1. Introduction
Pain and pain chronification are incompletely understood and unresolved medical problems that continue to have a high prevalence.14 It has been accepted that pain is a complex phenomenon.2,32,72 Contemporary methods of computational science51 can use complex clinical and experimental data to better understand the complexity of pain. Among data science techniques, machine learning is referred to as a set of methods (Fig. 1) that can automatically detect patterns in data and then use the uncovered patterns to predict or classify future data, to observe structures such as subgroups in the data, or to extract information from the data suitable to derive new knowledge.11,43 Together with (bio)statistics, artificial intelligence and machine learning aim at learning from data.
Figure 1.
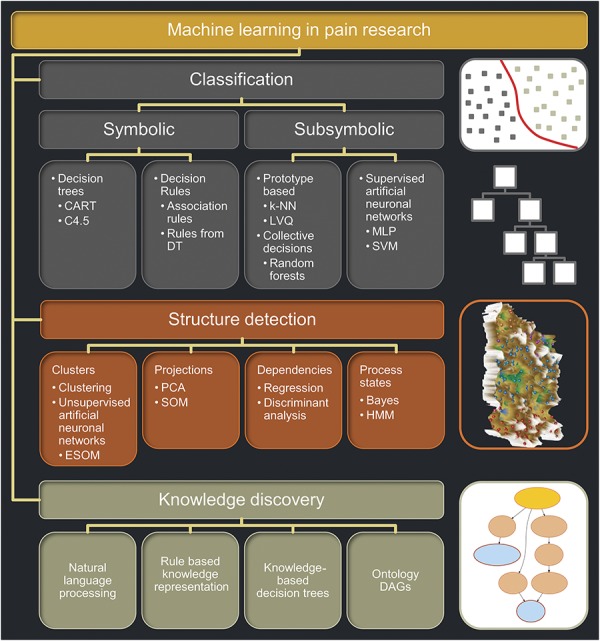
Overview and classification of machine learning methods, selected for their use in a pain research context. The figure structures machine learning for its main uses comprising (1) classification tasks used for example to obtain a clinical diagnosis, (2) data structure detection including the identification of clusters, and (3) knowledge discovery in experimental or clinical data or in large databases structured hierarchically such as ontologies. Short descriptions of key methods are provided in Box 1. The icons at the right of each main application field symbolize respective typical machine learning methods, that is, from top to bottom: (1) SVM where the grouping (classification) is obtained by placing a border (hyperplane) between classes (subsymbolic classifier), (2) a decision tree where the classification is obtained through hierarchical rules (symbolic classifier), (3) an emergent self-organizing maps as an unsupervised machine learning method able to find an interesting structure in high-dimensional data such as clusters. In this figure, the map was colored using a geographical analogy with brown (up to snow-covered) heights and green valleys, on which clusters can be separated (from Ref. 36). Finally, (4) a directed acyclic graph is drafted depicting the polyhierarchy of, for example, the functions of pain-relevant genes (from Ref. 69). CART, classification and regression tree; DAG, directed acyclic graph; DT, decision tree; ESOM, emergent self-organizing map; HMM, hidden Markov models; k-NN, k nearest neighbor; LVQ, learning vector quantization; MLP, multilayer perceptron; PCA, principal component analysis; SVM, support vector machine.
Although statistics can be regarded as a branch of mathematics, artificial intelligence and machine learning have developed from computer science (Ref. 58; see also https://en.wikipedia.org/wiki/Artificial_intelligence). The initial definition of artificial intelligence originates from Alan Turing who proposed an experiment where 2 players, who can either be human or artificial, try to convince a human third player, that they are also humans.68 The test of artificial intelligence is passed if the third player cannot tell who is the machine. Important steps in the development of machine learning were the first creation of the computer learning program, which was a checker game,54 and the first neural network called the perceptron.53 Statistics uses mathematical equations to model probability relationships between data variables, whereas machine learning learns from data without the necessity of previous knowledge. It aims at optimization and performance of an algorithm rather than on the analysis of the probabilities of observations, given a known underlying data distribution. Nevertheless, both machine learning and statistics techniques are working in concert for pattern recognition, knowledge discovery, and data mining and share partly the same methods such as regression, which is used widely in statistics but is also considered as a classification method in machine learning (Fig. 1).
In the present research context, when provided with pain-related data, machine-learned methods are able to learn a mapping of complex features to a known class, that is, to predict a pain phenotype class from a complex pattern of acquired parameters. After the machine has learned the prediction of a pain-related phenotype, the algorithm can subsequently be used on new data from which the class membership of a novel yet unclassified subject can be identified. However, machine learning methods can also be used for pattern recognition in complex pain-related data to reveal traces of an underlying molecular background or for knowledge discovery in big data in a drug discovery or repurposing context. The increasing use of contemporary methods of computational science is reflected in the rising number of reports using machine learning for pain research (Table 1). This review is focused on machine-learned technologies applied to general pain research that allow one to analyze and predict pain phenotypes and to obtain knowledge from experimental and clinical pain-related data.
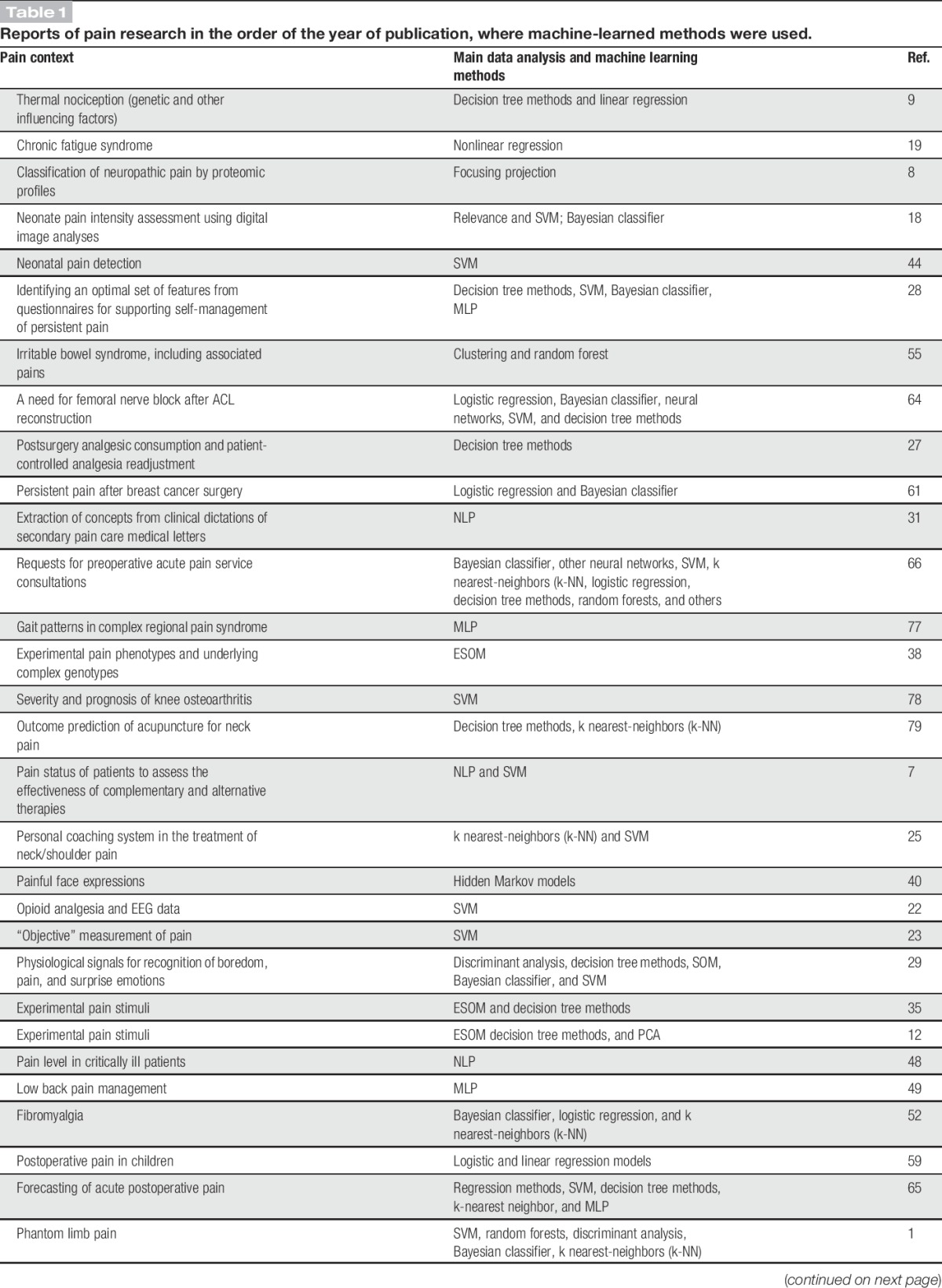
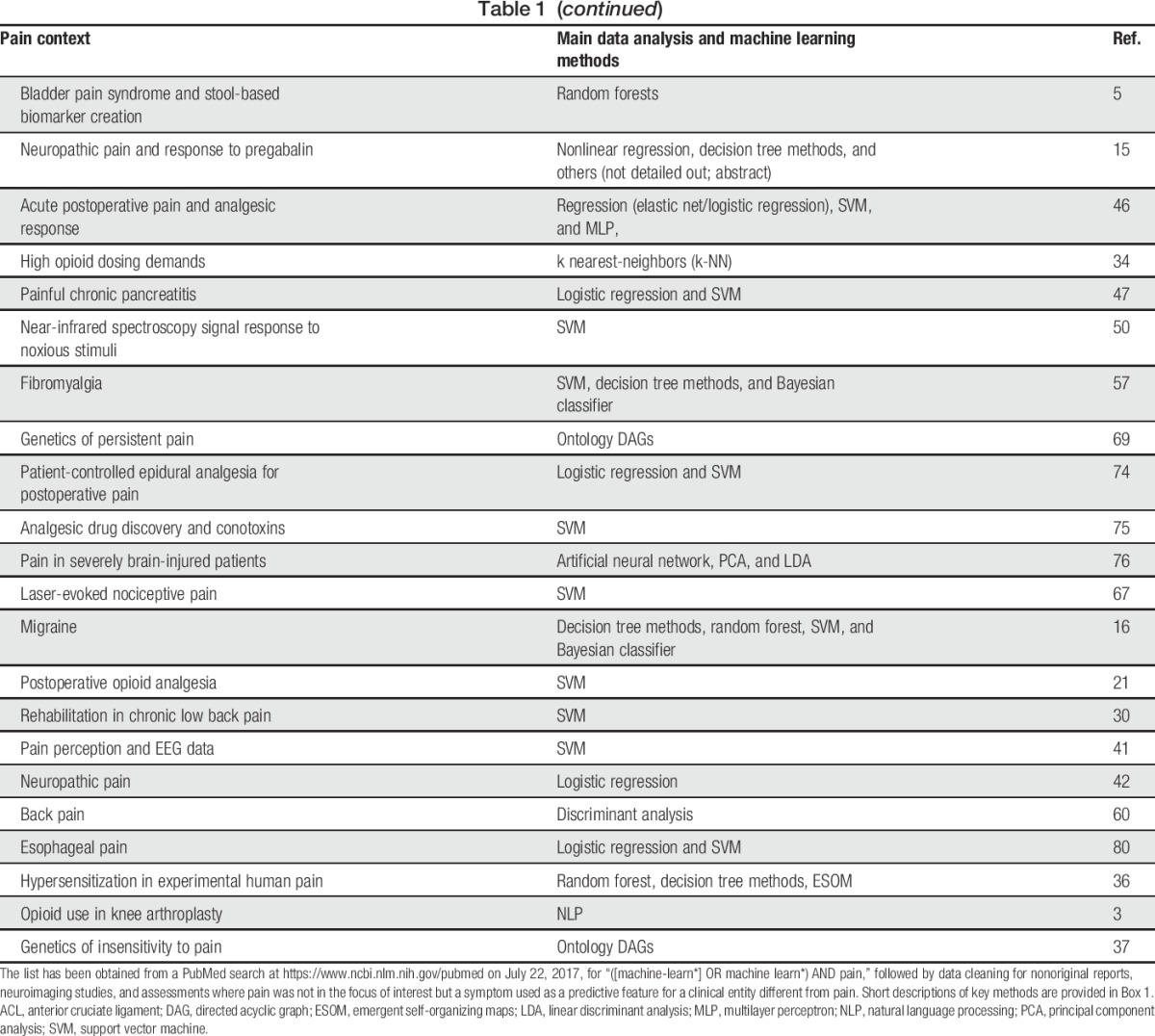
Table 1.
Reports of pain research in the order of the year of publication, where machine-learned methods were used.
2. Pain research involving machine learning
A literature search was conducted using PubMed at https://www.ncbi.nlm.nih.gov/pubmed on July 22, 2017 for “([machine-learn*] OR machine learn*) AND pain.” One hundred ten results published between 2002 and 2017 with an increasing number of publications over time were obtained (Table 1), and a few more reports were obtained from reference tracking. After elimination of editorials, reviews, repeated reports of the same machine-learned analysis, 88 original reports of the use of machine learning in a pain context were identified. Twenty-two articles that regarded pain only as a symptom interesting in another context such as chest pain as a diagnostic criterion for pneumonia10 or coronary syndromes4 or phantom limb pain as an indicator or prosthesis functioning1 were excluded as well as 14 reports about neuroimaging of pain, a topic that has been reviewed separately.26,39 This resulted in 52 reports that were analyzed for the use of several different methods of machine learning in pain research (Table 1). For a short description of the mentioned machine learning methods, please refer to Box 1.
Text box 1. Definitions and descriptions of key methods of machine learning most frequently used so far in the pain research context (Table 1 and Fig. 1). For a detailed description of these and further machine learning methods, see, for example, Ref. 11,43.
(1) Classification solves the problem of identifying to which category (diagnosis) a new case belongs, based on a training data set of data containing cases whose category is known.
(2) A Bayes classifier minimizes the probability of misclassification, given the prerequisites of the theorem of Bayes, that is, distributions and (conditional) probabilities.
(3) Decision tree methods output a tree-structured graph consisting of variables (features) in the decisions nodes (points of split) and conditions in the edges.
(4) Random forests use a multitude of simple decision trees usually based on a random selection of a small set of features.
(5) Projection methods represent the data space in a lower-dimensional space with the aim of conserving important structural properties.
(6) Focusing projection methods are learned using a function of the neighborhood of points in the data space.
(7) K nearest-neighbors (k-NN) methods use k (classified) prototypes to which new cases are assigned depending on their distances to all prototypes.
(8) Artificial neural networks (ANNs) are computer programs that operate a multitude of simple processing elements (neurons), which are connected to each other by (weighted) synapses.
(9) Multilayer perceptrons are ANN, where the connections are structured in layers. The neurons are of the McCulloch-Pitts type, that is, nonlinear decisions using hyperplanes are used.
(10) Support vector machines are multilayer perceptron with 1 layer using McCulloch-Pitts type of neurons, where the input data are projected into a (possible infinite dimensional) vector space in which (1) scalar products are easily computable and (2) the decision surface can be more complex than simple hyperplanes.
(11) A self-organizing map (SOM) is an unsupervised learning ANN producing a 2-dimensional discretized representation of the data space through a focusing projection.
(12) Emergent SOM are SOMs able to show emergent structures in the form of (U-, P-, and U*-) matrix representations, which display structural features of the data space using a geographical map metaphor.
(13) Knowledge in data science is a symbolic representation of taxonomic categorizations and decisions using an algorithmic treatable (ie, decidable or provable) part of natural human language, such as (a subset of) predicate logic, with the requirement to generalize to unseen data.
(14) Ontologies use data science knowledge in the form of a naming and definition of the terms and semantic interrelationships of the entities that really or fundamentally exist for a particular domain of discourse.
(15) Ontology directed acyclic graphs are graph-based representations of a polyhierarchy of terms of an ontology.
3. Pain phenotype prediction from complex case data
Machine learning addresses the so-called data space including an input space X comprising vectors xi = <xi,1,…xi,d> with d > 0 different parameters (variables and features24) acquired from n > 0 cases. In supervised machine learning, algorithms enable a mapping of the input parameters xi to the output classes yi in the data space 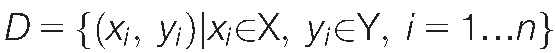 . The information consisting of several biomedical parameters is used to derive a mapping that allows assigning future cases to the right class (prediction and generalization Ref. 11), for example, the pain phenotype group or a clinical diagnosis. The main types of classifiers provided by supervised machine learning are symbolic45 or subsymbolic63 classifiers.
. The information consisting of several biomedical parameters is used to derive a mapping that allows assigning future cases to the right class (prediction and generalization Ref. 11), for example, the pain phenotype group or a clinical diagnosis. The main types of classifiers provided by supervised machine learning are symbolic45 or subsymbolic63 classifiers.
In symbolic classifiers, the decision how a classification is obtained can be interpreted by a domain expert as a combination of conditions on the features. For example, a symbolic45 classifier composed of a decision tree was created to predict patient-controlled analgesia consumption from approximately 30 acquired features including demographic (age, sex, and weight), biomedical (eg, blood pressure, diabetes, and arterial hypertension), surgery-related therapy (eg, type of surgery, duration, and details of anesthesia) and analgesic-related therapy (eg, consumption of analgesics before the surgery and dose demands during the first 24 hours after surgery) parameters.27 Importantly, for each of the parameters, the value range underlying the decision with respect to analgesics demands remained accessible (see Tables 1 and 11 in Ref. 27). In decision trees, the features are also weighted according to their importance (most important first). Another example of a symbolic classifier is the creation of a Bayesian diagnostic tool from demographic-, pain-, and surgery-related parameters for the prediction of persistence of pain in a breast cancer surgery context. It provided a sensitivity and specificity of 33% and 95%, respectively.61 Again, the classification procedure was accessible to direct interpretation through the Bayesian decision limits calculated for the single parameters.
In subsymbolic classifiers, a better performance of a machine learned algorithm is sought by waiving the possibility of understanding the details, that is, it is impossible to obtain biomedical explanations for the functioning of the algorithm. For example, random forests use hundreds or thousands of simple decision trees that escape interpretation; the classification is obtained through the complete set of trees, that is, the “forest.”6 Such a classifier was created from various stool-based markers to diagnose a bladder pain syndrome.5 Similarly, a projection method for high-dimensional data, specifically, minimum curvilinear embedding, was applied to obtain from complex proteomics data a clustering of patients with neuropathic pain from controls and a further separation of different types of neuropathy such as neuropathy associated with amyotrophic lateral sclerosis and peripheral neuropathy with or without or pain.8 Machine-learned algorithms were further applied to predict thermal pain sensitivity from bioresponses acquired through electromyography, skin conductance level, and electrocardiography.23 Specifically, using support vector machines (SVMs56), individual pain threshold and tolerance to thermal stimulation could be predicted from the noninvasive measurements at accuracies of >91% and 79%, respectively.23 This aimed at obtaining information about pain in subjects with verbal and/or cognitive impairments in whom queries of pain such as standard visual rating scales cannot be applied. Moreover, predicting which patients required high opioid doses for analgesia, based on a next-generation sequencing–derived opioid receptor genotype, was achieved with a subsymbolic classifier based on k-nearest neighbors calculations.34 Another application of subsymbolic classifiers has been implemented as neural networks. The so-called elastic net regression models and SVMs predicted pain scores measured between 40 and 120 minutes after the administration of 10 mg oxycodone from interpolated pain score values before drug administration.46 The elastic net regression model provided pain scores that had a correlation coefficient of 0.6 with the observed scores.
4. Structure detection in complex pain-related data
Detecting structures in the d-dimensional data space 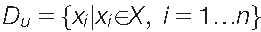 pointing at patterns or subgroups accessible to biomedical interpretation is a typical application of unsupervised machine learning. In contrast to the supervised learning setting, the class information Y is absent or ignored; the task is to find “interesting” data structures that can be interpreted as subgroups (clusters and strata) in the studied cases or made accessible for biomedical interpretation by domain experts, including the discovery of new knowledge in data-driven research approaches.
pointing at patterns or subgroups accessible to biomedical interpretation is a typical application of unsupervised machine learning. In contrast to the supervised learning setting, the class information Y is absent or ignored; the task is to find “interesting” data structures that can be interpreted as subgroups (clusters and strata) in the studied cases or made accessible for biomedical interpretation by domain experts, including the discovery of new knowledge in data-driven research approaches.
For example, in a data matrix comprising several quantitative sensory testing (QST) parameters acquired from healthy subjects, a pattern was detected allowing one to identify a subgroup of healthy subjects who reacted to hypersensitization with topical capsaicin with a shift in QST parameters that resembled the parameter pattern observed in patients with neuropathic pain.35 Similarly, in a set of pain phenotype data comprising responses to experimental heat, cold, mechanical, and electrical pain stimuli applied in 125 healthy subjects, structures were detected using unsupervised machine learning implemented as emergent self-organizing maps.71 These data structures could be associated with a complex genotype composed of 30 reportedly pain relevant variants in 10 genes, which was able to correctly identify 80% of the subjects as belonging to an extreme pain phenotype in an independent and prospectively assessed cohort of 89 other subjects.38
5. Knowledge discovery and exploration of pain-related data
Machine learning methods can be used to explore data sets by reversing the analytical focus of classifier building and pattern detection. Supervised machine learning methods qualify for data exploration under the assumption that if a biomedical parameter qualifies for inclusion in a classifier, then it is probably important for the addressed pain-related problem. In contrast to classic statistical methods, where knowledge or at least presumptions about the distributions and/or functional dependencies of the data are necessary, machine learning methods allow for data-driven research approaches. Hence, techniques of feature selection, which are common in machine learning, enable one to identify relevant modulators of pain-related outcomes in data-driven and hypothesis-free explorative research approaches. For example, a machine-learned analysis identified, among hundreds of biomedical parameters, demographic-, psychological-, and pain-related parameters as the most relevant for explaining the persistence of pain in women who underwent breast cancer surgery.61
Moreover, unsupervised machine learning methods can be used to assess, at a whole-study level, whether the acquired biomedical parameters demonstrate the efficacy of a treatment applied during a research project. The rationale is to detect data structures that are congruent with a known preclassification such as the presence of a modulator of the pain phenotype. For example, after treatment of 82 subjects with local UV-B irradiation or capsaicin application and assessing the pain phenotype using 10 different QST parameters, a 246 × 10-sized data matrix was obtained in a human experimental pain study.36 Using unsupervised machine learning implemented as emergent self-organizing maps,71 data structures were detected that coincided with applied known treatments indicating that a modulation of the complex pain phenotype had been obtained.36
A machine learning algorithm consisting of a classification and regression tree analysis was applied to 8034 independent observations of baseline thermal nociceptive sensitivity in mice.9 The analysis identified the mouse genotype as predictive of the pain phenotype; however, it also revealed that the experimenter performing the test and additional laboratory factors including season/humidity, cage density, time of day, sex, and within-cage or order of testing modulated the pain phenotypes.9 Finally, natural language progressing methods,73 which combine linguistics with computer science to analyze human language in speech or written text, were used to extract signs from clinical notes using, such as the occurrence of terms, for example, keywords that hint at a clinically incident, in a document.48 Prediction accuracy of this method for the patient's pain level was reported to be better than 99%.
6. Limitations of machine learning in pain research
Machine learning is vulnerable to overfitting and may end up in describing noise or irrelevant relationships rather than the true relationship between features and classes. In that case, only the actual data on which the mapping has been learned are successfully classified, but the algorithm fails to classify new data. This can be addressed by building the classifier on a training data set and testing its performance on a test data set obtained in a separate experiment or through splitting the available data, and/or by cross validation using creating data subsets randomly resampled from the original data sets. Furthermore, machine learning may be fooled by data sets containing dominant but irrelevant features. A classic example is the training of a neuronal network to recognize camouflaged tanks hidden in trees.13 The network was apparently successfully trained with a set of photographs of tanks in trees and just trees without tanks. However, in a new set of photographs of trees with or without tanks hidden among them, the neuronal network failed. It turned out that in the training set, photographs of camouflaged tanks had been taken on cloudy days, whereas photographs of trees without tanks had been taken on sunny days. The neural network had learned to recognize the weather rather than distinguishing tanks among trees. In the new set of photographs, forests with and without tanks had been photographed in the same weather; hence, a neuronal network merely able to distinguish the weather was unable to identify tanks.
Furthermore, applications of machine learning in pain research may be limited by the availability and quality of data; it depends on the maintenance of knowledge bases or on the success of enrolling the necessary large number of subjects in clinical studies. The latter has become easier, thanks to funding activities of concerted large-scale pain research projects.33 However, even the analysis of apparently large data sets can quickly be confronted with small sample problems when data structure detection results in many subgroups of small sizes. Then, the rather typical setting of many more features than cases poses challenges on a valid data analysis. Possibly, generative machine learning methods17 are able to reduce this problem. Such models range from Gaussian mixture models as a simple form of a generative model up to more complex approaches such as generative adversarial networks,20 generative restricted Boltzman machines,62 or generative emergent self-organizing neuronal networks.70
7. Conclusions
The emerging discipline of computational pain research provides contemporary tools to understand pain. This discipline uses computer-based processing of complex pain-related data and relies on “intelligent” learning algorithms. By extracting information from complex pain-related data and generating knowledge from this, information will be facilitated. Therefore, machine learning has the ability to influence the study and treatment of pain profoundly. Indeed, the application of machine learning for pain research–related nonimaging problems has been mentioned in publications in scientific journals since 2002 (Table 1). Among machine learning methods,11,43 a subset has so far been applied to pain research–related problems (Fig. 1), SVMs, regression models, and several kinds of neural networks so far most frequently mentioned in the pain literature. Machine learning receives increasing general interest and appears to penetrate many parts of daily life and natural sciences. This tendency is likely to extend to pain research. The present review aims to acquaint pain domain experts with the methods and current applications of machine learning in pain research, possibly facilitating the awareness of the methods in current and future projects.
Conflict of interest statement
This work has been funded by the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 602919 (“GLORIA”, J.L.) and by the Landesoffensive zur Entwicklung wissenschaftlich-ökonomischer Exzellenz (LOEWE), LOEWE-Zentrum für Translationale Medizin und Pharmakologie (J.L.).
The authors have declared that no further conflicts of interest exist.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
References
- [1].Atzori M, Gijsberts A, Castellini C, Caputo B, Hager AGM, Elsig S, Giatsidis G, Bassetto F, Müller H. Effect of clinical parameters on the control of myoelectric robotic prosthetic hands. J Rehabil Res Dev 2016;53:345–58. [DOI] [PubMed] [Google Scholar]
- [2].Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell 2009;139:267–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ben-Ari A, Chansky H, Rozet I. Preoperative opioid use is associated with early revision after total knee arthroplasty: a study of male patients treated in the Veterans Affairs System. J Bone Joint Surg Am 2017;99:1–9. [DOI] [PubMed] [Google Scholar]
- [4].Berikol GB, Yildiz O, Özcan IT. Diagnosis of acute coronary syndrome with a support vector machine. J Med Syst 2016;40:84. [DOI] [PubMed] [Google Scholar]
- [5].Braundmeier-Fleming A, Russell NT, Yang W, Nas MY, Yaggie RE, Berry M, Bachrach L, Flury SC, Marko DS, Bushell CB, Welge ME, White BA, Schaeffer AJ, Klumpp DJ. Stool-based biomarkers of interstitial cystitis/bladder pain syndrome. Sci Rep 2016;6:26083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Breiman L. Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- [7].Bui DDA, Zeng-Treitler Q. Learning regular expressions for clinical text classification. J Am Med Inform Assoc 2014;21:850–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cannistraci CV, Ravasi T, Montevecchi FM, Ideker T, Alessio M. Nonlinear dimension reduction and clustering by Minimum Curvilinearity unfold neuropathic pain and tissue embryological classes. Bioinformatics 2010;26:i531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chesler EJ, Wilson SG, Lariviere WR, Rodriguez-Zas SL, Mogil JS. Identification and ranking of genetic and laboratory environment factors influencing a behavioral trait, thermal nociception, via computational analysis of a large data archive. Neurosci Biobehav Rev 2002;26:907–23. [DOI] [PubMed] [Google Scholar]
- [10].DeLisle S, Kim B, Deepak J, Siddiqui T, Gundlapalli A, Samore M, D'Avolio L. Using the electronic medical record to identify community-acquired pneumonia: toward a replicable automated strategy. PLoS One 2013;8:e70944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dhar V. Data science and prediction. Commun ACM 2013;56:64–73. [Google Scholar]
- [12].Dimova V, Oertel BG, Kabakci G, Zimmermann M, Hermens H, Lautenbacher S, Ultsch A, Lötsch J. A more pessimistic life orientation is associated with experimental inducibility of a neuropathy-like pain pattern in healthy individuals. J Pain 2015;16:791–800. [DOI] [PubMed] [Google Scholar]
- [13].Dreyfus HL, Dreyfus SE. What artificial experts can and cannot do. AI Soc 1992;6:18–26. [Google Scholar]
- [14].Elzahaf RA, Tashani OA, Unsworth BA, Johnson MI. The prevalence of chronic pain with an analysis of countries with a Human Development Index less than 0.9: a systematic review without meta-analysis. Curr Med Res Opin 2012;28:1221–9. [DOI] [PubMed] [Google Scholar]
- [15].Emir B, Kuhn M, Johnson K, Parsons B. Predictors of response to pregabalin for broad neuropathic pain: results from 11 machine learning methods from a 6-week German observational study. J Pain 2016;17:S78. [Google Scholar]
- [16].Garcia-Chimeno Y, Garcia-Zapirain B, Gomez-Beldarrain M, Fernandez-Ruanova B, Garcia-Monco JC. Automatic migraine classification via feature selection committee and machine learning techniques over imaging and questionnaire data. BMC Med Inform Decis Mak 2017;17:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Generative learning. In: Sammut C, Webb GI, editors. Encyclopedia of machine learning. Boston: Springer US, 2010. p. 455–6. [Google Scholar]
- [18].Gholami B, Haddad WM, Tannenbaum AR. Relevance vector machine learning for neonate pain intensity assessment using digital imaging. IEEE Trans Biomed Eng 2010;57:1457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Goertzel BN, Pennachin C, de Souza Coelho L, Maloney EM, Jones JF, Gurbaxani B. Allostatic load is associated with symptoms in chronic fatigue syndrome patients. Pharmacogenomics 2006;7:485–94. [DOI] [PubMed] [Google Scholar]
- [20].Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial networks. Ithaca, NY: Cornell University, ArXiv e-prints, 2014. [Google Scholar]
- [21].Gram M, Erlenwein J, Petzke F, Falla D, Przemeck M, Emons MI, Reuster M, Olesen SS, Drewes AM. Prediction of postoperative opioid analgesia using clinical-experimental parameters and electroencephalography. Eur J Pain 2017;21:264–77. [DOI] [PubMed] [Google Scholar]
- [22].Gram M, Graversen C, Olesen AE, Drewes AM. Machine learning on encephalographic activity may predict opioid analgesia. Eur J Pain 2015;19:1552–61. [DOI] [PubMed] [Google Scholar]
- [23].Gruss S, Treister R, Werner P, Traue HC, Crawcour S, Andrade A, Walter S. Pain Intensity recognition rates via biopotential feature patterns with support vector machines. PLoS One 2015;10:e0140330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res 2003;3:1157–82. [Google Scholar]
- [25].Hermens H, op den Akker H, Tabak M, Wijsman J, Vollenbroek M. Personalized coaching systems to support healthy behavior in people with chronic conditions. J Electromyogr Kinesiol 2014;24:815–26. [DOI] [PubMed] [Google Scholar]
- [26].Hu L, Iannetti GD. Painful issues in pain prediction. Trends Neurosci 2016;39:212–20. [DOI] [PubMed] [Google Scholar]
- [27].Hu YJ, Ku TH, Jan RH, Wang K, Tseng YC, Yang SF. Decision tree-based learning to predict patient controlled analgesia consumption and readjustment. BMC Med Inform Decis Mak 2012;12:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Huang Y, Zheng H, Nugent C, McCullagh P, Black N, Vowles KE, McCracken L. Feature selection and classification in supporting report-based self-management for people with chronic pain. IEEE Trans Inf Technol Biomed 2011;15:54–61. [DOI] [PubMed] [Google Scholar]
- [29].Jang EH, Park BJ, Park MS, Kim SH, Sohn JH. Analysis of physiological signals for recognition of boredom, pain, and surprise emotions. J Physiol Anthropol 2015;34:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jiang N, Luk KDK, Hu Y. A machine learning based surface electromyography topography evaluation for prognostic prediction of functional restoration rehabilitation in chronic low back pain. Spine (Phila Pa 1976) 2017;42:1635–42. [DOI] [PubMed] [Google Scholar]
- [31].Juckett D. A method for determining the number of documents needed for a gold standard corpus. J Biomed Eng Inform 2012;45:460–70. [DOI] [PubMed] [Google Scholar]
- [32].Julius D, Basbaum AI. Molecular mechanisms of nociception. Nature 2001;413:203–10. [DOI] [PubMed] [Google Scholar]
- [33].Kringel D, Lötsch J. Pain research funding by the European Union Seventh Framework Programme. Eur J Pain 2015;19:595–600. [DOI] [PubMed] [Google Scholar]
- [34].Kringel D, Ultsch A, Zimmermann M, Jansen JP, Ilias W, Freynhagen R, Griessinger N, Kopf A, Stein C, Doehring A, Resch E, Lötsch J. Emergent biomarker derived from next-generation sequencing to identify pain patients requiring uncommonly high opioid doses. Pharmacogenomics J 2017;17:419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lötsch J, Dimova V, Hermens H, Zimmermann M, Geisslinger G, Oertel BG, Ultsch A. Pattern of neuropathic pain induced by topical capsaicin application in healthy subjects. PAIN 2015;156:405–14. [DOI] [PubMed] [Google Scholar]
- [36].Lötsch J, Geisslinger G, Heinemann S, Lerch F, Oertel BG, Ultsch A. QST response patterns to capsaicin- and UV-B-induced local skin hypersensitization in healthy subjects: a machine-learned analysis. PAIN 2017;159:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lötsch J, Lippmann C, Kringel D, Ultsch A. Integrated computational analysis of genes associated with human hereditary insensitivity to pain. A drug repurposing perspective. Front Neurosci 2017;10:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lötsch J, Ultsch A. A machine-learned knowledge discovery method for associating complex phenotypes with complex genotypes. Application to pain. J Biomed Inform 2013;46:921–8. [DOI] [PubMed] [Google Scholar]
- [39].Mayer EA, Gupta A, Kilpatrick LA, Hong JY. Imaging brain mechanisms in chronic visceral pain. PAIN 2015;156(suppl 1):S50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Meng H, Bianchi-Berthouze N. Affective state level recognition in naturalistic facial and vocal expressions. IEEE Trans Cybernetics 2014;44:315–28. [DOI] [PubMed] [Google Scholar]
- [41].Misra G, Wang WE, Archer DB, Roy A, Coombes SA. Automated classification of pain perception using high-density electroencephalography data. J Neurophysiol 2017;117:786–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mohan A, Moreno N, Song JJ, De Ridder D, Vanneste S. Evidence for behaviorally segregated, spatio-temporally overlapping subnetworks in phantom sound perception. Brain Connect 2017;7:197–210. [DOI] [PubMed] [Google Scholar]
- [43].Murphy KP. Machine learning: a probabilistic perspective. Cambridge, MA: The MIT Press, 2012. [Google Scholar]
- [44].Nanni L, Lumini A, Brahnam S. Local binary patterns variants as texture descriptors for medical image analysis. Artif Intell Med 2010;49:117–25. [DOI] [PubMed] [Google Scholar]
- [45].Newell A, Simon HA. Computer science as empirical inquiry: symbols and search. Commun ACM 1976;19:113–26. [Google Scholar]
- [46].Nickerson P, Tighe P, Shickel B, Rashidi P. Deep neural network architectures for forecasting analgesic response. Conf Proc IEEE Eng Med Biol Soc 2016;2016:2966–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Olesen SS, Graversen C, Bouwense SA, Wilder-Smith OHG, van Goor H, Drewes AM. Is timing of medical therapy related to outcome in painful chronic pancreatitis? Pancreas 2016;45:381–7. [DOI] [PubMed] [Google Scholar]
- [48].Patterson OV, Jones M, Yao Y, Viernes B, Alba PR, Iwashyna TJ, DuVall SL. Extraction of vital signs from clinical notes. Stud Health Technol Inform 2015;216:1035. [PubMed] [Google Scholar]
- [49].Pesteie M, Abolmaesumi P, Ashab HAD, Lessoway VA, Massey S, Gunka V, Rohling RN. Real-time ultrasound image classification for spine anesthesia using local directional Hadamard features. Int J Comput Assist Radiol Surg 2015;10:901–12. [DOI] [PubMed] [Google Scholar]
- [50].Pourshoghi A, Zakeri I, Pourrezaei K. Application of functional data analysis in classification and clustering of functional near-infrared spectroscopy signal in response to noxious stimuli. J Biomed Opt 2016;21:101411. [DOI] [PubMed] [Google Scholar]
- [51].President's Information Technology Advisory C. Report to the president: computational science: ensuring America's competitiveness. Marc R. Benioff, Edward D. Lazowska. (Eds.). Arlington, VA: National Coordination Office for Information Research and Development, 2005. [Google Scholar]
- [52].Robinson ME, O'Shea AM, Craggs JG, Price DD, Letzen JE, Staud R. Comparison of machine classification algorithms for fibromyalgia: neuroimages versus self-report. J Pain 2015;16:472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev 1958;65:386–408. [DOI] [PubMed] [Google Scholar]
- [54].Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev 1959;3:210–29. [Google Scholar]
- [55].Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011;141:1782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Schölkopf B, Smola AJ. Learning with Kernels: support vector machines, regularization, optimization, and beyond. Cambridge, MA: MIT Press, 2002. [Google Scholar]
- [57].Sevel L, Letzen J, Boissoneault J, O'Shea A, Robinson M, Staud R. MRI based classification of chronic fatigue, fibromyalgia patients and healthy controls using machine learning algorithms: a comparison study. J Pain 2016;17:S60. [Google Scholar]
- [58].Shalev-Shwartz S, Ben-David S. Understanding machine learning: from theory to algorithms. Cambridge, United Kingdom: Cambridge University Press, 2014. [Google Scholar]
- [59].Sikka K, Ahmed AA, Diaz D, Goodwin MS, Craig KD, Bartlett MS, Huang JS. Automated assessment of children's postoperative pain using computer vision. Pediatrics 2015;136:e124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sing DC, Metz LN, Dudli S. Machine learning-based classification of 38 years of spine-related literature into 100 research topics. Spine (Phila Pa 1976) 2017;42:863–70. [DOI] [PubMed] [Google Scholar]
- [61].Sipilä R, Estlander AM, Tasmuth T, Kataja M, Kalso E. Development of a screening instrument for risk factors of persistent pain after breast cancer surgery. Br J Cancer 2012;107:1459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Smolensky P. Information processing in dynamical systems: foundations of harmony theory. In: David ER, James LM; CPR Group, editors. Parallel distributed processing: explorations in the microstructure of cognition. Vol. 1: MIT Press, 1986. p. 194–281. [Google Scholar]
- [63].Smolensky P. On the proper treatment of connectionism. Behav Brain Sci 2010;11:1–23. [Google Scholar]
- [64].Tighe P, Laduzenski S, Edwards D, Ellis N, Boezaart AP, Aygtug H. Use of machine learning theory to predict the need for femoral nerve block following ACL repair. Pain Med 2011;12:1566–75. [DOI] [PubMed] [Google Scholar]
- [65].Tighe PJ, Harle CA, Hurley RW, Aytug H, Boezaart AP, Fillingim RB. Teaching a machine to feel postoperative pain: combining high-dimensional clinical data with machine learning algorithms to forecast acute postoperative pain. Pain Med 2015;16:1386–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tighe PJ, Lucas SD, Edwards DA, Boezaart AP, Aytug H, Bihorac A. Use of machine-learning classifiers to predict requests for preoperative acute pain service consultation. Pain Med 2012;13:1347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tu Y, Tan A, Bai Y, Hung YS, Zhang Z. Decoding subjective intensity of nociceptive pain from pre-stimulus and post-stimulus brain activities. Front Comput Neurosci 2016;10:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Turing A. Computing machinery and intelligence. Mind 1950;LIX:433–60. [Google Scholar]
- [69].Ultsch A, Kringel D, Kalso E, Mogil JS, Lötsch J. A data science approach to candidate gene selection of pain regarded as a process of learning and neural plasticity. PAIN 2016;157:2747–57. [DOI] [PubMed] [Google Scholar]
- [70].Ultsch A, Lötsch J. Generative learning with emergent self-organizing neuronal networks. Conference of the International Federation of Classification Societies. IFCS-2017 Organizing Team, Tokyo, Japan, 2017:266.
- [71].Ultsch A, Lötsch J. Machine-learned cluster identification in high-dimensional data. J Biomed Inform 2017;66:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].von Hehn CA, Baron R, Woolf CJ. Deconstructing the neuropathic pain phenotype to reveal neural mechanisms. Neuron 2012;73:638–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Winograd T. Understanding natural language. Cogn Psychol 1972;3:1–191. [Google Scholar]
- [74].Wu HY, Gong CSA, Lin SP, Chang KY, Tsou MY, Ting CK. Predicting postoperative vomiting among orthopedic patients receiving patient-controlled epidural analgesia using SVM and LR. Sci Reps 2016;6:27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wu Y, Zheng Y, Tang H. Identifying the types of ion channel-targeted conotoxins by incorporating new properties of residues into pseudo amino acid composition. Biomed Res Int 2016;2016:3981478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yang L, Wang S, Jiang X, Cheng S, Kim HE. PATTERN: pain Assessment for paTients who can't TEll using Restricted Boltzmann machiNe. BMC Med Inform Decis Mak 2016;16(suppl 3):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yang M, Zheng H, Wang H, McClean S, Hall J, Harris N. A machine learning approach to assessing gait patterns for Complex Regional Pain Syndrome. Med Eng Phys 2012;34:740–6. [DOI] [PubMed] [Google Scholar]
- [78].Yoo TK, Kim SK, Choi SB, Kim DY, Kim DW. Interpretation of movement during stair ascent for predicting severity and prognosis of knee osteoarthritis in elderly women using support vector machine. Conf Proc IEEE Eng Med Biol Soc 2013;2013:192–6. [DOI] [PubMed] [Google Scholar]
- [79].Zhang G, Liang Z, Yin J, Fu W, Li GZ. A similarity based learning framework for interim analysis of outcome prediction of acupuncture for neck pain. Int J Data Min Bioinform 2013;8:381–95. [DOI] [PubMed] [Google Scholar]
- [80].Zifan A, Jiang Y, Mittal RK. Temporal and spectral properties of esophageal mucosal blood perfusion: a comparison between normal subjects and nutcracker esophagus patients. Neurogastroenterol Motil 2017;29. 10.1111/nmo.12917. [DOI] [PMC free article] [PubMed] [Google Scholar]


