Abstract
Purpose:
Several studies have examined the potential role of p16 protein expression as a diagnostic and prognostic biomarker in various cancers. However, it remains unclear whether p16 protein expression is a prognostic and diagnostic factor for colorectal cancer. Therefore, this meta-analysis is conducted to evaluate the associations of p16 protein expression with overall survival (OS) and clinicopathological characteristics of colorectal cancer.
Methods:
According to PRISMA guideline, relevant literatures were identified by searching Medicine, Web of Science, WanFang, and CNKI databases. The pooled hazard ratios (HRs) with 95% confidence intervals (CIs) were extracted from included studies to assess the association between p16 protein expression and OS of patients with colorectal cancer. Other relevant data were extracted to evaluate the correlations of p16 protein expression with risk and clinicopathological characteristics of colorectal cancer. Stata 12.0 software was applied to calculate the strength of association between p16 protein expression and colorectal cancer.
Results:
Forty-one studies were included to evaluate the association between p16 protein expression and colorectal cancer. Nine studies involving 1731 patients with colorectal cancer found that there was no association between p16 protein expression and OS of colorectal cancer in the overall analysis (HR = 0.78, 95% CI: 0.55–1.10). However, p16 protein overexpression was significantly associated with a better prognosis in patients with colorectal cancer when cut-off value of p16 protein expression was <10% (HR = 0.23, 95% CI: 0.08–0.66). The results of subgroup analysis based on ethnicity indicated that p16 protein overexpression was a risk factor for the occurrence of colorectal cancer in Caucasians (odds ratio = 28.95, 95% CI: 6.08–137.89), but not in Asians. Furthermore, p16 protein overexpression was significantly associated with the Dukes stage, lymph node metastasis, tumor location, and Tumor Lymph Node Metastasis-stage of colorectal cancer.
Conclusions:
p16 protein overexpression might be a useful biomarker to predict the clinicopathological progress and prognosis of colorectal cancer.
Keywords: colorectal neoplasms, expression, meta-analysis, p16 protein
1. Introduction
Colorectal cancer (CRC) is the third most common cancer with the estimated 150,000 new cases and 693,900 deaths worldwide in 2012.[1,2] A majority of patients with CRC are sporadic cases, and only 5% to 10% are familial CRC, which is largely caused by inherited mutations. However, 25% of patients with CRC have family history, which reveals that many relevant genes are needed to be discovered in these patients.[3] In addition, sporadic CRC is a somatic genetic disease, which is influenced by colonic environment and individual's genetic background.[4] CRC may result from transformation of normal colonic mucosa cells to tumor cells through accumulation of genetic alterations such as adenomatous polyposis coli and tumor protein 53.[5] Genomic instability and chromosomal instability play important roles in colorectal carcinogenesis, and a number of gene mutations are detected in the hereditary nonpolyposis colon cancer/Lynch syndrome and MUTYH-associated polyposis.[6,7] Chromosomal instability commonly leads to gene deletions, duplications, and chromosomal rearrangements, which are observed in 70% to 85% of patients with CRC.[7] Furthermore, microsatellite instability is another risk factor for the occurrence of CRC. Currently, many studies have focused on the influence of instability of CpG island methylator phenotype in tumor suppressor genes and aberrant promoter methylation of relevant genes in the development of CRC.[8] Although we have found a lot of environmental factors and genetic factors, there are still many patients with CRC every year.
It has been reported that 5-year survival of early stage tumor reached 90%, but a 5-year survival of <10% is detected in advanced tumor.[9] Currently, many CRC cases in advanced stage are observed due to a lack of disease-specific symptoms and disease markers. Therefore, early detection of CRC in local region or preinvasive form may be the better approach to the early treatment of CRC and the decline of death rate in CRC. Although fecal occult blood testing and colonoscopy are very effective detection means to the early finding of CRC, the corresponding difficulties also present in these detection means. Hence, biomarkers that are extracted in peripheral blood, stool, or tissue sample may be beneficial as a more reliable and specific marker for the early detection of CRC.[10] Clinical applicable biomarkers had better be significantly associated with occurrence, progress, and prognosis of cancers. However, this kind of biomarker is hardly to detect; hence, several biomarkers may be used to supervise one of the occurrence, progress, and prognosis of cancer. In addition, several studies have focused on epigenetic changes of CRC in which DNA sequence is not changed. But methyl group is transferred to DNA, and expression of genes or proteins is restrained. Many studies have reported that promoter region hypermethylation in tumor suppressor genes had a strong relationship with diagnosis and prognosis of CRC.[11] For example, aberrant promoter methylation of p14, p15, and p16 gene are associated with occurrence, progress, and prognosis of many cancers such as gastric cancer, breast cancer, ovarian cancer, CRC, and lung cancer.[12–15] p16 (cyclin-dependent kinase inhibitor 2A [CDKN2A]), one of the most frequently altered genes, consisting of 3 exons and 2 introns, is located on chromosome 9p21 where its overexpression and hypermethylation are commonly detected in various cancers.[16] p16 protein binds cyclin-dependent kinase 4 protein to produce a complex, and further inhibits the interaction of CDK4 and cyclin D1 to control the cell cycle and prevent cancer.[17] Loss of p16 function, which is commonly resulted from gene sequence changes and promoter aberrant methylation, might lead to aberrant cells proliferation and further produce cancer cells. Numerous studies based on molecular biology have found that aberrant methylation of p16 gene could restrain expression of p16 protein.[18,19] The inactivation of p16 gene leads to uncontrolled proliferation of cells, which is one of reasons of tumorigenesis.[20] According to previous reports, p16 protein expression might be associated with the occurrence, progress, or prognosis of CRC. However, accurate conclusions are not obtained due to several reasons such as clinical stage of patients with CRC, sample size, detection method, or ethnicity. Hence, we performed this meta-analysis to explore the association of p16 protein expression with risk, clinicopathological characteristics, and prognosis of CRC.
2. Materials and methods
2.1. Search strategy
Relevant literature searching in Medicine, Web of Science, WanFang, and CNKI databases was conducted with the following keywords: “p16,” “CDKN2A,” “p16INK4a,” “expression,” “colorectal cancer,” “CRC,” “colorectal carcinoma,” “colorectal neoplasm,” “prognosis,” “overall survival,” “OS,” “survival,” and “outcome.” The articles concerning the association of p16 protein expression with CRC were identified before May 2017 (NZ and QG). The references of the eligible articles and relevant reviews were hand searched to find other relevant original researches (NZ and QG). Titles, abstracts, and full text of retrieved articles were gradually scanned to identify eligible studies, which met the relevant inclusion criteria (NZ and QG).
2.2. Selection criteria
Articles were included if they met the following criteria: original articles with English or Chinese language; articles should focus on the p16 protein expression and prognosis, risk, or clinicopathological features of CRC; articles should contain available data or curves; and p16 protein expression was examined in the normal tissue, adjacent tissue, or CRC tissue. The following exclusion criteria was used to exclude irrelevant articles: reviews and other nonoriginal articles; studies did not contain sufficient data; and studies on cells or animals.
2.3. Data collection and methodology quality assessment
Two investigators independently checked the relevant literatures and extracted the data to improve the reliability of included studies (NZ and QG). The following characteristics of included studies were extracted: first author, country, ethnicity, publication year, hazard ratio (HR) and 95% confidence interval (CI) of overall survival (OS), survival curve, cut-off value, the frequency of p16 protein negative and positive expression in control and case group, disease type, sample type, and method of p16 protein expression detection. In addition, 2 reviewers evaluated the methodology quality of included studies independently according to Newcastle-Ottawa Scale table. If any divergence existed between 2 reviewers, a third investigator would join the discussion.
2.4. Statistical analysis
Odds ratio (OR) and 95% CI were used to evaluate the association of p16 protein expression with risk and clinicopathological features of CRC, whereas HR and 95% CI were extracted and calculated to assess the correlation between p16 protein expression and prognosis of patients with CRC. Forest map was drawn to merge the extracted data from included studies. A value of P < .05 indicated that p16 protein positive expression might be significantly associated with the risk, clinicopathological features, or prognosis of CRC. Heterogeneity among studies was assessed by the Cochran Q test and I2 statistic.[21] If P < .05 or I2 > 50%, significant heterogeneity between studies existed and a random effects model was needed, whereas significant heterogeneity was not detected, the meta-analysis used a fixed effects model.[22,23] Moreover, Begg regression test and Egger test were used to evaluate the publication bias among studies.[24,25] The stability of overall results was validated through sensitivity analysis in this meta-analysis. In the sensitivity analysis, each included study was omitted one by one, and remaining studies were pooled to calculate the overall OR and 95% CI. If significant changes were found in the overall OR and 95% CI, it means that the removed study should not be put together with other relevant studies. Or we can say, this study resulted in significant heterogeneity in the overall analysis. Maybe the used statistical approach may be not appropriate to the study. And different detection methods, populations, or sample size often contributed a lot to the difference of the study. In addition, meta-regression was conducted to explore the source of heterogeneity (NZ). If HR and 95% CI were not directly provided in eligible studies, Engauge Digitizer 4.1 was used to extract HR and 95% CI from survival curves. STATA 13.0 was applied to summarize and analyze the extracted data (NZ and QG).
2.5. Ethnical statement
This study did not need consent of ethics committee or institutional review board, because the research was a meta-analysis which belonged to a review article.
3. Results
3.1. Search results
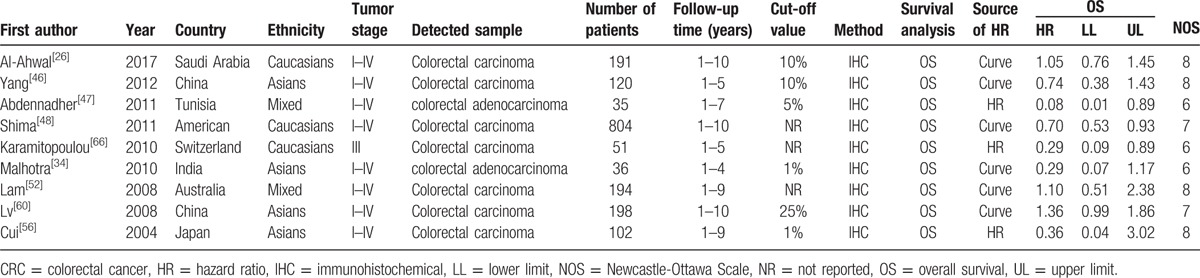
According to the PRISMA statement, the literatures search flow is shown in Figure 1. Based on the search strategy, 835 articles were obtained. Eighty-six articles were remained after titles and abstracts were read, whereas 749 studies including duplicate, meta-analysis, reviews, and other articles, which were not involved in p16 protein expression and CRC were excluded. Then, the full text of remained articles was scanned. In finally, a total of 41 articles were included to explore the association of p16 protein expression with the risk, clinicopathological features, and OS of CRC. Among the 41 studies, 9 studies with 1731 patients were performed to analyze the correlation between p16 protein expression and OS of CRC, and 20 studies with 755 controls and 1733 cases were included for the analysis of risk of CRC.[26–66] In addition, clinical information was extracted to analyze the association between p16 protein expression and clinicopathological features of CRC. These characteristics of eligible studies are presented in Tables 1 and 2. In the 9 studies for OS, 3 studies directly provided HR and 95% CI, whereas other 6 studies provided survival curves. Only 1 study used western blotting, whereas other studies applied immunohistochemistry to detect the level of p16 expression in CRC tissue.
Figure 1.
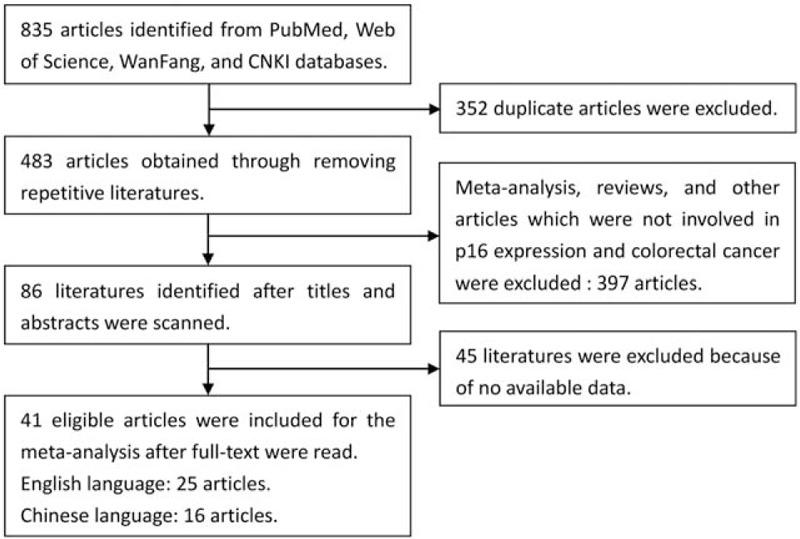
Flow chart of eligible literatures searching.
Table 1.
Main characteristics of 40 eligible studies evaluating the association between p16 protein expression and clinicopathologic characteristics of colorectal cancer.
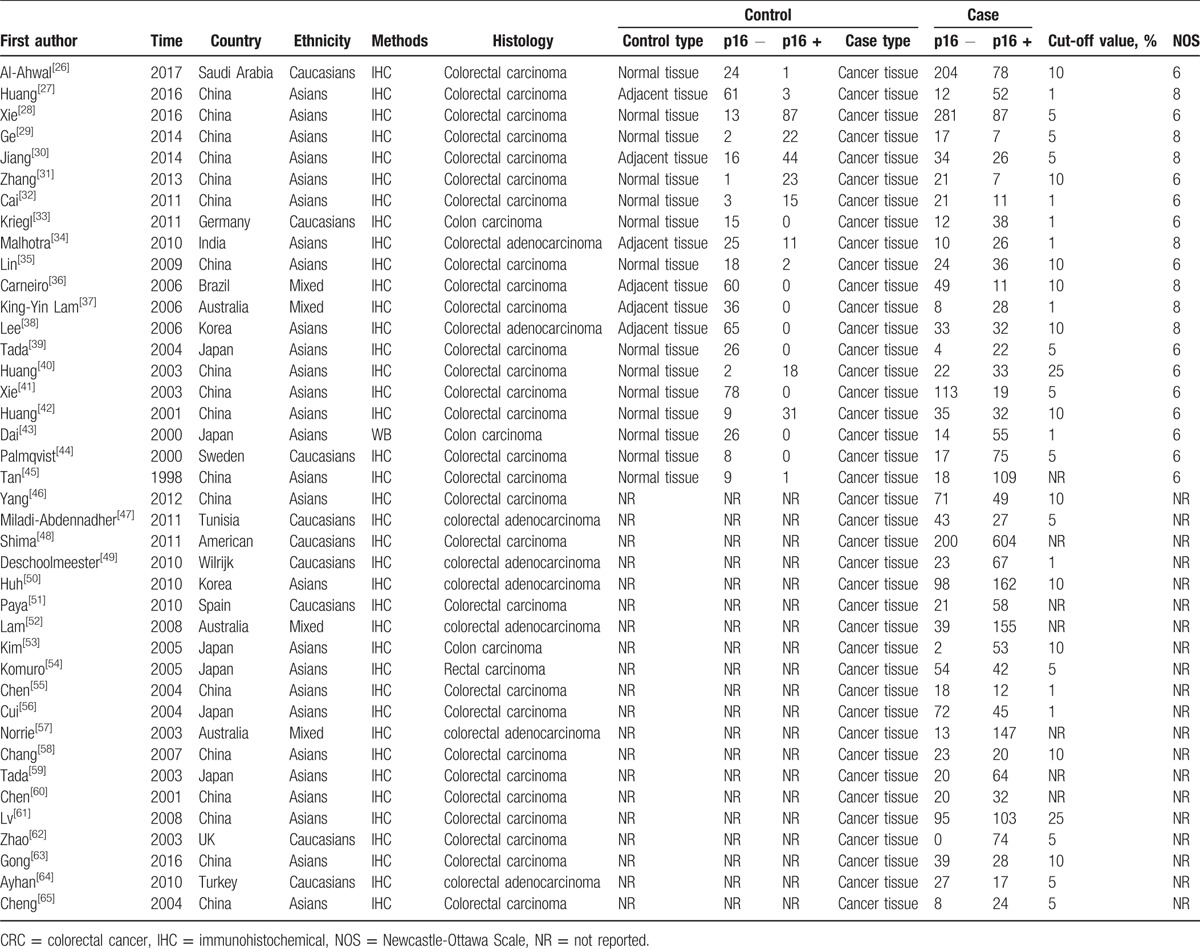
Table 2.
Main characteristics of the included studies which evaluated the correlation of p16 protein expression with overall survival of colorectal cancer.
3.2. Correlation of p16 protein expression with clinicopathological parameters of CRC
Twenty studies compared the p16 protein expression status between control and case group, and random effects model was applied since significant heterogeneity was found. The results indicated that p16 protein overexpression was a significant risk factor for the risk of CRC in Caucasians (OR = 28.95, 95% CI: 6.08–137.89). Interestingly, no significant association between p16 protein overexpression and CRC risk was found in Asians. And in the included studies for Asians, many studies got opposite result which indicated unstable results, which are shown in the Figure 2. The subgroups analysis were performed based on sex, differentiation, Tumor Lymph Node Metastasis (TNM)-stage, lymph node metastasis, location of tumor, vascular invasion, and Dukes stage to explore the correlation between p16 protein expression and clinicopathologic characteristics of CRC. Notably, p16 protein overexpression was significantly associated with Dukes stage, lymph node metastasis, and TNM stage of CRC. However, significant association between p16 protein expression and TNM stage of CRC was only found in Caucasians (HR = 0.68, 95% CI: 0.50–0.92). To be specific, p16 protein overexpression was a protective factor for the lymph node metastasis of CRC (OR = 0.52, 95% CI: 0.32–0.86). In addition, p16 protein overexpression was also a protective factor for the advanced TNM stage of CRC in Caucasians (TNM-stage, OR = 0.68, 95% CI: 0.50–0.92) (Figs. 3–5, Table 3).
Figure 2.
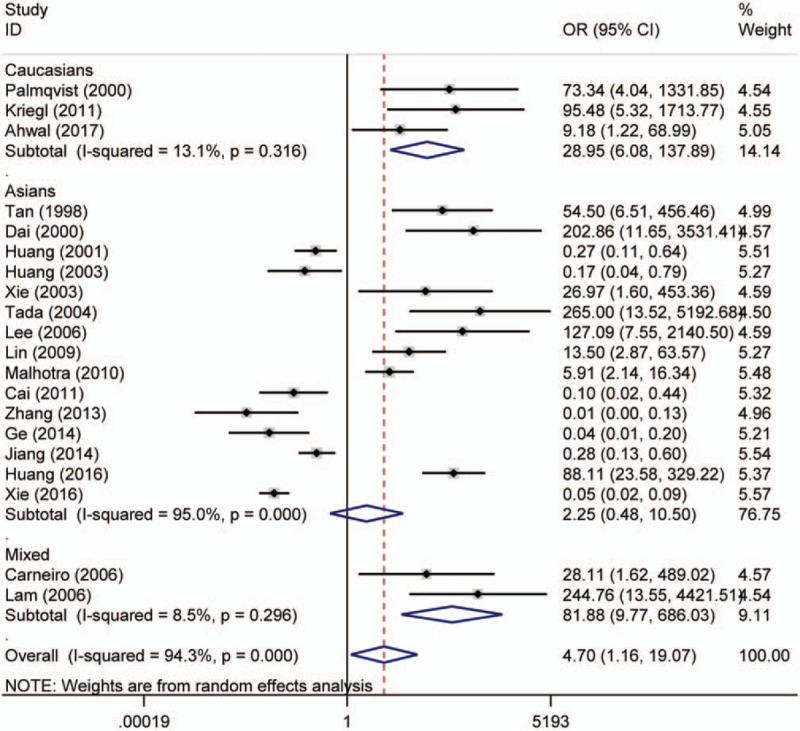
Forest plot for the association between p16 protein expression and colorectal cancer (CRC) risk. CI = confidence interval, OR = odds ratio.
Figure 3.
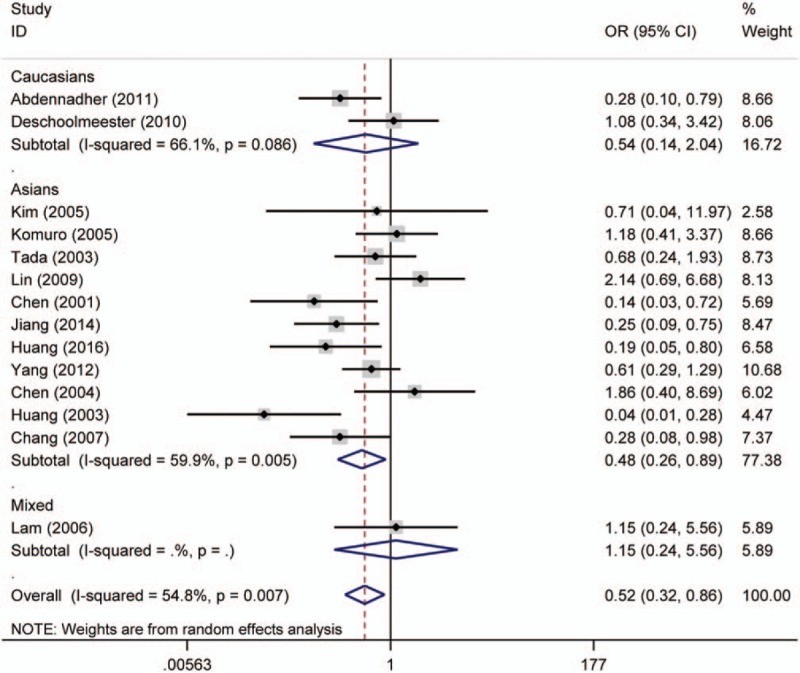
Forest plot for the association of p16 protein expression with lymph node metastasis of colorectal tumor. CI = confidence interval, OR = odds ratio.
Figure 5.
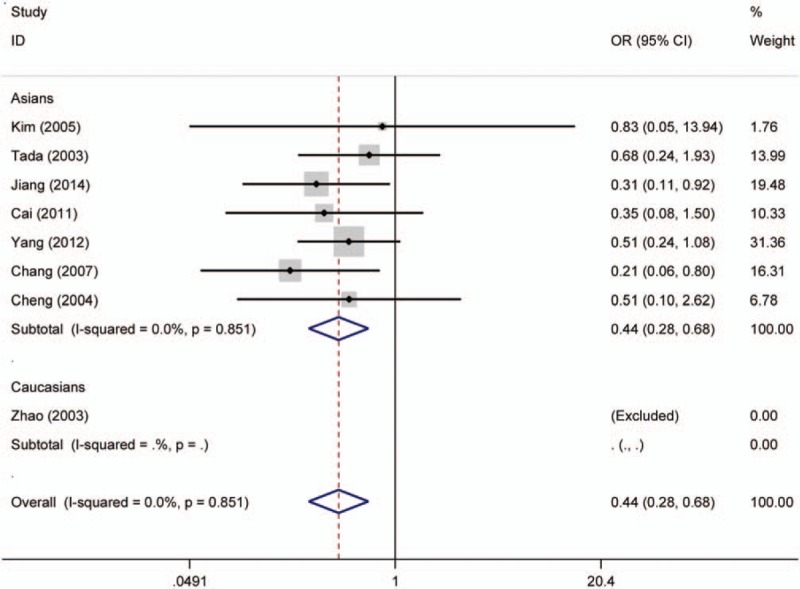
Forest plot for the association of p16 protein expression with Dukes stage of colorectal tumor. CI = confidence interval, OR = odds ratio.
Table 3.
Association between p16 protein expression and clinicopathological characteristics of patients with colorectal cancer (p16 [−] vs p16 [+]).
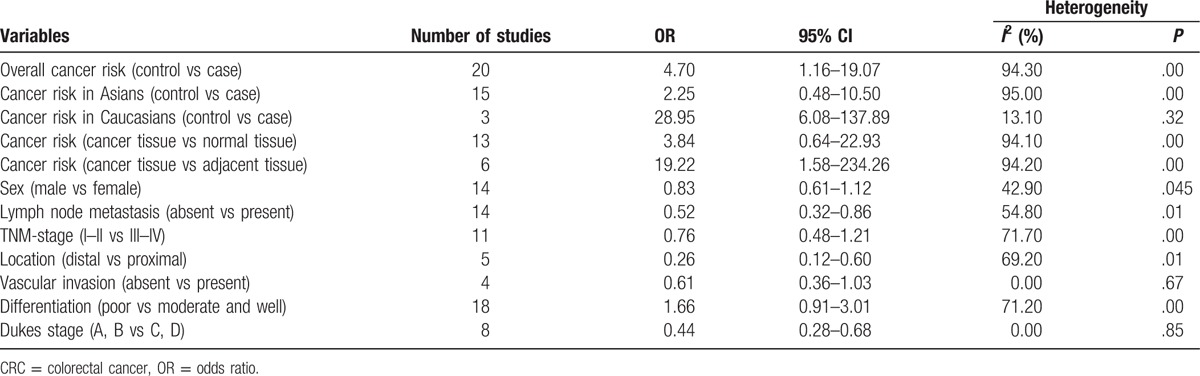
Figure 4.
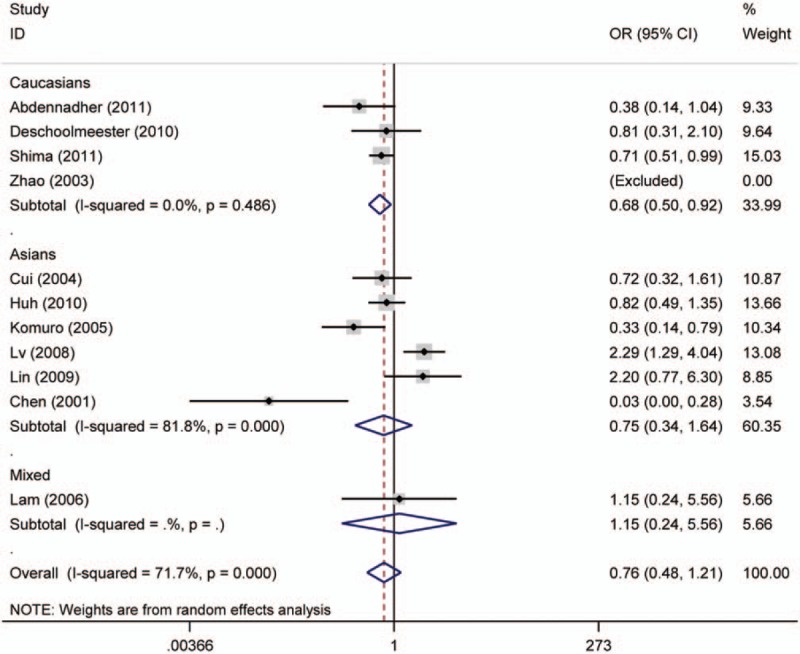
Forest plot for the association of p16 protein expression with TNM-stage of colorectal tumor. CI = confidence interval, OR = odds ratio.
3.3. Association between p16 protein expression and OS of CRC
Nine studies investigating OS were pooled to analyze the association between p16 protein expression and OS of CRC in the meta-analysis. The overall results demonstrated that p16 protein expression was not significantly associated with the OS of CRC (HR = 0.78, 95% CI: 0.55–1.10). When grouped by ethnicity, no significant association was found in Caucasians and Asians (Asians, HR = 0.79, 95% CI: 0.40–1.54; Caucasians, HR = 0.75, 95% CI: 0.47–1.18). When stratified analysis based on cut-off values was conducted, significant association was observed in the subgroup analysis with cut-off value of <10% (cut-off <10%, HR = 0.23, 95% CI: 0.08–0.66). Interestingly, the results revealed that p16 protein overexpression was helpful for the OS of CRC patients. In addition, p16 protein overexpression was associated with the OS of ORC in the subgroup with sample size <100 (HR = 0.24, 95% CI: 0.11–0.56). Finally, no significant heterogeneity was detected through Cochran Q test and I2 statistic; thus, fixed effects model was used to calculate the pooled HR and 95% CI (Fig. 6, Table 4). Moreover, in the analysis on CRC risk and TNM stage of CRC, the results of metaregression indicated that country was a significant influence factor in which the result of China had an obvious heterogeneity (for cancer risk P = .002, for TNM stage of CRC P = .049), compared with results of other countries. Furthermore, Asians and Caucasians caused the main heterogeneity in the analysis of Dukes of CRC (P = .011) (Table 5). In order to lower the heterogeneity and acquire more accurate result in Asians and Caucasians, the subgroup analysis based on ethnicity has been conducted. Therefore we could compare the results in Asians with Caucasians on the forest plot. Furthermore, we could observe the results of China on the forest plots.
Figure 6.
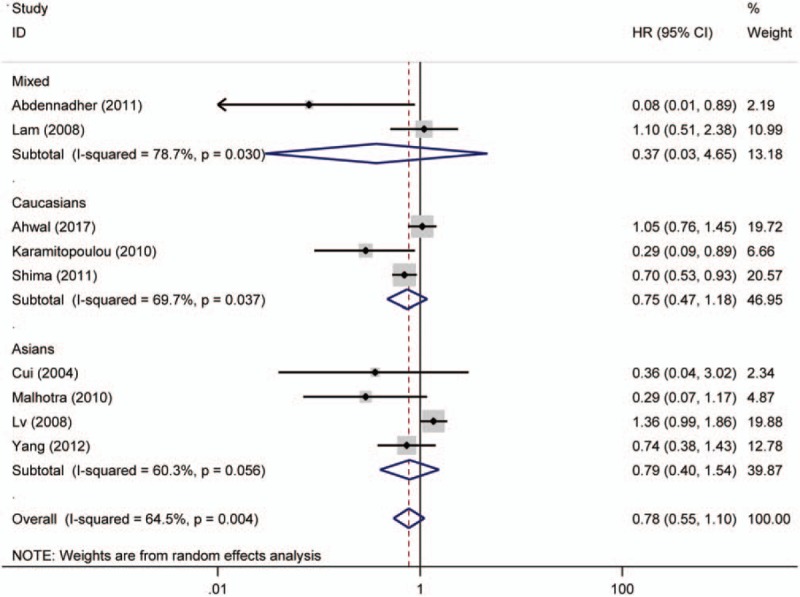
Forest plot for the association of p16 protein expression with overall survival of colorectal cancer (CRC). CI = confidence interval, HR = hazard ratio.
Table 4.
Associations between p16 protein overexpression and overall survival of patients with colorectal cancer.
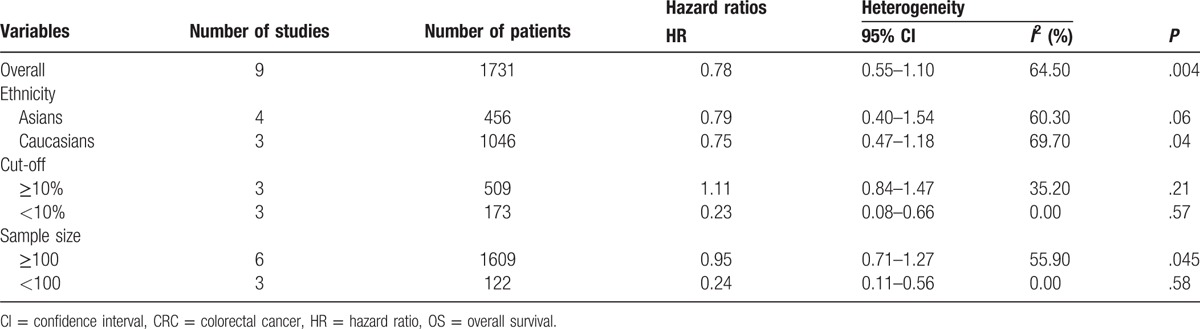
Table 5.
Results of meta-regression for the meta-analysis (P value).
3.4. Publication bias analysis
Publication bias of included studies was evaluated by Begg test and Egger test, in which significant publication bias was found for the OS of CRC (OS, Begg test P = .21, Egger test P = .28). However, small publication bias was observed through Egger test in included studies for the risk of CRC (CRC risk, Begg test P = .09, Egger test P = .002). In order to seek the source of publication bias, meta-regression was performed to analyze the effect of ethnicity, control tissue type, and publication year in publication bias. But no significant factors were found for the pooled HR and 95% CI, which indicated that other factors may bring significant impact on the publication bias among studies (Table 5). Furthermore, the results of sensitivity analysis indicated that the overall HR and OR were not significantly changed by removing each study at a time (Figures 7–9).
Figure 7.
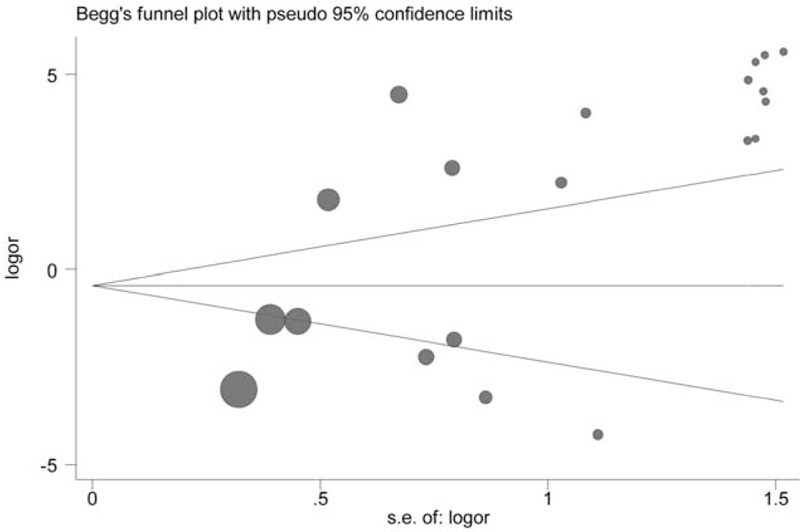
Begg funnel plot for publication bias test for the role of p16 protein expression in colorectal cancer (CRC) risk.
Figure 9.
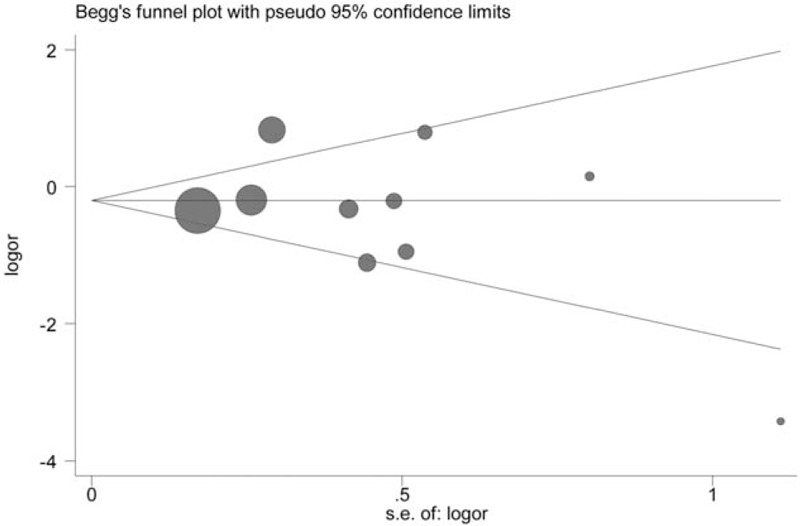
Begg funnel plot for publication bias test for the role of p16 protein expression in TNM stage of colorectal cancer (CRC).
Figure 8.
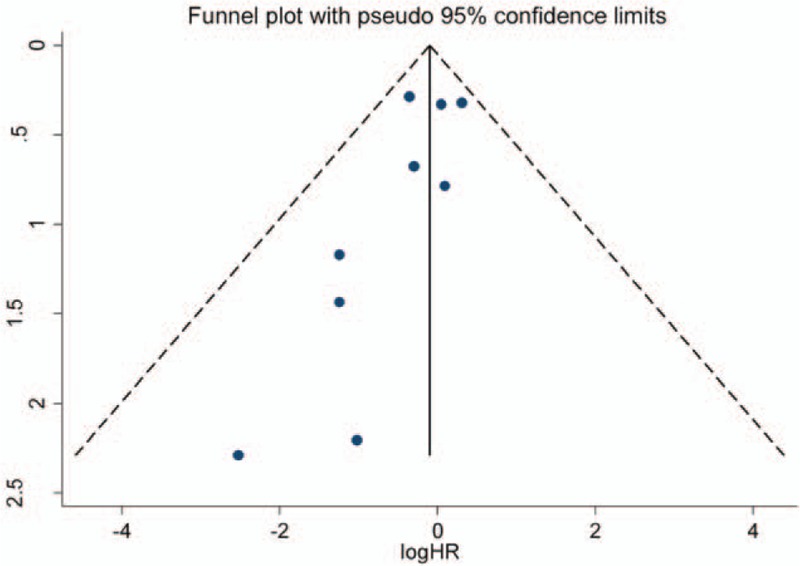
Begg funnel plot for publication bias test for the role of p16 protein expression in overall survival of patients with colorectal cancer (CRC). HR = hazard ratio.
4. Discussion
In the present study, the associations of p16 protein expression with prognosis and clinicopathological features of CRC were comprehensively assessed with 41 publications. According to the result of literature retrieval, this is the first meta-analysis aimed to explore the role of p16 protein expression in prognosis and clinicopathological characteristics of patients with CRC. Previous study have shown that p16 protein expression was changed in the development of CRC through gene mutation or promoter region methylation.[16] Furthermore, many studies have demonstrated that p16 gene promoter aberrant methylation would repress the transcription of p16 gene, and therefore lead to the inactivation of p16 protein.[18,67,68] Other studies have shown that overexpression of p16 protein would lead to the combination of CDK4/6-p16 complexes and the loss of CDK4/6-cyclin D.[69] And elevated levels of p16 protein expression could prevent cell cycle pass the G1/S restriction point and repress cells proliferation.[70] Previous clinical study have also revealed that p16 was inactivated by point mutation, promoter hypermethylation, or homozygous deletion which was commonly found in many human cancers.[71] The genetic variation of p16 gene was commonly found in patients with cancer, and p16 had a crucial role in the process of cell growth. For instance, the status of p16 gene promoter methylation was commonly studied and significantly associated with the development of bladder cancer, lung cancer, brain cancer, and esophagus cancer.[72–75] At the same time, the study of Chen et al[76] has suggested that promoter hypermethylation of the p16 gene might be significantly associated with the clinicopathologic features of CRC. However, an accurate expression level of p16 gene was not detected in these studies despite the significant impact of p16 promoter hypermethylation on p16 protein expression. So relevant studies were searched and included to explore the role of p16 protein expression in the development and prognosis of CRC in this study.
The results showed that p16 protein overexpression was correlated with the Dukes stage, lymph node metastasis, and TNM stage (only in Caucasians) of CRC, suggesting that p16 protein overexpression might play a crucial effect in the development of CRC. The frequency of p16 protein negative expression in advanced stage CRC patients was higher than that in early stage patients with CRC. And the patients with CRC with p16 protein negative expression would have a bigger chance of lymph node metastasis in tumor cells. And these results were consistent with the founding of Chen et al.[76] They also found that p16 gene promoter hypermethylation was significantly associated with the TNM stage and Dukes stage of patients with CRC. Therefore, p16 gene promoter hypermethylation might repress the activity of p16 protein, which promoted differentiation of tumor cells and aggravated the progression of CRC.[77] However, interestingly, no significant association between p16 protein expression and CRC differentiation was found. According to the results of included studies in this meta-analysis, 6 studies got opposite results and only 1 study obtained a significant result for the differentiation of colorectal tumor.[27,35,36,51,55,59] The sample size of each study in the 6 studies was <100 which suggested that results of the 6 studies might be debatable. Thus, studies with larger sample size were still needed to assess the role of p16 expression in differentiation of CRC. Several studies have been investigated to explore the prognostic relevance of p16 protein expression in CRC; however, the sample size of included studies was very small and conclusions were conflicting. In this study, 9 studies with 1731 patients with CRC were included to evaluate the association of p16 protein expression with OS of patients with CRC. It was described that p16 protein expression was not correlated with OS of patients with CRC (HR = 0.78, 95% CI: 0.55–1.10) in the overall analysis. Subgroup analysis based on ethnicity was conducted due to significant heterogeneity in the overall analysis (I2 = 64.5%, P = .004), but the results still indicated that there was no significant association between p16 protein expression and OS of patients with CRC in Asians and Caucasians (Asians, HR = 0.79, 95% CI: 0.40–1.54; Caucasians, HR = 0.75, 95% CI: 0.47–1.18). In order to get accurate result, subgroup analysis based on sample size and cut-off values was performed, in which significant correlation between p16 protein expression and OS was exhibited in the subgroup with cut-off values of <10% or sample size of <100. Therefore, if cut-off value was set lower, different results might be showed in the future studies. However, we believed that the results in the subgroup with sample size <100 might not be accurate due to the small size of included studies. Moreover, 3 studies observed positive results and the sample size of the 3 studies was not small for the analysis of the association between p16 protein expression and OS of CRC.[47,48,66] Therefore, other factors might affect the overall results such as tumor stage, medication use, country, cut-off value, and sex. According to these results, studies with accurate tumor TNM stage should be performed to assess the effect of p16 protein expression on the prognosis of patients with CRC. At the same time, the frequency of p16 protein overexpression in case group was significantly higher than those in normal colorectal tissue or adjacent tissue in Caucasians but not in Asians. It needs to be emphasized that the results in the included studies were very unstable and several opposite results were obtained. Moreover, some studies believed that this result might attribute to cell heterogeneity, in which some cancer cells showed high methylation of p16 gene promoter region and inactivation of p16 protein, whereas other cancer cells had an elevated level of p16 protein.[19,78] In addition, Ohhara et al[79] have reported that activation of p16 protein was significantly correlated with primary CRC. Therefore, the level of p16 protein expression might have a change along with the development of CRC.
In order to get a more accurate result, subgroup analysis was conducted and no significant results were observed in Asians and Caucasians. However, many opposite results were found in the included Asians studies and heterogeneity was very obvious. Hence, the result of Asians might be unstable and more studies with larger sample size, more accurate clinical stage, and same cut-off value were still urgently needed. Finally, no significant associations of p16 protein expression with sex, location, and vascular invasion of CRC were detected in this meta-analysis.
From these results, we might conclude that the expression of p16 protein was low in normal cells. However, when cells proliferation was out of control, p16 protein expression would be activated to restrain the cells proliferation. Hence, from these results, although p16 protein overexpression might increase the risk of CRC, p16 protein overexpression might prevent the development of CRC.
From the test of publication bias, there existed significant publication bias in the analysis of the correlation of p16 expression with risk of CRC. Metaregression was applied to analyze the source of publication bias, and the factors such as: country, publication year, ethnicity, and control sample type were included to calculate relevant P value. However, no evident source was found other than China, which revealed that some factors might lead to the publication bias such as the instability of results in Asians studies. Moreover, the results of sensitivity analysis did not exhibit very prominent study. Thus, the overall results were relatively stable based on the present study.
Nevertheless, some limitations were also shown in this meta-analysis. First, no accurate reasons could explain why so many studies in Asians obtained opposite results in the analysis of association between p16 protein expression and risk of CRC, and this phenomenon was not found in Caucasians. Second, the sample size was relatively small to evaluate the relationship of p16 protein expression with clinicopathologic features of CRC such as location of colorectal tumor, vascular invasion of colorectal tumor cells, and Dukes stage of CRC. Third, subject selection bias might exist in the retrospective studies. Fourth, inclusion criterion was not very clear and consistent, which might affect final conclusions. Although included studies might declare that most patients were examined in detail in hospital, we could not get the diagnostic clinical information. However, this study was the first comprehensive meta-analysis, which assessed the role of p16 protein expression in the prognosis, occurrence, and progression of CRC. Thus, to some extent, it might point out the direction of future research about CRC for us.
5. Conclusion
In conclusion, our meta-analysis shows that p16 protein overexpression is significantly associated with the risk of CRC. p16 protein overexpression may be a useful predicted biomarker for the progression of colorectal tumor such as: TNM-stage, Dukes stage, and lymph node metastasis. Based on these data of the study, p16 protein overexpression may be beneficial to improve the OS of patients with CRC. However, more investigations should be made to further identify the influence of p16 protein expression in the prognosis and development of CRC.
6. Author contributions
Conceptualization: N. Zhou.
Data curation: N. Zhou, Q. Gu.
Formal analysis: N. Zhou.
Funding acquisition: N. Zhou.
Investigation: N. Zhou.
Methodology: N. Zhou.
Software: N. Zhou.
Validation: Q. Gu.
Writing – original draft: N. Zhou, Q. Gu.
Writing – review & editing: N. Zhou, Q. Gu.
Footnotes
Abbreviations: APC = adenomatous polyposis coli, CDKN2A = cyclin-dependent kinase inhibitor 2A, CI = confidence interval, CNKI = China National Knowledge Infrastructure, CRC = colorectal cancer, HR = hazard ratio, OR = odds ratio, OS = overall survival.
The authors have no conflicts of interest to disclose.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014;64:104–17. [DOI] [PubMed] [Google Scholar]
- [3].Rustgi AK. The genetics of hereditary colon cancer. Genes Dev 2007;21:2525–38. [DOI] [PubMed] [Google Scholar]
- [4].Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology 2015;149:1177.e3–90.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- [6].Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet 2009;10:353–8. [DOI] [PubMed] [Google Scholar]
- [7].Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology 2010;138:2059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 1999;96:8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [10].Coghlin C, Murray GI. Progress in the identification of plasma biomarkers of colorectal cancer. Proteomics 2013;13:2227–8. [DOI] [PubMed] [Google Scholar]
- [11].Jia Y, Yang Y, Brock MV, et al. Epigenetic regulation of DACT2, a key component of the Wnt signalling pathway in human lung cancer. J Pathol 2013;230:194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhao R, Choi BY, Lee MH, et al. Implications of genetic and epigenetic alterations of CDKN2A (p16(INK4a)) in cancer. EBioMedicine 2016;8:30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rasmussen SL, Krarup HB, Sunesen KG, et al. Hypermethylated DNA as a biomarker for colorectal cancer: a systematic review. Colorectal Dis 2016;18:549–61. [DOI] [PubMed] [Google Scholar]
- [14].Wang F, Li H, Long J, et al. Clinicopathological significance of p14ARF expression in lung cancer: a meta-analysis. Onco Targets Ther 2017;10:2491–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yang X, Yang L, Dai W, et al. Role of p14ARF and p15INK4B promoter methylation in patients with lung cancer: a systematic meta-analysis. Onco Targets Ther 2016;9:6977–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Okamoto A, Demetrick DJ, Spillare EA, et al. Mutations and altered expression of p16INK4 in human cancer. Proc Natl Acad Sci U S A 1994;91:11045–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sherr CJ. Cancer cell cycles. Science 1996;274:1672–7. [DOI] [PubMed] [Google Scholar]
- [18].Li Y, Nichols MA, Shay JW, et al. Transcriptional repression of the D-type cyclin-dependent kinase inhibitor p16 by the retinoblastoma susceptibility gene product pRb. Cancer Res 1994;54:6078–82. [PubMed] [Google Scholar]
- [19].Gonzalez-Zulueta M, Bender CM, Yang AS, et al. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res 1995;55:4531–5. [PubMed] [Google Scholar]
- [20].Pajcini KV, Corbel SY, Sage J, et al. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell 2010;7:198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Der Simonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [23].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [24].Song F, Gilbody S. Bias in meta-analysis detected by a simple, graphical test. Increase in studies of publication bias coincided with increasing use of meta-analysis. BMJ 1998;316:471. [PMC free article] [PubMed] [Google Scholar]
- [25].Peters JL, Sutton AJ, Jones DR, et al. Comparison of two methods to detect publication bias in meta-analysis. JAMA 2006;295:676–80. [DOI] [PubMed] [Google Scholar]
- [26].Al-Ahwal M, Gomaa W, Emam E, et al. p16 protein is upregulated in a stepwise fashion in colorectal adenoma and colorectal carcinoma. Saudi J Gastroenterol 2016;22:435–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang XZ, Guo R, Huang Y, et al. Expression and significance of P16INK4a protein in colorectal carcinoma. J Chin Pract Diagn Ther 2016;30:1227–9. [Google Scholar]
- [28].Xie XP, Lin AF, Chen XF. Association between p16 protein expression and colorectal cancer with III–IV stage. J Yunnan Univ Trad Chin Med 2016;39:84–6. [Google Scholar]
- [29].Ge C, Wang LP, Xu CW, et al. p16 gene promoter methylation and protein expression in colorectal serrated lesions. Chin J Diagn Pathol 2014;21:169–74. [Google Scholar]
- [30].Jiang H. Expression and implication of P16, cyclin D1 and CDK4 in colonic carcinoma. Med Philos 2014;35:56–9. [Google Scholar]
- [31].Zhang YP, Wang LP, Fang Y. Clinicopathologic and immunohistochemical features of colorectal sessile serrated adenoma/polyp and traditional serrated adenoma. J Diag Pathol 2013;20:150–6. [Google Scholar]
- [32].Cai J. Correlation between p16 expression and biological behavior of colorectal cancer. Chin J Urban Rural Ind Hygiene 2011;4:75–6. [Google Scholar]
- [33].Kriegl L, Neumann J, Vieth M, et al. Up and downregulation of p16(Ink4a) expression in BRAF-mutated polyps/adenomas indicates a senescence barrier in the serrated route to colon cancer. Mod Pathol 2011;24:1015–22. [DOI] [PubMed] [Google Scholar]
- [34].Malhotra P, Kochhar R, Vaiphei K, et al. Aberrant promoter methylation of p16 in colorectal adenocarcinoma in North Indian patients. World J Gastrointest Oncol 2010;2:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lin MX, Wen ZF, Chen YM, et al. Expression and significance of Bmi-1 and P16 in colorectal carcinoma. China Trop Med 2009;9:2112–4. [Google Scholar]
- [36].Carneiro FP, Ramalho LN, Britto-Garcia S, et al. Immunohistochemical expression of p16, p53, and p63 in colorectal adenomas and adenocarcinomas. Dis Colon Rectum 2006;49:588–94. [DOI] [PubMed] [Google Scholar]
- [37].King-Yin Lam A, Ong K, Ho YH. Colorectal mucinous adenocarcinoma: the clinicopathologic features and significance of p16 and p53 expression. Dis Colon Rectum 2006;49:1275–83. [DOI] [PubMed] [Google Scholar]
- [38].Lee M, Sup HW, Kyoung KO, et al. Prognostic value of p16INK4a and p14ARF gene hypermethylation in human colon cancer. Pathol Res Pract 2006;202:415–24. [DOI] [PubMed] [Google Scholar]
- [39].Tada T, Watanabe T, Kanazawa T, et al. Genetic characterization of colorectal cancers in young patients based on chromosomal loss and microsatellite instability. Scand J Gastroenterol 2004;39:1134–40. [DOI] [PubMed] [Google Scholar]
- [40].Huang SM, Yi SB. Clinical effect of p16 protein expression in the development of colorectal cancer. Clin Med 2003;23:32–3. [Google Scholar]
- [41].Xie D, Sham JS, Zeng WF, et al. Heterogeneous expression and association of beta-catenin, p16 and c-myc in multistage colorectal tumorigenesis and progression detected by tissue microarray. Int J Cancer 2003;107:896–902. [DOI] [PubMed] [Google Scholar]
- [42].Huang WB, Peng DF, Qi Q. Clinical effect of p16 and cyclin D1 protein expression in the development of colorectal cancer. Chin J Gastrointest Surg 2001;4:123–4. [Google Scholar]
- [43].Dai CY, Furth EE, Mick R, et al. p16(INK4a) expression begins early in human colon neoplasia and correlates inversely with markers of cell proliferation. Gastroenterology 2000;119:929–42. [DOI] [PubMed] [Google Scholar]
- [44].Palmqvist R, Rutegard JN, Bozoky B, et al. Human colorectal cancers with an intact p16/cyclin D1/pRb pathway have up-regulated p16 expression and decreased proliferation in small invasive tumor clusters. Am J Pathol 2000;157:1947–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tan WL, Wang YH, Dong JR, et al. Study on the expression of MTS1 gene in different pathological types of colorectal neoplasm. Chin J Exp Surg 1998;15:499–500. [Google Scholar]
- [46].Yang YF, Hu XQ, Liang XB, et al. Expression of p16 associated with survival time in colorectal cancer. Modern Preventive Medicine 2012;39:5911–7. [Google Scholar]
- [47].Miladi-Abdennadher I, Abdelmaksoud-Damak R, Ayadi L, et al. Expression of p16INK4a, alone or combined with p53, is predictive of better prognosis in colorectal adenocarcinoma in Tunisian patients. Appl Immunohistochem Mol Morphol 2011;19:562–8. [DOI] [PubMed] [Google Scholar]
- [48].Shima K, Nosho K, Baba Y, et al. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: cohort study and literature review. Int J Cancer 2011;128:1080–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Deschoolmeester V, Van Marck V, Baay M, et al. Detection of HPV and the role of p16INK4A overexpression as a surrogate marker for the presence of functional HPV oncoprotein E7 in colorectal cancer. BMC Cancer 2010;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Huh JW, Lee JH, Kim HR. Expression of p16, p53, and Ki-67 in colorectal adenocarcinoma: a study of 356 surgically resected cases. Hepatogastroenterology 2010;57:734–40. [PubMed] [Google Scholar]
- [51].Paya A, Alenda C, Perez-Carbonell L, et al. Utility of p16 immunohistochemistry for the identification of Lynch syndrome. Clin Cancer Res 2009;15:3156–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lam AK, Ong K, Giv MJ, et al. p16 expression in colorectal adenocarcinoma: marker of aggressiveness and morphological types. Pathology 2008;40:580–5. [DOI] [PubMed] [Google Scholar]
- [53].Kim BN, Yamamoto H, Ikeda K, et al. Methylation and expression of p16INK4 tumor suppressor gene in primary colorectal cancer tissues. Int J Oncol 2005;26:1217–26. [PubMed] [Google Scholar]
- [54].Komuro Y, Watanabe T, Tsurita G, et al. Evaluating the combination of molecular prognostic factors in tumor radiosensitivity in rectal cancer. Hepatogastroenterology 2005;52:666–71. [PubMed] [Google Scholar]
- [55].Chen YQ, Lin MB, Cai D, et al. Expression of P19ARF and P16INK4a and their relationships to tumor biological behaviors in human pancreatic cancer and colorectal cancer. Clin Med J China 2004;11:378–82. [Google Scholar]
- [56].Cui X, Shirai Y, Wakai T, et al. Aberrant expression of pRb and p16(INK4), alone or in combination, indicates poor outcome after resection in patients with colorectal carcinoma. Hum Pathol 2004;35:1189–95. [DOI] [PubMed] [Google Scholar]
- [57].Norrie MW, Hawkins NJ, Todd AV, et al. Inactivation of p16INK4a by CpG hypermethylation is not a frequent event in colorectal cancer. J Surg Oncol 2003;84:143–50. [DOI] [PubMed] [Google Scholar]
- [58].Chang XH, Jiang AM, Zhang C, et al. The expression and clinical significance of survivin, PTEN and p16 in colorectal cancer. Clin Med J China 2007;14:31–2. [Google Scholar]
- [59].Tada T, Watanabe T, Kazama S, et al. Reduced p16 expression correlates with lymphatic invasion in colorectal cancers. Hepatogastroenterology 2003;50:1756–60. [PubMed] [Google Scholar]
- [60].Chen Q, Zheng SG, Tan JL. The expression of p16 and P-gp protein in colorectal cancer. Guangxi Med J 2001;23:1061–3. [Google Scholar]
- [61].Lv Y, Zhang HL, Li YZ, et al. Clinicopathological significance of expressions of DNA dependent protein kinase catalytic subunit and P16 in colorectal carcinoma. Natl Med J China 2008;88:2025–9. [PubMed] [Google Scholar]
- [62].Zhao P, Hu YC, Talbot IC. Expressing patterns of p16 and CDK4 correlated to prognosis in colorectal carcinoma. World J Gastroenterol 2003;9:2202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gong L, Chen P, Liu XJ, et al. Correlation of the expression of Ki-67 and p16 protein with KRAS mutations. Chin J Diag Pathol 2016;23:701–3. [Google Scholar]
- [64].Ayhan S, Isisag A, Saruc M, et al. The role of pRB, p16 and cyclin D1 in colonic carcinogenesis. Hepatogastroenterology 2010;57:251–6. [PubMed] [Google Scholar]
- [65].Cheng SQ, Cao J, Liu JW, et al. Study on the aberrant methylation and protein of p16 gene in patients with colorectal cancer. Chin J Lab Med 2004;27:307–10. [Google Scholar]
- [66].Karamitopoulou E, Zlobec I, Koumarianou A, et al. Expression of p16 in lymph node metastases of adjuvantly treated stage III colorectal cancer patients identifies poor prognostic subgroups: a retrospective analysis of biomarkers in matched primary tumor and lymph node metastases. Cancer 2010;116:4474–86. [DOI] [PubMed] [Google Scholar]
- [67].Kondo E, Gu Z, Horii A, et al. The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16(INK4a) and hMLH1 genes. Mol Cell Biol 2005;25:4388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dai W, Zeller C, Masrour N, et al. Promoter CpG island methylation of genes in key cancer pathways associates with clinical outcome in high-grade serous ovarian cancer. Clin Cancer Res 2013;19:5788–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Semczuk A, Jakowicki JA. Alterations of pRb1-cyclin D1-cdk4/6-p16(INK4A) pathway in endometrial carcinogenesis. Cancer Lett 2004;203:1–2. [DOI] [PubMed] [Google Scholar]
- [70].Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol 2006;24:1770–83. [DOI] [PubMed] [Google Scholar]
- [71].Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res 2001;264:42–55. [DOI] [PubMed] [Google Scholar]
- [72].Hibi K, Taguchi M, Nakayama H, et al. Molecular detection of p16 promoter methylation in the serum of patients with esophageal squamous cell carcinoma. Clin Cancer Res 2001;7:3135–8. [PubMed] [Google Scholar]
- [73].Fukushima N, Sato N, Ueki T, et al. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol 2002;160:1573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hasegawa M, Nelson HH, Peters E, et al. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene 2002;21:4231–6. [DOI] [PubMed] [Google Scholar]
- [75].Liu Y, Lan Q, Siegfried JM, et al. Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking lung cancer patients. Neoplasia 2006;8:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chen YZ, Liu D, Zhao YX, et al. Relationships between p16 gene promoter methylation and clinicopathologic features of colorectal cancer: a meta-analysis of 27 cohort studies. DNA Cell Biol 2014;33:729–38. [DOI] [PubMed] [Google Scholar]
- [77].Yi J, Wang ZW, Cang H, et al. p16 gene methylation in colorectal cancers associated with Duke's staging. World J Gastroenterol 2001;7:722–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Simpson DJ, Bicknell JE, McNicol AM, et al. Hypermethylation of the p16/CDKN2A/MTSI gene and loss of protein expression is associated with nonfunctional pituitary adenomas but not somatotrophinomas. Genes Chromosomes Cancer 1999;24:328–36. [PubMed] [Google Scholar]
- [79].Ohhara M, Esumi M, Kurosu Y. Activation but not inactivation of the MTS1 gene is associated with primary colorectal carcinomas. Biochem Biophys Res Commun 1996;226:791–5. [DOI] [PubMed] [Google Scholar]


