Abstract
Recent technological breakthroughs in image acquisition have enabled single-particle cryo-electron microscopy (cryo-EM) to achieve near-atomic resolution structural information for biological complexes. The improvements in image quality coupled with powerful computational methods for sorting distinct particle populations now also allow the determination of compositional and conformational ensembles, thereby providing key insights into macromolecular function. However, the inherent instability and dynamic nature of biological assemblies remain a tremendous challenge that often requires tailored approaches for successful implementation of the methodology. Here, we briefly describe the fundamentals of single-particle cryo-EM with an emphasis on covering the breadth of techniques and approaches, including low- and high-resolution methods, aiming to illustrate specific steps that are crucial for obtaining structural information by this method.
Keywords: single-particle EM, cryo-EM, negative-stain EM, macromolecular structure
Introduction
Delineating the molecular mechanisms underlying cellular processes necessitates structural characterization of the participating components, such as protein complexes and their assemblies with RNA or DNA. Broadly, structural biology seeks to identify how these molecules work by obtaining snapshots of functional complexes at the highest resolution possible. These assemblies can be helical polymers such as microtubules and actin filaments, complex asymmetric machines such as the ribosome, or symmetric oligomers such as the proteosome. Invariably however, biological processes involve a heteromeric and dynamic collection of macromolecular components that undergo cycles of distinct conformational rearrangements associated with various types of association or dissociation events. Thus, macromolecular complexes are not static; biological function dictates that they contain moving parts, flexible domains and multiple binding interactions. The presence of such a dynamic compositional and conformational ensemble presents challenges for obtaining molecular-level snapshots of biological complexes in defined states.
NMR and X-ray crystallography are long-established techniques for determining structures at atomic resolution. NMR is advantageous in yielding an ensemble of models, potentially identifying flexible arrangements in solution, but it is limited by the size of the macromolecule that can be studied and typically requires molecular weights of <50 kDa for conventional applications. X-ray crystallography has been widely successful, but obtaining crystals is a major bottleneck, especially for large and conformationally variable macromolecular assemblies. For the past 20 years single-particle cryo-electron microscopy (cryo-EM) has been steadily gaining popularity due to its ability to visualize large, multi-subunit complexes without the need for crystallization or large amounts of sample. With this method, 3D reconstructions are calculated by combining 2D EM projections of macromolecules in different orientations. However, due to limitations stemming from both the inherent characteristics of biological specimen as well as adverse effects associated with data acquisition, the recorded cryo-EM images contain limited signal compared to noise. For single-particle cryo-EM 3D reconstructions, which necessitate the determination of angular orientations for every particle projection, the low signal has imposed major limitations in alignment accuracy. As a result, only large (>1 MDa) or symmetric macromolecules could readily reach a resolution better than 10 Å (see for example [1–3]), whereas near-atomic resolution could only be obtained for the highly symmetric icosahedral viruses [4–6]. However, the recent development of direct electron detectors for recording EM data has enabled a quantum leap in single-particle cryo-EM [7]. Direct electron detectors provide images with much-improved signal and contrast that enable the calculation of near-atomic resolution 3D cryo-EM structures [8–11]. With these technological breakthroughs complemented by advances in image-processing methodologies, single-particle cryo-EM is entering a new frontier for the structural characterization of challenging macromolecular assemblies. The goal of this review is to provide the non-expert with a concise description of the methodology and scope of work required for single-particle EM analysis of biological complexes, the current challenges and limitations of the technique, and its future development.
General principles
For cryo-EM, a small amount of specimen in buffer solution is applied on an EM grid and rapidly frozen, resulting in particles suspended in a layer of non-crystalline, glass-like ice that is termed vitreous ice (Fig. 1a). The vitrified sample can withstand the vacuum of the electron microscope, while imaging at very low temperatures reduces the effect of radiation damage [12]. This preparation enables visualization of macromolecular complexes under near-physiological conditions and preserves high-resolution information, making it the method of choice for 3D structure determination. In contrast, the traditional specimen preparation, termed negative staining, involves embedding and drying the specimen in a layer of heavy metal salt [13]. The areas occupied by the specimen exclude the stain and are relatively electron transparent, resulting in high reverse contrast images. Negative-stain EM is a low-resolution technique, providing at best 3D reconstructions with a resolution of ∼2 nm, based on the average grain size of the dried heavy metal salt. In addition, the preparation induces specimen dehydration, which results in a variable degree of particle collapse or distortions. Nevertheless, the high-contrast images obtained by this method make it a valuable and versatile tool for various stages of sample characterization [14].
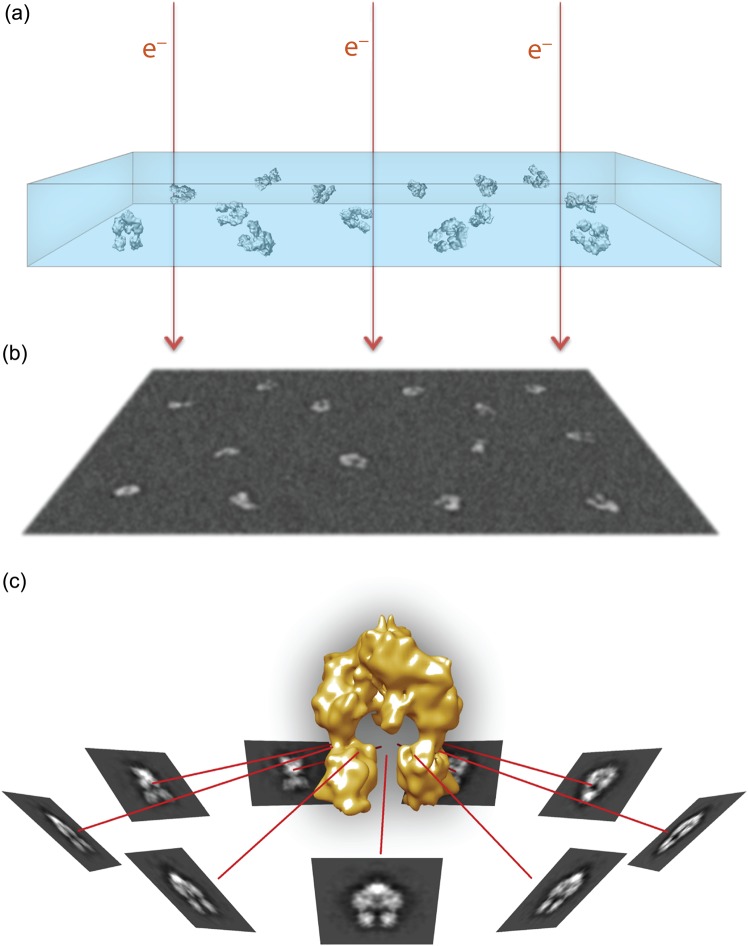
Fig. 1.
Overview of single-particle cryo-EM visualization. (a) Purified macromolecular complexes in solution are rapidly frozen in a thin layer of vitreous ice, ideally in random orientations. (b) Electrons passing through the sample in a transmission electron microscope are recorded as 2D projection images on a recording medium, such as a CCD or a direct electron detector. The particle projections have low signal-to-noise ratio due to the relatively weak electron scattering and the low-dose imaging requirements. (c) Similar particle projections are combined to form class averages that can be assigned with experimentally determined angular relationship. The projection averages are back-projected along their assigned angles to generate a 3D density map representing the particles.
In the transmission electron microscope, emitted electrons travel through the relatively thin sample and become scattered due to electrostatic interactions with the specimen atoms. The interference pattern between scattered and unscattered electrons results in the formation of 2D projections that can be recorded below the specimen level (Fig. 1b). The EM images contain convoluted structural information from all levels of the 3D object as projected on a 2D plane from one direction or orientation. For structure determination, a set of 2D projections encompassing views of the sample from different orientations are combined and ‘back-projected’ to recreate a model of the object (Fig. 1c) [15]. The orientation parameters are determined experimentally by comparing particle projections to angularly defined re-projections of a model that is an initial rough approximation of the overall 3D object architecture [16]. The assigned angles are used to generate a new model from the corresponding experimental projection images, and the process is iterated to gradually produce improved 3D reconstructions until the solution becomes stable. The accuracy of the orientation assigned to each experimental projection is one of the main determinants of the achievable resolution for a 3D reconstruction. However, biological specimens scatter electrons weakly, providing images of very low contrast between the molecule of interest and the surrounding medium. This problem is exacerbated by the need to limit the electron dose to <20–40 e−/Å2 in order to record projections of the sample before extensive radiation damage occurs [17]. To increase the specimen visibility, a variable extent of defocus is applied for imaging, which provides a modest increase in phase contrast. Nevertheless, the raw EM images have a significant amount of noise, which is the major limiting factor for accurate determination of angular orientation parameters in single-particle cryo-EM. The low signal-to-noise ratio also explains the challenges with applying single-particle cryo-EM analysis to complexes smaller than 200–300 kDa, given that the limited features that they display are usually insufficient for accurate determination of particle projection orientations.
Sample preparation for single-particle EM
The complexity of macromolecular assemblies and their variable stability during in vitro preparations requires adequate characterization before embarking on single-particle EM analysis. In most cases, an extensively heterogeneous sample would require further biochemical optimization or engineering before cryo-EM analysis. Attempts with ‘in silico’ particle purification, whereby the heterogeneity is sorted during EM image processing, may not always work well and can produce misleading results. Therefore, the experimentalist should always strive for an optimal biochemical preparation and not start by applying cryo-EM analysis on a heterogeneous sample. Of course, this raises the question of what is deemed to be acceptable homogeneity or heterogeneity for successful cryo-EM analysis. The answer depends on the particle sample characteristics, such as size, pronounced features and symmetry. In general, however, the larger a particle is the relatively easier it is to use computational approaches for identifying subpopulations because of the adequate signal for accurate projection alignment and sorting.
An important test for the applicability of single-particle cryo-EM is the behavior of complexes during size-exclusion chromatography (SEC), which enables separation of sample subpopulations based on the particle's hydrodynamic radius [18]. With this method a complex can be purified from unbound components and elution peaks can be analyzed to probe for the presence of all anticipated subunits or the existence of incomplete complexes. Nevertheless, it is important to note that conformational heterogeneity can be present within a sample elution volume even if it is characterized by a narrow peak distribution. Purification of target complexes by SEC is optimally performed immediately prior to negative-stain or cryo-EM grid preparations allowing one to avoid potential instability issues over time. In addition, SEC facilitates a relatively rapid exchange into the buffer deemed more optimal for cryo-EM visualization. Certain complexes may be refractory to SEC methods due to interactions with the column matrix or dissociation during the run. However, disassembly of a complex in the process of SEC is a good indicator that the complex is not stable enough for EM and that additional optimization is required.
Complex instability also becomes pronounced at the relatively low concentrations that are required for single-particle EM. This is especially true for negative-stain techniques, for which protein concentrations are typically very low (∼0.01 mg ml−1), requiring affinities stronger than 100 nM to withstand the necessary dilution. Weak binding can be a challenge for investigations of an interaction partner that transiently interacts with a larger, more stable complex. For these instances covalently linking complexes together, for example by engineering cysteine mutations to form a disulfide bridge, or the use of chemical cross-linkers, may improve stability and homogeneity. For complex stabilization through disulfide bond formation, significant structural information must already be available, such as the crystal structure of the interacting subdomains and a well-informed location of the binding interface, so that mutations can be made with the goal of positioning the thiol groups within a few angstroms [19]. Chemical cross-linking can be performed with a variety of commercially available compounds with specific bifunctionality, or simple lysine reactive compounds including N-hydroxysuccinimide esters or glutaraldehyde [20]. Because many compounds react readily with surface accessible residues, typically lysines, careful experimental conditions are required to optimize concentrations and avoid over-cross-linking intermolecular sites or aggregation. By nature of this reactivity, cross-linking alters the surface charge of macromolecules and can thereby reduce or change the preferred orientation of particles, which can be an advantage if additional views are needed. Analysis of cross-linked samples by gel electrophoresis and SEC is important to validate and purify the correct molecular weight species. Alternatively, cross-linking methods, involving glycerol/cross-linker gradient centrifugation [21], or ‘on-column’ glutaraldehyde cross-linking during SEC runs [22] have been shown to be effective at enriching for specific cross-linked complexes for single-particle EM. While these methods are beneficial for improving overall homogeneity or trapping specific conformations, the results need to be analyzed with caution for potential cross-linking artifacts while it is also important to show that the stabilized conformation occurs in the native sample. In addition, 3D EM reconstructions of cross-linked complexes may be limited to more modest resolutions (8–15 Å) [23–25] most likely due to variability in the cross-linked sites between particles or the persistence of flexibility among the targets.
EM visualization as an analytical tool to evaluate the effect of different biochemical conditions and purification approaches is an extremely useful component of the sample optimization process (Fig. 2). This is ideally achieved by negative-stain EM, which can often allow for more immediate results compared with cryo-EM. Due to the higher contrast provided by the heavy metal stain, the overall homogeneity of the sample and features of the individual particles can often be assessed with only a few images. Detailed methods describing the negative-stain procedure are available [13,14,26]. While many stains have been used in the field, for macromolecules smaller than 500 kDa uranyl formate is an ideal choice due to its fine granularity that allows for improved stain penetration and excellent contrast compared with other stains [27]. Although uranyl stains are acidic (the pH of uranyl formate stain is adjusted to 5.0–5.5 before use), they have a beneficial rapid fixative effect before the negative stain preparation dehydrates and compresses the particle [28]. Alternatively, neutral stains such as vanadate and molybdate stains can also be used and compared with uranyl formate. Care should be taken to obtain the appropriate stain thickness, optimally as thick as the particle diameter, in order to obtain full stain embedding and produce well-defined particle projections.
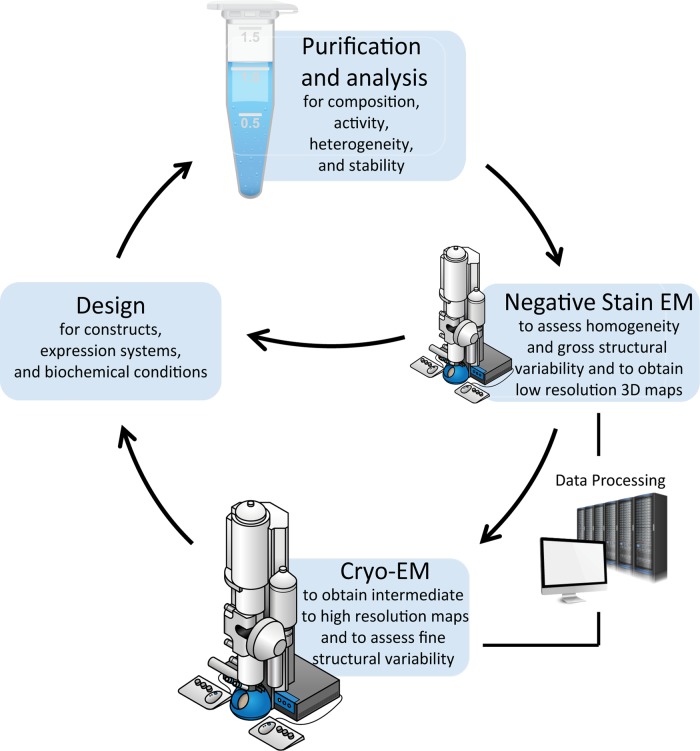
Fig. 2.
Overview of sample optimization and analysis for cryo-EM. For many samples iterative processes are necessary for obtaining a structure, improving resolution, identifying conformations or validating structural models. Following purification, initial negative-stain EM screening can provide a check-point regarding the suitability of the specimen for cryo-EM studies. Evaluation of samples as heterogeneous or unstable should lead to the redesign of constructs, buffer conditions and purification strategies to facilitate improvement. Significant computational analysis is required throughout EM steps for 2D/3D classification and map refinement. Following an initial cryo-EM characterization, additional construct designs or biochemical conditions are often necessary to probe conformational changes under different biochemical sates (e.g. inactive or activated enzymes or receptors) or validate domain localization in maps at intermediate to low-resolution.
For cryo-EM, a small amount of sample is applied on a holey carbon-coated grid, which is then plunge-frozen in liquid ethane. With this method, developed in the 1980s by Dubochet et al. [29], the water molecules freeze rapidly to form vitreous ice while preserving the sample fully hydrated in a buffer of choice. As with negative staining, several reviews are available for preparing samples by plunge-freezing techniques [30,31]. Optimization in order to minimize artifacts, such as crystalline ice and surface contamination, and to achieve thin ice with low background noise and evenly spread, intact particles is critical for imaging. Sample concentrations are typically around 10-fold higher compared with negative-stain preparations, but preferential absorption onto the surrounding carbon support and potential denaturation or aggregation at the air–water interface can add significant variability. The proximity of the air–water interface can also lead to preferred orientations of the particles, which hinders structure determination. The inclusion of detergents at low concentrations (below the CMC), or, more recently, amphipathic polymers (amphipols), has been shown to improve particle orientation in thin ice [10,32]. The addition of additives such as detergents, however, can increase the background scattering and reduce contrast.
Initial stages of single-particle EM data analysis and the need for classification
Negative-stain EM, besides its validity as a simple and high-contrast technique for assessing the sample quality, offers additional advantages during preliminary structural characterization. The carbon support of the EM grids used for negative staining often imposes preferred particle orientations, which is particularly useful when seeking to obtain additional insights into the sample during initial stages of characterization. This is accomplished with the application of 2D classification, a very important projection sorting process for both negative-stain EM and cryo-EM analysis. With 2D classification, particle projections are aligned and grouped based on their similarity and are subsequently used to calculate 2D averages for each group. 2D classification procedures are typically based on K-means clustering, a widely applicable methodology for identifying variations in a population [33]. In the process of 2D classification for single-particle EM analysis, every particle image is aligned, compared and classified according to its agreement against a number of templates [34]. In the first step the templates can be randomly selected particle projections from the dataset, and thus the process can be free of externally specified references. After the first clustering iteration, the group of particles in each class is used to calculate an average image, and all averages serve as the new templates for comparing every particle projection in the next iteration of the process. The improved signal present in the averages will gradually improve particle classification and the resulting class averages, and the process will be iterated until the partitioning is stable. While most programs include a variation in the 2D clustering routines, the recently developed Iterative Stable Alignment and Classification (ISAC) procedure in SPARX includes several validation steps that result in reliable classification and reproducible averages [35]. The limited particle orientations often present in negatively stained preparations make it more straightforward to compare features of particle averages from the same orientation in order to evaluate compositional homogeneity or regional flexibility [14]. Therefore, a relatively quick 2D classification and averaging of negatively stained samples can be used as a robust quality control test before attempting more sophisticated cryo-EM experiments.
As with 2D classification, the often assumed preferred orientation of negatively stained particles can be an advantage for the application of geometrically constrained 3D reconstruction approaches involving tilt pair images, such as the most commonly used random conical tilt (RCT) technique [36] or the recently proposed orthogonal tilt reconstruction (OTR) method [37]. With these techniques, particle projections categorized in a single class can be used to obtain a low-resolution 3D reconstruction. The ability to compare multiple independent 3D maps from similar views of the molecule can facilitate a more straightforward assessment of stable or variable features in the particle. Considering that the resolution of 3D reconstructions obtained with negatively stained samples is limited to ∼20 Å, the missing information due to the limits in EM stage tilting is of less concern, though attention should be given to the extent of particle collapse due to drying of the specimen [13,14]. Nevertheless, the advantage of obtaining multiple geometrically constrained, and thus robust 3D reconstructions from preferentially oriented negatively stained particles lies in the ability to confidently deduce large-scale compositional and conformational variations that may inform further structural work.
Cryo-EM analysis
The recent development of direct electron detectors has led to a remarkable improvement in the quality of recorded cryo-EM images. These detectors can record high-resolution data with superior signal-to-noise ratio compared with film or CCD cameras [38–40]. Importantly, the high sensitivity and speed of the detectors allow the recording of many consecutive low-dose ‘movie’ frames instead of a single frame, with the total electron dose fractionated over several seconds of recording [41,42]. This feature makes it possible to effectively correct for beam-induced motion and specimen drift encountered during image acquisition, which results in blurring and loss of high-resolution information. The individual movie frames can be aligned to each other to correct for the drift and summed to generate a single, total dose image that is used for extracting the individual particles. Additionally, individual image frames that are of poor quality, due to high drift and/or cumulative electron dose, can be removed at various stages of the refinement to improve resolution.
A prerequisite for any step in single-particle EM analysis, whether 2D averaging or 3D reconstruction, is the correction of the raw images for an electron-optical shortcoming termed contrast transfer function (CTF) [43]. The CTF modulates the phase and amplitude of the contrast in the recorded image as a function of spatial frequency. Amongst other parameters, most of which are characteristic and well-defined for each microscope, image modulation by the CTF depends on the defocus value that is used to enhance phase contrast in the images. Therefore, to determine the CTF and restore images for its effects requires accurate determination of defocus values for each image. CTF correction can be applied in a fully automated or interactive fashion and is implemented as part of all the main software platforms for image processing, such as SPIDER [44], EMAN2 [45], RELION [46], FREALIGN [47], XMIPP [48] and SPARX [49].
Single-particle cryo-EM analysis and 3D reconstruction involve a number of distinct steps of data processing (Fig. 3). After collecting a sufficient number of good-quality cryo-EM images, as judged by examination of the image power spectrum, a careful choice needs to be made in regards to particle selection. Whereas different kinds of automated particle selection programs work well for large complexes, such as ribosomes or the proteasome, their application may be problematic for low-contrast images of complexes smaller than 300 kDa, leading to many falsely picked image regions. A strategy that often works well is to manually select ∼5,000–10,000 particle projections and generate class averages, which can then be used as templates to guide the automated particle extraction routine [50,51]. In any case, a considerable user effort is usually needed for interactive examination of selected projections and the elimination of false particles. This is particularly important because including large numbers of falsely selected particles in a dataset may compromise the effectiveness of 2D classification of the particle projections. The unsupervised 2D classification routine, same as outlined above for negative-stain EM, is a necessary step that provides crucial information on the cryo-EM particle dataset. The ability to obtain well-defined class averages from a small dataset (∼5,000–10,000 projections) without any reference bias is usually a good indicator that the projections can be properly aligned, often facilitating an accurate 3D reconstruction. Another advantage is that the 2D classification will serve as a second seeding step to remove projections corresponding to false or damaged particles or a different species that could not be removed by biochemical purification. In addition, the classification will reveal whether there are strong preferred orientations assumed by the particles in the vitreous ice layer, which may significantly limit the quality of a cryo-EM 3D reconstruction and needs to be dealt with. Besides exploring biochemical conditions mentioned earlier (e.g. different detergents), the visualization of the specimen in slightly thicker vitreous ice may help improve the availability of different particle orientations. Another alternative, if cryo-EM is performed on a high-end microscope with very stable specimen stage, is to acquire additional images after tilting the sample ∼30°–45°, thereby supplementing the overall particle dataset with less frequent views.
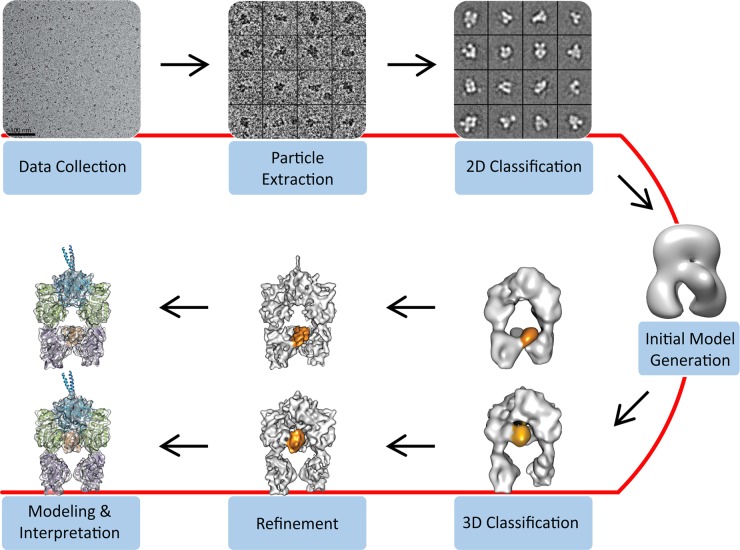
Fig. 3.
Overview of steps for the determination of single-particle cryo-EM 3D reconstructions. Cryo-EM images are acquired and individual particles are extracted manually or with automation, ideally followed by user examination to exclude falsely picked projections. The particle projections are subjected to 2D classification and averaging, aiming to group the projections according to their similarity. An initial 3D model may be generated ab initio using class averages representing different particle orientations, by geometrically constrained 3D reconstruction methods such as by RCT, or by utilizing available structural information. The initial 3D model can serve as a reference for 3D classification of the data, a useful step for identifying distinct conformations or macromolecular compositions. The 3D classification also facilitates identification of particle subsets that are amenable to higher resolution refinement. Depending on the achieved resolution, the final maps can be used for de novo structure building or the modeling of available structures based on various criteria.
Initial model generation
Well-defined 2D class averages representing several different views of the particle can be used to generate an initial low-resolution 3D reconstruction by back-projection. This ab initio model generation attempt takes advantage of the higher signal-to-noise ratio present in the class averages and follows the so-called common lines approach, as first implemented with 3D reconstruction by ‘angular reconstitution’ in IMAGIC [52]. The ‘common lines’ method aims to identify where pairs of projections intersect and to assign them angular orientations for the back-projection operation. In an iterative fashion, the experimental averages will then be compared with re-projections of the model, be assigned new orientations, and combined to create a new 3D model until the solution converges. There are different implementations of this routine in various image-processing programs. In our recent experience, the VIPER algorithm in SPARX has proven to be a robust method for the calculation of a first model from class averages as it incorporates critical validation steps [53]. However, especially in the case of small and asymmetric complexes with limited features for projection alignment, ab initio model generation may not deliver reliable solutions. In those cases it would be useful to obtain a first model from negatively stained sample using a geometrically constrained 3D reconstruction approach, such as the RCT method mentioned earlier [36]. Another option might be to create informed models from complete or even partial crystal structures if available. Especially in the case of small complexes the establishment of a first model that closely approximates the true structure is very important for further 3D reconstruction steps toward higher resolution. While projections of relatively large complexes can converge to the right architecture even by employing a starting model with very different shape, the limited features of complexes smaller than 300 kDa make it challenging to avoid getting trapped in local minima and to overcome reference bias. It is therefore useful to validate the obtained low-resolution model. This should be done in at least a few of several ways, such as by comparing results across different ab initio reconstruction programs, employing different starting references to test whether the refinement converges to the same overall architecture (see, for example, [54]), applying tilt pair parameter tests [55], comparing model re-projections with reference free averages, examining the regional correspondence with the shape of available crystal structures, and including invariably some common sense.
3D Classification
Prior to the refinement of a cryo-EM reconstruction, the particle projections can be further classified with 3D classification routines that attempt to iteratively categorize the particles according to their similarities with one or multiple references (Fig. 3). Essentially, 3D classification aims to differentiate between different views and different conformations of the particle, thereby producing projection subsets reflecting the different conformational sates. The process may employ a single or multiple reference models that supervise the initial partitioning of projections. A more recent variation of 3D clustering incorporates a maximum likelihood approach [56], which integrates over probability distributions for particle categorization, and is currently implemented in RELION [46] and FREALIGN [47]. In general, the 3D classification step can be useful for identifying dataset subpopulations that can facilitate high-resolution structure calculation, or exhibit a unique structural conformation of the macromolecule, or are simply of much lower quality, necessitating removal prior to high-resolution structure refinement. Again, the effectiveness of 3D classification is limited in the case of relatively small particles, and care must be taken to avoid producing erroneous results. In addition, the process should be executed with variable numbers of classes in an effort to test consistency and identify low occupancy particle categories that might otherwise get lost within other dominant classes. The number of projections needed to successfully tackle the variability in the particle ensemble depends on many factors, including the size and features of the target particle, the scale of the variability (large or small changes) and the number of different states. It is not uncommon to assemble datasets in excess of 300,000 projections in order to effectively deal with structural or compositional heterogeneity [57–59]. A key parameter for defining the required size for a dataset is whether the number of particles in a given state is high enough to facilitate a reconstruction with the desired resolution, assuming of course that this resolution signal is available in the raw data. It is currently the case for direct detector imaging that near-atomic resolution 3D reconstructions of asymmetric particles require a final dataset in the range of ∼50,000–150,000 projections (see, for example, [58,60]).
Cryo-EM structure refinement
Provided that a homogeneous particle subset has been defined and a starting reference model is available, the next step is the actual refinement of the structure (Fig. 3). In this process, the angular and translational positioning of a particle projection is iteratively refined through comparison to re-projections of the 3D map calculated in the previous step. Structure refinement can be executed using the entire particle dataset or after splitting it into two independent halves followed by independent refinement and comparison of the ‘half’ reconstructions. In either case, it is usually a good practice to refine cryo-EM structures with a resolution limited approach, in an effort to avoid excess noise build-up in the structure through noise fitting and to be able to define resolution more accurately [61]. Such approaches would include filtering the resulting 3D model from each round according to its resolution estimation before being used as a reference for the next round of refinement and also limiting the resolution of refinement of the raw projections. Resolution estimates can be obtained through Fourier shell correlation comparisons between two ‘half’ maps [62], where each map is generated from half the dataset projections refined together or independently of the other half [63]. The resolution numbers usually reported come from the overall map and provide no indication of local resolution, which can vary significantly depending on the positional stability of the corresponding segment. This characteristic is pronounced in cryo-EM as central densities are usually more stable and better defined compared to flexible peripheral densities in solution. Therefore, it is useful to estimate local resolution, such as with programs like ResMap [64], and also carefully examine and compare the map quality to provide a justified interpretation of features. It is also important to note that the alignment of cryo-EM projections against a reference map is dominated by the larger and more stable portion of the complex, leading to lower resolution for sub-regions assuming different relative conformations. In such cases, it is often possible to focus the refinement on the flexible portion by masking out the density of the larger stable region in the reference map [59,65] or even in the aligned projections. This strategy requires that the density of the target area is adequate for facilitating accurate alignment on its own, such as, for example, in the case of the small ribosomal subunit that can assume different ratcheting configurations in relation to the large subunit [66].
In the case of direct electron detector movie data, particularly when the resolution of the refined map is higher than ∼6 Å, further improvement may be achieved through the alignment of the movie frames for each individual particle [67]. This leads to better summed particle projections that can contribute higher resolution information towards the 3D map. Furthermore, this process can be coupled with different types of weighting of the sub-frames according to their signal-to-noise ratio and a model for resolution-dependent radiation damage [68]. These routines, though computationally demanding, have recently enabled final 3D maps with overall resolution improvements that may be critical for the interpretation of particular regions of interest. It should also be noted that when such weighted projections have been obtained, it might be useful to repeat previous classification steps aiming at improving 2D and 3D particle partitioning.
3D map interpretation
The interpretation of a cryo-EM 3D map is inherently dependent on the resolution achieved for the reconstruction. Resolutions of ∼15 Å are required to outline subunit boundaries within a map, better than 10 Å is needed to observe rod-like densities corresponding to α-helices, ∼4 Å to trace peptide backbone and closer to 3 Å to discern the side chains for most amino-acids or resolve stacked bases for double-stranded DNA or RNA (Fig. 4). Maps at near-atomic resolution can be used directly to build and refine atomic models with approaches that are largely established for X-ray crystallography. For maps with resolutions in the range of 4.5–10 Å, the docking of available high-resolution structures for individual domains can be invaluable for the creation of pseudo-atomic models. The modeling is usually performed computationally and aims to identify and optimize the positioning of the structure model within the map, either as a rigid body [70] or through different extents of flexible fitting. Computational methods for flexible fitting are particularly important for intermediate resolution maps with distinct secondary structure features and can be very useful for the identification of important interfaces. Flexible docking operations are accomplished primarily through variations of either normal modes flexible fitting (NMFF) [71,72] or molecular dynamics flexible fitting (MDFF) [73,74]. NMFF is driven by correlation optimization of the model against the 3D map, whereas in MDFF the Coulomb potentials of the 3D map are described as an additional force field driving the MD simulation. The development of these approaches brought very valuable insights to molecular transitions associated with function. However, flexible-docking routines should be controlled by strict constraints depending on the resolution of the map, with higher constraints employed for lower-resolution maps [75]. These conditions can ensure that the results are not prone to overfitting and are conservatively interpreted.
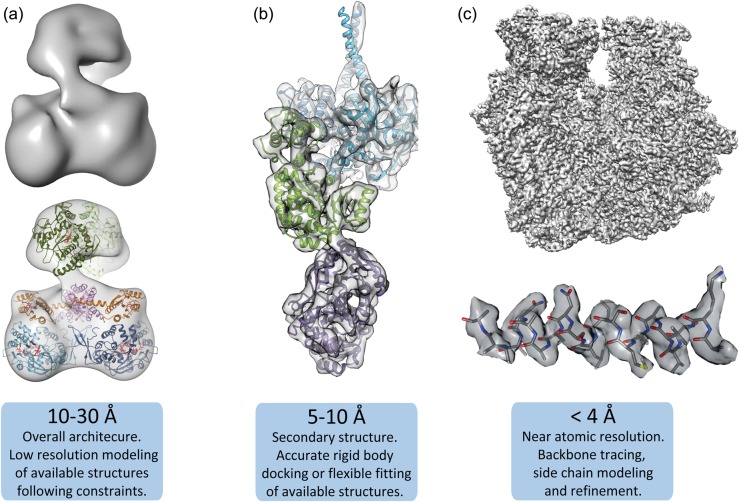
Fig. 4.
Molecular modeling at different resolution ranges. (a) Low resolution maps (10–30 Å), e.g. from a nitric oxide synthase (NOS) complex [69], can be used to dock large domains or protomer subunits and often require significant additional biochemical or structural information to provide constraints and validate fitting. (b) Intermediate resolution 3D reconstructions (5–10 Å), such as the one shown for a polyketide synthase (PKS) module [54], identify secondary structure elements, like α-helices, and subdomain densities that are reliably docked within the density map. Structure docking in this resolution range may benefit from flexible fitting procedures to identify novel conformational arrangements. (c) High-resolution maps (<4 Å), such as from a 70S ribosome (unpublished data), resolve backbone and side-chain densities, enabling de novo structure building.
While obtaining high-resolution cryo-EM maps is always the objective, it is often the case that a complex under investigation might prove refractory to structure characterization at a resolution better than 10 Å. Such limits may be posed due to the absence of enough features to facilitate accurate alignment of projections, as is the case with small particles, or due to extended conformational and compositional variability in the particle population. However, low-resolution maps can often be very informative, especially in the case of previously uncharacterized macromolecular complexes [69,76,77]. The interpretation of a low-resolution map can be challenging, particularly in the absence of known crystal structures. To this end, modifying the macromolecular complex in order to test predictions about the location of individual components is critical for developing structural models. Peripheral subunit localization can be achieved by performing subunit deletion or depletion experiments, assuming that this does not compromise the integrity of the macromolecular complex [78]. For macromolecules that have few subunits or are easily destabilized, the use of protein tags can be very useful for localizing specific domains. Soluble, structurally defined proteins, such as SH3 domain [79], green fluorescent protein (GFP) [80], maltose binding protein (MBP) [81] or T4 lysozyme [77] can be added to the N- or C-terminus or between domain boundaries. Similarly, biotinylation of specific residues can be used to decorate a particle with monomeric avidin [82]. By comparison with the untagged complex in 2D projection averages or 3D reconstructions, ideally through the use of difference maps, the density corresponding to the additional proteins is identified. Flexibility of the protein tag can be problematic, and thus limiting the linker length or truncating the target protein may be necessary.
Antibody labeling is another approach for localizing specific regions of a protein complex. This is ideally achieved through the use of a monovalent fragment antigen-binding (Fab) region, isolated by papain digestion of a preferably monoclonal antibody. Due to the high-affinity of the antigen-binding site, the inclusion of Fab fragments may increase complex stability and improve 2D projection alignments for reconstructions of small complexes [83]. Small gold clusters, such as 1.4 nm Nanogold (Nanoprobes) derivatized with Ni-NTA, Fab or cross-linker for protein attachment can also enable domain localization [84]. However, gold labeling may be less ideal for 2D classification or 3D reconstruction purposes because its high scattering power may negatively affect the alignment of projections as well as mask the protein densities. It should be emphasized that the interpretation of low-resolution maps requires the orthogonal integration of various types of data (biochemical or biophysical) that will enable the creation of testable models for the target complexes. To this end, chemical cross-linking can be coupled with EM structural analysis and combined with mass spectrometry to identify neighboring intermolecular sites in a complex [85,86]. Distance constraints, based on the length of the cross-linker, are applied and used to refine models as a powerful first step in localizing specific protein regions [22].
Thoughts and future perspectives
Owing to steady developments over the last 20 years, and recently propelled by breakthroughs in image acquisition hardware, single-particle cryo-EM has been elevated to a major tool for structure determination alongside X-ray crystallography and NMR. The methodology is particularly powerful for relatively large, multi-component macromolecular complexes that can now be characterized at high resolution. Near-atomic resolution reconstructions have also been obtained in cases of complexes as small as 200–300 kDa [10,58], although the results in this size category may vary depending on whether the particle geometry and features can facilitate accurate projection alignment. However, even low-resolution reconstructions combined with data from other techniques to produce testable models can also be very informative in the process of characterizing challenging complexes. This becomes all the more important, considering that this process necessitates a stepwise and often iterative approach that gradually identifies specific issues that need to be resolved in order to obtain a high-resolution structure. Furthermore, even though conformational variability poses challenges for high-resolution cryo-EM, especially for smaller targets, visualizing this variability is crucial for understanding large-scale dynamics that often underlie biological mechanisms. Such dynamics would include inherent flexibility of certain regions or precise structural rearrangements involving two or more biochemical states, such as those defining the steps of a catalytic mechanism [54,87]. Thus, such information would provide valuable insights on how components interact in the system, even in the absence of high-resolution information. It is perhaps this characteristic of providing both the architecture but also dynamics from the same sample population that elevates single-particle cryo-EM to a unique biophysical methodology for the characterization of biological mechanisms. Whether it is a simple negative-stain EM experiment to evaluate the quality of the biochemical preparation or a sophisticated cryo-EM analysis yielding near-atomic resolution, single-particle EM provides a comprehensive toolbox for various types of sample characterization.
The current realization of the advantages of this powerful, multipronged approach is already attracting more researchers into applying various levels of single-particle EM analysis. As the procedures become more standardized, the technique will become increasingly accessible, enhancing the research of not only the structural biologist but also the cell biologist or biochemist. Consequently, there is an intense need for additional EM instrumentation resources, which should be satisfied by both institutional and national centers, with the latter providing perhaps higher end instrumentation for established projects and also advice on sophisticated applications. This also underlines the need for more formal training and workshops in single-particle EM, in an effort to provide a solid and systematic background for the application toward the non-expert. Nevertheless, it is important to note that recognizing problems in cryo-EM analysis is not always straightforward and expert advice is highly recommended for any type of project.
Notwithstanding these challenges, the widespread interest in single-particle EM will accelerate further method development and user-friendly solutions tailored to different kinds of experimental needs. The next generation of direct electron detectors is expected to provide even higher signal-to-noise, which will enable the calculation of high-resolution reconstructions from smaller datasets and more robust classification of different particle populations. Importantly, the improved detectors will enable high-quality cryo-EM 3D reconstructions for smaller complexes. In the same direction, the rapidly developing phase plate technology for recording cryo-EM images will provide an additional tool for recording high-contrast images of relatively small macromolecules [88,89]. In regards to cryo-EM grid preparation, there is clearly a need to develop trouble-shooting procedures and systematically screen the conditions or chemical reagents for improving the dispersion of macromolecules within the holey grids and increase the efficiency of data collection. Along the same lines, the development of different types of grids and specimen supports [90,91] is expected to reduce the number of failed attempts and improve the quality of data recorded. Finally, there will continue to be significant improvements in the analysis software, with more refined 2D and 3D classification tools to efficiently address various levels of heterogeneity in the specimen and lead to high-resolution reconstructions. Collectively, these avenues will push the application of single-particle cryo-EM to full maturity, expanding its repertoire in both width and depth and rendering it more accessible and widely applicable. However, the inherent character of biological complexes will remain a limiting factor for single-particle cryo-EM—the dynamic and flexible nature of macromolecules often requires tailored approaches to resolve information at any resolution. Therefore, as is the case for X-ray crystallography, extensive biochemical control and protein engineering will also play an increasingly important role for obtaining high-quality and targeted structural information. In any case, modern single-particle cryo-EM is unique in its ability to provide both high-resolution structure determination as well as a characterization of the conformational ensemble adopted by the biological complexes under near-physiological conditions. This combination of information of both the structure and conformational dynamics will lead to unprecedented insights into the molecular mechanisms underlying crucial biological processes.
Acknowledgements
We thank Thomas Walz for critical reading of this manuscript and Alpay B. Seven for assistance with figure preparation.
References
- 1.Armache J P, Jarasch A, Anger A M, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Söding J, Westhof E, Wilson D N, Beckmann R (2010) Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc. Natl Acad. Sci. USA 107: 19748–19753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ludtke S J, Baker M L, Chen D H, Song J L, Chuang D T, Chiu W (2008) De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure 16: 441–448. [DOI] [PubMed] [Google Scholar]
- 3.Rabl J, Smith D M, Yu Y, Chang S C, Goldberg A L, Cheng Y (2008) Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30: 360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H, Jin L, Koh S B S, Atanasov I, Schein S, Wu L, Zhou Z H (2010) Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329: 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X, Jin L, Zhou Z H (2008) 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature 453: 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Jin L, Fang Q, Hui W H, Zhou Z H (2010) 3.3 Å cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141: 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kühlbrandt W. (2014) The resolution revolution. Science 343: 1443–1444. [DOI] [PubMed] [Google Scholar]
- 8.Bai X C, Fernandez I S, McMullan G, Scheres S H W (2013b) Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife 2:e00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao E, Liao M, Cheng Y, Julius D (2013) TRPV1 structures in distinct conformations reveal mechanisms of activation. Nature 504: 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao M, Cao E, Julius D, Cheng Y (2013) Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504: 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Mooney P, Zheng S, Booth C, Braunfeld M B, Gubbens S, Agard D A, Cheng Y (2013) Electron counting and beam-induced motion correction enable near atomic resolution single particle cryoEM. Nat. Methods 10: 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark H, Zemlin F, Boettcher C (1996) Electron radiation damage to protein crystals of bacteriorhodopsin at different temperatures. Ultramicroscopy 63: 75–79. [Google Scholar]
- 13.Ohi M, Li Y, Cheng Y, Walz T (2004) Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proc. Online 6: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peisley A, Skiniotis G (2015) 2D projection analysis of GPCR complexes by negative stain electron microscopy. Methods Mol. Biol. 1335: 29–38. [DOI] [PubMed] [Google Scholar]
- 15.Penczek P A. (2010a) Fundamentals of three-dimensional reconstruction from projections. Methods Enzymol 482: 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penczek P A, Grassucci R A, Frank J (1994) The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy 53: 251–270. [DOI] [PubMed] [Google Scholar]
- 17.Bammes B E, Jakana J, Schmid M F, Chiu W (2010) Radiation damage effects at four specimen temperatures from 4 to 100 K. J. Struct. Biol. 169: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duong-Ly K C, Gabelli S B (2014) Gel filtration chromatography (size exclusion chromatography) of proteins. Methods Enzymol. 541: 105–114. [DOI] [PubMed] [Google Scholar]
- 19.Dombkowski A A, Sultana K Z, Craig D B (2014) Protein disulfide engineering. FEBS Lett. 588: 206–212. [DOI] [PubMed] [Google Scholar]
- 20.Wong S S, Wong L J (1992) Chemical crosslinking and the stabilization of proteins and enzymes. Enzyme Microb. Technol. 14: 866–874. [DOI] [PubMed] [Google Scholar]
- 21.Kastner B, Fischer N, Golas M M, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, Uchtenhagen H, Urlaub H, Herzog F, Peters J M, Poerschke D, Lührmann R, Stark H (2008) GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5: 53–55. [DOI] [PubMed] [Google Scholar]
- 22.Shukla A K, Westfield G H, Xiao K, Reis R I, Huang L Y, Tripathi-Shukla P, Qian J, Li S, Blanc A, Oleskie A N, Dosey A M, Su M, Liang C R, Gu L L, Shan J M, Chen X, Hanna R, Choi M, Yao X J, Klink B U, Kahsai A W, Sidhu S S, Koide S, Penczek P A, Kossiakoff A A, Woods V L Jr, Kobilka B K, Skiniotis G, Lefkowitz R J (2014) Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512: 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Sielaff B, Lee J, Tsai F T (2010) CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc. Natl Acad. Sci. USA 107: 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Southworth D R, Agard D A (2011) Client-loading conformation of the Hsp90 molecular chaperone revealed in the cryo-EM structure of the human Hsp90:Hop complex. Mol. Cell 42: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark H. (2010) GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481: 109–126. [DOI] [PubMed] [Google Scholar]
- 26.Booth D S, Avila-Sakar A, Cheng Y (2011) Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. J. Vis. Exp. 58: pii3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takagi J, Strokovich K, Springer T A, Walz T (2003) Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J. 22: 4607–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao F Q, Craig R (2003) Capturing time-resolved changes in molecular structure by negative staining. J. Struct. Biol. 141: 43–52. [DOI] [PubMed] [Google Scholar]
- 29.Dubochet J, Booy F P, Freeman R, Jones A V, Walter C A (1981) Low temperature electron microscopy. Annu. Rev. Biophys. Bioeng. 10: 133–149. [DOI] [PubMed] [Google Scholar]
- 30.Dobro M J, Melanson L A, Jensen G J, McDowall A W (2010) Plunge freezing for electron cryomicroscopy. Methods Enzymol. 481: 63–82. [DOI] [PubMed] [Google Scholar]
- 31.Grassucci R A, Taylor D J, Frank J (2007) Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc. 2: 3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flotenmeyer M, Weiss H, Tribet C, Popot J L, Leonard K (2007) The use of amphipathic polymers for cryo electron microscopy of NADH:ubiquinone oxidoreductase (complex I). J. Microsc. 227: 229–235. [DOI] [PubMed] [Google Scholar]
- 33.Duda R O, Hart P E, Stork D G (2001) Pattern classification, (Wiley, NewYork: ). [Google Scholar]
- 34.van Heel M, Stöffler-Meilicke M (1985) Characteristic views of E. coli and B. stearothermophilus 30S ribosomal subunits in the electron microscope. EMBO J. 4: 2389–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, Fang J, Chittuluru J, Asturias F J, Penczek P A (2012) Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radermacher M, Wagenknecht T, Verschoor A, Frank J (1987) Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 146: 113–136. [DOI] [PubMed] [Google Scholar]
- 37.Leschziner A E, Nogales E (2006) The orthogonal tilt reconstruction method: an approach to generating single-class volumes with no missing cone for ab initio reconstruction of asymmetric particles. J. Struct. Biol. 153: 284–299. [DOI] [PubMed] [Google Scholar]
- 38.McMullan G, Chen S, Henderson R, Faruqi A R (2009) Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy 109: 1126–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMullan G, Faruqi A R, Clare D, Henderson R (2014) Comparison of optimal performance at 300 keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy 147: 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruskin R S, Yu Z, Grigorieff N (2013) Quantitative characterization of electron detectors for transmission electron microscopy. J. Struct. Biol. 184: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brilot A F, Chen J Z, Cheng A, Pan J, Harrison S C, Potter C S, Carragher B, Henderson R, Grigorieff N (2012) Beam-induced motion of vitrified specimen on holey carbon film. J. Struct. Biol. 177: 630–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell M G, Cheng A, Brilot A F, Moeller A, Lyumkis D, Veesler D, Pan J, Harrison S C, Potter C S, Carragher B, Grigorieff N (2012) Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure 20: 1823–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penczek P A. (2010b) Image restoration in cryo-electron microscopy. Methods Enzymol 482: 35–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116: 190–199. [DOI] [PubMed] [Google Scholar]
- 45.Tang G, Peng L, Baldwin P R, Mann D S, Jiang W, Rees I, Ludtke S J (2007) EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157: 38–46. [DOI] [PubMed] [Google Scholar]
- 46.Scheres S H W. (2012) RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180: 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grigorieff N. (2007) FREALIGN: High-resolution refinement of single particle structures. J. Struct. Biol. 157: 117–125. [DOI] [PubMed] [Google Scholar]
- 48.Sorzano C O, Marabini R, Velazquez-Muriel J, Bilbao-Castro J R, Scheres S H, Carazo J M, Pascual-Montano A (2004) XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148: 194–204. [DOI] [PubMed] [Google Scholar]
- 49.Hohn M, Tang G, Goodyear G, Baldwin P R, Huang Z, Penczek P A, Yang C, Glaeser R M, Adams P D, Ludtke S J (2007) SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157: 47–55. [DOI] [PubMed] [Google Scholar]
- 50.Chen J Z, Grigorieff N (2007) SIGNATURE: a single-particle selection system for molecular electron microscopy. J. Struct. Biol. 157: 168–173. [DOI] [PubMed] [Google Scholar]
- 51.Sigworth F J. (2004) Classical detection theory and the cryo-EM particle selection problem. J. Struct. Biol. 145: 111–122. [DOI] [PubMed] [Google Scholar]
- 52.Van Heel M. (1987) Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21: 111–123. [DOI] [PubMed] [Google Scholar]
- 53.Penczek P A. (2014) SPARX Wiki.
- 54.Dutta S, Whicher J R, Hansen D A, Hale W A, Chemler J A, Congdon G R, Narayan A R, Hakansson K, Sherman D H, Smith J L, Skiniotis G (2014) Structure of a modular polyketide synthase. Nature 510: 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henderson R, Chen S, Chen J Z, Grigorieff N, Passmore L A, Ciccarelli L, Rubinstein J L, Crowther R A, Stewart P L, Rosenthal P B (2011) Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J. Mol. Biol. 413: 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sigworth F J, Doerschuk P C, Carazo J M, Scheres S H W (2010) An introduction to maximum-likelihood methods in cryo-EM. Methods Enzymol 482: 263–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amunts A, Brown A, Bai X C, Llácer J L, Hussain T, Emsley P, Long F, Murshudov G, Scheres S H W, Ramakrishnan V (2014) Structure of the yeast mitochondrial. Science 343: 1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bai X C, Yan C, Yang G, Lu P, Ma D, Sun L, Zhou R, Scheres S H, Shi Y (2015) An atomic structure of human gamma-secretase. Nature 525: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen T H, Galej W P, Bai X C, Savva C G, Newman A J, Scheres S H, Nagai K (2015) The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 523: 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voorhees R M, Fernandez I S, Scheres S H, Hegde R S (2014) Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell 157: 1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen S, McMullan G, Faruqi A R, Murshudov G N, Short J M, Scheres S H, Henderson R (2013) High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Penczek P A. (2010c) Resolution measures in molecular electron microscopy. Methods Enzymol 482: 73–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheres S H, Chen S (2012) Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9: 853–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kucukelbir A, Sigworth F J, Tagare H D (2014) Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11: 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan C, Hang J, Wan R, Huang M, Wong C C, Shi Y (2015) Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- 66.Valle M, Sengupta J, Swami N K, Grassucci R A, Burkhardt N, Nierhaus K H, Agrawal R K, Frank J (2002) Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 21: 3557–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bai X-C, Fernandez I S, McMullan G, Scheres S H W (2013a) Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife 2: e00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheres S H. (2014) Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife 3: e03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yokom A L, Morishima Y, Lau M, Su M, Glukhova A, Osawa Y, Southworth D R (2014) Architecture of the nitric-oxide synthase holoenzyme reveals large conformational changes and a calmodulin-driven release of the FMN domain. J. Biol. Chem. 289: 16855–16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wriggers W, Milligan R A, McCammon J A (1999) Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125: 185–195. [DOI] [PubMed] [Google Scholar]
- 71.Tama F, Miyashita O, Brooks C L 3rd (2004a) Flexible multi-scale fitting of atomic structures into low-resolution electron density maps with elastic network normal mode analysis. J. Mol. Biol. 337: 985–999. [DOI] [PubMed] [Google Scholar]
- 72.Tama F, Miyashita O, Brooks C L 3rd (2004b) Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J. Struct. Biol. 147: 315–326. [DOI] [PubMed] [Google Scholar]
- 73.Trabuco L G, Villa E, Mitra K, Frank J, Schulten K (2008) Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trabuco L G, Villa E, Schreiner E, Harrison C B, Schulten K (2009) Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vashisth H, Skiniotis G, Brooks C L 3rd (2012) Using enhanced sampling and structural restraints to refine atomic structures into low-resolution electron microscopy maps. Structure 20: 1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takahashi Y H, Westfield G H, Oleskie A N, Trievel R C, Shilatifard A, Skiniotis G (2011) Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc. Natl Acad. Sci. USA 108: 20526–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Westfield G H, Rasmussen S G, Su M, Dutta S, DeVree B T, Chung K Y, Calinski D, Velez-Ruiz G, Oleskie A N, Pardon E, Chae P S, Liu T, Li S, Woods V L Jr, Steyaert J, Kobilka B K, Sunahara R K, Skiniotis G (2011) Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc. Natl Acad. Sci. USA 108: 16086–16091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Strunk B S, Loucks C R, Su M, Vashisth H, Cheng S, Schilling J, Brooks C L 3rd, Karbstein K, Skiniotis G (2011) Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333: 1449–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skiniotis G, Surrey T, Altmann S, Gross H, Song Y H, Mandelkow E, Hoenger A (2003) Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J. 22: 1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choy R M, Kollman J M, Zelter A, Davis T N, Agard D A (2009) Localization and orientation of the gamma-tubulin small complex components using protein tags as labels for single particle EM. J. Struct. Biol. 168: 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Z, Speck C, Wendel P, Tang C, Stillman B, Li H (2008) The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 105: 10326–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lau P W, Potter C S, Carragher B, MacRae I J (2012) DOLORS: versatile strategy for internal labeling and domain localization in electron microscopy. Structure 20: 1995–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu S, Avila-Sakar A, Kim J, Booth D S, Greenberg C H, Rossi A, Liao M, Li X, Alian A, Griner S L, Juge N, Yu Y, Mergel C M, Chaparro-Riggers J, Strop P, Tampe R, Edwards R H, Stroud R M, Craik C S, Cheng Y (2012) Fabs enable single particle cryoEM studies of small proteins. Structure 20: 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ackerson C J, Powell R D, Hainfeld J F (2010) Site-specific biomolecule labeling with gold clusters. Methods Enzymol. 481: 195–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kalisman N, Adams C M, Levitt M (2012) Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc. Natl Acad. Sci. USA 109: 2884–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold C B, Lakomek K, Aebersold R, Beckmann R, Hopfner K P (2013) Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154: 1207–1219. [DOI] [PubMed] [Google Scholar]
- 87.Whicher J R, Dutta S, Hansen D A, Hale W A, Chemler J A, Dosey A M, Narayan A R, Hakansson K, Sherman D H, Smith J L, Skiniotis G (2014) Structural rearrangements of a polyketide synthase module during its catalytic cycle. Nature 510: 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang W H, Chiu M T K, Chen C Y, Yen C F, Lin Y C, Weng Y P, Chang J C, Wu Y M, Cheng H, Fu J, Tu I P (2010) Zernike phase plate cryoelectron microscopy facilitates single particle analysis of unstained asymmetric protein complexes. Structure 18: 17–27. [DOI] [PubMed] [Google Scholar]
- 89.Danev R, Buijsse B, Khoshouei M, Plitzko J M, Baumeister W (2014) Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl Acad. Sci. USA 111: 15635–15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Russo C J, Passmore L A (2014a) Controlling protein adsorption on graphene for cryo-EM using low-energy hydrogen plasmas. Nat. Methods 11: 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Russo C J, Passmore L A (2014b) Ultrastable gold substrates for electron cryomicroscopy. Science 346: 1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]