Abstract
Previously, VLPs bearing JR-FL strain HIV-1 Envelope trimers elicited potent neutralizing antibodies (nAbs) in 2/8 rabbits (PLoS Pathog 11(5): e1004932) by taking advantage of a naturally absent glycan at position 197 that borders the CD4 binding site (CD4bs). In new immunizations, we attempted to improve nAb responses by removing the N362 glycan that also lines the CD4bs. All 4 rabbits developed nAbs. One targeted the N197 glycan hole like our previous sera. Two sera depended on the N463 glycan, again suggesting CD4bs overlap. Heterologous boosts appeared to reduce nAb clashes with the N362 glycan. The fourth serum targeted a N362 glycan-sensitive epitope. VLP manufacture challenges prevented us from immunizing larger rabbit numbers to empower a robust statistical analysis. Nevertheless, trends suggest that targeted glycan removal may improve nAb induction by exposing new epitopes and that it may be possible to modify nAb specificity using rational heterologous boosts.
Keywords: HIV-1, Env, trimer, gp120, neutralization, vaccine, VLP, antibody, glycan
INTRODUCTION
Eliciting broadly neutralizing antibodies (bnAbs) is a major goal of HIV-1 vaccine development. NAbs block infection by binding to native envelope glycoprotein (Env) spikes on virus surfaces, thereby preventing transmission (Burton and Hangartner, 2016). The compact, sequence-diverse, and heavily glycosylated nature of these spikes allows the virus to largely evade neutralization (Kwong and Mascola, 2012; Pancera et al., 2014). Nevertheless, a growing number of monoclonal antibodies (mAbs) have been isolated from HIV-infected individuals that can potently and broadly neutralize tier 2 strains (Burton and Hangartner, 2016). These bnAbs can protect animal models against viral challenge (Shingai et al., 2014), suggesting that if sufficient titers of similar bnAbs can be induced by vaccination, they could prevent HIV infection.
Autologous nAbs typically develop within a few months during natural infection, and invariably precede any breadth development. The appearance of bnAbs is frequently associated with increased viral diversity as the virus escapes autologous nAbs, often by the acquisition of new glycans (Bhiman et al., 2015; Liao et al., 2013; Moore et al., 2012; Moore et al., 2009; Murphy et al., 2013; Wibmer et al., 2013). A logical vaccine strategy is to mimic the bnAb development processes observed in natural infection by first developing a platform to consistently elicit potent autologous tier 2 nAbs as a foundation, then using heterologous boosts to broaden nAb cross-reactivity to shared epitopes (Derdeyn et al., 2014; Moore et al., 2012; Ping et al., 2013; Wibmer et al., 2013).
Until recently, most vaccine candidates largely elicited weak or undetectable autologous tier 2 nAbs, let alone any breadth (Burton and Hangartner, 2016). This may be because these vaccine candidates insufficiently resemble native spikes and consequently elicit largely “off target” responses. These problems have, to some extent, been recently solved by the development of “near native” soluble SOSIP gp140 trimers that can elicit potent autologous nAbs in most vaccinated rabbits (Cheng et al., 2015; de Taeye et al., 2015; Sanders et al., 2015). However, remaining challenges include the order of magnitude differences in nAb titers among immunized rabbits, the markedly differing propensities of SOSIP trimers prepared from different strains to induce nAbs and the relatively modest nAb titers in vaccinated non-human primates (NHPs). The helix breaking I559P mutation and/or the lack of a natural, membrane setting could also be problematic. The reduction in VRC01 and PGT151 bnAb recognition caused by the I559P mutation (Alsahafi et al., 2015) suggests a conformational impact. Furthermore, Env ectodomain conformation differs when expressed in situ with transmembrane and cytoplasmic domains (Abrahamyan et al., 2005; Chen et al., 2015; Montero et al., 2012). Thus, although all BG505 SOSIP.664 gp140 trimers-vaccinated animals may generate potent binding titers, not all may cross react with native membrane-expressed trimers and neutralize (Hu et al., 2015). This problem could be exacerbated in NHPs where a more sophisticated antibody repertoire may generate antibodies that can better distinguish the modest differences between SOSIP trimers and their native membrane counterparts.
Considering the above concerns, it is reasonable to pursue vaccine strategies in which Env trimers are presented in a native, membrane setting. To this end, we have been developing virus-like particles (VLPs) (Crooks et al., 2007; Crooks et al., 2015; Tong et al., 2014). We previously suggested that non-functional Env present on VLP surfaces may interfere with their ability to induce nAbs (Tong et al., 2014). To address this problem, we showed that a protease blast can selectively remove non-functional Env from VLP surfaces, leaving trimers intact (Crooks et al., 2011; Tong et al., 2012). Immunizations with high doses of the resulting “trimer VLPs” induced remarkably potent serum nAbs in 2 out of 8 rabbits that targeted a strain-specific hole in the trimer’s carbohydrate shell left by the natural absence of the N197 glycan in the C2 domain of JR-FL gp120 (Crooks et al., 2015). These sera neutralized other clade B tier 2 isolates when the N197 glycan was removed, suggesting that they target a conserved site to which access is usually regulated by a glycan.
In a more recent study, strain-specific glycan holes were also found to be targeted by autologous nAbs in rabbits immunized with BG505 SOSIP.664 gp140 trimers (McCoy et al., 2016). Similar glycan holes are also frequently targeted by autologous nAbs during natural infection (Derdeyn et al., 2014; Moore et al., 2012; Moore et al., 2009; Wibmer et al., 2016). These findings are in line with known antibody preference for binding protein over glycan, with the latter specificities being largely eliminated from the repertoire at tolerance checkpoints (Haynes et al., 2012). The carbohydrate shell encasing native Env trimers (Julien et al., 2013; Lee et al., 2016; Lyumkis et al., 2013; Pancera et al., 2014; Stewart-Jones et al., 2016) appears to dampen germline antibody triggering (McGuire et al., 2014; McGuire et al., 2013; Ota et al., 2012; Ping et al., 2013; Zhou et al., 2010). Nevertheless, bnAbs targeting the V2 apex, V3-glycan and gp120/gp41 interface epitope clusters make protein and glycan contacts (McLellan et al., 2011; Pejchal et al., 2011; Walker et al., 2011; Walker et al., 2009). In contrast, bnAbs targeting the CD4 binding site (CD4bs) (Burton and Hangartner, 2016) largely contact protein epitopes, but must also either avoid or make contacts with glycans that line the CD4bs (Huang et al., 2016; Pancera et al., 2014; Stewart-Jones et al., 2016).
The fence of glycans lining the CD4bs has evolved to leave a gap sufficient to accommodate the single immunoglobulin-like domain of CD4 but insufficient to grant easy access to a pair of immunoglobulin domains, i.e. antibody heavy and light chains (Heydarchi et al., 2016; McCoy et al., 2012). Despite this problem, the CD4bs remains an attractive vaccine target in part due to the many paradigms provided by monoclonal antibodies (mAbs) recovered from naturally infected donors. These include germline-encoded specificities like that of the VRC01 mAb, in which binding is dominated by the HCDR2 domain, and those in which binding is dominated by a long CDRH3 domain (Huang et al., 2016; Kwong and Mascola, 2012; Stewart-Jones et al., 2016; Zhou et al., 2010; Zhou et al., 2015).
Here, we tried to improve the frequency of nAb responses to JR-FL trimer VLPs by enlarging the CD4bs glycan hole. We found that 4 out of 4 animals (100%) developed autologous nAbs, in contrast to 25% in our previous study (Crooks et al., 2015) - a difference that appeared to stem from the exposure of new epitopes by removing the N362 glycan. Boosting with VLPs in which the N362 glycan was reinstated appeared to enable sera to navigate past this glycan and neutralize the parent virus. In a second study, plasmid DNA expressing VLP components appeared augment nAb induction by VLP protein immunogens. Although our VLP production challenges precluded a larger analysis that would be needed for full statistical powering, our data provide some leads that might benefit vaccine development.
RESULTS
An enlarged glycan hole improves trimer access to CD4 binding site mAbs
Our previous immunogenicity studies suggested that VLP efficacy may be improved by i) eliminating antigenic interference by non-functional Env, ii) by using higher VLP doses and iii) by using rabbit models (Crooks et al., 2015; Tong et al., 2014). So far, at best, potent tier 2 nAbs developed in only 25% of rabbits, suggesting a need for further improvements (Crooks et al., 2015). These sera targeted the space vacated by the naturally absent N197 glycan on the JR-FL vaccine strain, situated adjacent to the CD4bs. Figure 1A shows the fence of glycans (magenta) through which antibodies must navigate to contact the underlying CD4bs protein site (yellow). MAb VRC01 docking (Figure 1B) reveals how this ligand must exquisitely circumnavigate this fence to contact underlying protein.
Fig 1.

N362 glycan removal improves antibody access to the CD4 binding site. A) The ectodomain of native, full-length cleaved JR-FL Env trimers in pdb 5FUU (Lee et al., 2016) is shown in side and top views. Gp41 is shown in grey and gp120 protomers are shown in different shades of green. The CD4 binding loop (residues 360–370) and putative CD4 contact positions 427 and 477 are shaded yellow. Since complex glycans may exhibit branches that are difficult to predict and/or unequivocally model (Bonomelli et al., 2011), all glycans were modeled as Man8GlcNAc2. Those that form a fence around the CD4bs are colored in magenta. Residue 197 is colored in red. This position bears a glycan in most strains that forms part of the glycan fence, but is naturally absent in the JR-FL strain. Close glycan spacing may limit their differentiation, presumably by limiting access to glycan processing enzymes (Behrens et al., 2016; Stewart-Jones et al., 2016; Zhou et al., 2013). However, since a carbohydrate-free gap is necessary to admit CD4 binding, it is logical that some of the surrounding glycans are relatively well differentiated (Behrens et al., 2016). Since complex glycans usually have a greater mass than Man8GlcNAc2, the CD4bs is probably even better protected than shown here. Glycans were added to all NXS/T sequons except for N189 and N637. Due steric clashes with glycans at proximal sites, these sites may not be occupied. Furthermore, regions of structural disorder in the C1, V1 and V4 and any associated glycans (see Figure S1) are not shown. B) Fab VRC01 (3NGB), rendered in aqua ribbon format, is docked onto the trimer. C) The N362 glycan is removed, as in our JR-FL N362Q mutant vaccine strain and the underlying residue is colored red. D) Sensitivity of JR-FL glycan fence mutants to VRC01 neutralization. E) Sensitivity of the JR-FL SOS E168K parent and N362Q mutants to various mAbs. F) Binding of mAbs VRC01 and 2G12 to VLPs bearing the same Env trimers by ELISA.
In the JR-FL strain, the fence occurs at positions N276, N362, N386, N392 and N463 (Figure 1A). The precise positioning of fence glycans differs between strains. Indeed, one or more fence glycans may be missing in some strains (as for the N197 glycan of the JR-FL strain). In this case, partial masking of underlying protein may still be achieved by rearrangement of the interlocking network of remaining glycans to partially fill the vacated space (Gristick et al., 2016; Stewart-Jones et al., 2016; Zhou et al., 2016). The remaining glycans may also undergo more complex differentiation, adding mass to partially fill glycan holes (Behrens et al., 2016). Indeed, despite being a nAb target in our previous study (Crooks et al., 2015), the N197 glycan hole was only targeted by 2 out of 8 animals, suggests that is only partially accessible.
In this follow up study, we reasoned that enlarging the glycan-free CD4bs hole by removing another glycan (in addition to the already absent N197 glycan) might improve nAb induction. To inform our selection of a glycan knockout mutant, we first analyzed the effects of knocking out each of the 5 JR-FL strain fence glycans on JR-FL E168K mutant virus sensitivity to mAb VRC01. The E168K mutant was previously used to partially knock in V2 apex bnAb sensitivity and fortuitously improves trimer expression. The VRC01 mAb was selected to characterize these mutants because it exhibits a germline-encoded CDRH2-dominated binding mode that is almost exactly replicated by other “VRC01 class” bnAbs isolated from other donors, thus making it a useful prototype for vaccine researchers (Kwong and Mascola, 2012; Zhou et al., 2016; Zhou et al., 2010; Zhou et al., 2015; Zhou et al., 2013). The N276A and N362Q mutants were approximately 3- and 8-fold more VRC01-sensitive than the parent, respectively (Figure 1D), whereas N386, N392 and N463 knockout mutants all had little or no effect. Conversely, a glycan knock-in at position 197 markedly reduced VRC01 sensitivity (Figure 1D). We previously observed that this mutation reduces mAb b12 sensitivity by ~100-fold, demonstrating its key role in regulating CD4bs nAb access (Crooks et al., 2015).
Based on the above findings, we selected the N362Q mutant for follow up (Figure 1C). In further neutralization assays, we examined the effects of this mutant on sensitivity to other mAbs. MAb 8ANC131, which also employs a CDRH2-dominated binding interface but encoded by a different germline (Zhou et al., 2015) was also 2-fold more potent on the N362Q mutant. MAb b12, that uses a long CDRH3-dominated CD4bs binding mode, neutralized the N362Q mutant with similar potency to the parent virus (Figure 1E). As mentioned above, b12 sensitivity is instead heavily regulated by the N197 glycan. VRC13, another CDRH3-dominated prototype mAb, was 2–3-fold more effective against the mutant. A broadly neutralizing HIV-positive donor serum (1688) that targets the CD4bs was 3-fold more potent on the mutant, whereas little change was noted in sensitivity to a serum that targets the gp41 membrane proximal ectodomain region (MPER) (Figure 1E; (Binley et al., 2008)). MAbs HJ16 and 179NC75, both of which depend on the N276 glycan, were unable to neutralize either the parent or the mutant virus (Figure 1E and not shown). The neutralizing activities of various other mAbs were essentially unchanged (Figure 1E). Non-neutralizing mAbs 15e and CO11, directed to CD4bs and V3 epitopes, respectively, did not neutralize either the parent or the N362Q mutant, suggesting that the mutant did not overtly impact trimer conformation. PG16 did not reach an IC50 against this virus at the concentrations tested, as full sensitivity requires an additional N189A mutation of this strain to eliminate a competing glycosylation site (Doores and Burton, 2010). VLP ELISAs mirrored the neutralization data: VRC01 bound the mutant 10-fold better than the parent VLPs, whereas 2G12 bound both VLPs equivalently (Figure 1F). Collectively, this data shows that the N362Q mutant is more sensitive to the VRC01 mAb and certain other CD4bs mAb and was therefore selected for immunization studies to test whether a larger glycan hole might improve nAb induction.
Effects of an enlarged glycan hole on immunogenicity
Figure 2 provides an overview of Study 1. The “SOS” mutant was used to introduce a gp120-gp41 disulfide bond (Binley et al., 2000) and also improves trimer expression compared to the non-mutated wild type (WT) trimer (Crooks et al., 2005; Moore et al., 2006). We previously showed that E168K, SOS mutations and gp41 tail truncation all have only marginal effects on neutralization sensitivity, justifying their use as immunogens (Crooks et al., 2005). VLPs were produced by co-expression of Env, ERV (MuLV) Gag and HIV-1 Rev plasmids. MuLV Gag modestly improved VLP Env trimer yield, as compared to SIV-derived p55 Gag used previously (Crooks et al., 2015).
Fig 2.

Overview of rabbit immunizations. 4 rabbits were immunized with N362Q trimer VLPs and were later boosted with a mixture of these VLPs with parent VLPs (N362 glycan intact). VLPs were produced in 293T cells and were formulated in AS01B. The day of each immunization is indicated along with bleeds (B1–B5), taken two weeks thereafter.
Prior to immunizations, VLPs were treated with proteases to substantially remove non-functional Env, leaving native Env trimers intact, resulting in “trimer VLPs” (Crooks et al., 2011; Tong et al., 2012). We immunized a group of 4 rabbits, starting with 4 shots of N362Q mutant trimer VLPs (our “index” immunogen) at 0, 1, 3 and 6 months. VLP doses were high and matched those used in our previous study (~150μg gp120 equivalents/kg) (Crooks et al., 2015). None of the VLP-immunized animals exhibited any clear ill effects or toxicity related to immunizations. However, rabbit 742 died unexpectedly 168 days after the 4th shot for reasons that did not appear to be related to immunizations. The 3 surviving animals were later boosted with parent VLPs (Figure 2) to try to train rabbit nAbs to navigate the N362 glycan. This was mixed with smaller dose of the N362Q index immunogen to ensure the reactivation of nAb memory B cell clones primed by the preceding 4 N362Q VLP shots.
We did not immunize a concurrent control group with parent trimer VLPs because the high VLP doses needed to induce nAbs that we reported in our previous studies require significant labor to produce (Crooks et al., 2015; Tong et al., 2014). Indeed, we estimate that, using equivalent expression systems, per unit time, >100-fold more doses of SOSIP trimer can be produced compared to VLPs. This may relate to the relative inefficiency of multi-plasmid transfection, low Env spike density on VLP surfaces and the relatively constrained (i.e. less immunogenic) Env conformation in a membrane setting, that collectively mandate high VLP doses. Therefore, we decided to bridge the current immunized rabbits to the rabbits we previously immunized with parent trimer VLPs (termed reference groups A and B). The latter received equivalent high VLP doses as in the new study, were administered in the same adjuvant and were analyzed in the same way.
A profile of peak sera from the new rabbits along with reference sera is shown in Figure 3. “Peak” time points are those with the highest neutralizing ID50 against either the JR-FL parent or N362Q virus (a kinetic analysis of nAb development follows below). The bleed numbers shown (i.e. B3, B4 and B5) correspond to those depicted in Figure 2. Data for bleed 4 sera from reference groups A and B is shown except for rabbit 347 that died before the 4th shot (Crooks et al., 2015). We also included plasmas from an HIV-1 donor (1702) and an uninfected human control (210) (Binley et al., 2008; Crooks et al., 2005).
Fig 3.

Study 1 serum binding and neutralization profiles. Peak time points for each N362Q trimer VLP-immunized rabbits were compared with two groups of 4 sera from our previous study, termed here as reference groups A and B (Crooks et al., 2015), along with human HIV-1-seropositive (1702) and uninfected (210) human control plasmas. A) ELISA titers against recombinant JR-FL gp120 monomer and bald-VLPs. Data are means of at least two repeats. Neutralizing activity against B) the tier 1 phenotype mutant JR-FL WT A328G (Tong et al., 2014) and SIVmac239, and C) the JR-FL SOS E168K+N362Q index strain and parent JR-FL SOS E168K measured in CF2Th.CD4.CCR5 cells. All neutralization assays were performed at least three times in duplicate. Mean ID50s and standard deviations are shown. P values for Mann-Whitney tests are shown only when there were significant differences (i.e. <0.05) between groups. For these tests, undetectable <1:4 neutralizing ID50s were assigned as 1:4.
Serum titers to parent JR-FL gp120 monomer (i.e. carrying the N362 glycan) are shown as green circles in Figure 3A. Gp120 titers ranged from 1:1,520–1:54,192 (Crooks et al., 2015). Titers between groups were compared by Mann-Whitney tests. Reference group B titers were not significantly higher than those in group B (P=0.0571). However, Study 1 titers were significantly higher than those in group A (P=0.0286) but were not higher than those in group B (P=0.4857). Overall, despite the presence of the N362 glycan on the gp120 monomer antigen used in these assays, the new rabbit sera exhibited equivalent or better titers than previous sera but they were weaker than human plasma 1702 (although the need for different conjugates precludes a formal comparison of the latter).
Serum titers to bald VLPs are shown as blue triangles in Figure 3A. High titers were observed in all Study 1 animals. Differences between groups A and B were not significantly different (P=0.2), nor were they for Study 1 and group A (P=0.6857). However, Study 1 titers were significantly higher than those in group B (P=0.0286). Again, this data supports the equivalent or better immunogenicity of the new VLP immunogens. The apparently high bald VLP titer also observed in plasma 1702 may in part be explained by lipid-reactive antibodies that are benign and ubiquitous in nature (Alving, 2008; Tong et al., 2014).
Serum neutralization was assessed using CF2 target cells (Crooks et al., 2015; Tong et al., 2014). The JR-FL A328G mutant (Tong et al., 2014) was used to assay tier 1 nAbs, shown as magenta diamonds in Figure 3B. This mutant causes a folding defect that results in V3 loop exposure, similar to some other reported Env mutants (Blish et al., 2008) and can therefore detect “off target” tier 1 nAbs that matches the JR-FL vaccine strain Env sequence. Titers in groups A and B were comparable (P=0.7429), as were those of group A and Study 1 (P=0.2286) and group B and study 1 (P=0.3429). As expected, none of the sera neutralized the control virus SIVmac239 (black triangles in Figure 3B).
Unlike the reference groups, all Study 1 rabbits developed nAbs to the tier 2 index N362Q virus (orange triangles in Figure 3C). By Mann-Whiney test, there were no differences between groups A and B (P>0.99). Neither were there significant differences between Study 1 sera and the two reference groups (P=0.2 in both cases). The difference between Study 1 sera and groups A and B combined was also not quite significant (P=0.0687).
Three Study 1 sera also neutralized the parent virus (red circles in Figure 3C). Sera 740 and 742 titers were comparable to sera 347 and 613. As for the N362 titers, there was no difference in parent virus nAb responses by groups A and B. The differences between Study 1 and each of groups A and B were not significant (P= 0.4857 and 0.3714, respectively) and were also not significant when groups A and B were combined (P=0.2788). Overall, although a trend that suggests positive nAb responses in the new N362Q VLP-immunized rabbits, the group differences did not reach statistical significance. It should be emphasized that until recently, most vaccine sera have failed to generate any detectable autologous tier 2 nAb responses, as exemplified by sera 611, 612, 614, 345, 346 and 348 of the control groups A and B (Figure 3C) (Sanders et al., 2015).
Three Study 1 sera neutralized the N362Q index virus more effectively than the JR-FL parent, whereas serum 742 neutralized the parent marginally more effectively (Figure 3C). We next analyzed neutralizing ID50 kinetics over the course of immunizations (Figure 4). To detect subtle changes, we used 4 JR-FL mutants: WT parent, SOS parent, WT N362Q and SOS N362Q (Figure 4). Rabbits 740 and 741 both exhibited an early strong preference for SOS N362Q, matching the immunogen, at bleeds 3 and 4. However, this changed dramatically after rabbit 740 was boosted with a mixture of parent and N362Q VLPs, whereupon it gained an ability to neutralize all 4 variants equally. Similar mixed boosting of rabbit 741 also led to an increase in neutralization against the SOS parent virus, with a concomitant lowering of titers against the two N362Q mutants (Figure 4).
Fig 4.

Longitudinal analysis of neutralizing responses in vaccinated rabbits. Changes in serum ID50 titers against four JR-FL variants were followed over the course of immunization of all 4 Study 1 rabbits that generated tier 2 nAbs. The four variants analyzed were JR-FL WT (open symbols) and SOS versions (solid symbols) of the N362Q index (orange) and parent (red) strains. The preceding immunogen boost for each bleed is indicated by arrows. The color scheme matches that used in Figures 2 and 3.
Serum 742 exhibited strong initial titers, particularly to SOS mutant viruses (regardless of N362Q mutation) at bleed 3, and had lower titers against the respective WT versions of these viruses. Upon further boosting, titers against these N362Q viruses improved while those against parent viruses dropped. Thus, it appears that repeated boosting led to increased N362 glycan sensitivity. Nevertheless, the general ability of this serum to neutralize all variants effectively suggests a conserved epitope. Unfortunately, this animal died before heterologous boosting.
Serum 743 developed nAbs against the N362Q index immunogen at bleed 5. This was somewhat surprising, considering that the preceding boost predominantly consisted of parent VLPs (Figure 4). We infer that serum 743 overlaps the protein surface revealed by the missing N362 glycan, otherwise referred to as a N362 glycan-sensitive.
Potent nAbs recognize native trimers in a largely D368-independent manner
We previously showed that neutralization correlates with antibody binding to native trimers as measured in BN-PAGE-Western blot shifts (Binley et al., 2008; Crooks et al., 2007; Crooks et al., 2005; Moore et al., 2006; Tong et al., 2012). Peak serum IgG (Figure 3) from animals 740–743 of Study 1 all bound to native SOS E168K+N362Q trimer, as evidenced by depletion of the unliganded trimer (Fig 5A, upper panel). Serum 613 from reference group A also bound these trimers, as did mAbs b12 and 2G12. Neutralizing IgG from rabbits 773 and 776 of Study 2 (described later below) also bound to trimers but non-neutralizing serum IgG from rabbit 770 (also from Study 2) did not bind trimers. Study 1 sera also bound SOS E168K trimers (Figure 5A, middle panel). Notably, although neutralization was not detected in Figure 4, serum 743 IgG shifted this trimer, suggesting some weak activity. Serum IgG from rabbits 740, 741, 743 and 613 all bound to triple mutant SOS E168K+N362Q+D368R trimers (Figure 5A, lower panel), consistent with D368-independent trimer binding. Interestingly, rabbit 742 IgG failed to fully deplete this mutant trimer, thus indicating some D368-dependency. As expected, b12 binding was eliminated by the D368R mutation, but 2G12 binding was retained.
Fig 5.

Serum binding to native trimers and quaternary epitope dependency. A) Antibody binding to the native Env trimer was assessed in BN-PAGE shift assays. Briefly, mAbs b12 (3μg/ml) and 2G12 (30μg/ml) or protein A-purified IgG from rabbit serum “peak” bleeds (see Figure 3) were incubated with JR-FL SOS E168K+N362Q, JR-FL SOS E168K, or JR-FL SOS E168K+N362Q+D368R trimer VLPs, as indicated. VLPs were then washed and lysed, and Env was resolved by BN-PAGE-Western blot. As we observed previously, IgG-trimer complexes were rarely visible (Tong et al., 2012), perhaps due to the formation of higher order complexes. In marked contrast, mAb 2G12 exhibits a ladder of bound trimer species owing to monovalent binding. B) The effect of adding 10μg/ml of purified N362Q+D368R JR-FL gp120 monomer on serum and mAb neutralization of JR-FL gp160ΔCT WT E168K and JR-FL gp160ΔCT WT A328G is shown.
Quaternary epitope dependency of serum neutralization
Interference neutralization assays in which monomeric gp120 is pre-incubated with virus-serum mixtures can reveal whether sera target epitopes that found on the gp120 monomer or, alternatively, may be exclusively accessible on the native trimer. Above, we verified that all our vaccine sera can bind to E168K+N362Q+D368R mutant trimers, albeit weakly for the 742 serum (Figure 5A, lower panel). We therefore used E168K+N362Q+D368R gp120 monomer in interference assays using SOS E168K+N362Q and globally sensitive tier 1 phenotype WT A328G viruses (Figure 5B).
For rabbits 740 and 741, we examined bleed 4 and 5 sera to investigate any differences that might associate with those we observed in Figure 4. Neutralization of both viruses by bleed 4 of rabbit 740 was highly gp120 sensitive (Figure 5B). However, some gp120 monomer resistance was acquired at bleed 5. Thus, following mixed VLP boosting, nAbs became more gp120-resistant, suggesting quaternary epitope dependency (i.e. exclusive trimer binding) while also gaining an ability to cross-neutralize JR-FL variants. To a lesser extent, neutralization of the A328G mutant virus also became gp120-resistant at bleed 5. Although this tier 1 variant is expected to be neutralized predominantly by V3 loop-specific antibodies that can be adsorbed by gp120, this suggests that quaternary nAbs may also neutralize this variant. In contrast, serum 741 neutralized all viruses in a gp120 monomer-sensitive manner (Figure 5B), perhaps reflecting the less dramatic changes observed in Figure 4. Neutralization by sera 742 and 743 was also largely sensitive to gp120. The modest gp120-resistance of serum 742 neutralization of the index virus may be explained by its modest D368R-sensitivity observed in BN-PAGE shifts (Figure 5A). As expected, gp120 adsorbed 2G12 activity. Overall, although some sera (740 B5 and 742 B4) exhibited some gp120-resistance, most vaccine sera targeted epitopes that were largely contained within gp120. This contrasted with our previous study in which sera were insensitive to gp120 (see Figure S7 in (Crooks et al., 2015)). As described below, this could reflect the somewhat different epitopes targeted by the new sera.
Neutralizing sera target the CD4bs glycan hole
Study 1 rabbit sera failed to significantly neutralize other tier 2 viruses aside from the vaccine strain – as has been consistently true all HIV vaccine studies to date. This allowed us to map the 740, 741 and 742 sera by checking their activities against a panel of chimeric JR-FL Env pseudoviruses with JR-CSF domain swaps (Narayan et al., 2013) (Figure 6). We did not map the 743 as it did not significantly neutralize the JR-FL parent virus (Figure 4). As we reported previously, most chimeras were infectious and resistant to the V3 loop-specific mAb CO11 (Crooks et al., 2015; Tong et al., 2014), suggesting that they retain a tier 2 phenotype. All chimeras bearing JR-CSF strain V5 domains were resistant to the 740 and 741 sera. A V5 sequence comparison reveals a glycan at positions 461 and 463 in the JR-CSF and JR-FL strains, respectively (sequons are boxed in red in Figure 7A). Further analysis revealed that serum 740 neutralized a JR-CSF chimera with an engrafted JR-FL V5 region, but did not neutralize the JR-FL N463Q mutant (Figure 7B). Together, this suggests that the N463 glycan is either part of the serum epitope or imparts a local conformational effect that is important for recognition. For convenience, we henceforth term this N463 glycan-dependent. Notably, the infectivities of all 740 serum “resistant” viruses were nevertheless somewhat reduced (but did not reach an IC50; Figure 7B). We speculate that serum 740 may recognize protein sequence common to these related variants, but requires the glycan to fully neutralize. Incomplete neutralization is in fact a common feature of several bnAbs (McCoy et al., 2015). Serum 741 also depended on the N463 glycan (Figure 7C). An analysis of 4,265 Env sequences in the Los Alamos HIV sequence database (www.hiv.lanl.gov) reveals that this glycan is ~20% conserved. However, the 740 serum did not cross-neutralize 6 other strains that naturally carry a glycan at this position (Figure 7D), perhaps due to sequence variation. Given that the N463 glycan is part the CD4bs fence (at least in the JR-FL strain), we wondered if removing other fence glycans might impact virus sensitivity to serum 740 (or, in the case of D197N, knocking in a glycan). However, these mutants had no impact (Figure 7E).
Fig 6.

Neutralization of JR-FL-JR-CSF chimeras. Chimeras comprising of a JR-FL Env background (violet) with JR-CSF domain swaps (salmon) were evaluated for infectivity (in relative light units; RLU) and sensitivity to vaccine sera and mAb CO11 and b12 neutralization. N.D. denotes not done due to low infectivity.
Fig 7.
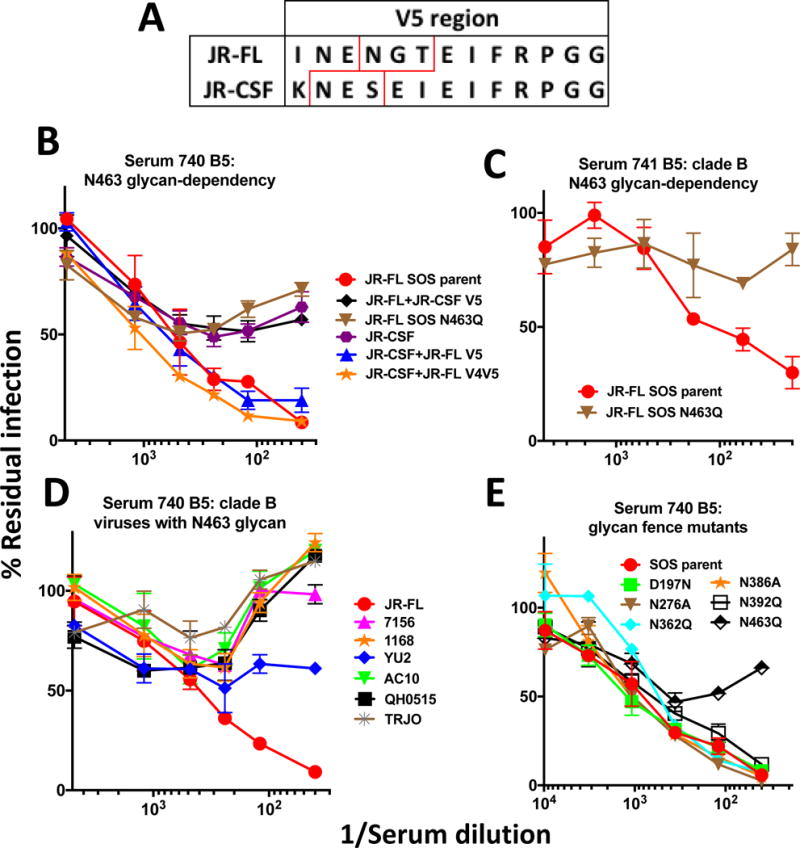
Fine mapping of the sera from rabbits 740 and 741. A) Alignment of the JR-FL and JR-CSF V5 sequences. Glycan sequons are boxed in red. B) Serum 740 (bleed 5) activity against various JR-FL and JR-CSF viruses, including those carrying V4V5 or V5 domain swaps and a JR-FL N463Q knockout mutant. C) Serum 741 (bleed 5) activity against the JR-FL parent and N463Q glycan knockout mutant. D) Serum 740 neutralization of various clade B viruses that carry a N463 glycan. E) Effect of glycan fence modifications on serum 740 neutralization.
We next investigated the effects of viral glycan modification on sensitivity to sera 740 and 741. Approximately 2/3 of the glycans on native trimers are of high mannose type and 1/3 are of complex type (Bonomelli et al., 2011). Several lines of evidence suggest that the N463 glycan is complex in nature. First, previous studies of various forms of Env derived from various strains have consistently described the glycan at this position as being complex (Behrens et al., 2016; Go et al., 2008; Leonard et al., 1990). Second, high mannose glycans tend to occur at well-conserved gp120 positions (e.g. N156 and N276) and may therefore have a structural role in trimer folding. Thus, the relatively poor conservation of the N463 glycan is in keeping with a complex glycan. Third, the proximity of the N463 glycan to the CD4 binding site may facilitate its processing and maturation (Behrens et al., 2016). Thus, assuming the N463 glycan is complex in JR-FL native trimers, we next determined the effect of adding kifunensine during transfection on the neutralization sensitivity. Kifunensine blocks α-mannosidase I trimming, thereby rendering all Env glycans as untrimmed high mannose type. Unexpectedly, this significantly increased virus sensitivity to these sera but had little effect on the 742 and 613 sera (Figure 8). As expected, PG9 sensitivity was knocked out by this treatment (Walker et al., 2011), while mAb VRC01 was marginally more effective. This data is consistent with the idea that the 740 and 741 sera recognize an epitope dependent on the N463 glycan stem (or a structure influenced by it).
Fig 8.
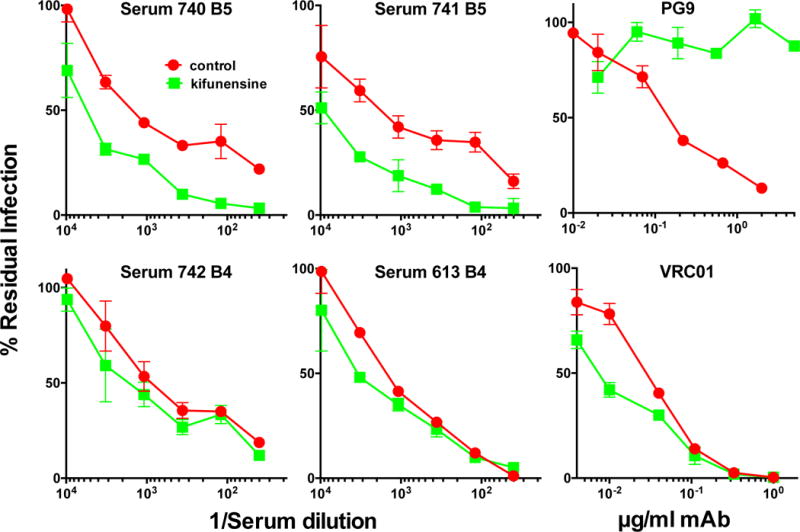
N463 glycan-dependent sera target the glycan core. Rabbit vaccine sera and mAb controls PG9 and VRC01 were evaluated for neutralization of the N362 index virus prepared in the presence or absence of kifunensine to inhibit high mannose glycan trimming.
We next attempted to map serum 742 which neutralized all the functional chimeras in Figure 6. However, chimeras in which the JR-CSF C2 region was substituted into the JR-FL background were not functional ((Crooks et al., 2015), Figure 6). We suspected that serum 742 might recognize a N197 glycan-sensitive epitope like that recognized by the 347 and 613 sera (Crooks et al., 2015). The N197 glycan is present in 98.7% of circulating strains (Crooks et al., 2015). Its absence in the JR-FL isolate is therefore highly unusual. To investigate this idea, we examined the sensitivity of viruses with mutations at this position to the 742 serum. A D197N glycan knock-in mutant of the parent JR-FL strain was resistant (Figure 9A). Reciprocally, a JR-CSF N197D mutant was sensitive. This suggests that the N197 glycan regulates this serum (Figure 9A). Further analysis of N197 glycan knockout mutants revealed that serum 742 cross-neutralized N197D mutants of the ADA isolate but no other clade B strains (Figure 9A). Glycan fence mutants revealed no other changes in neutralization sensitivity aside from that at position 197 (Figure 9B). Thus, the 742 serum exhibits modest tier 2 breadth within clade B by recognizing the conserved protein surface protected by the N197 glycan. Notably, the identities of the N197 glycan-lacking viruses sensitive to the 742 serum were the same as those that were sensitive to serum 347, and were among the larger number of N197 glycan-lacking viruses sensitive to the 613 serum (Crooks et al., 2015). As also coincidentally occurred with the 347 rabbit, serum 742 breadth might have been limited by the fact that this rabbit died before boosting was complete.
Fig 9.

Fine mapping of serum 742. A) Serum 742 was assayed for cross-neutralization of various clade B N197 knockout mutants. Global changes in sensitivity of these mutants was ruled out previously (Crooks et al., 2015), suggesting that any serum cross-neutralization observed is genuine and is not due to overt changes in folding related to the mutation. B). Effect of JR-FL glycan fence modifications on sensitivity to serum 742.
Effects of modifying VLP production and immunization strategies
To try to alleviate our VLP production challenges, in Study 2 (outlined in Figure S2), VLPs were prepared in 293 freestyle (293F) cells and were used to immunize 5 groups of 4 rabbits to attempt to investigate different immunization variables. First, we investigated the effects of two different adjuvants (AS01B and Adjuplex in groups 2A and 2B, respectively) on nAb induction to parent VLPs. Second, we investigated the effects of administering plasmids encoding the same Env, Gag and Rev used to make N362Q VLPs in prime-boost (group 2D) or co-administration formats (group 2E), as compared to a N362Q VLP only control group (group 2C). A comparison of groups 2A and 2C should allow us to determine the effects of removing the N362 glycan. Groups 2C–2E were later boosted with parent VLPs mixed with the N362Q VLP at day 190. Due to some evidence of nAb development (see below), groups 2D and 2E were boosted again at day 267.
Day 190 sera from study 2 were analyzed in Figure S3. To facilitate comparisons of responses to VLPs produced in 293T cells, sera from Study 1 and groups A and B are included. We mined this data by Mann-Whitney analyses of paired groups (Figure S4). To investigate the effect producer cell line, we compared Study 1 and group 2C (both used N362Q VLPs in AS01B) and groups [A and B] (combined) and 2A (both used parent VLPs formulated in AS01B). One caveat, however, is that the precise regime in Study 2 and previous studies differ somewhat (Figure 2 and S2 and ref (Crooks et al., 2015)). Nevertheless, we hoped to document any significant leads by analyzing samples in parallel using the same assays. By similar reasoning, we compared other groups to evaluate other variables (see Figure S4).
Gp120 titers were not significantly different between most group pairings (Figure S4). A direct comparison of groups 2A and 2B that received identical regimens aside from the adjuvant revealed no significant difference. However, a comparison of group 2A and all the other Study 2 sera reached significance (P=0.0157 in Figure S4). A caveat, however, is that these regimens differed by more than just the adjuvant.
Bald VLP titers were significantly higher in rabbits immunized with 293F VLPs than those produced in 293T cells (P=0.0286 and 0.004 in Figure S4). We return to this point later below. The higher bald-VLP titers in group 2B were not quite significantly higher than those in group 2A. However, they were significant when group 2B was compared to the other Study 2 sera (P=0.0074). This latter comparison carries weight, because the differences in Env between groups are largely irrelevant for inducing bald-VLP titers and all rabbits received identical doses of VLPs that are known to induce bald VLP titers. Indeed, there were no significant differences in bald VLP titers between the other Study 2 groups. This was true even for group 2D (DNA prime, VLP boost group) that did not receive protein immunogen until the 4th shot and bald VLP titers were measured after only one additional protein boost (Figure S2). Taken together, 293F production dramatically increases bald VLP binding titers and there is a possible slight improvement in binding responses with Adjuplex.
We next compared the neutralizing responses of the same groups. Tier 1 responses did not differ significantly between groups (Figure S4). However, there was significant individual variation. Although the titers in group 2E appeared to be the lowest, they were not significantly different from those in groups 2D and 2E. Tier 2 nAbs, where measureable, were not different in most cases, except that N362Q titers were significantly higher in Study 1 sera than in group 2C (where no nAbs were detected). This trend was also true for parent virus neutralization, but did not reach significance in part due to the lack of response in rabbit 743. In fact, tier 2 nAb responses were poor throughout Study 2. Only 2 out of 20 Study 2 animals exhibited measurable ID50s. Serum 773 neutralized N362Q and parent viruses and serum 776 neutralized only the N362Q virus. It is perhaps no coincidence that both neutralizing Study 2 sera came from groups D and E (see Fig. 2) that, unlike the other groups, received plasmid DNA as well as VLP immunogens.
Serum IgG from the 773 and 776 rabbits bound to native SOS E168K+N362Q trimer (Fig 5A, upper panel). However, non-neutralizing IgG from rabbit 770 did not (Figure S3). Similar patterns were observed using SOS E168K trimers, except that serum 776 failed to bind effectively (Figure 5A, middle panel), consistent with its lack of parent virus neutralization (Figure S3). These sera also recognized D368-independent epitopes (Figure 5A, lower panel).
The data shown in Figure S3 used Study 2 sera taken 2 weeks after the day 90 boost (Figure S2). Due to the nAbs detected in rabbits 773 and 776, groups 2D and 2E were boosted once more with a mixture of parent and N362Q VLPs. However, improved nAbs titers did not develop in rabbits 773 and 776 (not shown) and nAbs did not develop in the other rabbits.
The general failure of Study 2 suggests that 293F-produced VLPs, unlike those produced in 293T cells are ineffective at inducing nAbs. We attempted to understand this result so that similar problems might be avoided in the future. In Studies 1 and 2, VLP doses were prepared from the same volumes of supernatants derived from 293T or 293F transfections. Although these cell lines derive from the same origin, their VLPs products exhibited qualitative differences. 293F VLP pellets were noticeably larger than those produced by 293T cells (Figure S5A). Furthermore, trimer staining was less intense for undigested 293F VLPs compared to 293T VLPs (Figure S5B, lanes 1–6). However, upon protease digestion prior to VLP lysis and BN-PAGE, the trimer staining was comparable (Figure S5B, lanes 7–12). VLPs coated on ELISA wells at 20× concentration and measured for antigenicity revealed significantly weaker mAb binding to 293F VLPs (Figure S5C). However, mAb 2F5 bound equivalently probably due to its previously reported ability to cross-react with lipid, as indicated by its ability to recognize bald VLPs (Tong et al., 2012). Finally, pseudovirus produced in 293F cells was very poorly infectious compared to the cognate 293T virus (Figure S2D).
DISCUSSION
Here, we set out with two objectives. First, we sought to improve the frequency of VLP-induced nAb responses by increasing access to the CD4bs. Second, we attempted to increase the cross-reactivity of initial autologous nAbs using modified VLP boosts. Regarding the first goal, we removed the N362 glycan to try to minimize potential clashes with nascent antibodies that overlap the CD4bs and thereby relax roadblocks to nAb induction (Bonsignori et al., 2016; Kong et al., 2016; Li et al., 2008; McGuire et al., 2013). Tier 2 isolates frequently lack some glycans that are present on most other circulating strains (McCoy et al., 2016). Glycan holes are also a feature of strains associated with nAb breadth in natural infection (Moore et al., 2015; van den Kerkhof et al., 2013). Modestly glycan-depleted tier 2 viruses can also transmit infection, perhaps because they function well in settings where nAb resistance is unimportant (Chohan et al., 2005; Derdeyn et al., 2004; Ping et al., 2013). Here we found that enlarging the CD4bs glycan hole led to the induction of tier 2 nAbs in 4 out of 4 (100%) of animals in Study 1 (Figure 3). However, perhaps inevitably, the bigger hole exposed additional antibody targets.
In Figure 10, we modeled the targets of the sera of the present and previous studies. All our mapped sera appeared to target epitopes close to the CD4bs. Serum 742 targeted an N197 glycan-sensitive epitope (“epitope cluster 1” in Figure 10) that was akin those of sera 347 and 613 (Crooks et al., 2015). Although the vaccine group sizes (n=4) are small, it is perhaps no coincidence that 1/4 rabbits in each group targeted this epitope (i.e., 25%; 3 out of 12 rabbits in total). The frequency of responses to this site was apparently not improved by N362 glycan removal. Indeed, the 742 serum depends heavily on the glycan hole at position 197, but is impartial to the additional space provided by the N362 glycan vacancy (Figures 1E and 9B). Like the 347 and 613 sera (Crooks et al., 2015), serum 742 cross-neutralized N197 glycan knockout mutants of both the JR-CSF and ADA strains. This may reflect the limited IgG repertoire solutions able to take advantage of the N197 glycan hole. The fact that the cross-neutralized N197 glycan-deficient viruses retain a tier 2 phenotype provides hope that inducing tier 2 breadth may be feasible.
Fig 10.

Epitope footprints of neutralizing sera. The model shown in Fig. 1C was reproduced with putative epitope footprint clusters for trimer VLP vaccine sera of the current study and two potently neutralizing rabbit sera 613 and 347 from our previous study (Crooks et al., 2015). Serum from animal 773 was not mapped because the titer was too low. Footprint sizes are intended as a guide only and may be larger than shown, based on VRC01 docking in Figure 1B.
Our attempt to improve the frequency of nAb responses to the N197 glycan hole by removing the adjacent N362 glycan had the unintended consequence of leading to responses to other, less conserved sites. Sera 740 and 741 were dependent on the N463 glycan - a fence glycan in the V5 loop (modeled as “epitope cluster 2” in Figure 10). The V5 loop has in fact been frequently reported as a target of low titer tier 2 vaccine sera (Law et al., 2007; Leaman et al., 2015; Narayan et al., 2013). The N463 glycan is present in only 20.3% of circulating viruses (analyzed from 4,265 sequences in www.hiv.lanl.gov). Other poorly conserved glycans appear at adjacent positions in the V5 loop that serve to limit access to CD4bs-specific nAbs by introducing steric clashes. In the case of VRC01 class bnAbs, V5 glycan-light chain clashes are a major mechanism of resistance (Huang et al., 2016). Although these sera also appear to clash with complex glycan head groups (Figure 8), they were distinct in that they required the N463 glycan rather than avoided it.
This is not necessarily at odds with the growing evidence that glycan holes are preferred nAb targets (Crooks et al., 2015; McCoy et al., 2016). Despite the apparent N463-dependency, these sera initially targeted the N362 glycan hole. As mentioned above, we can not rule out the possibility that the sera recognize a structure created by the glycan rather than the glycan itself. Fence glycans N362 and N463 are spatially close and it is logical that removal of the former might expose epitopes that impinge somewhat on the latter (Figure 10), especially given that CD4bs bnAbs appear to be unable to completely avoid making some glycan contacts (Stewart-Jones et al., 2016). The same is also true for certain autologous nAbs in natural infection (Wibmer et al., 2016). The 743 and 776 sera also targeted N362 glycan hole. However, unlike the 740 and 741 sera, they did not gain an ability to navigate the N362 glycan upon boosting. As a result, we classified these sera as N362Q sensitive and did not attempt to map them further (epitope cluster 3). We note however the likely overlap between clusters 2 and 3 in Figure 10. The low titer of the N362Q-insensitive serum of rabbit 773 precluded any attempt at mapping (Figure 10).
Despite the N463 glycan-dependency, the 740 serum incompletely neutralized an N463 glycan-lacking JR-FL mutant (Figure 7B) suggesting that it may contact the underlying protein. Given V5 sequence variability, it is perhaps not surprising that the 740 and 741 sera did not neutralize other strains that carry the N463 glycan (Figure 7D). Overall, the apparent frequent targeting of this site by vaccine nAbs suggests that it is relatively exposed and may be used by the virus to attract strain-specific responses that are easy to escape (Bradley et al., 2016; Wibmer et al., 2016) making it a poor target for any attempt to evolve nAb breadth.
In addition to exposing new nAb epitopes, our data hint that N362 glycan removal may have had other subtle effects. Unlike our previous study, our new sera targeted epitopes that were largely contained within gp120 monomers (Figure 5B). Furthermore, serum 742, unlike the 613 and 347 sera, was partially sensitive to the D368R mutation in the CD4 binding loop. However, it is important to point out that the small numbers of responders to each epitope preclude any firm statement regarding the consistency of these findings. Overall, our data suggest that improving the frequency of nAbs to the N197 glycan hole will require a different strategy. Rather than improving lateral access to this site by N362 glycan removal, one approach may be to minimize glycan size at priming to eliminate any potential glycan clashes, perhaps by preparing trimer VLPs in GnTI- cells (Binley et al., 2010).
Our second major goal was to improve nAbs cross-reactivity. In both vaccination and natural infection settings, initial nAbs are invariably highly strain-specific and so there may be little hope of eliciting bnAbs directly. If so, it might be possible to evolve breadth from initial autologous nAbs using modified boosts to change nAb specificity. A recent study showed that multi-strain SOSIP gp140 trimer vaccines induce very limited breadth against strains not included in the regimen (Klasse et al., 2016). Thus, it may be important to design vaccines that promote nAb development to conserved targets. Natural infection studies provide relevant paradigms of the events leading up to bnAb development (Derdeyn et al., 2014; Doria-Rose et al., 2014; Gao et al., 2014; Liao et al., 2013; Moore et al., 2012; Murphy et al., 2013; Ping et al., 2013). In some cases, breadth develops after the virus escapes autologous nAbs, as a newly introduced glycan creates a new broad site of vulnerability (Derdeyn et al., 2014; Moore et al., 2012; Ping et al., 2013; Wibmer et al., 2013). In some cases, bnAbs avoid glycan clashes by altering their angle of approach (Garces et al., 2015; Tran et al., 2014). In other cases, rather by necessity rather than preference, bnAbs acquire glycans contacts (Stewart-Jones et al., 2016; Zhou et al., 2015; Zhou et al., 2013). This contrasts with other viruses such as influenza where, despite quite high glycan content (albeit not as high as for HIV Env), nAbs rarely contact glycans (Pancera et al., 2014; Stewart-Jones et al., 2016).
In a vaccine setting, breadth may be improved simply by additional boosts, as occurred for rabbit 613, where boosting expanded nAb breadth from neutralizing 3 of 18 (like the 347 and 742 sera) to 9 of 18 clade B N197 glycan-lacking viruses (Crooks et al., 2015). However, the natural infection paradigms above suggest that targeted boosting strategies might be important. The need to broaden cross-reactivity using boosts is underlined by our sera’s preference for the precise immunogen presentation even within the vaccine strain. For example, in Figure 4, serum 742 preferentially neutralizes SOS mutant viruses over their WT counterparts, despite the negligible effect of this mutation on trimer conformation. The 4th N362Q VLP shot of this rabbit promoted an overlap of the N362 glycan hole, suggesting that boosting can narrow as well as broaden cross-reactivity.
The acute effects of marginal antigenic differences on antibody targeting we discussed above together the inability of multi-strain prime boost approaches to induce breadth, suggest that expanding nAb cross-reactivity may at least initially require a subtler, rationally designed boosting approach. One approach is to select prospective boosts based on their recognition by interim sera. Unlike the multi-strain strategies, this “affinity drop” concept (Steichen et al., 2016) should ensure that boosts will reactivate pre-existing clones. Incremental changes induced by this approach might lead nAbs on a rational path to breadth.
In keeping with the affinity drop concept, Study 1 rabbits were boosted with parent VLPs mixed with the index N362Q VLPs. A kinetic analysis of rabbits 740 and 741 (Figure 4) suggested that cross-reactivity with JR-FL variants may have been promoted by these boosts, especially in rabbit 740. However, as mentioned elsewhere, VLP production challenges precluded the larger rabbits that would be needed to identify consistent patterns.
Given its conserved epitope, had rabbit 742 survived, the affinity drop approach might have been more valuable. In this case, potential boosts could be designed to i) increase cross-reactivity with N197 knockout mutants of other Env strains and ii) navigate the N197 glycan. Clearly, the value of these ideas need to be determined experimentally, with the only stipulation that boosts are recognized preceding sera. A complete strategy may also require an initial sensitive prime (e.g. GnTI- VLPs) to promote consistent nAb responses in all vaccinees.
Glycan holes are likely to remain a focus of vaccine development in the forseeable future. However, several factors need to be considered. One is that they should target conserved sites where a glycan is normally present in most strains. Thus, N197 hole (98.7% conserved) is preferable to one at position 463 (20.3% conserved). It may also be useful to plug holes at highly variable holes with glycan to prevent unwanted responses. However, it may be no coincidence that the glycan holes targeted by autologous vaccine nAbs reported so far usually occur at reasonably conserved positions, for example N197 (98.7%), N230 (32.7%) N241 (97%), N289 (69.4%), N611 (98.4%) (Crooks et al., 2015; McCoy et al., 2016). Glycan absences at these sites appear to have greater consequences than those at more variable sites. A second factor is antigenic context. For example, while JR-FL strain VLPs induced nAbs to the 197 glycan hole, a JR-FL gp160 DNA prime, protein boost regimen targeted the N230 glycan hole (Crooks et al., 2015), perhaps due to the subtle antigenic differences between these antigens. A third factor is that when Env glycans are absent, neighboring glycans may reposition and/or differentiate into larger multi-branched glycans that may interlock in new ways to partially occupy the vacated space, thereby complicating their ability to induce nAbs (Behrens et al., 2016; Stewart-Jones et al., 2016; Zhou et al., 2016).
Previous studies showed that removing the N276 glycan improves VRC01 class germline priming (Jardine et al., 2013; Jardine et al., 2016; McGuire et al., 2013). However, a high sensitivity to the mature VRC01 bnAb (Figure 1D) led us to choose the N362Q mutant here. In a study by another group, removing four CD4bs fence glycans from BG505 SOSIP trimers (Zhou et al., 2016) led to dramatically improved (>1,000 fold) neutralizing ID50s to the CD4bs of the glycan-depleted parent strain that could, to some extent, neutralize other isolates if the fence was similarly dismantled. However, as in our study, a drawback is that these nAbs do not effectively navigate the intact glycans of the parent virus. For these reasons, the subtler approaches we describe above may be more effective.
The need for high VLPs doses raises questions about their prospects for process development. Our current VLP platform is largely intended for proof of concept. Over the last decade, we have optimized plasmid combinations to improve trimer output by at least 10-fold per unit culture (Crooks et al., 2007; Crooks et al., 2015; Tong et al., 2014). Our investigation of 293F VLPs was intended to eliminate this production bottleneck, but was unsuccessful, apparently due to contaminating material, possibly vesicles. An alternative platform to investigate membrane trimers that we are now investigating is to extract membrane trimers, then reconstitute them into liposomes or nanodiscs (Frauenfeld et al., 2016; Nakatani-Webster et al., 2015). In the meantime, the consistently positive nAb responses to N362Q mutant VLPs observed here should facilitate immediate efforts to formally re-evaluate the VLP dose required to induce nAbs, which could temporarily alleviate our current challenges in evaluating concepts with full concurrent controls (current doses are high in part to err on the side of caution).
Overall, our study provides some encouraging leads for vaccine development. The frequency of autologous tier 2 nAbs appeared to be improved by enlarging the CD4bs glycan hole. Targeted glycan holes at conserved positions might be most effective if used as part of a composite strategy with other glycan modifications and heterologous boosts selected by the affinity drop concept. A continued commitment to iterative, rational improvements may in future bring us closer to our goal of inducing tier 2 breadth.
MATERIALS and METHODS
Anti-HIV-1 Env monoclonal antibodies
Monoclonal antibodies (mAbs) were obtained from their producers, the NIH AIDS reagent repository, or from commercial suppliers. Information on these mAbs can be found at the web link: (www.hiv.lanl.gov). Our mAb panel included the following (originators given in parentheses): 2G12 (Katinger), directed to a unique glycan-dependent epitope of gp120 (Scanlan et al., 2002); 39F and CO11 (J. Robinson), directed to the gp120 V3 loop (Crooks et al., 2005; Tong et al., 2012; Tong et al., 2014); b12 (Burton), VRC01 and VRC13 (Mascola), 8ANC131 (Nussenzweig), HJ16 (Lanzavecchia), 15e (J.Robinson), directed to epitopes that overlap the CD4bs (Scheid et al., 2011; Tong et al., 2012; Wu et al., 2010; Zhou et al., 2015); PGT121 (Burton) directed to epitopes involving the base of the V3 loop of gp120 and the N332 glycan (Walker et al., 2011); PG9 and PG16 (Burton); directed to quaternary, glycan-dependent epitopes that involve the V2 loop (Doria-Rose et al., 2014; Walker et al., 2009); 7B2 and 2.2B (J. Robinson), directed to the gp41 cluster I and II epitopes, respectively (Moore et al., 2012); 4E10 and 2F5 (Katinger), directed to the gp41 membrane-proximal ectodomain region (MPER) (Huang et al., 2012).
Recombinant gp120 monomer, gp41, soluble CD4 and CD4-IgG
Recombinant monomeric JR-FL gp120 and mutants thereof were produced in 293T cells and purified from supernatants over Galanthus nivalis agarose. Soluble CD4 (sCD4) consisting of its 4 outer domains was a gift from Progenics Pharmaceuticals (Tarrytown, NY).
Plasmids and mutagenesis
Plasmid pCAGGS was used to express JR-FL gp160ΔCT on VLP surfaces (Moore et al., 2006). Gp160ΔCT is truncated at amino acid 709, leaving a 3 amino acid gp41 cytoplasmic tail. This increases native trimer expression and produces pseudoviruses with similar neutralization sensitivity profiles compared to their full-length gp160 counterparts (Crooks et al., 2005). The use of the JR-FL Env strain has several advantages, including efficient expression and gp120/gp41 processing (Crooks et al., 2005; Moore et al., 2006). Mutants were generated by Quikchange (Agilent Technologies) and were numbered per the HXB2 reference strain (Binley et al., 2008). “SOS” mutations (A501C and T506C) introduce an intermolecular disulfide bond between gp120 and gp41 (Binley et al., 2000). The E168K mutation knocks in the broadly neutralizing “PG” epitopes that are normally absent in the JR-FL isolate and increases trimer expression (Tong et al., 2012). Mutants D197N, N276A, N362Q, N386A, N392Q and N463Q knock-in or knockout glycans that line the CD4bs. The A328G mutation dramatically enhances neutralization sensitivity, otherwise known as “global” sensitivity (Tong et al., 2012). The D368R mutation abrogates CD4 binding capability.
Plasmids expressing gp160 from other strains were obtained from the NIH AIDS Reagent Repository. Mutant versions of these sequences in which the N197 glycan is eliminated were produced. Another set of plasmids encoded a series of domain-exchanged hybrid gp160s using sequences from the JR-FL and JR-CSF isolates (Narayan et al., 2013). Plasmids used to make VLPs included pMV-ERV Gag (expresses endogenous murine leukemia virus Gag under the control of a CMV promoter (Bashratyan et al., 2017) and pMV-Rev 0932 (expresses codon-optimized HIV-1 Rev under the control of a CMV promoter).
VLP production
VLPs were produced by co-transfecting 293T cells with an Env-expressing plasmid (typically pCAGGS JR-FL gp160ΔCT SOS E168K and mutants thereof and, in select cases, p0001 JR-CSF gp160ΔCT SOS), pMV-ERV Gag and pMV-0932, using polyethyleneimine (PEI Max), as described previously (Tong et al., 2012). Two days later, supernatants were collected, precleared by low speed centrifugation, filtered and pelleted at 50,000 × g in a Sorvall SS34 rotor. To remove residual medium, VLP pellets were diluted with 1ml of PBS, then re-centrifuged at 15,000 rpm and resuspended in PBS at 1,000 × the original concentration. VLPs were referred to as WT-VLPs or SOS-VLPs, depending on the form of Env displayed on their surfaces or as bald-VLPs, bearing no Env, produced by transfecting pNL4-3.Luc.R-E- alone (Crooks et al., 2007). “Trimer-VLPs” were made by digestion using a cocktail of proteases including proteinase K, subtilisin, trypsin and chymotrypsin, as previously described (Crooks et al., 2011; Tong et al., 2012). VLPs were inactivated using aldrithiol (AT-2) (Crooks et al., 2007). VLPs were also made in 293 Expi freestyle cells. Following transfection, cells were harvested when they reached ~60% viability, at which point, cells were pre-cleared by low speed centrifugation. The supernatant was then filtered with 1.2 μm MWCO dead-end filter and concentrated by tangential flow filtration (100 kDa filter) to approximately 100–200× the initial volume.
Animal immunizations
Species
New Zealand white rabbits were housed and immunized at the Pocono Rabbit Farm (Canadensis, PA) – a site that has been approved by the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC). All animals were fed and handled in strict accordance with the recommendations of the NIH Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act.
All immunization protocols were approved (protocol PRF2A) by the Explora Biolabs Animal Care and Use Committee (IACUC). Explora Biolabs’ animal welfare assurance (AWA) number is A4487-01. Pocono Rabbit Farm is approved for rabbit and guinea pig immunizations by the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC). The AWA number Pocono Rabbit Farm is A3886-01. For all immunization and bleed protocols, pain and distress was slight and momentary and did not affect animal health. Discomfort and injury to animals was limited to that which is unavoidable in the conduct of scientifically valuable research. Analgesics, anesthetics, and tranquilizing drugs were used as necessary by veterinary staff. After the completion of all immunizations and bleeds, rabbits were euthanized by injection of an overdose of anesthesia per NIH guidelines. Ketamine and xyalzine were administered intramuscularly at 35mg/kg and 5mg/kg, respectively, followed by exsanguination via cardiac puncture.
Procedures for adverse reactions to immunization followed established practices that include measures for dealing with unexpected outcomes. For example, if an animal had an adverse reaction at the injection site, depending on the severity, the response would be either i) none in the case of mild reactions, ii) anesthetic in the case of medium reaction, and iii) veterinary staff would be brought in for severe cases, in which it may be necessary to euthanize via an overdose of pentobarbital (100mg/kg i.v.), per guidelines established in 1993 by the American Veterinary Medical Association Panel on Euthanasia. However, in this study, reactions to all procedures were only mild and no animals were euthanized over the course of immunizations and bleeds. Two rabbits, however (742 and 743 in Study 1) died unexpectedly, but this was unrelated to the immunization and bleed procedures.
Immunizations
Six groups of 4 female rabbits were immunized in two studies, as outlined in Figures 2 and S2. In all groups, each dose of VLPs was harvested from a total of 1L transfection supernatant for initial shots and 500mL for boosts. Standard serum volumes were drawn on the day of each immunization and two weeks thereafter. Study 1. One group of 4 rabbits were immunized intramuscularly with VLPs formulated in AS01B (Glaxo SmithKline, consisting of liposomes containing deacylated monophosphoryl lipid A and QS-21). VLP doses were determined by documenting their relative antigenicity compared with JR-FL gp120 by ELISA, using various mAbs, as we described previously (Tong et al., 2014). As depicted in Fig. 2, initial shots consisted of JR-FL SOS N362Q trimer VLPs, but later shots included mixtures of VLPs.
Study 2
Five groups of 4 rabbits were immunized to try to evaluate adjuvant and plasmid DNA immunizations. Groups A and B were immunized intramuscularly with JR-FL SOS E168K parent trimer VLPs formulated in either AS01B or Adjuplex, respectively. Adjuplex was obtained from Advanced BioAdjuvants, and consists of purified lecithin and carbomer homopolymer. Group C was a protein only immunogen control group for DNA immunization Groups D and E. Group C rabbits received 3 shots of JR-FL SOS N362Q trimer VLPs formulated in AS01B, followed by boost containing the same index immunogen mixed with JR-FL SOS parent trimer VLPs also formulated in AS01B. Group D rabbits received 3 shots of plasmid DNA, consisting of 227μg pCAGGS JR-FL SOS E168K+N362Q, 227μg pMV-ERV Gag and 46μg of pMV-Rev 0932 that was administered intramuscularly without adjuvant using a BTX ECM 830 electroporation device. Protein boosts consisted of VLPs mixed with AS01B. Group E rabbits received concomitant priming immunizations of plasmid DNA (as above) and VLPs. These immunogens were administered separately, one in each hind leg. Rabbits were later boosted with trimer VLP mixtures in AS01B, as indicated in Fig. S2.
Reference sera and plasmas
Reference controls included a pooled serum generated from six rabbits that were immunized with monomeric gp120 in AS01B, as described previously (Tong et al., 2014). We also included rabbit sera from our previous study (Crooks et al., 2015). HIV-1-infected donor plasma 1702 and uninfected control plasma 210 were also described previously (Binley et al., 2008; Tong et al., 2014).
ELISAs using recombinant gp120 and VLPs
ELISAs were used to measure serum binding to various antigens and were also used to determine their specificities (Tong et al., 2012; Tong et al., 2014; Tong et al., 2013). Briefly, Immulon II plates were coated with 20× concentrated VLPs, recombinant gp120 at 5μg/ml overnight at 4°C. Following a PBS wash and blocking, sera were titrated against each antigen in blocking buffer. Species-specific alkaline phosphatase anti-Fc conjugates (Accurate, Westbury, NY) and SigmaFAST p-nitrophenyl phosphate tablets (Sigma) were then used to detect binding. Plates were read at 405nm. Titers are taken when ELISA signals exhibited an optical density of 0.5 (approximately 3× background).
Blue Native PAGE (BN-PAGE)-Western blots
Blue native PAGE (BN-PAGE) was performed as described previously (Crooks et al., 2007; Crooks et al., 2005; Moore et al., 2006). Briefly, VLPs were solubilized in 0.12% Triton X-100 in 1 mM EDTA. An equal volume of 2× sample buffer (100 mM morpholinepropanesulfonic acid (MOPS), 100 mM Tris HCl, pH 7.7, 40% glycerol, and 0.1% Coomassie blue) was added. Samples were then loaded onto a 4–12% Bis-Tris NuPAGE gel (Invitrogen) and separated at 4°C for 3 hours at 100V. The gel was then blotted onto polyvinylidene difluoride membrane, destained, immersed in blocking buffer (4% nonfat milk in PBS) and probed with an anti-gp120 cocktail (mAb b12 and 39F at 1μg/ml) and/or an anti-gp41 cocktail (mAb 2F5, 4E10, 7B2, 2.2B at 1μg/ml). Blots were then probed by an anti-human Fc alkaline phosphatase conjugate (Accurate Chemicals) and developed using SigmaFast BCIP/NBT substrate (Sigma).
BN-PAGE “shift” assays were used to measure the ability of antibodies to bind and deplete the unliganded trimer (Crooks et al., 2007; Crooks et al., 2005; Moore et al., 2006; Tong et al., 2012). VLPs were incubated with mAb or protein A-purified serum IgG for 1h at 37°C, then washed with PBS and resolved by BN-PAGE-Western blot, as above.
Neutralization assays
Heat-inactivated sera were analyzed for neutralization of various pseudoviruses produced by co-transfecting either 293T cells with an Env plasmid and pNL4-3.Luc.R-E- (CF2 assays). Data is representative of at least 3 repeat assays performed in duplicate. Neutralization assays using CF2 cell targets.
Neutralization assays using canine CF2 cells expressing CD4 and CCR5 receptors (CF2Th.CD4.CCR5) have been described previously (Crooks et al., 2005). Briefly, virus was incubated with graded dilutions of mAb or serum for 1 h at 37°C. The mixture was then added to CF2 cells, spinoculated at 300 × g for 15 min, incubated at 37°C. For WT viruses, cells were cultured for a further 3 days, then luciferase activity was measured. For SOS mutant viruses, after 2h, the medium was removed and infection was activated by adding 5mM DTT for 5 minutes, followed by a wash (Crooks et al., 2005) and culturing for a further 3 days, after which luciferase activity was measured.
Monomeric gp120 interference of neutralization
In modified neutralization assays, fixed concentrations (~1μg/ml) of recombinant JR-FL gp120 monomer trimer carrying N362Q and D386R mutations, was added to the virus-antibody mixture to test for its ability to interfere with the neutralizing activity in the test serum.
Statistical analyses
Immunization groups were compared using Mann-Whitney tests. For neutralization titer comparisons, undetectable titers were given an arbitrary titer of 4 for calculations.
Molecular modeling
The recently reported full-length JR-FL Env structure of (PDB: 5FUU) (Lee et al., 2016) was used to generate a visual model for the distribution of glycans on the native spike. First, atomic clashes present in the 5FUU structure were relieved and missing side-chains rebuilt by executing 1,000 symmetric ROSETTA-fixbb simulations, selecting the lowest scoring model, and then running a constrained ROSETTA-relax simulation. Each N-linked glycosylation motif was decorated with a Man8GlcNAc2 glycan. The glycan at position N637 in gp41 is absent, per evidence that one or other glycans at N625 and N637 remain unoccupied (Go et al., 2008). For any overlapping sequons, for example those at N188 and 189, only the first sequon is occupied. GlycanRelax (Pancera et al., 2010) was used to approximate the conformational behavior of glycans in a glycoprotein context. For each model, 10 separate GlycanRelax trajectories of 10,000 cycles of MonteCarlo trials were carried out. Each gp120 glycan could move independently throughout the GlycanRelax minimization. A single low energy model was chosen and figures were generated in PyMOL Molecular Modeling Software (Version 1.5.0.4 Schrödinger, LLC).
Ethics Statement
The archived adult human plasmas used in this study have previously been described (Binley et al., 2008; Tong et al., 2014). All donors provided written consent for the use of these samples. Institutional Review Board (IRB) approval for this project was obtained through the San Diego Biomedical Research Institute IRB Committee (approval number: IRB-14-04-JB; Federal Wide Assurance number: 00021327).
All immunization protocols for rabbits were approved (protocol PRF2A) by the Explora Biolabs Animal Care and Use Committee (IACUC). Explora Biolabs’ animal welfare assurance (AWA) number is A4487-01. Pocono Rabbit Farm are approved for rabbit immunizations by the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC). The AWA number Pocono Rabbit Farm is A-3886-01. All animals were fed, housed and handled in strict accordance with the recommendations of the NIH Guide for the Care and Use of Laboratory Animals, the Animal Welfare Act and Regulations and guidelines established in 1993 by the American Veterinary Medical Association Panel on Euthanasia.
Supplementary Material
Figure S1: Comparison of structural model sequences and the full JR-FL Env sequence. The 3 Env subunit protomers of our model (A, B and C derived from pdb 5FUU) were aligned with the complete JR-FL Env sequence (shown at the top) (http://www.uniprot.org/uniprot/Q6BC19). Regions of disorder in the structure in the C1, V1 and V2 regions shown as dashes in the alignment are not rendered in the models. Besides the missing amino acids, this also entails the loss of one or two potential glycans in the V1 and the V4 regions of each protomer. The sequons at positions 189 and 637 (HXB numbering, highlighted in magenta) do not carry a glycan in the models shown in Figures 1 and 10, reflecting natural overlap or competition with neighboring glycans at these sites (Go et al., 2008).
Figure S2: Overview of Study 2 rabbit immunizations. 5 groups of 4 rabbits were immunized. Details or immunization type (VLPs or plasmid DNA), route (intramuscular injection) and dose are provided in Materials and Methods. VLPs were produced in 293F cells and formulated in adjuvants AS01B or Adjuplex. The timeline shows the days of each immunization and bleeds (B1–B6) taken two weeks thereafter.
Figure S3: Study 2 serum binding and neutralization profiles. Peak time points for Study 2 sera were compared to replots of Study 1 sera and groups A and B from our previous study (Crooks et al., 2015). For clarity and to facilitate comparisons, the VLP Env (parent or N362Q), regimen, adjuvant and producer cells are indicated. A) ELISA titers against recombinant JR-FL gp120 monomer and bald-VLPs. Data are means of at least two repeats. Neutralizing activity against B) the tier 1 mutant JR-FL WT A328G (Tong et al., 2014) and SIVmac239, and C) the JR-FL SOS E168K+N362Q index strain and parent JR-FL SOS E168K measured in CF2Th.CD4.CCR5 cells. Neutralization assays were performed at least three times in duplicate. Mean ID50s and standard deviations are shown.
Figure S4: Mann-Whitney analysis of Study 2 data and reference samples. Mann-Whitney tests were performed on pairs of groups as listed in the first column. The concept these comparisons sought to test are shown in the second column. P values for each kind of data in each column are shown in the remaining columns, where significant differences (i.e. <0.05) between groups are highlighted in red.
Figure S5: Comparison of VLPs produced from 293T and 293F cells. A) Size of 1,000× concentrated pellet from a 25ml culture of the two cell types. Note that ~12-fold higher numbers of non-adherent 293F cells are used to seed a given volume than adherent 293T cells. Each cell type was transfected with plasmids expressing JR-FL SOS E168K using MuLV Gag and Rev. Photographs of VLP pellets harvested at day 3 and 7 after transfection are shown. B) Env expression in the two VLP preparations (harvested at day 3) was compared at various dilutions by BN-PAGE-Western blot. C) Antigenicity of trimer 293T and 293F VLPs was assessed by ELISA using serial dilutions of mAbs. D) Infectivity of JR-FL WT parent pseudovirus supernatants produced in both cell types in a single round assay using CF2 target cells.
Acknowledgments
This work was supported by grants RO1AI00790, RO1AI93278 and R33AI84714 (J.M.B.). We thank IAVI and NIH AIDS Reagent Programs and the individual suppliers for providing mAbs, Clarisse Lorin for providing AS01B, Emily Carrow (Advanced BioAdjuvants) for providing Adjuplex; Progenics Pharmaceuticals, Inc. for providing gp120 and sCD4; Scott Conklin (Pocono Rabbit Farm) for assistance with rabbit immunizations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahamyan LG, Mkrtchyan SR, Binley J, Lu M, Melikyan GB, Cohen FS. The Cytoplasmic Tail Slows the Folding of Human Immunodeficiency Virus Type 1 Env from a Late Prebundle Configuration into the Six-Helix Bundle. J Virol. 2005;79:106–115. doi: 10.1128/JVI.79.1.106-115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsahafi N, Debbeche O, Sodroski J, Finzi A. Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer. PLoS One. 2015;10:e0122111. doi: 10.1371/journal.pone.0122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alving CR. 4E10 and 2F5 monoclonal antibodies: binding specificities to phospholipids, tolerance, and clinical safety issues. AIDS. 2008;22:649–651. doi: 10.1097/QAD.0b013e3282f51922. [DOI] [PubMed] [Google Scholar]
- Bashratyan R, Regn D, Rahman J, Marquardt K, Fink EA, Hu WY, Elder JH, Binley J, Sherman LA, Dai YD. Type 1 diabetes pathogenesis is modulated by spontaneous autoimmune responses to endogenous retrovirus antigens in NOD mice. European Journal of Immunology accepted. 2017 Jan 3; doi: 10.1002/eji.201646755. 2017. [DOI] [PubMed] [Google Scholar]
- Behrens AJ, Vasiljevic S, Pritchard LK, Harvey DJ, Andev RS, Krumm SA, Struwe WB, Cupo A, Kumar A, Zitzmann N, Seabright GE, Kramer HB, Spencer DI, Royle L, Lee JH, Klasse PJ, Burton DR, Wilson IA, Ward AB, Sanders RW, Moore JP, Doores KJ, Crispin M. Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhiman JN, Anthony C, Doria-Rose NA, Karimanzira O, Schramm CA, Khoza T, Kitchin D, Botha G, Gorman J, Garrett NJ, Abdool Karim SS, Shapiro L, Williamson C, Kwong PD, Mascola JR, Morris L, Moore PL. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat Med. 2015;21:1332–1336. doi: 10.1038/nm.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Ban YE, Crooks ET, Eggink D, Osawa K, Schief WR, Sanders RW. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. Journal of virology. 2010;84:5637–5655. doi: 10.1128/JVI.00105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion- associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blish CA, Nguyen MA, Overbaugh J. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 2008;5:e9. doi: 10.1371/journal.pmed.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, Alam SM, Bradley T, Gladden MA, Hwang KK, Iyengar S, Kumar A, Lu X, Luo K, Mangiapani MC, Parks RJ, Song H, Acharya P, Bailer RT, Cao A, Druz A, Georgiev IS, Kwon YD, Louder MK, Zhang B, Zheng A, Hill BJ, Kong R, Soto C, Program NCS, Mullikin JC, Douek DC, Montefiori DC, Moody MA, Shaw GM, Hahn BH, Kelsoe G, Hraber PT, Korber BT, Boyd SD, Fire AZ, Kepler TB, Shapiro L, Ward AB, Mascola JR, Liao HX, Kwong PD, Haynes BF. Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell. 2016;165:449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley T, Fera D, Bhiman J, Eslamizar L, Lu X, Anasti K, Zhang R, Sutherland LL, Scearce RM, Bowman CM, Stolarchuk C, Lloyd KE, Parks R, Eaton A, Foulger A, Nie X, Karim SS, Barnett S, Kelsoe G, Kepler TB, Alam SM, Montefiori DC, Moody MA, Liao HX, Morris L, Santra S, Harrison SC, Haynes BF. Structural Constraints of Vaccine-Induced Tier-2 Autologous HIV Neutralizing Antibodies Targeting the Receptor-Binding Site. Cell Rep. 2016;14:43–54. doi: 10.1016/j.celrep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B. HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science. 2015;349:191–195. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Pancera M, Bossert A, Schmidt SD, Chen RE, Chen X, Druz A, Narpala S, Doria-Rose NA, McDermott AB, Kwong PD, Mascola JR. Immunogenicity of a Prefusion HIV-1 Envelope Trimer in Complex with a Quaternary-Structure-Specific Antibody. J Virol. 2015;90:2740–2755. doi: 10.1128/JVI.02380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Moore PL, Franti M, Cayanan CS, Zhu P, Jiang P, de Vries RP, Wiley C, Zharkikh I, Schulke N, Roux KH, Montefiori DC, Burton DR, Binley JM. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology. 2007;366:245–262. doi: 10.1016/j.virol.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Moore PL, Richman D, Robinson J, Crooks JA, Franti M, Schulke N, Binley JM. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum Antibodies. 2005;14:101–113. [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O’Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site. PLoS Pathog. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Tong T, Osawa K, Binley JM. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol. 2011;85:5825–5839. doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye S, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bonjier I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, Labranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Moore PL, Morris L. Development of broadly neutralizing antibodies from autologous neutralizing antibody responses in HIV infection. Curr Opin HIV AIDS. 2014;9:210–216. doi: 10.1097/COH.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Burton DR. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J Virol. 2010;84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Program NCS, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfeld J, Loving R, Armache JP, Sonnen AF, Guettou F, Moberg P, Zhu L, Jegerschold C, Flayhan A, Briggs JA, Garoff H, Low C, Cheng Y, Nordlund P. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods. 2016;13:345–351. doi: 10.1038/nmeth.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, Lynch RM, Alam SM, Moody MA, Ferrari G, Berrong M, Kelsoe G, Shaw GM, Hahn BH, Montefiori DC, Kamanga G, Cohen MS, Hraber P, Kwong PD, Korber BT, Mascola JR, Kepler TB, Haynes BF. Cooperation of B Cell Lineages in Induction of HIV-1-Broadly Neutralizing Antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, Sanders RW, Ward AB, Wilson IA. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao HX, Sutherland LL, Alam SM, Haynes BF, Desaire H. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes’ accessibility. J Proteome Res. 2008;7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristick HB, von Boehmer L, West AP, Jr, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, Bjorkman PJ. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydarchi B, Center RJ, Gonelli C, Muller B, Mackenzie C, Khoury G, Lichtfuss M, Rawlin G, Purcell DF. Repeated Vaccination of Cows with HIV Env gp140 during Subsequent Pregnancies Elicits and Sustains an Enduring Strong Env-Binding and Neutralising Antibody Response. PLoS One. 2016;11:e0157353. doi: 10.1371/journal.pone.0157353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, de Taeye SW, Sok D, Ozorowski G, Deresa I, Stanfield R, Ward AB, Burton DR, Klasse PJ, Sanders RW, Moore JP, Crotty S. Murine Antibody Responses to Cleaved Soluble HIV-1 Envelope Trimers Are Highly Restricted in Specificity. J Virol. 2015;89:10383–10398. doi: 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, O’Dell S, Chuang GY, Druz A, Georgiev IS, Schramm CA, Zheng A, Joyce MG, Asokan M, Ransier A, Darko S, Migueles SA, Bailer RT, Louder MK, Alam SM, Parks R, Kelsoe G, Von Holle T, Haynes BF, Douek DC, Hirsch V, Seaman MS, Shapiro L, Mascola JR, Kwong PD, Connors M. Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity. 2016;45:1108–1121. doi: 10.1016/j.immuni.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereno-Orbea J, Kalyuzhniy O, Deresa I, Hu X, Spencer S, Jones M, Georgeson E, Adachi Y, Kubitz M, deCamp AC, Julien JP, Wilson IA, Burton DR, Crotty S, Schief WR. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, Ringe RP, Golabek M, van Gils MJ, Guttman M, Lee KK, Wilson IA, Butera ST, Ward AB, Montefiori DC, Sanders RW, Moore JP. Sequential and Simultaneous Immunization of Rabbits with HIV-1 Envelope Glycoprotein SOSIP.664 Trimers from Clades A, B and C. PLoS Pathog. 2016;12:e1005864. doi: 10.1371/journal.ppat.1005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Ju B, Chen Y, He L, Ren L, Liu J, Hong K, Su B, Wang Z, Ozorowski G, Ji X, Hua Y, Chen Y, Deller MC, Hao Y, Feng Y, Garces F, Wilson R, Dai K, O’Dell S, McKee K, Mascola JR, Ward AB, Wyatt RT, Li Y, Wilson IA, Zhu J, Shao Y. Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor. Immunity. 2016;44:939–950. doi: 10.1016/j.immuni.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human Antibodies that Neutralize HIV-1: Identification, Structures, and B Cell Ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M, Cardoso RM, Wilson IA, Burton DR. Antigenic and immunogenic study of membrane-proximal external region-grafted gp120 antigens by a DNA prime-protein boost immunization strategy. J Virol. 2007;81:4272–4285. doi: 10.1128/JVI.02536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman DP, Lee JH, Ward AB, Zwick MB. Immunogenic Display of Purified Chemically Cross-Linked HIV-1 Spikes. J Virol. 2015;89:6725–6745. doi: 10.1128/JVI.03738-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- Li Y, Cleveland B, Klots I, Travis B, Richardson BA, Anderson D, Montefiori D, Polacino P, Hu SL. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol. 2008;82:638–651. doi: 10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Program NCS, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM Structure of a Fully Glycosylated Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013 doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy LE, Falkowska E, Doores KJ, Le K, Sok D, van Gils MJ, Euler Z, Burger JA, Seaman MS, Sanders RW, Schuitemaker H, Poignard P, Wrin T, Burton DR. Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies. PLoS Pathog. 2015;11:e1005110. doi: 10.1371/journal.ppat.1005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy LE, Quigley AF, Strokappe NM, Bulmer-Thomas B, Seaman MS, Mortier D, Rutten L, Chander N, Edwards CJ, Ketteler R, Davis D, Verrips T, Weiss RA. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J Exp Med. 2012;209:1091–1103. doi: 10.1084/jem.20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, Kulp DW, Macauley MS, Sok D, Pauthner M, Menis S, Cottrell CA, Torres JL, Hsueh J, Schief WR, Wilson IA, Ward AB, Sanders RW, Burton DR. Holes in the Glycan Shield of the Native HIV Envelope Are a Target of Trimer-Elicited Neutralizing Antibodies. Cell Rep. 2016;16:2327–2338. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Glenn JA, Lippy A, Stamatatos L. Diverse Recombinant HIV-1 Envs Fail To Activate B Cells Expressing the Germline B Cell Receptors of the Broadly Neutralizing Anti-HIV-1 Antibodies PG9 and 447–52D. J Virol. 2014;88:2645–2657. doi: 10.1128/JVI.03228-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, Schief WR, Stamatatos L. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O’Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero M, Gulzar N, Klaric KA, Donald JE, Lepik C, Wu S, Tsai S, Julien JP, Hessell AJ, Wang S, Lu S, Burton DR, Pai EF, Degrado WF, Scott JK. Neutralizing epitopes in the membrane-proximal external region of HIV-1 gp41 are influenced by the transmembrane domain and the plasma membrane. J Virol. 2012;86:2930–2941. doi: 10.1128/JVI.06349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. Nature of Nonfunctional Envelope Proteins on the Surface of Human Immunodeficiency Virus Type 1. J Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, Lambson BE, Ranchobe N, Ping L, Ngandu N, Abdool Karim Q, Abdool Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Ranchobe N, Lambson BE, Gray ES, Cave E, Abrahams MR, Bandawe G, Mlisana K, Abdool Karim SS, Williamson C, Morris L. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009;5:e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Williamson C, Morris L. Virological features associated with the development of broadly neutralizing antibodies to HIV-1. Trends Microbiol. 2015;23:204–211. doi: 10.1016/j.tim.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MK, Yue L, Pan R, Boliar S, Sethi A, Tian J, Pfafferot K, Karita E, Allen SA, Cormier E, Goepfert PA, Borrow P, Robinson JE, Gnanakaran S, Hunter E, Kong XP, Derdeyn CA. Viral escape from neutralizing antibodies in early subtype A HIV-1 infection drives an increase in autologous neutralization breadth. PLoS Pathog. 2013;9:e1003173. doi: 10.1371/journal.ppat.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani-Webster E, Hu SL, Atkins WM, Catalano CE. Assembly and characterization of gp160-nanodiscs: A new platform for biochemical characterization of HIV envelope spikes. J Virol Methods. 2015;226:15–24. doi: 10.1016/j.jviromet.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan KM, Agrawal N, Du SX, Muranaka JE, Bauer K, Leaman DP, Phung P, Limoli K, Chen H, Boenig RI, Wrin T, Zwick MB, Whalen RG. Prime-boost immunization of rabbits with HIV-1 gp120 elicits potent neutralization activity against a primary viral isolate. PLoS One. 2013;8:e52732. doi: 10.1371/journal.pone.0052732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Doyle-Cooper C, Cooper AB, Huber M, Falkowska E, Doores KJ, Hangartner L, Le K, Sok D, Jardine J, Lifson J, Wu X, Mascola JR, Poignard P, Binley JM, Chakrabarti BK, Schief WR, Wyatt RT, Burton DR, Nemazee D. Anti-HIV B Cell Lines as Candidate Vaccine Biosensors. Journal of immunology. 2012;189:4816–4824. doi: 10.4049/jimmunol.1202165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping LH, Joseph SB, Anderson JA, Abrahams MR, Salazar-Gonzalez JF, Kincer LP, Treurnicht FK, Arney L, Ojeda S, Zhang M, Keys J, Potter EL, Chu H, Moore P, Salazar MG, Iyer S, Jabara C, Kirchherr J, Mapanje C, Ngandu N, Seoighe C, Hoffman I, Gao F, Tang Y, Labranche C, Lee B, Saville A, Vermeulen M, Fiscus S, Morris L, Karim SA, Haynes BF, Shaw GM, Korber BT, Hahn BH, Cohen MS, Montefiori D, Williamson C, Swanstrom R, Study, C.A.I., the Center for, H.I.V.A.V.I.C. Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol. 2013;87:7218–7233. doi: 10.1128/JVI.03577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of a1–>2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, Nason MC, Montefiori D, Moldt B, Poignard P, Diskin R, Bjorkman PJ, Eckhaus MA, Klein F, Mouquet H, Cetrulo Lorenzi JC, Gazumyan A, Burton DR, Nussenzweig MC, Martin MA, Nishimura Y. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, Briney B, Schiffner T, Garces F, Freund NT, Gitlin AD, Menis S, Georgeson E, Kubitz M, Adachi Y, Jones M, Mutafyan AA, Yun DS, Mayer CT, Ward AB, Burton DR, Wilson IA, Irvine DJ, Nussenzweig MC, Schief WR. HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, AJ B, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CC, Wu CY, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong CH, Mascola JR, Kwong PD. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T, Crooks ET, Osawa K, Binley JM. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J Virol. 2012;86:3574–3587. doi: 10.1128/JVI.06938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T, Crooks ET, Osawa K, Robinson JE, Barnes M, Apetrei C, Binley JM. Multi-parameter exploration of HIV-1 virus-like particles as neutralizing antibody immunogens in guinea pigs, rabbits and macaques. Virology. 2014:456–457. doi: 10.1016/j.virol.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T, Osawa K, Robinson JE, Crooks ET, Binley JM. Topological analysis of HIV-1 glycoproteins expressed in situ on virus surfaces reveals tighter packing but greater conformational flexibility than for soluble gp120. J Virol. 2013;87:9233–9249. doi: 10.1128/JVI.01145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, Karlsson Hedestam GB, Wyatt RT. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A. 2014;111:E738–747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Kerkhof TL, Feenstra KA, Euler Z, van Gils MJ, Rijsdijk LW, Boeser-Nunnink BD, Heringa J, Schuitemaker H, Sanders RW. HIV-1 envelope glycoprotein signatures that correlate with the development of cross-reactive neutralizing activity. Retrovirology. 2013;10:102. doi: 10.1186/1742-4690-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol GPI, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. Viral Escape from HIV-1 Neutralizing Antibodies Drives Increased Plasma Neutralization Breadth through Sequential Recognition of Multiple Epitopes and Immunotypes. PLoS Pathog. 2013;9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer CK, Gorman J, Anthony CS, Mkhize NN, Druz A, York T, Schmidt SD, Labuschagne P, Louder MK, Bailer RT, Abdool Karim SS, Mascola JR, Williamson C, Moore PL, Kwong PD, Morris L. Structure of an N276-Dependent HIV-1 Neutralizing Antibody Targeting a Rare V5 Glycan Hole Adjacent to the CD4 Binding Site. J Virol. 2016;90:10220–10235. doi: 10.1128/JVI.01357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chambers M, Chuang GY, Druz A, Geng H, McKee K, Kwon YD, O’Dell S, Sastry M, Schmidt SR, Xu K, Chen L, Chen RE, Louder M, Pancera M, Wanninger TG, Zhang B, Zheng A, Foulds KE, Georgiev IS, Joyce MG, Lemmin T, Narpala S, Soto C, Todd JP, Shen C-H, Tsybovsky Y, Zhao P, Haynes BF, Stamatatos L, Tiemeyer M, Wells L, Scorpio D, Shapiro L, McDermott A, Mascola JR, Kwong PD. Impact of the HIV-1-glycan shield on antibody elicitation. Cell submitted. 2016 doi: 10.1016/j.celrep.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O’Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng HP, Yang AS, Program NCS, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP, Jr, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, Do Kwon Y, Longo NS, Louder MK, Luongo T, McKee K, Schramm CA, Skinner J, Yang Y, Yang Z, Zhang Z, Zheng A, Bonsignori M, Haynes BF, Scheid JF, Nussenzweig MC, Simek M, Burton DR, Koff WC, Program NCS, Mullikin JC, Connors M, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Comparison of structural model sequences and the full JR-FL Env sequence. The 3 Env subunit protomers of our model (A, B and C derived from pdb 5FUU) were aligned with the complete JR-FL Env sequence (shown at the top) (http://www.uniprot.org/uniprot/Q6BC19). Regions of disorder in the structure in the C1, V1 and V2 regions shown as dashes in the alignment are not rendered in the models. Besides the missing amino acids, this also entails the loss of one or two potential glycans in the V1 and the V4 regions of each protomer. The sequons at positions 189 and 637 (HXB numbering, highlighted in magenta) do not carry a glycan in the models shown in Figures 1 and 10, reflecting natural overlap or competition with neighboring glycans at these sites (Go et al., 2008).
Figure S2: Overview of Study 2 rabbit immunizations. 5 groups of 4 rabbits were immunized. Details or immunization type (VLPs or plasmid DNA), route (intramuscular injection) and dose are provided in Materials and Methods. VLPs were produced in 293F cells and formulated in adjuvants AS01B or Adjuplex. The timeline shows the days of each immunization and bleeds (B1–B6) taken two weeks thereafter.
Figure S3: Study 2 serum binding and neutralization profiles. Peak time points for Study 2 sera were compared to replots of Study 1 sera and groups A and B from our previous study (Crooks et al., 2015). For clarity and to facilitate comparisons, the VLP Env (parent or N362Q), regimen, adjuvant and producer cells are indicated. A) ELISA titers against recombinant JR-FL gp120 monomer and bald-VLPs. Data are means of at least two repeats. Neutralizing activity against B) the tier 1 mutant JR-FL WT A328G (Tong et al., 2014) and SIVmac239, and C) the JR-FL SOS E168K+N362Q index strain and parent JR-FL SOS E168K measured in CF2Th.CD4.CCR5 cells. Neutralization assays were performed at least three times in duplicate. Mean ID50s and standard deviations are shown.
Figure S4: Mann-Whitney analysis of Study 2 data and reference samples. Mann-Whitney tests were performed on pairs of groups as listed in the first column. The concept these comparisons sought to test are shown in the second column. P values for each kind of data in each column are shown in the remaining columns, where significant differences (i.e. <0.05) between groups are highlighted in red.
Figure S5: Comparison of VLPs produced from 293T and 293F cells. A) Size of 1,000× concentrated pellet from a 25ml culture of the two cell types. Note that ~12-fold higher numbers of non-adherent 293F cells are used to seed a given volume than adherent 293T cells. Each cell type was transfected with plasmids expressing JR-FL SOS E168K using MuLV Gag and Rev. Photographs of VLP pellets harvested at day 3 and 7 after transfection are shown. B) Env expression in the two VLP preparations (harvested at day 3) was compared at various dilutions by BN-PAGE-Western blot. C) Antigenicity of trimer 293T and 293F VLPs was assessed by ELISA using serial dilutions of mAbs. D) Infectivity of JR-FL WT parent pseudovirus supernatants produced in both cell types in a single round assay using CF2 target cells.


