Supplemental Digital Content is available in the text
Keywords: carotid atherosclerosis, diagnosis, gene, microRNA, protein–protein interaction network, regulatory network
Abstract
This study was aimed to explore the crucial genes and microRNAs (miRNAs) associated with the carotid atherosclerosis (CA).
Two public datasets GSE28829 and GSE43292 were obtained from Gene Expression Omnibus databases to analyze the differentially expressed genes (DEGs) between primary and advanced atherosclerotic plaque tissues. The Gene Ontology (GO) terms, pathways, and protein–protein interactions (PPIs) of these DEGs were analyzed. miRNAs and transcription factor (TF) were predicted.
A total of 112 upregulated and 179 downregulated intersection DEGs were identified between 2 datasets. In the PPI network, HSP90AB1 (degree = 19), RAP1A (degree = 14), and integrin subunit beta 1 (ITGB1) had higher degrees. A total of 23 miRNAs were predicted, such as miR-126, miR-155, miR-19A, and miR-19B. Four TFs were associated with upregulated DEGs, while 10 TFs were identified to be associated with downregulated genes.
Our study suggests the important roles of HSP90AB1, RAP1A, and integrins proteins of ITGB1, ITGA11, ITGA9, and ITGB2 in the progression of CA plaque. Additionally, miR-126, miR-155, miR-19B, and miR-19A may be considered as biomarkers of CA.
1. Introduction
Cardiovascular disease is the prime cause of death among the elderly and is a major determinant of chronic disability.[1] Carotid atherosclerosis (CA), a common type of cardiovascular disease, appears to be highly prevalent in the ageing population. According to a recent epidemiological survey, up to 12% of men and 5% of women over the age of 80 suffer from asymptomatic moderate atherosclerotic carotid artery stenosis, while 3% and 1%, respectively, suffer from severe carotid stenosis.[2] CA has become one of the main causes leading to morbidity and death around the world and has been predicted to be “the number one killer” by 2020.[3] Recent studies have revealed that CA plays a major role in cerebral stroke induced by ischemic in senior people.[4] The first characterization in CA pathogenesis is intima-media thickness (IMT) in the middle membrane of cervical artery, and then gradually formation of atherosclerotic plaque. Although the previous studies have showed that IMT in carotid artery is an independent risk factor to predict the cerebral stroke, its potential relationships in stroke evaluation remained unclear.[5,6]
IMT can directly reflect the extent of atherosclerosis, which, together with carotid atherosclerotic plaque, could serve as good clinical markers of atherosclerosis.[7] It has been reported that the formation of carotid IMT and plaque is regulated by many genes.[8,9] For instance, alanine-glyoxylate aminotransferase 2 gene (AGXT2)[10] was recently reported to have a closely relationship with arteriosclerotic changes through carotid IMT in Japanese subjects, indicating that AGXT2 genotype may be an important factor underlying atherosclerosis. Tanja et al[11] showed that the promoter variants of 3 specific candidate genes, matrix metalloproteinase-3, interleukin-6, and hepatic lipase gene were respectively related to matrix deposition, inflammation, and lipid metabolism, all of which could lead to an increased carotid IMT. Moreover, microRNAs (miRNAs) have also been revealed to play a major role in atherosclerosis, such as miR-320b, let-7, and miR-29b.[12–14] Although numerous genes and miRNAs associated with the pathogenesis of CA have been identified, it is far from enough for the development of novel therapeutic targets.
Recently, it is possible to analyze the genome-wide mRNA or miRNAs expression thanks to the development of microarrays.[15] In this study, 2 public gene expression profiles (GSE28829 and GSE43292) were downloaded from Gene Expression Omnibus databases. Both of 2 dataset contained atherosclerotic plaque tissues of primary and advanced stages, which may help to uncover the biomarkers of CA develop from primary to advanced stages. The differentially expressed genes (DEGs) between primary and advanced atherosclerotic plaque tissues were identified. The gene functional enrichment analyses as well as protein–protein interaction (PPI) network analysis were separately performed for the DEGs. Additionally, the miRNAs of DEGs were predicted. We aimed to explore more crucial genes and miRNAs associated with the CA progression.
2. Materials and methods
2.1. Data sources
The mRNA expression profiles data, GSE28829 and GSE43292, were downloaded from the Gene Expression Omnibus database. GSE28829, which included 13 primary atherosclerotic plaques and 16 advanced atherosclerotic plaques, was sequenced on the platform of GPL570-Affymetrix Human Genome U133 plus 2.0 Array [HG-U133_Plus_2]. GSE43292, including 32 atheroma plaque and 32 macroscopically intact samples, was sequenced on the platform of GPL6244-Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] [HuGene-1_0-st].
2.2. Data preprocessing
The R software package affy (version 1.50.0, http://www.bioconductor.org/packages/release/bioc/html/affy.html)[16] was used to read the microarray data. The robust multiarray business method[17,18] was used for data preprocessing, including background correction, normalization, and expression calculation. Platform annotations files were used to annotate the probes. The probes without matching genes (gene symbol) were deleted. For the conditions that multiple probes mapped to the same gene, we took the average of these probes as the ultimate value of gene expression.
2.3. DEGs screening
For the 2 datasets, limma[19] package was used to screen DEGs. Adjusted t test was used to calculate the P values, and Benjamini and Hochberg method was used to adjust the P values. The DEG with adjusted P value of less than 0.05 was selected. Finally, the intersection DEGs between GSE2889 and GSE43292 were obtained.
2.4. Functional analysis of DEGs
For the identified DEGs, we performed Gene Ontology (GO)[20] and Kyoto Encyclopedia of Genes and Genomes (KEGG)[21] pathway enrichment analyses using DAVID (version 6.8).[22]P < .05 was used as threshold for the identification of significant GO terms and KEGG pathways. To visually observe the functions of DEGs, we use the ClueGO plugin (version 2.2.6)[23] in Cytoscape software (version 3.4.0)[24] to draw the cross-linked enrichment graph between GO terms and KEGG pathways. P < .05 was set as threshold for the identification of significant enrichment.
2.5. PPI network analysis
Based on the Search Tool for the Retrieval of Interacting Genes/Proteins,[25] we performed PPI network analysis. PPI network can reveal the interactions between proteins. The parameter of PPI score (medium confidence) was set to 0.4. The obtained PPI pairs were then visualized to construct the PPI network using Cytoscape. The degree value was counted and the closeness connection between nodes was observed.
2.6. Network module analysis
The MCODE[26] method was used to analyze the significant module in PPI network. MCODE algorithm could calculate the score of each module, and the higher score meant that the interaction relations inside the module were more closed. According to the analysis results, the module with score ≥5 and node ≥5 was selected. After that, we performed GO and KEGG pathway enrichment analyses for the DEGs in module.
2.7. miRNA-target gene regulatory relations analysis
The overrepresentation enrichment analysis method in Webgestalt[27] (http://www.webgestalt.org/) tool was used to predict the miRNAs of DEGs that involved in PPI network. The key miRNAs and genes were identified based on the network topological property analysis. The genes count ≥he and Benjamini and Hochberg method adjusted P < .05 were set as thresholds.
2.8. Transcription factor (TF) prediction and network integration
Based on the TF regulatory network data in the databases of ITFP (http://itfp.biosino.org/itfp) and TRANSFAC (http://www.gene-regulation.com/pub/databases.html), we predicted the TFs that related to DEGs and further screened out DEGs which were regulated by these TFs. Finally, we integrated the TF-target, miRNA-target genes, and PPI networks. The network diagram was mapped by the Cytoscape software.
3. Results
3.1. DEGs between advanced and early atherosclerotic plaque patients
In total, 233 up- and 486 downregulated DEGs were identified from GSE28829. Additionally, 4183 up- and 4156 downregulated DEGs were identified from GSE43292. Subsequently, we analyzed the 2 datasets and calculated the intersection DEGs. Total 291 intersection DEGs (112 upregulated and 179 downregulated) were obtained (Supplementary data S1).
3.2. Functional and pathway enrichment analyses
GO and KEGG pathway enrichment analyses were performed for both up- and downregulated DEGs. The top 5 GO and KEGG pathway terms enriched by the upregulated DEGs are shown in Table 1, such as leukocyte migration (GO:0050900), regulation of immune response (GO:0050776), innate immune response (GO:0045087P), and rheumatoid arthritis (hsa05323). The downregulated DEGs enriched GO and pathway terms included cardiac muscle fiber development (GO:0048739), smooth muscle contraction (GO:0006939), arrhythmogenic right ventricular cardiomyopathy (hsa05412), and Focal adhesion (hsa04510) (Table 1). The crosslinking of GO terms and pathways is shown in Fig. 1. The complete output of functional analysis is shown in Supplementary data S1.
Table 1.
The GO and KEGG pathway enrichment results of differentially expressed genes.
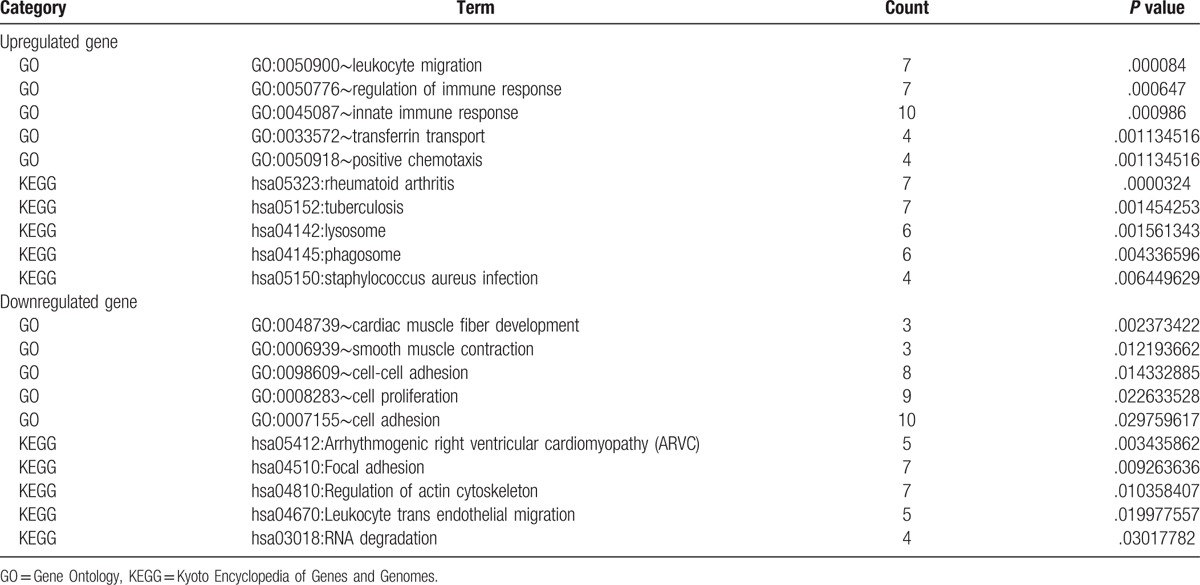
Figure 1.
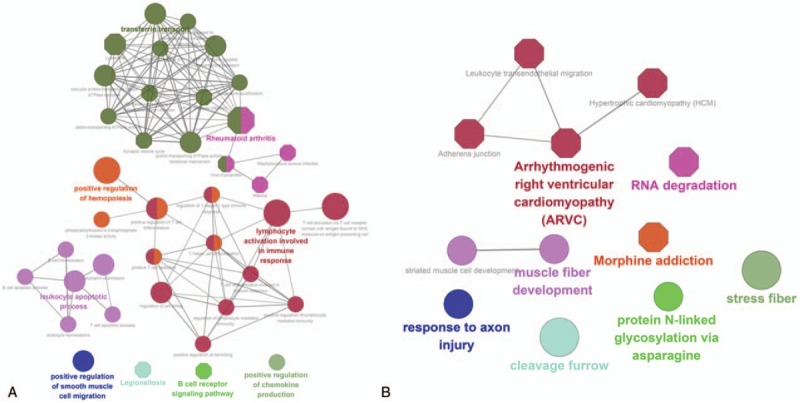
The enrichment analysis of up- and downregulated DEGs in GO biological process and KEGG pathway. (A) Upregulated genes, (B) downregulated genes. Roundness nodes represent GO biological process classification; hexagon nodes represent KEGG pathway classification; different color nodes represent different pathway/GO biological process classification. DEG = differentially expressed gene, GO = Gene Ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes.
3.3. PPI network and module analyses
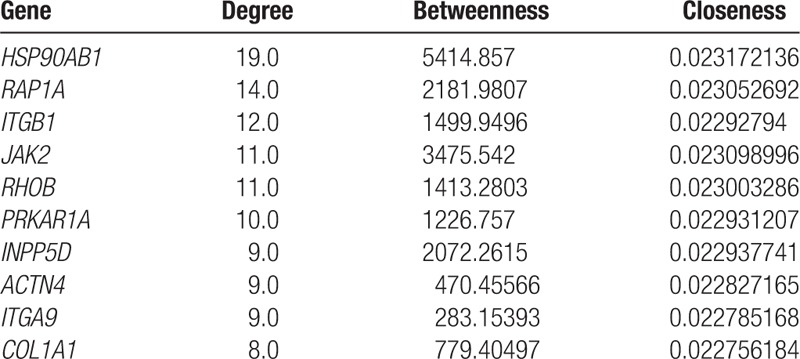
The PPI network was established with 162 nodes and 225 interaction pairs (Fig. 2). Analysis of nodes in the PPI network revealed that heat shock protein 90 kDa alpha, class B member 1 (HSP90AB1), ras-related protein (RAP1A), and integrin subunit beta 1 (ITGB1) had the top 3 highest degrees. The top 10 nodes with higher topological property scores (regarded as hub proteins) are shown in Table 2.
Figure 2.
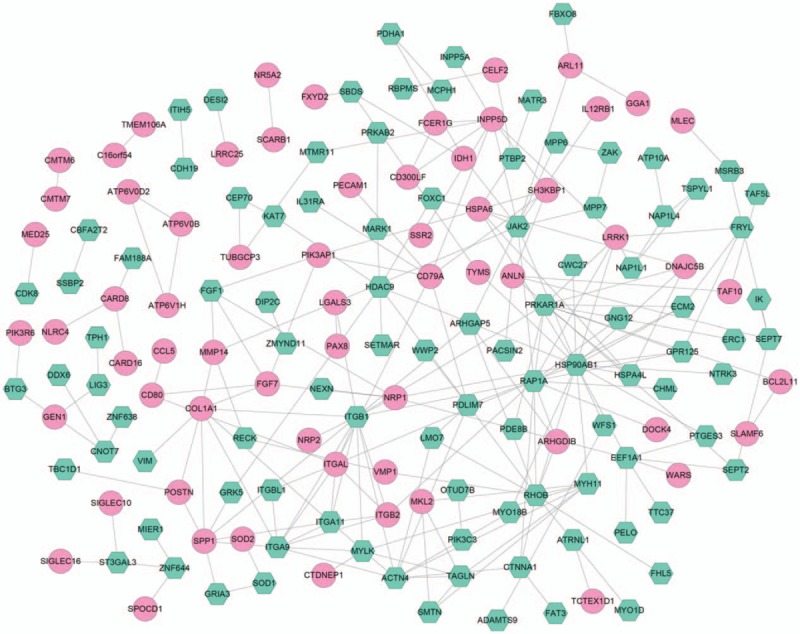
The construction of protein–protein interaction network that based on the differentially expressed genes. The pink round represents upregulated gene, green hexagon represents downregulated gene.
Table 2.
The top 10 nodes with high degrees.
From the PPI network, only 1 module with score more than 5 (5.667) was identified. As shown Fig. 3, this module was made up of 17 edges and 7 nodes, such as integrin subunit alpha 11 (ITGA11), ITGA9, ITGB1, and ITGB2. The 7 genes were significantly enriched in ECM-receptor interaction (hsa04512), Focal adhesion (hsa04510), Regulation of actin cytoskeleton (hsa04810), PI3K-Akt signaling pathway (hsa04151), and cell adhesion molecules (hsa04514).
Figure 3.
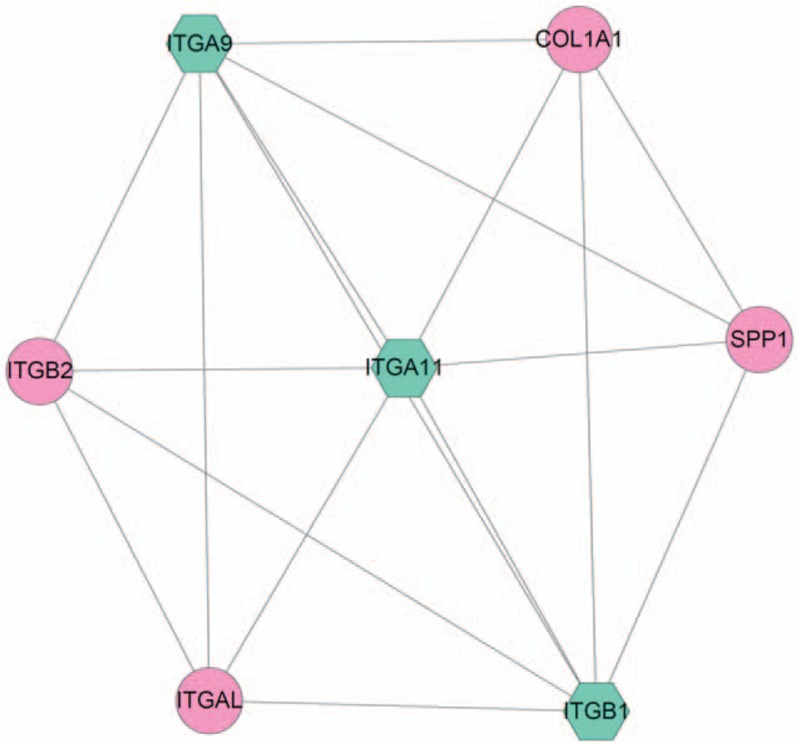
The module with highest scores in protein–protein interaction network. The pink round represents upregulated gene, green hexagon represents the downregulated gene.
3.4. miRNA prediction and analysis
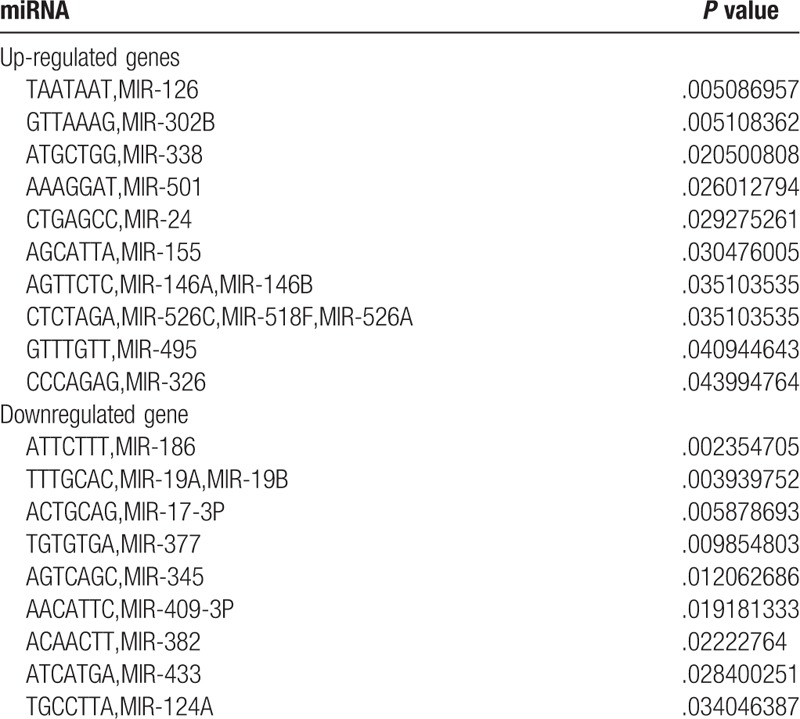
Based on the threshold of P < .05, 13 miRNAs were predicted in the upregulated genes, such as miR-126, miR-302B, and miR-338. Additionally, total 10 miRNAs were predicted in the downregulated genes, such as miR-186, miR-19A, miR-19B, and miR-17-3P. The results are shown in Table 3.
Table 3.
The miRNA prediction results in upregulated genes.
3.5. TF prediction and network integration
TFs were predicted from the DEGs. From the upregulated DEGs, 4 TFs were predicted, including ANLN, KLHL6, PAX8, and SAMSN1, among which SAM domain, SH3 domain, and nuclear localization signals 1 (SAMSN1) was a TF of ITGB2, a gene in module. In the downregulated DEGs, 10 TFs were identified, including CNOT7, INPP5A, LMO7, matrin 3 (MATR3), NFIA, PTBP2, TAF5L, ZMYND11, zinc finger protein 638 (ZNF638), and ZNF644. Besides, MATR3, ZNF638, and ZNF644 were TFs of HSP90AB1.
Based on the obtained PPI, TF-target, and miRNA-target pairs, we constructed an integrated network using Cytoscape software. This network contained 218 nodes, and HSP90AB1, RAP1A, miR-19B, miR-19A, miR-124A, and RHOB had degrees more than 12. The network is shown in Fig. 4.
Figure 4.
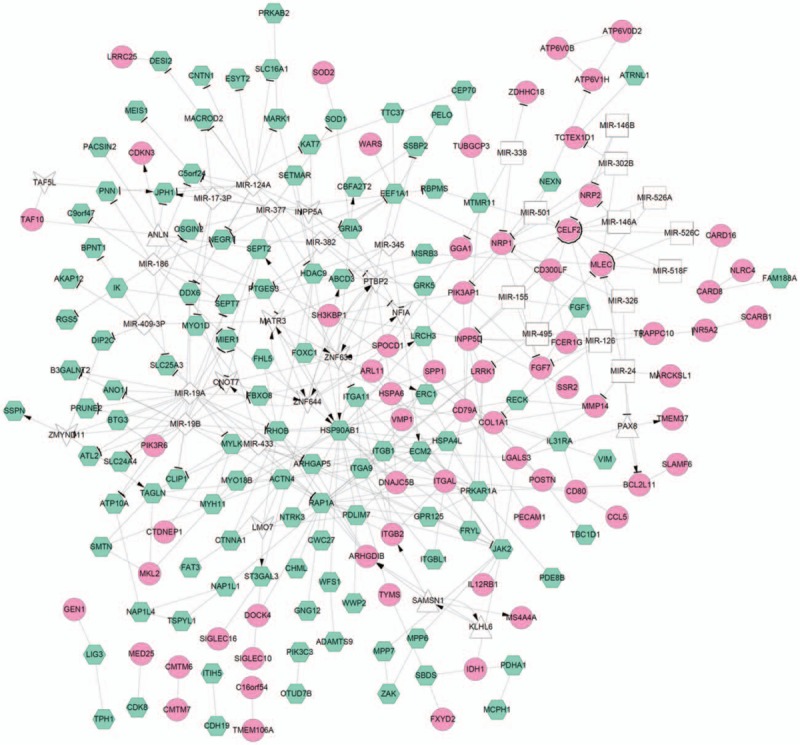
The integration network of protein–protein interaction pairs, transcription factor-target, and miRNA-target. The pink round represents upregulated gene, green hexagon represents downregulated gene; white triangle represents transcription factor predicted in upregulated genes, white V shape represents transcription factor predicted in downregulated genes; white diamond shape represents miRNA predicted in downregulated genes, white square shape represents miRNA predicted in upregulated genes; line with arrow represents regulation relation between transcription factor and target genes, T shape represents regulation relation between the miRNA and target genes; lines without arrow represents the interaction relation between proteins.
4. Discussion
In our study, we identified 291 intersection DEGs in 2 databases. HSP90AB1, RAP1A, and ITGB1 were hub proteins in the PPI network. ITGA11, ITGA9, ITGB1, and ITGB2 involved in the module, which were involved in several pathways, including Regulation of actin cytoskeleton (hsa04810). Based on these DEGs, 23 miRNAs were predicted, such as miR-126, miR-186, and miR-155. Furthermore, HSP90AB1, RAP1A, miR-19B, and miR-19A had high degrees in the integrated network.
The PPI network contains a small number of highly connected protein nodes (known as hub proteins) and many poorly connected nodes. Genome-wide studies have shown that deletion of a hub protein is more likely to be lethal than deletion of a nonhub protein, which is known as the centrality-lethality rule.[28] Therefore, in this study, the hub proteins in the PPI network may play a critical role in CA progression. In the constructed PPI network, we identified several hub proteins, such as HSP90AB1 (degree = 19.0) and RAP1A (degree = 14.0). HSP90AB1, as 1 key member of heat shock protein 90 family, was observed to downregulate in ischemic human heart tissue; and the decreased expression of HSP90AB1 may potentially demonstrate a decreased defense mechanism against ischemic injury of myocardial cells, which finally leads to decreased inflammation and atherosclerosis.[29] Jean et al[30] evaluated the differentially expressed networks of genes in 10-week-old Cx37−/−ApoE−/− mice and found that HSP90AB1 was downregulated, which indicated that HSP90AB1 may interact with Cx37 to regulate the progression of atherosclerosis. Interestingly, several other heat shock proteins such as HSP70 and HSP90 have also been proposed to serve as potential biomarkers in cardiovascular diseases or other specific diseases, such as heart failure and arrhythmogenic right ventricular cardiomyopathy.[31–33] Moreover, HSP90AB1 was predicted to have 3 TFs (MATR3, ZNF638, and ZNF644). MATR3 is strongly expressed in the developing heart, cardiac outflow tract valves, and aorta. MATR3 disruption appears to cause similar cardiac left ventricular outflow tract defects in human and mouse,[34] suggesting the role of MATR3 in cardiovascular disease. Although the roles of ZNF638 and ZNF644 in cardiovascular disease or CA have not been investigated, given their transcriptional regulation in HSP90AB1, we speculated that they may play a role in CA. RAP1A is an important isoforms of RAP1A (RAP) family, which is the most abundant GTPases in platelets.[35] Rap1 was reported to overexpress in human atherosclerotic lesions and induce cytokine production in pro-inflammatory macrophages via NFκB signaling.[36] The results indicated that RAP1A and HSP90AB1 may be involved in the progression of CA.[31,32]
In addition to HSP90AB1 and RAP1A, ITGB1 also had high degree (degree = 12.0). Besides, ITGB1 interacted with several other integrins proteins, like ITGA11, ITGA9, and ITGB2. Integrins are cell-surface heterodimers, which mediate interactions between the extracellular environment and platelets, inflammatory cells, and the vasculature.[37] Importantly, integrins have been reported to be associated with angiogenesis.[38] In addition, ITGA9, ITGA11, ITGB2, and ITGB1 were significantly enriched in Regulation of actin cytoskeleton (hsa04810). Study has reported that changes in the actin cytoskeleton underlie vascular contractility and remodeling.[39] Moreover, TF prediction analysis found that SAMSN1 was a TF of ITGB2. A recent study revealed that an rs2822693 that was located in an open chromatin region upstream of SAMSN1 was associated with the number of diseased coronary vessels.[40] We speculated that these integrins proteins may involved in the CA development via regulation of actin cytoskeleton.
Additionally, miRNA-target gene analysis predicted 23 miRNAs, such as miR-126 miR-155, miR-19B, and miR-19A. miR-126 was reported as one specifically expressed miRNA in endothelial cells and could accelerate the proliferation and angiogenesis of endothelial cell.[41] Zernecke et al[42] found that the intravenous injection of miR-126 can lead to the inhibition effect of atherosclerosis in mice and finally reduced the formation of the atherosclerotic plaques. Recently, Pan's report showed that expression of miR-126 was downregulated significantly in the model mice with carotid atherosclerotic plaque, while its target gene vascular cell adhesion molecule-1 was significantly upregulated, which accelerated the progression of atherosclerosis.[43] Together with our results, these findings indicated that miR-126 was involved in the formation of atherosclerosis.
Li et al[44] reported that increased miR-155 led to relieve of chronic inflammation through a negative feedback loop, which played a protective role during atherosclerosis-related cell formation. The potential mechanism may be associated with the miR-155-CARHSP1-TNF-α signaling pathway, which repressed the expression of calcium-regulated heat stable protein 1. Moreover, macrophages play a central role in the initiation and development stages of atherosclerosis pathogenesis. Several evidences have indicated that, both as inducers and carriers of miR-155, low-density lipoprotein and its oxidized derivatives can regulate miR-155-mediated apoptotic and inflammatory responses in lesional macrophages at different stages of atherosclerosis.[45] Therefore, our data were in consistent with the previous study, which further suggested the importance of miRNAs in atherosclerosis.
miR-19B and miR-19A had higher degrees (degree = 16.0) in the integrated network. The two miRNAs belongs to the miR-17–92 cluster. Researchers have noted that the miR-17-92 cluster is significantly downregulated among patients with atherosclerosis.[46] A recent study suggested that the levels of miR-19B were significantly lower in patients with coronary artery diseases compared with healthy control subjects.[47] They elaborated that inflammatory factors can destroy the integrity of the endothelium, triggering the development of atherosclerosis. miR-19B plays a key role in the attenuation of TNF-α-induced endothelial cell apoptosis. Moreover, miR-19A was also found to play a key role in HIF-1α-induced endothelial inflammation in atherosclerosis.[48] Taken together, miR-19B and miR-19A may be 2 biomarkers of CA.
Although several genes or miRNAs were predicted and considered to play significant roles in CA progression, no biological experiments were carried out to further confirm their differential expression in CA tissue and their functions in CA. It is a limitation for this study. Therefore, our future study will focus on their functions in CA through animal and clinical experiments.
In conclusion, our study suggests the important roles of HSP90AB1, RAP1A, and integrins proteins of ITGB1, ITGA11, ITGA9, and ITGB2 in the progression of CA plaque. Additionally, miR-126, miR-155, miR-19B, and miR-19A may also play critical roles in pathogenesis of CA, therefore, these miRNAs may be considered as biomarkers of CA. Findings of this study would hopefully reveal new therapeutic targets for CA.
Author contributions
Data curation: Z. Mao, F. Wu.
Formal analysis: F. Wu.
Writing – original draft: Z. Mao.
Writing – review & editing: Y. Shan.
Supplementary Material
Footnotes
Abbreviations: CA = carotid atherosclerosis, DEG = differentially expressed gene, GO = Gene Ontology, HSP90AB1 = heat shock protein 90 kDa alpha, class B member 1, IMT = intima-media thickness, ITGA11 = integrin subunit alpha 11, ITGB1 = integrin subunit beta 1, KEGG = Kyoto Encyclopedia of Genes and Genomes, MATR3 = matrin 3, PPI = protein–protein interaction, RAP1A = ras-related protein, TF = transcription factor, ZNF638 = zinc finger protein 638.
Ethical approval: The article did not involve animals or clinical trials, so ethical approval was not necessary.
The authors have no funding and conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Thom TJ. International mortality from heart disease: rates and trends. Int J Epidemiol 1989;18:20–8. [PubMed] [Google Scholar]
- [2].De WM, Greving JB, Lorenz MW, et al. Prevalence of asymptomatic carotid artery stenosis in the general population an individual participant data meta-analysis. Stroke 2010;41:1294–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lopez AD, Mathers CD, Ezzati M, et al. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet 2006;367:1747–57. [DOI] [PubMed] [Google Scholar]
- [4].Barski L, Porath A, Novack V, et al. Prevalence, recognition and treatment of cardiovascular risk factors in outpatients with atherothrombosis in Israel. Isr Med Assoc J 2007;9:376–9. [PubMed] [Google Scholar]
- [5].Hermann DM, Gronewold J, Lehmann N, et al. Intima-media thickness predicts stroke risk in the Heinz Nixdorf Recall study in association with vascular risk factors, age and gender. Atherosclerosis 2012;224:84–9. [DOI] [PubMed] [Google Scholar]
- [6].Chien KL, Su TC, Jeng JS, et al. Carotid artery intima-media thickness, carotid plaque and coronary heart disease and stroke in Chinese. PLoS One 2008;3:e3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fox CS, Polak JF, Chazaro I, et al. Genetic and environmental contributions to atherosclerosis phenotypes in men and women: heritability of carotid intima-media thickness in the Framingham Heart Study. Stroke 2003;34:397–401. [DOI] [PubMed] [Google Scholar]
- [8].Sayed-Tabatabaei FA, van Rijn MJ, Schut AF, et al. Heritability of the function and structure of the arterial wall: findings of the Erasmus Rucphen Family (ERF) study. Stroke 2005;36:2351–6. [DOI] [PubMed] [Google Scholar]
- [9].Hunt KJ, Duggirala R, Goring HH, et al. Genetic basis of variation in carotid artery plaque in the San Antonio Family Heart Study. Stroke 2002;33:2775–80. [DOI] [PubMed] [Google Scholar]
- [10].Yoshino Y, Kohara K, Abe M, et al. Missense variants of the alanine: glyoxylate aminotransferase 2 gene correlated with carotid atherosclerosis in the Japanese population. J Biol Regul Homeost Agents 2014;28:605–14. [PubMed] [Google Scholar]
- [11].Rundek T, Elkind MS, Pittman J, et al. Carotid intima-media thickness is associated with allelic variants of stromelysin-1, interleukin-6, and hepatic lipase genes: the Northern Manhattan Prospective Cohort Study. Stroke 2002;33:1420–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen KC, Hsieh IC, Hsi E, et al. Negative feedback regulation between microRNA let-7 g and the oxLDL receptor LOX-1. J Cell Sci 2011;124(Pt 23):4115–24. [DOI] [PubMed] [Google Scholar]
- [13].Chen KC, Liao YC, Hsieh IC, et al. OxLDL causes both epigenetic modification and signaling regulation on the microRNA-29b gene: novel mechanisms for cardiovascular diseases. J Mol Cell Cardiol 2012;52:587–95. [DOI] [PubMed] [Google Scholar]
- [14].Zhang R, Qin Y, Zhu G, et al. Low serum miR-320b expression as a novel indicator of carotid atherosclerosis. J Clin Neurosci 2016;33:252–8. [DOI] [PubMed] [Google Scholar]
- [15].Liang RQ, Li W, Li Y, et al. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res 2005;33:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gautier L, Cope L, Bolstad BM, et al. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307–15. [DOI] [PubMed] [Google Scholar]
- [17].Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185–93. [DOI] [PubMed] [Google Scholar]
- [18].Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–64. [DOI] [PubMed] [Google Scholar]
- [19].Smyth GK. Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 2005;Berlin, Germany: Springer, 397–420. [Google Scholar]
- [20].Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [23].Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43(Database issue):D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson 2012;14:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005;33(Web Server issue):W741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet 2006;2:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hermansson C, Lundqvist A, Wasslavik C, et al. Reduced expression of NLRP3 and MEFV in human ischemic heart tissue. Biochem Biophys Res Commun 2013;430:425–8. [DOI] [PubMed] [Google Scholar]
- [30].Derouette JP, Wong C, Burnier L, et al. Molecular role of Cx37 in advanced atherosclerosis: a micro-array study. Atherosclerosis 2009;206:69–76. [DOI] [PubMed] [Google Scholar]
- [31].Li Z, Song Y, Xing R, et al. Heat shock protein 70 acts as a potential biomarker for early diagnosis of heart failure. PloS One 2013;8:e67964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wei Y-J, Huang Y-X, Shen Y, et al. Proteomic analysis reveals significant elevation of heat shock protein 70 in patients with chronic heart failure due to arrhythmogenic right ventricular cardiomyopathy. Mol Cell Biochem 2009;332:103. [DOI] [PubMed] [Google Scholar]
- [33].Dong R, Deng P, Huang Y, et al. Identification of HSP90 as potential biomarker of biliary atresia using two-dimensional electrophoresis and mass spectrometry. PloS One 2013;8:e68602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Quintero-Rivera F, Xi QJ, Keppler-Noreuil KM, et al. MATR3 disruption in human and mouse associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus. Hum Mol Genet 2015;24:2375–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stefanini L, Bergmeier W. RAP1-GTPase signaling and platelet function. J Mol Med (Berl) 2016;94:13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cai Y, Sukhova GK, Wong HK, et al. Rap1 induces cytokine production in pro-inflammatory macrophages through NFkappaB signaling and is highly expressed in human atherosclerotic lesions. Cell Cycle 2015;14:3580–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Weng S, Zemany L, Standley KN, et al. β3 integrin deficiency promotes atherosclerosis and pulmonary inflammation in high-fat-fed, hyperlipidemic mice. Proc Natl Acad Sci 2003;100:6730–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brooks PC, Montgomery AM, Rosenfeld M, et al. Integrin alpha v beta 3 antagonists promote tumor regression by inducing apoptosis of angiogenic blood vessels. Cell 1994;79:1157. [DOI] [PubMed] [Google Scholar]
- [39].Zhou Q, Gensch C, Liao JK. Rho-associated coiled-coil-forming kinases (ROCKs): potential targets for the treatment of atherosclerosis and vascular disease. Trends Pharmacol Sci 2011;32:167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Duke University, Ward-Caviness CK. Gene-Environment Interactions in Cardiovascular Disease. 2014. [Google Scholar]
- [41].Harris TA, Yamakuchi M, Ferlito M, et al. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A 2008;105:1516–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal 2009;2:ra81. [DOI] [PubMed] [Google Scholar]
- [43].Pan X, Hou R, Ma A, et al. Atorvastatin upregulates the expression of miR-126 in apolipoprotein E-knockout mice with carotid atherosclerotic plaque. Cell Mol Neurobiol 2017;37:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Li X, Kong D, Chen H, et al. miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Sci Rep 2016;6:21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang E, Wu Y. Dual effects of miR-155 on macrophages at different stages of atherosclerosis: LDL is the key? Med Hypotheses 2014;83:74–8. [DOI] [PubMed] [Google Scholar]
- [46].Fichtlscherer S, De Rosa S, Fox H, et al. Circulating microRNAs in patients with coronary artery diseasenovelty and significance. Circ Res 2010;107:677–84. [DOI] [PubMed] [Google Scholar]
- [47].Tang Y, Zhang Y-c, Chen Y, et al. The role of miR-19b in the inhibition of endothelial cell apoptosis and its relationship with coronary artery disease. Sci Rep 2015;5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Akhtar S, Hartmann P, Karshovska E, et al. Endothelial hypoxia-inducible factor-1α promotes atherosclerosis and monocyte recruitment by upregulating microRNA-19a. Hypertension 2015;66:1220–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


