1 (Fig 1). The HaRxL23 ortholog from Phytophthora sojae, PsAvh73 [51], overlaps structurally with the same domain of AvrE1 (S1 Fig). This domain of AvrE1 does not have the WxxxE and ER-MRS motifs, but corresponds to the double beta propeller domain of another AvrE-family T3E, DspA/E from Erwinia amylovora, for which mutational analysis has indicated functional importance [68].
Fig 1. Structural predictions of HaRxL23 and AvrE1.
(A) Predicted structure of AvrE1 by I-TASSER (B) Predicted structure of HaRxL23 by I-TASSER (C) Superimposed structure of HaRxL23 on AvrE1 by DaliLite. Structure of HaRxL23 is shown in red. (D) Structural alignment of DspA/E propeller domain and AvrE1 region using TM-aligned. ":" denotes aligned residue pairs of RMSD < 5.0 Angstroms, "." denotes other aligned residues.
HaRxL23 and AvrE1 induce cell death in young Arabidopsis plants when delivered by Pseudomonas phaseolicola (Pph) 3121
Because HaRxL23 has been confirmed as an effector [51] and also had the highest Z-score of 6.7 (S5 Table), we focused on functional comparisons of this protein with AvrE1. Previous studies with AvrE1 of PtoDC3000 and its ortholog in Pantoea stewartii, WstE, revealed that both proteins are capable of inducing a cell death response in Nicotiana tabacum and tomato plants when secreted by bacteria that are inoculated at a relatively high density [22, 24, 66]. As a first test of functional similarity between HaRxL23 and Pst AvrE1, we tested whether HaRxL23 was capable of inducing cell death in Arabidopsis when delivered from the bean pathogen, P. syringae pv. phaseolicola NPS3121 (Pph) at a high inoculum dose. We used Pph for our experiments because it is non-pathogenic on Arabidopsis, has a functional TTSS, and does not elicit cell death or any other symptoms. Thus, the endogenous AvrE homolog in Pph NPS3121 does not induce cell death in Arabidopsis. We engineered Pph to use the “effector detector vector (EDV)” to deliver HaRxL23 via the type III secretion system (TTSS) to the interior of plant cells [64]. The HaRxL23 coding region, beginning downstream of the cleavage site of the signal peptide, was cloned as a translational fusion to the promoter and N-terminal region of the P. syringae effector AvrRps4. Transgenic Pph strains containing this plasmid could translate the fusion protein and secrete it into plant cells via the Type III translocon, enabling the effects of HaRxL23 inside plant cells to be assayed experimentally. This strain was used in comparison with a previously described Pph3121 strain that delivers PstDC3000 AvrE1 from a plasmid vector [21].
In accordance with previous results, a typical HR cell death symptom of leaf collapse was observed when the P. syringae effector AvrRpt2 was delivered into the Arabidopsis ecotype Col-0 as a positive control (Fig 2) [64]. Delivery of either HaRxL23 or AvrE1 triggered leaf collapse symptoms comparable to AvrRpt2 in three-week old Arabidopsis plants (Fig 2). This response was not seen in five-week old plants (Fig 2). This experiment demonstrates that, when delivered by Pph, both AvrE1 and HaRxL23 can elicit cell death specifically in young Arabidopsis plants.
Fig 2. Both HaRxL23 and AvrE1 induce cell death in young Arabidopsis plants when delivered by Pseudomonas phaseolicola.
Images from the cell death test in leaves of Arabidopsis wild type plants (Col-0). Five-week old plants and three-week old plants were infiltrated with suspensions of P. phaseolicola expressing effectors (1 X 108 cfu/ml). Cell death was visually monitored over a period of 20 hours after inoculation. This experiment was repeated three times with similar results.
HaRxL23 and AvrE1 suppress callose deposition elicited in Arabidopsis by Pph
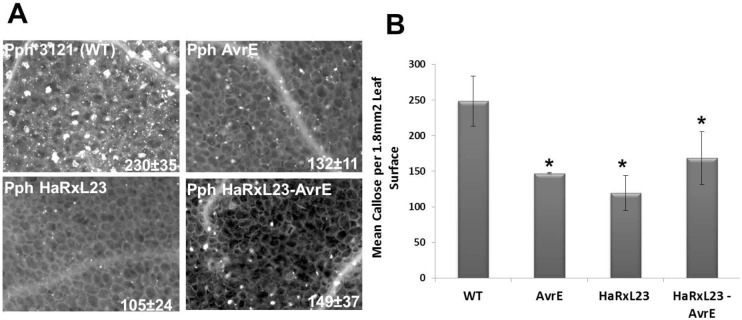
Another advantage of using Pph is that it also elicits robust defenses in Arabidopsis, including deposition of callose and accumulation of the defense marker protein Pathogenesis Related-1 (PR-1). Thus, the system can be used to study the ability of a heterologous effector to suppress the Pph-induced defenses. Suppression of cell wall-based defenses (e.g., callose deposition) is considered an important function of both bacterial and oomycete effectors. Callose is a polymer of β-1, 3 glucans that is deposited between the cell wall and cell membrane near the invading pathogen [69]. Such foci are easily visualized by fluorescence microscopy and can be quantified as a readout of host defense [64]. We compared the ability of HaRxL23 and AvrE1 to suppress callose deposition when delivered from Pph, by inoculating four-week old plants at a titer that does not induce the cell death response described in Fig 2. As expected, Arabidopsis Col-0 plants exhibit extensive callose deposition when syringe-infiltrated with wild type Pph 3121 (Fig 3A). Contrastingly, a reduction of close to 50% in callose deposits is observed in plants that were syringe-infiltrated with strains of Pph expressing either HaRxL23 or AvrE1 (Fig 3A and 3B). It is interesting to note that this suppression is not enhanced when a double transformant, Pph HaRxL23 + AvrE1, is used (Fig 3A and 3B). This lack of additivity indicates that both effectors likely interfere with the same regulatory pathway in the host.
Fig 3. AvrE and HaRxL23 suppress callose deposition in Arabidopsis when delivered by P. phaseolicola via the effector detector vector system.
(A) Four-week old Arabidopsis Col-0 plants were infiltrated with 5 X 107 cfu/ml Pph strains expressing either AvrE1or HaRxL23 individually, or a double transformant expressing HaRxL23 and AvrE1 together. Callose deposits were visualized by staining with aniline blue and (B) quantified using the Autospots software program. Four pictures per leaf from six leaves were analyzed per treatment. P-value * < 0.01; t-test comparisons representing significant differences with Col-0. Error bars represent Standard Error of six independent leaf samples tested at the same time. This experiment was repeated three times with similar results.
Neither HaRxL23 nor AvrE1 enhance Pph 3121 virulence
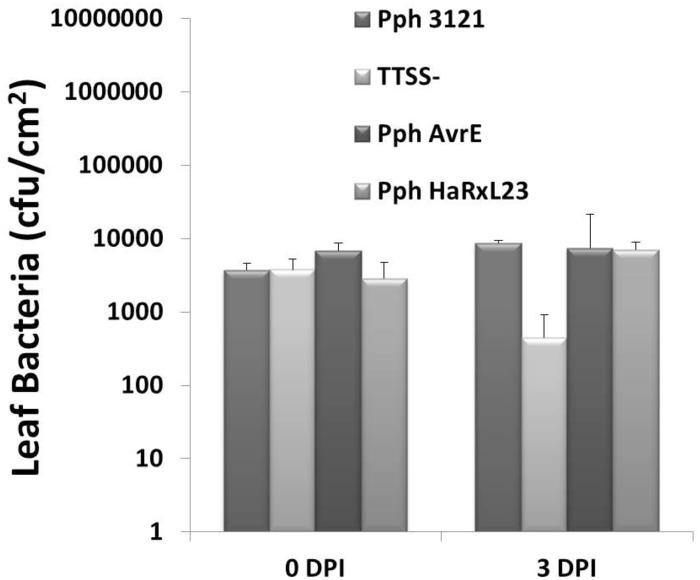
The assay in Fig 2 for macroscopic plant cell death was carried out with a high dose of bacterial inoculum (5 X 107 cfu/ml). To test whether HaRxL23 enhances or suppresses bacterial growth in planta, we infiltrated Arabidopsis leaves with a low dose of bacteria (1 X 105 cfu/ml) and then measured bacterial growth at three days after inoculation. We compared the growth, in Col-0, of Pph expressing HaRxL23 and AvrE1 to Pph with an empty vector control (Pph3121) and a non-pathogenic mutant deficient in Type III secretion (TTSS-). At three days post-infiltration, there was no enhancement of bacterial growth for either Pph HaRxL23 or Pph AvrE1 compared to Pph 3121 EV (Fig 4) in young Arabidopsis Col-0 plants. Thus, despite the ability of HaRxL23 and AvrE1 to suppress callose elicited by Pph 3121, there is no net enhancement of Pph virulence in Arabidopsis by either effector.
Fig 4. Pph 3121 virulence is not enhanced by transgenic HaRxL23 or AvrE.
Plants were infiltrated with a bacterial suspension of 1 X 105 cfu/ml. Bacterial populations were determined at day 0 and day 3 after inoculation. Error bars indicate Standard Error of six independent leaf samples tested at the same time. The experiment was repeated three times with similar results.
HaRxL23 can rescue the reduced virulence phenotype of the PtoDC3000ΔavrE1 strain in tomato Moneymaker
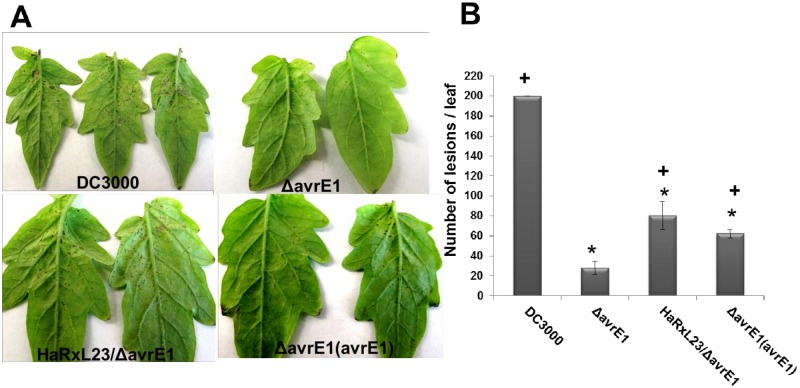
The most stringent genetic test for functional equivalence between HaRxL23 and AvrE1 is to assay whether HaRxL23 can rescue an AvrE loss-of-function mutant. However, this experiment is complicated by functional redundancy between AvrE and other Pst effectors, such that a ΔavrE1 mutant does not display a phenotype under most previously tested conditions. The only phenotype of an AvrE1 mutant was reported by Badel et al., who observed that P. syringae pv. tomato DC3000 ΔavrE1 deletion mutant was impaired in the formation of bacterial speck lesions in tomato (Solanum lycopersicum cv. Moneymaker) plants [66]. Thus, we hypothesized that if HaRxL23 is functionally similar to AvrE1, then HaRxL23 would be able to rescue the reduced bacterial speck lesion phenotype of the avrE1 mutant in tomato. Four-week old tomato Moneymaker plants were dip-inoculated with bacterial suspensions containing wild-type DC3000, the ΔavrE1 mutant, the ΔavrE1 mutant carrying HaRxL23, and the complemented ΔavrE1(avrE1) strain (Fig 5). Bacterial speck lesions ≥0.25 mm2 were quantified from infected leaves. Like AvrE1, HaRxL23 was able to significantly increase the number of lesions produced by the ΔavrE1 mutant strain (Fig 5). Interestingly, the virulence of the strain expressing HaRxL23 was slightly stronger than that observed in the complemented strain of ΔavrE1 [66], which is known to have a partial rescue phenotype, relative to the wild-type strain, presumably due to inefficient expression of the plasmid-encoded gene.
Fig 5. HaRxL23 rescues the reduced lesion phenotype of the ΔavrE1 strain in tomato Moneymaker plants.
(A) Disease symptoms (lesion production) on tomato cv. Moneymaker plants 8 days after dip inoculation of the indicated strains of Pto DC3000 at 1x108 cfu/ml bacterial culture. (B) Number of lesions (≥0.25mm2) per whole leaf appearing on plants 8 days after dipping inoculation with the respective bacterial strains. Values indicate mean and error bars indicate standard error on at least five whole leaves assayed for each treatment. P-value * < 0.01; t-test comparisons representing significant differences with DC3000; + < 0.01; t-test comparisons representing significant differences with ΔavrE1. This experiment was repeated three times with similar results.
Discussion
Over the last three decades, it has become clear that secreted effector proteins are critically important virulence or avirulence factors for essentially every category of plant pathogen and pest. Studies of bacterial effectors have provided seminal insights into the molecular mechanisms that underpin pathogen virulence and host plant resistance [2]. Moreover, bacterial effectors have been used as molecular probes to decipher unknown aspects of plant biology [70, 71]. This is well-exemplified by studies of AvrE-family T3Es, which have uncovered previously unknown plant immune regulators.
Similar studies of effectors from eukaryotic pathogens are progressing quickly, thanks in part to the falling cost of sequencing, enabling draft genomes and transcriptomes to be assembled for many fungi and oomycetes. These resources have been combined with computational tools to enable prediction of candidate effector genes [47]. This is well-exemplified by the use of Hidden Markov Models to identify RxLR genes in oomycete genomes [72]. However, such computational approaches typically yield hundreds or even thousands of candidate genes, particularly for fungal genomes in which no clear consensus motifs, other than a signal peptide, have emerged. Moreover, the primary sequences of effector proteins typically provide no clues to their biochemical functions or molecular targets in plant cells. Thus, a need exists for additional computation tools to enable prediction of molecular functions and prioritization of effector genes for time-consuming molecular studies [47].
With this bottleneck in mind, we connected two recent but disparate advances: The first is emergence of new computational methods for predicting structural similarity between proteins, in the absence of strong similarity in primary sequence. The second is the emerging understanding, from model plant pathosystems, that effectors from divergent pathogen lineages (e.g., bacteria and oomycetes) convergently evolve to target the same points of vulnerability in plant regulatory networks. Most often these targets comprise highly connected regulatory hubs, perturbation of which would have major effects on plant cell signaling [73]. We reasoned that this convergence provides an opportunity to exploit alignment-free methods to identify proteins that target the same protein via convergent evolution of protein interaction domains. Such convergence might not be evident in primary sequence but could be revealed by more sophisticated methods that probe physiochemical properties of amino acids such as mass, volume, surface area, hydrophilicity, hydrophobicity, isoelectric point, transfer of energy solvent to water, refractivity, non-polar surface area, and frequencies of alpha-helix, beta-sheet, and reverse turn. In this way, we could utilize information from bacterial effectors to accelerate understanding of effectors from organisms that contain more complex effectoromes and are less tractable from an experimental standpoint.
We approached this problem by testing whether a bioinformatics-driven approach could identify effector proteins from H. arabidopsidis that are similar to the well-studied AvrE1 protein. An alignment-free, PLS method was used because alignment-based methods, such as profile HMMs, become unreliable when sequence similarity drops below 40% [74]. Some families of effector proteins, such as the AvrE-family, are highly divergent and have low sequence identity even though they likely still share similar structures, biochemical properties, and functions. In such cases, obtaining reliable alignments among these protein sequences is extremely difficult, and alignment-based methods such as BLAST, PSI-BLAST (position specific iterative BLAST [75]), and profile HMMs would fail to identify these proteins from databases.
Using PLS alignment-free methods [56], we identified nine candidate effector genes from H. arabidopsidis predicted to be similar to AvrE-family type III effector proteins. The structures of these nine proteins were predicted using the I-TASSER server and compared with the AvrE1 predicted protein structure using DaliLite. Five of the nine Hpa proteins were predicted to share structural similarity with AvrE1. HaRxL23 was selected for further study because of its high Z-score (Fig 1, Table 1) and because it has been experimentally validated as a bona fide effector [51]. Moreover, we were intrigued that HaRxL23 is one of the most evolutionarily conserved RXLR effectors in the Hpa genome, it is one of only three Hpa effector candidates for which homologous genes are recognizable at syntenic loci in Phytophthora genomes, and it contributes to virulence in evolutionarily divergent species [39, 51, 66]. Similarly, AvrE-family effectors are among the most conserved bacterial Type III effectors, are broadly distributed among distinct genera of phytopathogenic genera, and contribute to the virulence of these diverse bacteria in divergent host plants [21–27]. Thus, we hypothesize that AvrE1 and HaRxL23 promote virulence by targeting evolutionarily conserved protein(s) in divergent plant species.
As a first step towards testing this hypothesis, we conducted genetic experiments to test for functional similarity between HaRxL23 and AvrE1. Importantly, we demonstrated that HaRxL23 could rescue the reduced bacterial speck lesion phenotype of the avrE1 mutant, demonstrating that HaRxL23 can genetically complement the avrE1 mutant. We also determined that, in older Arabidopsis plants, both effectors were successful in suppressing Pph 3121-induced callose deposition when delivered by Pph. PTI readouts, such as the callose deposition suppressed by AvrE1 and HopM1, are both SA-dependent [24] and SA-independent [76, 77]. It is possible that HaRxL23 targets similar conserved SA-mediated immunity pathways in the host as a possible virulence mechanism. Accordingly, Arabidopsis plants expressing transgenic HaRxL23 display reduced induction of the SA-responsive gene PR-1 during infection by Hpa [51].
Another set of experiments was directed towards understanding whether HaRxL23 and AvrE1 could induce cell death when delivered by Pph in Arabidopsis. It has been previously shown that AvrE1 induces cell death when delivered from a high dose of bacteria infiltrated into leaves of susceptible tobacco (Nicotiana tabacum) or tomato plants [66]. We delivered HaRxL23 and AvrE1 from Pseudomonas phaseolicola (Pph) 3121 and found that each induced cell death in the leaves of young Arabidopsis plants. Previous studies had hypothesized a strong correlation between suppression of basal defense and R protein-independent cell-death promotion by bacterial effectors AvrE1 and HopM1 [66], where both these effectors restored basal resistance suppression to the ΔCEL mutant and produced a delayed necrosis when transiently expressed in the host. The authors proposed that the recognition event triggered by AvrE1 could be explained by a mechanism through which the plant perceived strong suppression of PTI by high levels of AvrE1 and responded by triggering cell death. More recently, AvrE1 and HopM1 have been shown to promote water-soaking in the leaf, thereby converting the apoplast into a aqueous environment [16]. The promotion of water-soaking by HopM1 requires the Arabidopsis MIN7 protein that regulates vesicle trafficking and is targeted by HopM1. The mechanism through which AvrE1 promotes water soaking is not known. Water-soaking is thought to benefit bacteria by interfering with apoplastic defenses, facilitating nutrient flow, and/or promoting bacterial dissemination outside of the leaf. Considering that the vast majority of oomycete (i.e., hyphae) biomass resides in the apoplast during colonization, oomycetes could benefit similarly from an aqueous environment in the apoplast. Thus, it will be of interest to examine whether the virulence-promoting activities of HaRxL23 are linked to the water status of the apoplast.
As mentioned above, an important, emerging concept in effector biology is that effectors from pathogens as distantly related as bacteria, fungi, oomycetes, and nematodes can perturb the same target proteins/pathways in the host. One example of this is provided by the cysteine-rich protease RCR3 protein from tomato. RCR3 is targeted by effectors from three unrelated phytopathogens: Avr2 from the fungus Cladosporium fulvum, EPIC1 & EPIC2B from the oomycete Phytophthora infestans and VAP1 from the root nematode Globodera rostochiensis [78, 79]. Thus, RCR3 can be thought of as an Achilles’ Heel that fungi, oomycetes and nematodes have evolved independently to target as part of their virulence program.
In a similar vein, but on a much broader scale, Mukhtar et al. [49] undertook a yeast two-hybrid protein interaction study between a fraction of the Arabidopsis proteome and effectors from bacteria and Hpa. One goal of this screen was to identify effector targets. They identified a set of 18 “core” Arabidopsis proteins that were targeted by effectors from bacteria and Hpa, and presumably represent pathways that are important for the interaction with both pathogens [49]. A subsequent study extended this approach to identify Arabidopsis proteins convergently targeted by effectors from pathogens representing three kingdoms of life: The bacterium P. syringae, the oomycete Hpa, and Golovinomyces orontii, a powdery mildew fungus of Arabidopsis [48].
Based on these precedents and upon the data in this study, we propose that AvrE1 and HaRxL23 target the same (or similar) protein(s) in Arabidopsis. Considering that both effectors are amongst the most highly conserved in their respective pathogen lineages, their target(s) are likely to hold broad importance for plant-bacteria and plant-oomycete interactions. The next step will be directed and unbiased protein interaction assays to identify HaRxL23’s host protein target(s) and thereby directly test the hypothesis that HaRxL23 targets the same Arabidopsis proteins as do AvrE-family T3Es. Regulatory subunits of PP2A are one example of a possible shared target between AvrE1 and HaRxL23.
In a broader context, this study has provided an example of how alignment-free approaches can be applied to functional genomic characterization of effector protein, towards the ultimate goals of leveraging insight from model plant pathogens and identifying effectors that have convergently evolved to manipulate high-value host protein targets. The approaches in this paper are not restricted to any particular lineage of pathogen and could be applied to any species for which a set of effector genes has been delineated. Judicious application of these tools provides another approach towards the important and challenging goal of reducing a long list of effector protein candidates to a tractable list of high value leads and to illuminating shared activities and host targets of effectors from divergent pathogens.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Science Foundation Division of Integrative Organismal Systems Grant no. IOS-1353366 (JM) and by Agriculture and Food Research Initiative Competitive Grant nos. 2009-03008 (JM) and 2016-67013-24727 (DM) from the USDA National Institute of Food and Agriculture. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cook DE, Mesarich CH, Thomma BP. Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol. 2015;53:541–63. doi: 10.1146/annurev-phyto-080614-120114 . [DOI] [PubMed] [Google Scholar]
- 2.Bent AF, Mackey D. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol. 2007;45:399–436. Epub 2007/05/18. doi: 10.1146/annurev.phyto.45.062806.094427 . [DOI] [PubMed] [Google Scholar]
- 3.Katagiri F, Tsuda K. Understanding the plant immune system. Mol Plant Microbe Interact. 2010;23(12):1531–6. Epub 2010/07/27. doi: 10.1094/MPMI-04-10-0099 . [DOI] [PubMed] [Google Scholar]
- 4.Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Molecular Cell. 2014;54(2):263–72. doi: 10.1016/j.molcel.2014.03.028 . [DOI] [PubMed] [Google Scholar]
- 5.Toruno TY, Stergiopoulos I, Coaker G. Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol. 2016;54:419–41. doi: 10.1146/annurev-phyto-080615-100204 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–9. doi: 10.1038/nature05286 . [DOI] [PubMed] [Google Scholar]
- 7.Van der Biezen EA, Jones JDG. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;12:454–6. [DOI] [PubMed] [Google Scholar]
- 8.van der Hoorn RA, Kamoun S. From Guard to Decoy: a new model for perception of plant pathogen effectors. The Plant Cell. 2008;20(8):2009–17. Epub 2008/08/30. doi: 10.1105/tpc.108.060194 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012 . [DOI] [PubMed] [Google Scholar]
- 10.Khan M, Subramaniam R, Desveaux D. Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Current Opinion in Microbiology. 2016;29:49–55. doi: 10.1016/j.mib.2015.10.006 . [DOI] [PubMed] [Google Scholar]
- 11.Hein I, Gilroy EM, Armstrong MR, Birch PRJ. The zig-zag-zig in oomycete–plant interactions. Molecular Plant Pathology. 2009;10(4):547–62. doi: 10.1111/j.1364-3703.2009.00547.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468(7323):527–32. doi: 10.1038/nature09606 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohn M, Bart RS, Shybut M, Dahlbeck D, Gomez M, Morbitzer R, et al. Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector-mediated induction of a SWEET sugar transporter in cassava. Mol Plant Microbe Interact. 2014;27(11):1186–98. doi: 10.1094/MPMI-06-14-0161-R . [DOI] [PubMed] [Google Scholar]
- 14.Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013;200(3):808–19. doi: 10.1111/nph.12411 . [DOI] [PubMed] [Google Scholar]
- 15.Verdier V, Triplett LR, Hummel AW, Corral R, Cernadas RA, Schmidt CL, et al. Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol. 2012;196(4):1197–207. doi: 10.1111/j.1469-8137.2012.04367.x . [DOI] [PubMed] [Google Scholar]
- 16.Xin X-F, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, et al. Bacteria establish an aqueous living space in plants crucial for virulence. Nature. 2016;539:524 doi: 10.1038/nature20166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright CA, Beattie GA. Pseudomonas syringae pv. tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101(9):3269–74. doi: 10.1073/pnas.0400461101 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xin XF, He SY. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol. 2013;51:473–98. doi: 10.1146/annurev-phyto-082712-102321 . [DOI] [PubMed] [Google Scholar]
- 19.da Cunha L, Sreerekha MV, Mackey D. Defense suppression by virulence effectors of bacterial phytopathogens. Curr Opin Plant Biol. 2007;10(4):349–57. doi: 10.1016/j.pbi.2007.04.018 . [DOI] [PubMed] [Google Scholar]
- 20.Alfano JR, Charkowski AO, Deng WL, Badel JL, Petnicki-Ocwieja T, van Dijk K, et al. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci U S A. 2000;97(9):4856–61. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ham JH, Majerczak DR, Nomura K, Mecey C, Uribe F, He SY, et al. Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE require WxxxE motifs. Mol Plant Microbe Interact. 2009;22(6):703–12. Epub 2009/05/19. doi: 10.1094/MPMI-22-6-0703 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ham JH, Majerczak D, Ewert S, Sreerekha MV, Mackey D, Coplin D. WtsE, an AvrE-family type III effector protein of Pantoea stewartii subsp. stewartii, causes cell death in non-host plants. Mol Plant Pathol. 2008;9(5):633–43. Epub 2008/11/21. doi: 10.1111/j.1364-3703.2008.00489.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ham JH, Majerczak DR, Arroyo-Rodriguez AS, Mackey DM, Coplin DL. WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes disease-associated cell death in corn and requires a chaperone protein for stability. Mol Plant Microbe Interact. 2006;19(10):1092–102. Epub 2006/10/07. doi: 10.1094/MPMI-19-1092 . [DOI] [PubMed] [Google Scholar]
- 24.DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci U S A. 2004;101(26):9927–32. doi: 10.1073/pnas.0401601101 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogdanove AJ, Bauer DW, Beer SV. Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. Journal of Bacteriology. 1998;180(8):2244–7. Epub 1998/04/29. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudriault S, Malandrin L, Paulin JP, Barny MA. DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Molecular Microbiology. 1997;26(5):1057–69. Epub 1998/01/13. . [DOI] [PubMed] [Google Scholar]
- 27.Lorang JM, Keen NT. Characterization of AvrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol Plant Microbe Interact. 1995;8(1):49–57. Epub 1995/01/01. . [DOI] [PubMed] [Google Scholar]
- 28.Jin L, Ham JH, Hage R, Zhao W, Soto-Hernandez J, Lee SY, et al. Direct and Indirect Targeting of PP2A by Conserved Bacterial Type-III Effector Proteins. PLoS Pathog. 2016;12(5):e1005609 doi: 10.1371/journal.ppat.1005609 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xin XF, Nomura K, Ding X, Chen X, Wang K, Aung K, et al. Pseudomonas syringae Effector Avirulence Protein E Localizes to the Host Plasma Membrane and Down-Regulates the Expression of the NONRACE-SPECIFIC DISEASE RESISTANCE1/HARPIN-INDUCED1-LIKE13 Gene Required for Antibacterial Immunity in Arabidopsis. Plant Physiol. 2015;169(1):793–802. doi: 10.1104/pp.15.00547 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamoun S, Furzer O, Jones JD, Judelson HS, Ali GS, Dalio RJ, et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol. 2015;16(4):413–34. doi: 10.1111/mpp.12190 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thines M, Kamoun S. Oomycete-plant coevolution: recent advances and future prospects. Curr Opin Plant Biol. 2010;13:427–33. Epub 2010/05/08. doi: 10.1016/j.pbi.2010.04.001 . [DOI] [PubMed] [Google Scholar]
- 32.Jiang RH, Tyler BM. Mechanisms and evolution of virulence in oomycetes. Annu Rev Phytopathol. 2012;50:295–318. Epub 2012/08/28. doi: 10.1146/annurev-phyto-081211-172912 . [DOI] [PubMed] [Google Scholar]
- 33.Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461(7262):393–8. Epub 2009/09/11. doi: 10.1038/nature08358 . [DOI] [PubMed] [Google Scholar]
- 34.Raffaele S, Farrer RA, Cano LM, Studholme DJ, MacLean D, Thines M, et al. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010;330(6010):1540–3. Epub 2010/12/15. doi: 10.1126/science.1193070 . [DOI] [PubMed] [Google Scholar]
- 35.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313(5791):1261–6. doi: 10.1126/science.1128796 . [DOI] [PubMed] [Google Scholar]
- 36.Levesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biology. 2010;11(7):R73 Epub 2010/07/16. doi: 10.1186/gb-2010-11-7-r73 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biology. 2011;9(7):e1001094 doi: 10.1371/journal.pbio.1001094 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Links MG, Holub E, Jiang RH, Sharpe AG, Hegedus D, Beynon E, et al. De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics. 2011;12:503 doi: 10.1186/1471-2164-12-503 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, Kemen E, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330(6010):1549–51. Epub 2010/12/15. doi: 10.1126/science.1195203 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kale SD, Gu B, Capelluto DG, Dou D, Feldman E, Rumore A, et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142(2):284–95. Epub 2010/07/27. doi: 10.1016/j.cell.2010.06.008 . [DOI] [PubMed] [Google Scholar]
- 41.Tyler BM, Kale SD, Wang Q, Tao K, Clark HR, Drews K, et al. Microbe-independent entry of oomycete RxLR effectors and fungal RxLR-like effectors into plant and animal cells is specific and reproducible. Mol Plant Microbe Interact. 2013;26(6):611–6. doi: 10.1094/MPMI-02-13-0051-IA . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wawra S, Djamei A, Albert I, Nurnberger T, Kahmann R, van West P. In vitro translocation experiments with RxLR-reporter fusion proteins of Avr1b from Phytophthora sojae and AVR3a from Phytophthora infestans fail to demonstrate specific autonomous uptake in plant and animal cells. Mol Plant Microbe Interact. 2013;26(5):528–36. doi: 10.1094/MPMI-08-12-0200-R . [DOI] [PubMed] [Google Scholar]
- 43.Petre B, Kamoun S. How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biology. 2014;12(2):e1001801 doi: 10.1371/journal.pbio.1001801 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torto TA, Li S, Styer A, Huitema E, Testa A, Gow NA, et al. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Research. 2003;13(7):1675–85. doi: 10.1101/gr.910003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amaro TMMM, Thilliez GJA, Motion GB, Huitema E. A Perspective on CRN Proteins in the Genomics Age: Evolution, Classification, Delivery and Function Revisited. Frontiers in Plant Science. 2017;8:99 doi: 10.3389/fpls.2017.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson RG, Deb D, Fedkenheuer K, McDowell JM. Recent progress in RXLR effector research. Mol Plant Microbe Interact. 2015;28(10):1063–72. doi: 10.1094/MPMI-01-15-0022-CR [DOI] [PubMed] [Google Scholar]
- 47.Dalio RJD, Herlihy J, Oliveira TS, McDowell JM, Machado MAA. Effector biology in focus: a primer for computational prediction and functional characterization. Mol Plant Microbe Interact. 2017. doi: 10.1094/MPMI-07-17-0174-FI . [DOI] [PubMed] [Google Scholar]
- 48.Wessling R, Epple P, Altmann S, He Y, Yang L, Henz SR, et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host & Microbe. 2014;16(3):364–75. doi: 10.1016/j.chom.2014.08.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333(6042):596–601. Epub 2011/07/30. doi: 10.1126/science.1203659 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDowell JM. Hyaloperonospora arabidopsidis: A Model Pathogen of Arabidopsis In: Dean RA, Lichens-Park A, Kole C, editors. Genomics of Plant-Associated Fungi and Oomycetes: Dicot Pathogens: Springer; Berlin Heidelberg; 2014. p. 209–34. [Google Scholar]
- 51.Deb D, Anderson R, Kin THY, Tyler BM, McDowell JM. Conserved RxLR effectors from oomycetes Hyaloperonospora arabidopsidis and Phytophthora sojae suppress PAMP- and effector-triggered immunity in diverse plants. Mol Plant Microbe Interact. 2017. doi: 10.1094/MPMI-07-17-0169-FI . [DOI] [PubMed] [Google Scholar]
- 52.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–38. Epub 2010/04/03. nprot.2010.5 doi: 10.1038/nprot.2010.5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40 Epub 2008/01/25. doi: 10.1186/1471-2105-9-40 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77 Suppl 9:100–13. Epub 2009/09/22. doi: 10.1002/prot.22588 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–9. Epub 2010/05/12. doi: 10.1093/nar/gkq366 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Opiyo SO, Moriyama EN. Protein family classification with partial least squares. J Proteome Res. 2007;6(2):846–53. Epub 2007/02/03. doi: 10.1021/pr060534k . [DOI] [PubMed] [Google Scholar]
- 57.Opiyo S, Mackey D. Identification of proteins similar to AvrE type III effector proteins from Arabidopsidis thaliana genome with partial least squares. African Journal of Biotechnology. 2013;12:5804–8. [Google Scholar]
- 58.Geladi P, Kowalski BR. Partial least squares regression: A tutorial. Anal Chim Acta. 1986;185:1–7. [Google Scholar]
- 59.Wehrens R, Mevik B. pls: Partial Least Squares Regression(PLSR) and Principal Component Regression (PCR). R package version 1.2–1. 2007.
- 60.Karchin R, Karplus K, Haussler D. Classifying G-protein coupled receptors with support vector machines. Bioinformatics. 2002;18(1):147–59. Epub 2002/02/12. . [DOI] [PubMed] [Google Scholar]
- 61.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24(23):2780–1. Epub 2008/09/27. doi: 10.1093/bioinformatics/btn507 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Research. 2005;33(7):2302–9. doi: 10.1093/nar/gki524 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sohn KH, Lei R, Nemri A, Jones JD. The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. The Plant Cell. 2007;19(12):4077–90. doi: 10.1105/tpc.107.054262 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindgren PB, Peet RC, Panopoulos NJ. Gene cluster of Pseudomonas syringae pv. "phaseolicola" controls pathogenicity of bean plants and hypersensitivity of nonhost plants. Journal of Bacteriology. 1986;168(2):512–22. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Badel JL, Shimizu R, Oh HS, Collmer A. A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant Microbe Interact. 2006;19(2):99–111. Epub 2006/03/15. doi: 10.1094/MPMI-19-0099 . [DOI] [PubMed] [Google Scholar]
- 67.Cumbie JS, Pankow RC, Thomas WJ, Chang JH. AutoSPOTs: Automated Image Analysis for Enumerating Callose Deposition In: Akimitsu K, al e, editors. 10th Japan-US Seminar: Genome-Enabled Integration of Research in Plant Pathogen Systems; St. Paul, MN: APS press: 2010. [Google Scholar]
- 68.Siamer S, Gaubert S, Boureau T, Brisset MN, Barny MA. Mutational analysis of a predicted double beta-propeller domain of the DspA/E effector of Erwinia amylovora. FEMS Microbiology Letters. 2013;342(1):54–61. doi: 10.1111/1574-6968.12108 . [DOI] [PubMed] [Google Scholar]
- 69.Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J. Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact. 2011;24(2):183–93. Epub 2010/10/20. doi: 10.1094/MPMI-07-10-0149 . [DOI] [PubMed] [Google Scholar]
- 70.Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol. 2012;15(4):483–92. Epub 2012/04/10. doi: 10.1016/j.pbi.2012.03.008 . [DOI] [PubMed] [Google Scholar]
- 71.Feng F, Zhou JM. Plant-bacterial pathogen interactions mediated by type III effectors. Current Opinion in Plant Biology. 2012;15(4):469–76. doi: 10.1016/j.pbi.2012.03.004 . [DOI] [PubMed] [Google Scholar]
- 72.Tyler B, Gijzen M. The Phytophthora sojae Genome Sequence: Foundation for a Revolution In: Dean RA, Lichens-Park A, Kole C, editors. Genomics of Plant-Associated Fungi and Oomycetes: Dicot Pathogens: Springer; Berlin Heidelberg; 2014. p. 133–57. [Google Scholar]
- 73.Garbutt CC, Bangalore PV, Kannar P, Mukhtar MS. Getting to the edge: protein dynamical networks as a new frontier in plant-microbe interactions. Frontiers in Plant Science. 2014;5:312 doi: 10.3389/fpls.2014.00312 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petsko GA, Ringe D. Protein Structure and Function Primers in Biology. Sunderland (Massachusetts): Science Press; London, in association with Blackwell Publishing; Oxford, and Sinauer Associates; 2003. [Google Scholar]
- 75.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25(17):3389–402. Epub 1997/09/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gangadharan A, Sreerekha MV, Whitehill J, Ham JH, Mackey D. The Pseudomonas syringae pv. tomato type III effector HopM1 suppresses Arabidopsis defenses independent of suppressing salicylic acid signaling and of targeting AtMIN7. PLoS One. 2013;8(12):e82032 doi: 10.1371/journal.pone.0082032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geng X, Cheng J, Gangadharan A, Mackey D. The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. The Plant Cell. 2012;24(11):4763–74. doi: 10.1105/tpc.112.105312 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lozano-Torres JL, Wilbers RH, Gawronski P, Boshoven JC, Finkers-Tomczak A, Cordewener JH, et al. Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc Natl Acad Sci U S A. 2012;109(25):10119–24. doi: 10.1073/pnas.1202867109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song J, Win J, Tian M, Schornack S, Kaschani F, Ilyas M, et al. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci U S A. 2009;106(5):1654–9. Epub 2009/01/28. doi: 10.1073/pnas.0809201106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.