Abstract
This review, the first of an occasional series, tries to make sense of the concepts and uses of deep sequencing of polynucleic acids (DNA and RNA). Deep sequencing, synonymous with next-generation sequencing, high-throughput sequencing and massively parallel sequencing, includes whole genome sequencing but is more often and diversely applied to specific parts of the genome captured in different ways, for example the highly expressed portion of the genome known as the exome and portions of the genome that are epigenetically marked either by DNA methylation, the binding of proteins including histones, or that are in different configurations and thus more or less accessible to enzymes that cleave DNA. Deep sequencing of RNA (RNASeq) reverse-transcribed to complementary DNA is invaluable for measuring RNA expression and detecting changes in RNA structure. Important concepts in deep sequencing include the length and depth of sequence reads, mapping and assembly of reads, sequencing error, haplotypes, and the propensity of deep sequencing, as with other types of ‘big data’, to generate large numbers of errors, requiring monitoring for methodologic biases and strategies for replication and validation. Deep sequencing yields a unique genetic fingerprint that can be used to identify a person, and a trove of predictors of genetic medical diseases. Deep sequencing to identify epigenetic events including changes in DNA methylation and RNA expression can reveal the history and impact of environmental exposures. Because of the power of sequencing to identify and deliver biomedically significant information about a person and their blood relatives, it creates ethical dilemmas and practical challenges in research and clinical care, for example the decision and procedures to report incidental findings that will increasingly and frequently be discovered.
Keywords: ChipSeq, exome, methylome, next-generation sequencing, RNASeq
In 1995, Craig Venter reported the sequence of Hemophilus influenza, a bacterial genome containing 1.8 Mb (million DNA bases). Draft sequences of the human genome (3000 Mb) were published in 2001 by a public consortium led by Eric Lander and Francis Collins and a private corporation led by Venter. The sequencing technology that enabled these accomplishments was based on a primer extension method invented by Nobel Laureate Frederick Sanger et al. (1977), who used bacterial DNA polymerase, later modified, to trick DNA into synthesizing copies of itself in a test tube. By spiking the reaction mix with polymerization-defective, chain terminating dideoxynucleotides Sanger was able to stop the extension reaction at multiple points where the growing DNA chain randomly incorporated a dideoxynucleotide instead of the corresponding deoxy-A, G, C or T. It was thereby possible to ‘read’ the sequence by measuring sizes of fragments separated on a gel. For example an ‘A’ dideoxy sequencing reaction might generate a ladder of ‘bands’ with fragments 4, 7, 12 and 20 bases in size indicating that an ‘A’ was present at each of those positions. Initially it was necessary to perform a separate DNA synthesis for each nucleotide but after dye terminator dideoxy nucleotides of different colors were introduced one chemical reaction could be run, with the nucleotide at each position established by color. Sanger sequencing, and as should also be mentioned, Maxam–Gilbert sequencing which depended on cleavage of terminal nucleotides, revolutionized genetics by enabling the letters of the genetic code to be read.
Via Sanger sequencing, accurate sequences of ~500 to 1000 bases were produced in one go and this method as implemented on capillary sequencing machines remains widely available, highly accurate and applicable for small-scale, well-defined targets, for example portions of genes. However, even on the most powerful machines only a few hundred DNA fragments could be simultaneously sequenced, the process involved electrophoretic separation of DNA fragments and, worst of all, enough template DNA for the sequencing reaction had to be prepared. Preparing the template DNA required growing very large ‘libraries’ of DNA fragments in bacteria, cloning bacteria expressing specific fragments, and purifying DNA from each clone in the library. For genome sequencing projects that require sequencing millions of DNA fragments this was almost totally unworkable but by heroic effort, and expenditure, it was ‘made to work’ leading to the draft sequence of the human genome.
In 2001, Francis Collins said that the draft genome sequence was only the ‘end of the beginning.’ To understand the variation and functions of the human genome it was necessary to sequence genomes of many species and people and to obtain readouts of gene expression, DNA methylation and chromatin structure. A technological revolution was needed to make sequencing vastly more powerful and less expensive.
Massively parallel, ‘next-generation’ sequencing (NGS) technologies that were the product of this revolution have been transformative in two ways that are both strongly tied to throughput. First, NGS enables the sequencing of large regions of genomes and even whole genomes by the efforts of one person, and by one run on one machine rather than via the work of hundreds of people, hundreds of machines and thousands of machine runs. To sequence a human genome three billion nucleotides in length at 20-fold coverage (the reasons for which will be discussed later), some sixty billion nucleotides must be sequenced, representing 100–1000 million sequencing reads depending on read length. At the time the human genome was first sequenced this would have required more than a quarter million sequencing runs, if a single genome had been sequenced at 20×, which it actually was not because of the prohibitive cost. The introduction of NGS methods enabled whole genome sequencing but also stimulated the development of other methodologies such as exome capture and RNASeq that also require sequencing of millions of DNA fragments and with which the sequencing technology itself is now closely integrated. Second, NGS can be targeted to sequences and selected samples but owing to its capacity and low cost it can also be applied broadly or ‘omically’, deriving unexpected insights. As Francis Collins has also remarked, no longer do geneticists only have to search for the lost car keys directly under the lamp post. The low cost and high throughput of NGS also enable measurement of many individuals and samples. This led directly to the capture of the depth of human genetic variation, with for example 22 million common polymorphisms having been described, and finer grained studies of epigenetic regulation and gene expression, and for example studies that track such changes across time and in different tissues.
The first massively parallel ‘next-generation’ sequencing method, which used adapters linked to DNA to be sequenced on beads, was made commercially accessible in year 2000 by Lynx technologies. Subsequently and as speed of sequencing increased and price reciprocally dropped, it was frequently observed, perhaps irrelevantly, that sequencing technology advanced at a rate faster than Moore’s law, which states that the number of transistors on an integrated circuit board doubles every two years.
Relevantly, sequencing speed and capacity are continuing to grow exponentially and the cost is rapidly dropping. These advances in sequencing technology were made possible by fundamental breakthroughs that include the rapid generation of colonies of replicated DNA fragments to be sequenced on beads and other substrates removing the need to ever clone DNA, single molecule sequencing in real time and new bioinformatic techniques for fragment mapping, sequence assembly and feature identification. Because of the rapidity with which one generation of instruments replaces the previous, it is not easy to understand what is meant by next-generation sequencing, and it would be unhelpful in this article to explain the principles of each of the devices now in use but that will soon be outdated anyway, or to jump into the always controversial discussion of which technology is better and for what application. Therefore, the remainder of this review focuses on principles that are important and likely to remain so regardless of the particular way sequencing is performed.
Get ready to sequence. What is a sequencing library?
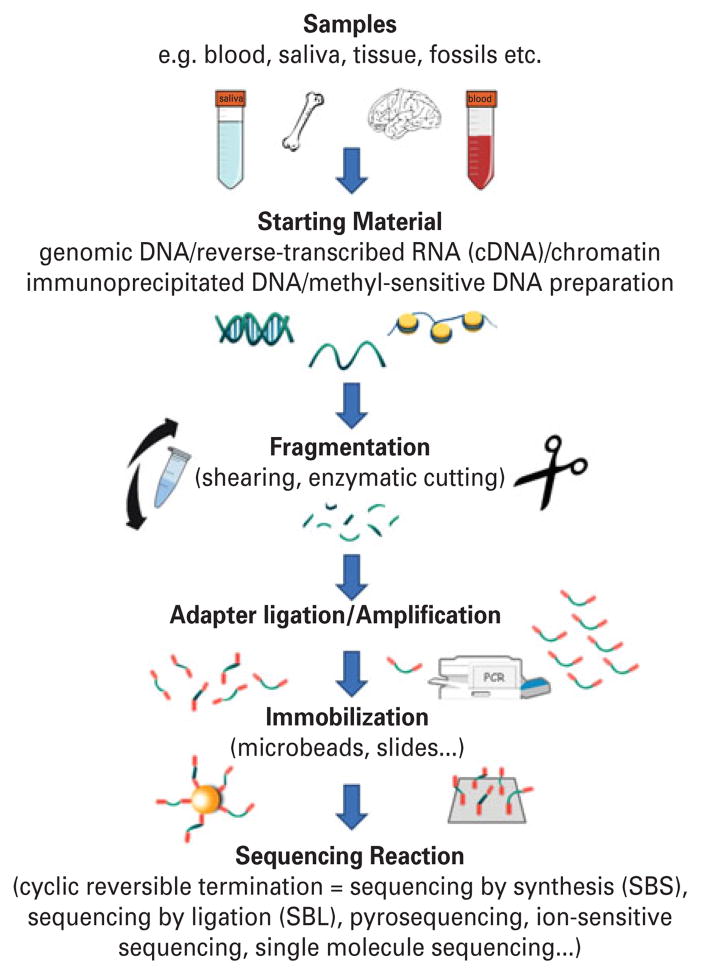
The starting material for sequencing is reverse-transcribed RNA or genomic DNA, either of which can be extracted from blood, saliva, another tissue or even fossils or forensic specimens (Fig. 1). The results of some analyses, for example epigenetic modifications of DNA and gene expression, but also some changes in DNA sequence and for example the mutations and DNA rearrangements that are important in cancer and may be important in other diseases, are tissue-specific. If a contract or core lab performs the sequencing, collecting the sample may be the last technical step prior to analyzing the results, but it is a critical step. DNA is relatively stable. RNA is not. To preserve DNA and RNA, tissue can be collected into media that neutralize nucleases that destroy them and the nucleic acid should also be shielded from physical shearing – a problem in freezers with a defrost cycle. The genome and large pieces of it such as the exome are too long to read in one go, so after sample handling steps in which the DNA was carefully protected it is deliberately fragmented at random locations by shearing or cut at specific locations using an endonuclease. To target DNA associated with specific proteins, chromatin (the DNA protein-complex) is isolated, which usually requires a different sample collection procedure. Probes can be used to capture specific DNA fragments by binding to those proteins or can recognize the DNA itself. For example fragments of genomic DNA containing methylcytosine, a regulatory nucleotide modification, can be captured using MECP2, a protein that recognizes methylcytosine and whose dysfunction causes Rett syndrome, a neurobehavioral disease. DNA fragments can also be made by simultaneous amplification (PCR) of up to tens of thousands of specific DNA targets, producing the mixture of targets one wants to sequence. Starting with these fragments, and as required by the specific technology, the DNA is put on microbeads, or immobilized on slides, or modified with nucleic acid adaptors, or bar-coded (using short DNA sequences –what else?), or combinations of these (Fig. 1). There are now many ways to avoid the need for cloning to obtain enough of the particular fragment to sequence. These include single molecule sequencing but more often used, at least for the time being, are methods that amplify single DNA fragments as pure colonies on beads or plates and leading to billions of colonies all waiting to be sequenced in a massively parallel fashion.
Fig. 1.
Schematic workflow of deep sequencing. RNA or genomic DNA is extracted from various sources including blood, saliva, tissue and forensic specimens. DNA or RNA reverse-transcribed to complementary DNA (cDNA) is physically fragmented at random locations or enzymatically cut at specific sites. Small synthetic DNAs are added to the ends of the fragments (‘adapter ligation’), which are subsequently clonally amplified by polymerase chain reaction (PCR), if not sequenced directly by single-molecule sequencing. Individual DNA fragments are spatially separated, including by use of oil–water emulsions, and in several procedures the fragments are immobilized by attachment to microbeads or slides. Finally, the fragments are sequenced in massively parallel fashion.
What is read length and why does it matter?
Accuracy degrades as read length increases, and varies between methods. Minimum lengths of 30 nucleotides or more are usually needed to map or assemble the sequences. Longer reads facilitate these steps and enable other insights – for example whether genetic variants at different loci travel apart (in trans) or together (in cis) on the same DNA fragment in which case they constitute a haplotype. Longer reads are useful to discover and define insertions and deletions and to accurately sequence regions that contain multiple tandem copies of a similar sequence or that are similar in sequence to other regions of the genome. On the other hand, the main goal in next-generation sequencing is massive amounts of accurate sequence, and tricks are often used to achieve some of the advantages of long reads.
What is a haplotype? How does sequencing reveal same-strand (in cis) genetic and regulatory interactions?
As just mentioned, a unique strength of sequencing is the ability to read whether two genetic elements are on the same DNA strand. Genotyping may not reveal those relationships. These same-strand relationships are important for identifying functional effects of genetic combinations involving two loci whose alleles may act differently if located on the same strand. For example, one DNA element might regulate the expression of another or the combination of two nucleotide substitutions in the messenger RNA could alter RNA folding, ultimately affecting processing, stability or translation of the RNA. The combinations themselves, called haplotypes, also can serve as barcodes to track or predict other elements on the same DNA strand. Imagine that an occasional kangaroo and mountain lion reside in an otherwise boring subdivision where all houses seem to be identical. Wouldn’t it be important to know if the kangaroo and mountain lion peacefully coexist in the same house? Like the nearly identical houses in that subdivision, each copy of a certain chromosome (for example Chromosome 1) is more or less the same but with minor variations (alleles) introduced as mutations or copy errors. Typically people carry some three million heterozygous loci, most owing to a genetic difference between the chromosome inherited from the mother and the father, and a few due to new mutations. Most genetic glitches do not alter the sense of the DNA message, but some do. Genotyping tells us which alleles are present at a genomic location (locus). However, if two nearby loci are heterozygous, with different alleles inherited from mother and father, genotyping does not directly tell us whether the alleles are on the same or opposite member of the chromosome pair. Especially with long sequencing reads, we can read whether the two alleles are present on the same fragment. There are also methods that cleverly link together sequences from different regions of DNA. These methods include mate-pair libraries in which larger DNA fragments are circularized and then cut, and the sequencing of both ends of a larger DNA fragment (paired-end reads).
By this same rationale, other measures of function such as differences in RNA expression or chromatin structure can be directly tied to a particular DNA sequence, and even if that functional element has not yet been identified. Imagine that a region transcribed into RNA contains a heterozygous locus. By sequencing different transcripts from this region it can determined if one copy of the chromosome is under-expressed relative to the other (differential allele expression) and by sequencing that DNA region that element can sometimes be identified. Frequently psychiatric geneticists speculate that unknown regulatory variants account for associations they observe to genetic markers. However, by a combination of DNA and RNA sequencing, including differential allele expression methods, it can be learned whether a nearby locus alters expression of the gene, the locus may be identified. Zhou et al. (2008) found, for example, that lower haplotype-driven neuropeptide Y (NPY) expression, attributable to a functional locus in the NPY promoter region, predicted higher emotion-induced activation of the amygdala, as well as diminished resiliency as assessed by pain/stress-induced activations of endogenous opioid neuro-transmission in various brain regions.
Why is sequencing redundant? What is read error?
Redundancy is multiple representations of a target nucleotide in many reads. Redundancy can overcome read errors, the rates of which are as high as several percent with some sequencing methods. A few misreads in a hundred can be dismissed. Redundancy is also necessary because of biology. RNA expression and epigenetic modifications are measured by counting sequence representations, quantitative accuracy depending on the number of representations. There is often wide variation in the number of reads expected for particular targets, so the overall coverage, or sequencing depth, a metric of the average number of times a nucleotide is read in the sequencing process, has to be adjusted for the rarer targets, for example low abundance RNAs. Frequently, variation in capture or amplification of different targets also leads to unevenness across targets, and the overall level of redundancy therefore has to be higher. Finally, human autosomes are diploid, requiring higher redundancy to accurately sequence both the maternally and paternally derived chromosome. Earlier whole genome sequencing at 4×–8× read depth is more often being replaced by 20×–40× depth, and higher.
What is DNA pooling?
Instead of sequencing samples individually they may be combined. An interesting observation made via pooling can be confirmed by analysis of individual samples as can be critical if the pooling process introduces error, for example because of uneven representation of samples. Furthermore, analysis of individual samples provides information on biological variability and actual genotypes (pooling usually only allows comparisons of allele frequencies, not genotypes or combinations of genotypes) and interactions of genes with mediators, including mundane ones such as sex and age. Several sequencing technologies now allow tagging of individual samples within pools.
How to analyze ‘big data’?
Although the major limitation of first-generation sequencing was sequence acquisition, the challenge for next-generation sequencing is data analysis and interpretation. In first-generation sequencing, specific fragments were usually cloned or amplified and then sequenced one at a time. One knew what one was sequencing. Most deep sequencing is conducted against mixtures of short fragments, many of which partially overlap. Frequently, billions of fragments are simultaneously sequenced. The ingenious idea that makes this possible is that – after initial sequence analysis (‘base calling’) – each fragment is identified post-hoc based on its sequence. For example, a sequence 20 nucleotides in length can have 420 combinations, more than enough to uniquely map it to some region of the human genome and even allowing for sequence errors and polymorphisms. Whenever possible, sequence reads are mapped to a known reference as a scaffold to which the generated sequence is aligned (‘resequencing’; ‘mapping’). However, reference sequences originally had to be constructed by de novo assembly of overlapping fragments, and often with the aid of longer sequence reads and reads derived from the ends of larger DNA fragments, thus bridging gaps between DNA regions that are difficult to read or to uniquely assign and forming large blocks of contiguous sequence.
After fragment mapping or assembly, the real bioinformatics challenges begin, with different methodologies tailored to the particular application (for example, sequence variant detection as in exome sequencing, RNA expression as in RNASeq, and cytosine methylation as in methylome studies). Advanced computational infrastructure, often involving parallel processing and analytic pipelines executed in UNIX environments, are applied to organize and make sense of the tremendously large data sets generated by NGS (cf. McPherson, 2009). Although it is beyond the scope of this review to describe these steps for any of the NGS applications it can be generally stated that the general flow of analysis is data parsing, cleanup, feature identification, feature quantification, and some type of globally based normalization and evaluation of features. For example, in exome or genome sequencing performed to find a disease mutation, sequence variants may be detected by search against a reference sequence. Sequence variants that are thus detected can be individually scored for quality (for example adequate sequence representation of both alleles), and the overall quality assessed in a variety of other ways: for example in males, X chromosomal loci are not expected to be heterozygous, and overall sequence variants tend to have a 2:1 transition:transversion ratio (the ratio of purine to purine and pyrimidine to pyrimidine substitutions vs. purine to pyrimidine substitutions) rather than the 1:2 ratio seen with random sequence errors. Furthermore, thousands of genotypes might already be precisely known, for example if the individual had been genotyped using an array method. Using such clues, the method for variant calling can be evaluated and if necessary subtly adjusted to yield a large panel of sequence calls that is highly, if not perfectly, accurate, and accuracy can be estimated. Next, all the sequence variants might be bioinformatically evaluated for probable functional significance, for example by using the evolutionary conservation of the nucleotide in question, or the amino acid if the variant is a missense variant causing an amino acid substitution. The availability of sequence information from multiple individuals within the same family and from multiple individuals with the same disease enables other types of analyses, both for quality control, and – as might be imagined –to home in on the causal variant.
For NGS analyses hinging on quantitation of genomic features the derivation and accurate background subtraction and normalization of features is key. Several types of global analyses and data representations often follow, and these are likely to include A–B plots comparing main conditions, volcano plots that represent both deviation in quantitative expression and p values, and Q–Q plots of the actual and randomly expected distributions of p values. Q–Q plots can rapidly convey whether there is global inflation of p values as can occur if there is some systematic (methodologic) bias, and on the other hand can help visualize that there is an excess of highly significant p values. When tens of thousands of features are tested some sort of correction is required. If there is no evidence of systematic bias leading to p value inflation it may be appropriate to use False Discovery Rate (FDR) correction, which hinges on the idea that if there is large excess of observations above a certain statistical threshold most of these are likely to be non-random (but note that this does not rule out methodologic bias). Did the ‘omic’ expression or epigenetic study succeed in distinguishing one condition or group from the other? One way to answer that question is to ask whether the samples of the same type cluster in unsupervised fashion, and as can be computed using several methods. For both genetic variation and quantitative NGS studies an increasingly valuable and widely applied tool is pathway analysis which relies on the idea that if a function is perturbed several genes in related pathways will show coordinate changes. Whereas a single change in structure or expression may not be characteristic, a coordinated series may be characteristic, and point to a common origin.
Why do genomic sequencing and other ‘big data’ produce artifacts and false positives?
Genomic technologies are powerful discovery tools but can also be factories for artifacts because of the very large number of measures, inherent methodologic and sampling errors, and the potential for small and unsuspected systematic errors in procedures or biologic variability between samples, for example there may be occult variation in ethnic origin of subjects. Random errors and sampling variation can be corrected statistically. However, although false discovery rate (FDR) statistical approaches are extremely helpful, they can mask or accentuate the problem of systematic errors and biologic biases. Some false results can replicate in new samples. As mentioned, to detect hidden biases, Q–Q plots comparing observed and randomly expected distributions of p values can reveal evidence of p value inflation. However, the value of genomic discoveries is ultimately established by predictive validity in other contexts and at functional levels.
What are the ethical implications of deep sequencing?
Privacy, necessitating standards for confidentiality, and disclosure of medically significant findings are the two main problems. The creation of databases of sequences and genotypes accessible to qualified investigators advances our understanding of genetic diseases and is a requirement for research funding. However, privacy issues are raised by the ability to link even partial sequence data to a person or their blood relatives. That linkage may establish their participation in a research study, or directly link them to findings predicting disease or other individual characteristics. On the other hand, deep sequencing performed in a medical context may create a ‘right to know’ incidentally observed medically significant findings. The American College of Medical Genetics has recommended that any of 56 incidental findings, a list that is likely to grow, should be reported. These recommendations are not without controversy. For example, should the sequence be reevaluated each time there is a new finding, what is a significant finding, who should report the finding to the patient, a physician or scientist when warranted for diagnosis/treatment purposes, how should it be reported, and what should be done if the person doesn’t want to know? Certain of these problems involve conflict between the principles of beneficence, autonomy, non-malfeasance, fidelity and justice that are foundational in clinical care. Moreover, additional issues are raised by deep sequencing performed in the context of human research studies, e.g. in many instances we do not know enough about the human genome to uncloak the meaning of private mutations and rare alleles, etc. (cf. Biesecker and Peay, 2013).
Applications and prospects
Applications of NGS vary widely as reflected by the different starting materials for NGS that include genomic DNA, reverse-transcribed RNA (complementary DNA (cDNA)) and immunoprecipitated DNA (see above). The massive capacity of NGS has led to a paradigm shift from gene by gene analyses enabled by first-generation sequencing to ‘omic’ analyses covering the whole genome, exome (expressed region of the genome), transcriptome (global sequencing of RNAs), and methylome. For genetic analyses, NGS has shifted the focus from candidate gene hypotheses and studies of known, usually common, variants to the survey of whole molecular pathways, discovery of novel pathways to disease, and the discovery of novel variants including deletions, duplications, insertions and inversions that may be unique to a person or a family, and thus generating novel hypotheses and disease concepts (cf. Biesecker and Peay, 2013). NGS permits methylome-wide association studies (MWAS) using methyl-CpG binding domain (MBD) protein-enriched genome sequencing (MBD-seq) and investigation of DNA-protein interactions via Chromatin ImmunoPrecipitation DNA-Sequencing (ChiP-Seq) identifying DNA binding sequences for proteins such as transcription factors or histones. Frequently, or even usually, candidate gene studies of DNA methylation, and chromatin structure had failed to interrogate the entire gene, leading to important omissions, and for example the importance of regulatory methyl CpGs in exons. At the mRNA level (RNA-Seq), provides a global view of the transcriptome but at the single gene level improves on candidate gene studies that frequently had not placed gene expression in the context of other co-regulated genes, had inconsistently captured differences in transcription start sites and RNA processing, had usually not detected opposite strand and overlapping transcripts, and had most often relied on one or two potentially inappropriate ‘housekeeping genes’ for normalization of expression levels. There are many obstacles to unraveling the systems relationships between genotypes, epigenetic modifications and gene expression that are beyond the scope of this review. However there is increasing evidence, for example from the ability to objectively identify whole sets of functionally related genes from interventional studies where RNA expression is globally measured, that the ‘omic’ views provided by NGS can be integrated within the reference framework of the human genome sequence. This can lead to a better understanding of the etiology of neuropsychiatric disorders and refined individual biomolecular profiles that together with other modalities of measurement could target preventive interventions and lead to innovative and personalized therapies.
In a general effort to provide a comprehensive resource on human genetic variation, in 2008 the 1000 Genomes Project was launched to sequence one thousand healthy subjects using a combination of low-depth (2–6×) genome sequence and high-depth deep (50–100×) exome sequence and high density SNP arrays. The recently published data from 1092 individuals sampled from 14 populations showed that this approach captured 99.7, 98 and 50% of accessible single nucleotide polymorphisms (SNPs) at a frequency of 5, 1 and ~0.1%, respectively. Private and rare variants were also captured to a great extent (60%) by the exome sequencing, but were mostly missed by the low coverage sequencing (1000 Genomes Project Consortium et al., 2012). The goal of the ENCODE project, launched 2003, is to identify the functional elements of the genome. In a series of landmark papers published 2013, ENCODE revealed that a large portion of the human genome is at least occasionally transcribed and defined motifs that powerfully predict DNA transcription.
Biomedically, deep sequencing has already had a major impact and several of its biggest successes have been in contexts where gene effects are most likely to be isolated: rare Mendelian Diseases transmitted in families (Ng et al., 2010; reviewed by Biesecker, 2010), cancer genomics where the sequence of the tumor can be compared to the non-tumor DNA from the same person to reveal somatic mutations that cause cancer or increase metastatic potential or treatment resistance (Ley et al., 2008).
Discoveries achieved with the aid of deep sequencing illustrate its power and scope, and suggest how it may be used to bridge gaps in knowledge. For psychiatric disorders, gene by gene sequencing and genome wide genotyping (genome wide association; GWA) failed to identify genetic variants that would explain most of the genetic liability to psychiatric disorders (the ‘missing heritability’). Deep sequencing can address the component of heritability that is attributable to rare and uncommon variants. In contrast to common variants, the uncommon variants are themselves unlikely or incapable of generating genetic associations either directly or via linkage disequilibrium with the common variants placed on large scale genotyping arrays. Although rarer variants are also being placed on these arrays as their capacity increases, next-generation sequencing identifies rare, previously unrecognized variants and even de novo mutations occurring in a single individual. An N of one may be insufficient to draw conclusions about the relationship of a gene to disease; however, it is increasingly feasible to score variants for functional significance, for example on the basis of evolutionary conservation, and to use their functional properties to combine them in linkage analyses.
An intriguing observation by the ENCODE consortium is that most of the human genome is at least occasionally transcribed into RNA. Furthermore, DNA sequences located thousands of DNA bases away can alter the transcription of genes. Yet we know little about the functional significance of these sequences, or even many of the sequences that are at or near genes. Working gene by gene, progress in understanding transcriptional control is slow. However, by deep sequencing, the ENCODE consortium and others have opened the first genome-wide views of DNA function. Understanding genome function requires a synthesis of interrelated pieces of information from different types of deep sequencing analyses. The ‘methylome,’ consisting of hundreds of thousands of cytosine methylations and hydroxymethylations informs us of regulatory DNA modifications. Protein/DNA interactions are elucidated by sequencing DNA fragments associated with specific proteins including many types of methylated and acetylated histones, thus telling us about changes in DNA structure that alter gene expression. Gene expression itself and the post-transcriptional processing of RNA is measured by sequencing RNA that has been reverse transcribed to cDNA (complementary DNA). DNA sequence (genotype) remains stable throughout life; however, exposures modify various aspects of genomic structure and gene expression.
Psychiatric diseases are moderately to highly heritable, but paradoxically the inception of most of them is strongly influenced, or dependent, on context and experience. Deep sequencing helps to measure the impact of the environment on the genome and the gene–environment interactions that are critical to psychiatry and neuropsychopharmacology. Along these lines, deep sequencing has already been powerfully applied in sporadic (non-familial) cases where de novo mutations can be found by comparison to parental DNA, studies of founder populations, in which variants may be common that on a worldwide basis are rare, and sequencing of model organisms, including ones that have been derived by artificial selection. For instance, exome sequencing parent–child trios of non-familial cases and twin pairs with autism spectrum disorders (ASD) has revealed an excess of de novo mutations in patients including plausibly functional variants at several candidate genes (O’Roak et al., 2011; Michaelson et al., 2012; Neale et al., 2012). Similarly, family-based sequencing of sporadic schizophrenia cases has also revealed de novo mutations, many of which alter protein function and some that are stop codons (terminating protein translation) in plausible candidate genes (Girard et al., 2011; Xu et al., 2011). In a Finnish founder population, an HTR2B (serotonin 2B receptor) stop codon leading to impulsivity and alcoholism was found by targeted deep sequencing serotonin and dopamine receptor genes, and although the stop codon is found in >100000 Finns it is absent in other populations (Bevilacqua et al., 2010). Via exome sequencing of the alcohol preferring rat, a stop codon was discovered in Grm2, the gene encoding a glutamate receptor (Zhou et al., 2013). This stop codon, which is also found in some Wistar rats (the parental strain of the rats), leads to widespread alterations in glutamate function and alters alcohol preference. Applying targeted deep sequencing to over 100 classical candidate genes for schizophrenia, Hu et al. (2013) discerned 5170 novel variants, with 70% of them being rare variants (RVs) with a minor allele frequency (MAF) <1%, and suggested enrichment of nonsense variants in schizophrenia particularly in the neurexin 1 (NRXN1) and transcription factor 4 (TCF4) genes. The first methylome-wide association study (MWAS) in 759 patients with schizophrenia and 738 healthy controls found 25 differentially methylated sites, with the top finding being located in a gene, FAM63B, linked to neuronal differentiation and dopaminergic gene expression (Aberg et al., 2014). The first specific interactions between environmentally induced DNA methylation and functional polymorphisms to alter behavior have recently been reported, both involving genes that modulate or alter stress response (e.g. Domschke et al., 2012; Klengel et al., 2013), foreshadowing the dissection of gene by environment interactions by sequencing at a genomic level. On the transcriptome level, 212 significant differential splicing (DS) events dependent on the neuronal specific splicing factor A2BP1 were identified in post-mortem brain tissue samples of patients with autism spectrum disorders using high-throughput RNA-sequencing (RNA-Seq) (Voineagu et al., 2011).
Conclusion
Deep sequencing, owing to its flexibility, completeness, quantitative properties and rapidly decreasing costs is encouraging ‘omic’ rather than gene-centric views of genotype, gene expression, and DNA structure and regulation. For many applications DNA sequencing can displace other technologies, such as arrays, that may genotype millions of genetic markers or measure the expression of thousands of RNA transcripts, but that have certain limitations. Potentially, many or most people in societies with advanced medical care may have their genome sequenced, providing benefits in terms of risk factors and treatment predictors, but raising bioethical problems including incidental findings, privacy, and findings with implications for blood relatives. New bio-informatic methods continue to enhance the power and variety of applications of deep sequencing and are also useful to detect and correct errors inherent to ‘big data.’ Finally, the ability of sequencing to define epigenetic changes in DNA opens new avenues to the effects of the environment on the genome and to understanding gene by environment interaction.
Acknowledgments
The present work was supported by the Deutsche Forschungsgemeinschaft (DFG), SFB-TRR-58, project C02 (to KD).
Glossary and frequently used abbreviations
- Assembly
Reconstruction of a longer, or even whole genome sequence, from smaller fragments.
- ChIP-Seq
Chromatin immunoprecipitation followed by sequencing of DNA fragments associated with proteins to identify regional differences in DNA conformation and regulation.
- Chromatin
DNA and its associated proteins.
- Differential allele expression
The detection of allele bias in RNA expression, indicative of a cis-acting functional locus.
- DNA library
A collection of different DNA fragments in some usable form.
- Epigenetics
The study of changes in DNA structure and expression that are secondary or in addition to the linear sequence of the genome.
- Exome
The portion of the genome that is most highly transcribed (expressed). Approximately 50 million base pairs in size.
- Haplotype
A DNA ‘bar code’ composed of a combination of alleles at two different loci on the same DNA strand.
- Heterozygous
Carrying two different alleles at a genetic locus, on opposite members of a chromosome pair.
- Mapping
Positioning a sequence fragment in a reference sequence.
- Methylome
The component of the genome containing methylcytosine, a regulator of DNA expression.
- Next-generation sequencing (NGS)
High throughput sequencing via any post-year-2000 technology.
- Read depth
The average number of representations of targeted nucleotides.
- RNA-Seq
Sequencing RNA to detect differences in levels of expression and structure of particular RNAs.
- Stop codon
Any one of three triplet codon sequences instructing the ribosome to end protein synthesis.
- Transcription
The synthesis of RNA complementary to DNA. The RNA transcript is often further processed and may encode protein, may have direct enzymatic or structural function, may be regulatory, or may be nonfunctional.
Footnotes
Conflict of Interest
Both authors state that they have no biomedical financial interests or potential conflicts of interest related to this manuscript to declare. All affiliations mentioned below have no relevance to the work covered in the manuscript: KD has received speaker fees from Pfizer, Lilly and Bristol-Myers Squibb, she was a consultant for Johnson & Johnson and has received funding by Astra Zeneca.
References
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W, Khachane AN, Xie L, Hudson A, Gao G, Harada A, Hultman CM, Sullivan PF, Magnusson PKE, van den Oord EJCG. Methylome-Wide association study of Schizophrenia identifying blood Biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–264. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua L, Doly S, Kaprio J, Yuan Q, Tikkanen R, Paunio T, Zhou Z, Wedenoja J, Maroteaux L, Diaz S, Belmer A, Hodgkinson CA, Dell’osso L, Suvisaari J, Coccaro E, Rose RJ, Peltonen L, Virkkunen M, Goldman D. A population-specific HTR2B stop codon predisposes to severe impulsivity. Nature. 2010;468:1061–1066. doi: 10.1038/nature09629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG. Exome sequencing makes medical genomics a reality. Nat Genet. 2010;42:13–14. doi: 10.1038/ng0110-13. [DOI] [PubMed] [Google Scholar]
- Biesecker BB, Peay HL. Genomic sequencing for psychiatric disorders: promise and challenge. Int J Neuropsychopharmacology. 2013;16:1667–1672. doi: 10.1017/S146114571300014X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domschke K, Tidow N, Kuithan H, Schwarte K, Klauke B, Ambrée O, Reif A, Schmidt H, Arolt V, Kersting A, Zwanzger P, Deckert J. Monoamine oxidase A gene DNA hypomethylation - a risk factor for panic disorder? Int J Neuropsychopharmacol. 2012;15:1217–1228. doi: 10.1017/S146114571200020X. [DOI] [PubMed] [Google Scholar]
- Girard SL, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Hu X, Zhang B, Liu W, Paciga S, He W, Lanz TA, Kleiman R, Dougherty B, Hall SK, McIntosh AM, Lawrie SM, Power A, John SL, Blackwood D, St Clair D, Brandon NJ. A survey of rare coding variants in candidate genes in schizophrenia by deep sequencing. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson JD. Next-generation gap. Nat Methods. 2009;6:2–5. doi: 10.1038/nmeth.f.268. [DOI] [PubMed] [Google Scholar]
- Michaelson JJ, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, Rieder MJ, Nickerson DA, Bernier R, Fisher SE, Shendure J, Eichler EE. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, et al. Genetic variation in human NPY expression affects stress response and emotion. Nature. 2008;452:997–1001. doi: 10.1038/nature06858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Karlsson C, Liang T, Xiong W, Kimura M, Tapocik JD, Yuan Q, Barbier E, Feng A, Flanigan M, Augier E, Enoch MA, Hodgkinson CA, Shen PH, Lovinger DM, Edenberg HJ, Heilig M, Goldman D. Loss of metabotropic glutamate receptor 2 escalates alcohol consumption. Proc Natl Acad Sci USA. 2013;110:16963–16968. doi: 10.1073/pnas.1309839110. [DOI] [PMC free article] [PubMed] [Google Scholar]