Abstract
With limited and low-genetic barrier drugs used for the prevention of mother-to-child transmission (PMTCT) of HIV in sub-Saharan Africa, vertically transmitted HIV-1 drug-resistance (HIVDR) is concerning and might prompt optimal pediatric strategies.
The aim of this study was to ascertain HIVDR and viral-tropism in majority and minority populations among Cameroonian vertically infected children.
A comparative analysis among 18 HIV-infected children (7 from PMTCT-exposed mothers and 11 from mothers without PMTCT-exposure) was performed. HIVDR and HIV-1 co-receptor usage was evaluated by analyzing sequences obtained by both Sanger sequencing and ultra-deep 454-pyrosequencing (UDPS), set at 1% threshold.
Overall, median (interquartile range) age, viremia, and CD4 count were 6 (4–10) years, 5.5 (4.9–6.0) log10 copies/mL, and 526 (282–645) cells/mm3, respectively. All children had wild-type viruses through both Sanger sequencing and UDPS, except for 1 PMTCT-exposed infant harboring minority K103N (8.31%), born to a mother exposed to AZT+3TC+NVP. X4-tropic viruses were found in 5 of 15 (33.3%) children (including 2 cases detected only by UDPS). Rate of X4-tropic viruses was 0% (0/6) below 5 years (also as minority species), and became relatively high above 5 years (55.6% [5/9], P = .040. X4-tropic viruses were higher with CD4 ≤15% (4/9 [44.4%]) versus CD4 >15% (1/6 [16.7%], P = .580); similarly for CD4 ≤200 (3/4 [75%]) versus CD4 >200 (2/11 [18.2%] cells/mm3, P = .077.
NGS has the ability of excluding NRTI- and NNRTI-mutations as minority species in all but 1 children, thus supporting the safe use of these drug-classes in those without such mutations, henceforth sparing ritonavir-boosted protease inhibitors or integrase inhibitors for the few remaining cases. In children under five years, X4-tropic variants would be rare, suggesting vertical-transmission with CCR5-tropic viruses and possible maraviroc usage at younger ages.
Keywords: children, coreceptor usage, HIV-1 drug resistance, next-generation sequencing, PMTCT, sanger sequencing
1. Introduction
Despite increasing coverage (to about 61%) in prevention of mother-to-child transmission (PMTCT), human immunodeficiency virus type 1 (HIV-1) vertical-transmission remains consistent in sub-Saharan Africa (SSA).[1] More so, although progress in PMTCT (from single-dose nevirapine [sd-NVP] to option-B+) has been reducing HIV-1 vertical-transmission, infected children stand at higher risks of HIV-1 drug resistance (HIVDR) to antiretrovirals administered pre-, per-, or post-partum.[1,2] This is particularly true in SSA because of wide use of low genetic-barrier drugs, recurrent stock-outs, impaired-adherence, inadequate monitoring, HIV-1 diversity and, importantly, limited pediatric highly active antiretroviral therapy (HAART) options.[3–5] All these factors lead to delayed detection of HAART failure and HIVDR accumulation even beyond 80%.[6,7]
As the footprint of long-term HAART depends largely on the effectiveness of first-line drugs in sustaining viral suppression, establishing adequacy between pediatric HAART and DR-mutations (DRMs) would be clinically relevant.[7,8] In this line, we earlier reported low- and high-HIVDR, respectively, in naïve and HAART-failing children, with successful switch to second-line.[9] From these observations, we postulated that minority DRMs in HAART-naïve children might grow-up through selective drug-pressure and populate plasma in a short-frame, herein justifying the rapidly emerging DRMs we observed at failure.[9] Although not yet clinically endorsed, pediatric minority DRMs might be more concerning in the context of PMTCT, henceforth underscoring an unmet clinical need.[10,11] Coupled to previous knowledge on the detection of DRMs by next-generation sequencing (NGS),[12–14] we thus hypothesized that using NGS to assess DRMs in vertically infected HAART-naïve children would contribute in designing long-term HAART strategies for SSA-children.
Current pediatric HAART-regimens consist of lamivudine (3TC), abacavir (ABC), or zidovudine (AZT), associated to ritonavir-boosted lopinavir (LPV/r) or NVP. LPV/r is recommended to overcome PMTCT-resulting non-nucleoside reverse transcriptase inhibitor (NNRTI) resistance, whereas NVP matches with postnatal prophylaxis.[15] As HAART would be reaching 1.5 million children by 2020, as high as 20% virological failure (VF) is expected, favored by high-viremia and poor adherence in children.[15,16] Without optimal strategies, VF would quickly overcome HAART success, maintaining children vulnerable.[17]
Moreover, pediatric HAART options are limited in SSA, urging the quest for a wider therapeutic portfolio.[3,8] Although not yet approved for under 16 years, the CCR5 antagonist—maraviroc—might represent a suitable antiretroviral alternative for children,[18] pending proof-of-concept towards relevant pediatric clinical trials. Particularly, there are limited evidence on the potential effectiveness of maraviroc for SSA-children in PMTCT, initial-HAART and/or following treatment-failure.[19–21] With rising concerns of minority variants on response to several classes of antiretrovirals,[14] a genuine delineation of HIV-1 tropism, considering both minority and majority quasi-species,[22,23] could rationalize maraviroc suitability for pediatric HAART-policies in SSA.
Based on these assumptions, we aimed to ascertain DRMs and HIV-1 co-receptor usage, in majority and minority viral populations, from children according to maternal PMTCT-exposure in a resource-limited setting (RLS).
2. Study design
2.1. Sampling and setting.
A comparative study was conducted in 2015 among 18 HIV-1 vertically infected Cameroonian children, all HAART-naïve, stratified according to maternal antiretroviral exposure during pregnancy: control-group (11 children from mothers without antiretroviral exposure) versus case-group (7 children from mothers exposed to reverse transcriptase inhibitors [RTIs]). For each child, a plasma sample was collected to perform both Sanger- and 454 ultra-deep pyrosequencing (UDPS).
2.2. Sanger sequencing.
Protease (PR)/RT Sanger sequencing was performed as previously described.[24] Briefly, viral RNA was extracted from plasma using QIAamp Viral RNA minikit (Qiagen, Milan, Italy), following manufacturer's instructions. PR/RT-containing region was then reverse-transcribed and amplified using SuperScript One-Step for long templates reverse transcriptase polymerase chain reaction (RT-PCR) of Invitrogen kit (Foster City, CA), with an eventual second-round seminested PCR. Direct sequencing was then performed using 7 overlapping primers.
V3 loop Sanger sequencing was performed as previously described.[25] Briefly, viral RNA containing the V3-loop region was reverse-transcribed and amplified using an RT/Taq mix, with an eventual second-round seminested PCR. Direct sequencing was then performed using 4 overlapping primers.
2.3. Amplification of PR/RT region for UDPS
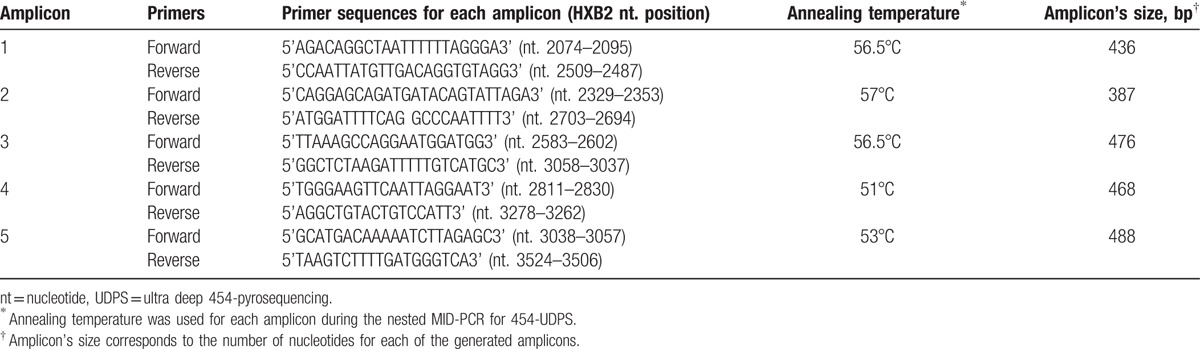
Ten milliliters of viral RNA was reverse transcribed and amplified using 1-step RT-PCR system containing 25 μL reaction mix (2×), 8 μL MgSO4 (5 mmol/L), 2.8 μL H2O DNase RNase free, 1 μL forward primer (10 μmol/L), 1 μL reverse primer (10 μmol/L), 1 μL RNase Out (40 U/μL Invitrogen) and 1.2 μL RT/TAQ, for a final volume of 50 μL. RT-PCR conditions were the following: 1 cycle 50°C, 30 minutes; 1 cycle 94°C, 2 minutes; 40 cycles (94°C, 30 seconds; 51°C, 30 seconds; 68°C, 2 minutes); a final extension 68°C, 10 minutes. Forward and reverse primers were respectively 5’GACAGGCTAATTTTTTAGGG3’ (2075–2094 bps, gag) and 5’GATAAATTTGATATGTCCATTG3’ (3555–3576bps, pol). Nested-mid PCR was then performed with the Fast Start HiFi PCR system (Roche Diagnostics, Mannheim, Germany) using 5 pairs of barcoded-modified forward and reverse primers for each amplicon (Table 1). Based on band's size from eurosafe (Euroclone) agarose gel, 31.1 μL in water diluted cDNA was mixed per tube with 3.75 μL PCR buffer (10×), 0.75 μL dNTPs (12.5%), 0.75 μL forward primer (10 μmol/L), 0.75 μL primer (10 μmol/L) and 0.4 μL Taq, under the following conditions: 1 cycle 94°C, 3 minutes; 30 cycles (94°C, 30 seconds; amplicon annealing temperature, 30 seconds; 72°C, 35 seconds); a final extension 72°C, 7 minutes.
Table 1.
UDPS primers, annealing temperatures, and amplicon's size.
2.4. Amplification of V3 loop region for UDPS.
Ten microliters viral RNA were reverse transcribed with 1-step RT-PCR system using forward (gp120, 5’CCAATTCCCATACATTATTGT3’; 49–669 bps) and reverse (gp120, 5’CTTCTCCAATTGTCCCTCA3’; 1421–1439 bps) primers, under the following conditions: 1 cycle 50°C, 30 minutes; 1 cycle 94°C, 2 minutes; 35cycles (94°C, 30 seconds; 51°C, 30 seconds; 68°C, 1 minute and 30 seconds); a final extension 68°C, 10 minutes. A nested mid-PCR was then performed with the Fast Start HiFi PCR system (Roche Diagnostics, Mannheim, Germany) as previously described.[26]
2.5. Amplicon purification and UDPS reaction.
PR/RT PCR products (5 fragments of 436, 387, 476, 468 and 488 bps) and V3 loop (one fragment of 367 bps) were purified using Agencourt AMPure PCR purification beads (Beckman Coulter, Brea, CA) and quantified with Quant-iT PicoGreen double-stranded DNA assay kit (Life Technologies, Eugene, OR) on a GloMax multidetection system (Promega, Madison, WI).
Pooled purified PCR products were clonally amplified by emulsion PCR and pyro-sequenced on the 454 GS junior platform (Roche Applied Science, Mannheimer Germany) as previously described.[26] Phylogenetic analyses excluded any possible sample contamination (data not shown).
2.6. Bioinformatics analyses of PR/RT and V3 sequences.
The entire PR (amino acid position: 1–99), RT (1–251) and the entire V3 loop (1–35) sequences obtained after 454-pyrosequencing were de-multiplexed and then quantified using the SFF tool Roche. Using a home-made Perl script and SHORAH package 0.5.1, sequences were filtered and corrected for homopolymeric region-associated errors and aligned against HIV-1 consensus B. Final alignments were manually checked for insertion or deletion in homopolymeric regions that could result in a frame shift. Nucleotidic/aminoacidic variants were evaluated and quantified by a home-made pearl script, and sequences were considered reliable when showed an intra-patient frequency ≥1% in both forward and reverse strands.
2.7. HIV drug resistance interpretation and viral-tropism determination.
PR/RT DRMs and HIV-1 co-receptor usage were interpreted using Stanford HIVdb list (updated March 9, 2015, available at http://hivdb.stanford.edu/pages/download/resistanceMutations_handout.pdf) and geno2pheno.v2.5 (http://coreceptor.geno2pheno.org/), respectively. Using a quantitative interpretation, viruses were considered CXCR4-tropic (X4-variants) by UDPS when ≥2% viral species had a false-positive rate (FPR) ≤3.5%,[27] or by Sanger sequencing when FPR was ≤10%, describing the probability of classifying an R5-virus falsely as an X4-variant.[25]
2.8. HIV-1 subtyping
Subtyping was performed through phylogenetic analysis, by aligning all PR/RT Sanger-sequences in Bio-Edit compared to reference sequences of HIV-1 subtypes and circulating recombinant forms (CRFs) available at http://www.hiv.lanl.gov as previously described.[28]
2.9. Statistical analysis
HIV-1 DRMs and coreceptor usage were compared between the two PMTCT-groups. Coreceptor results by Sanger sequencing and UDPS were considered concordant if viral-tropism was identical from both sequencing technologies. Viral-tropism was explored according to age and CD4 count.
All statistical analyses were performed using the statistical open source environment R.v.3.1.1. P values <.05 were considered statistically significant.
2.10. Ethical considerations.
Ethical clearance was obtained from the Cameroon National Ethics Committee (Ref. N°034/NEC/SE), proxy-informed consent was provided, unique identifiers were used for privacy and confidentiality, and a material transfer agreement was established.
3. Results
3.1. Characteristics of children analyzed.
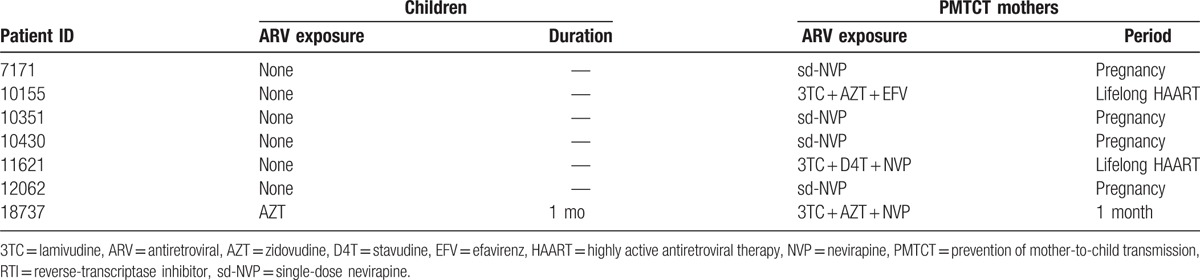
Overall, median (interquartile range [IQR]) age, viremia, and CD4 count were 6 (4–10) years, 5.5 (4.9–6.0) log10 copies/mL, and 526 (282–645) cells/mm3, respectively, without any significant difference between the 2 groups (data not shown). In the control, neither children nor their mothers had any antiretroviral exposure. Antiretroviral history of children belonging to the case-group, considered at higher risk of HIVDR, is described in Table 2.
Table 2.
Antiretroviral history of children with PMTCT exposure.
3.2. HIV-1 subtype distribution.
HIV-1 subtyping revealed 50% CRF02_AG (9/18), 33.3% F (6/18), 11.1% CRF01_AE (2/18), and 5.6% CRF11.cpx (1/18).
3.3. HIV-1 drug resistance in the children analyzed.
PR/RT sequences were successfully obtained both through Sanger sequencing and UDPS for 17/18 children. The median UDPS coverage was of 1642 (IQR: 1269–5193) reads. In the entire covered PR/RT regions, the 2 sequencing technologies showed total concordance in variants detection, and all UDPS variants with frequencies <20% were not detected by Sanger sequencing (Table 3).
Table 3.
HIV-1 DRMs according to sequencing technologies: 454 UDPS versus Sanger sequencing∗.
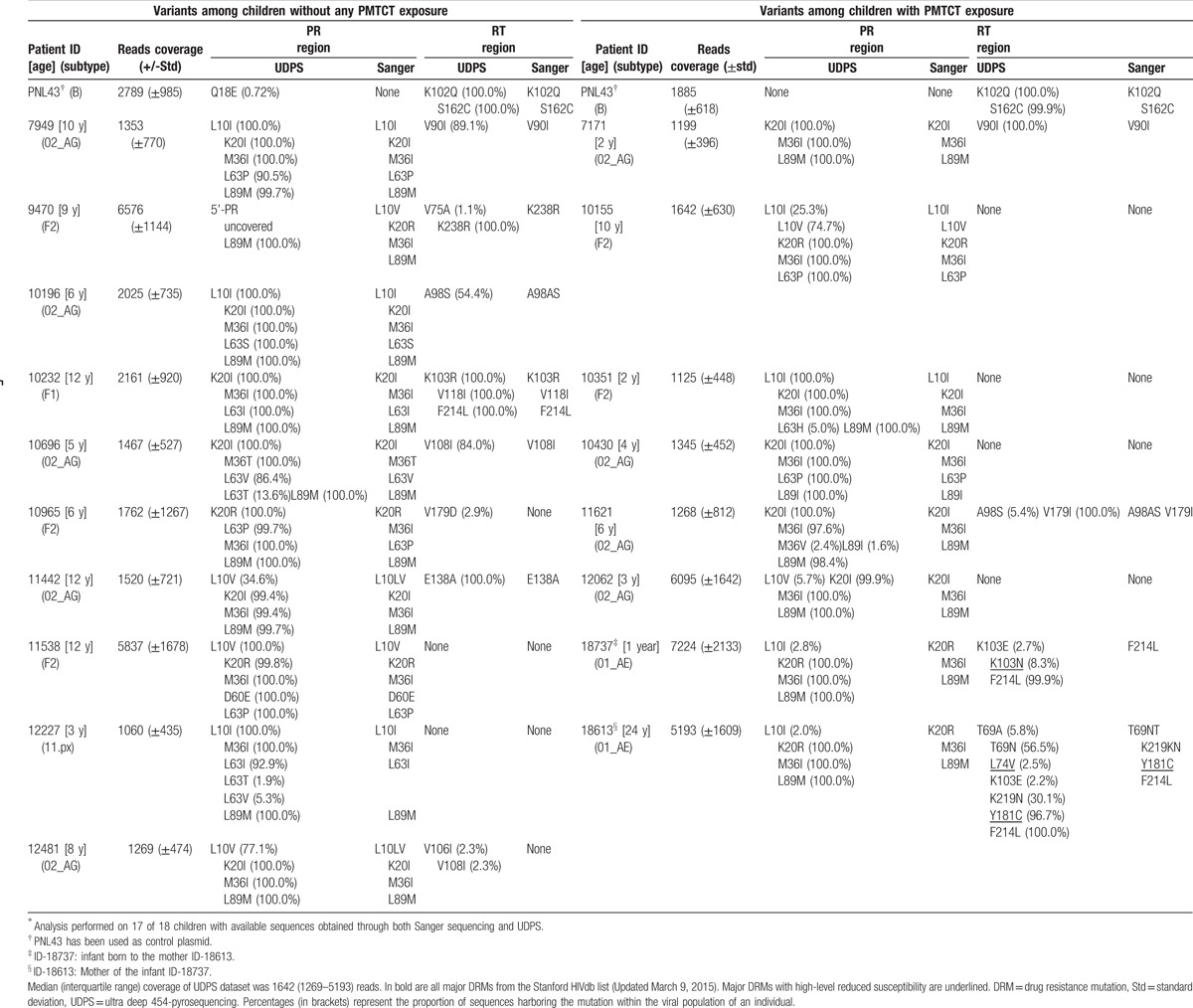
By using Sanger sequencing, all 17 children had a wild type virus. Only E138A (5.9%), an accessory polymorphism weakly selected under etravirine (ETR) and rilpivirine (RPV), was found in a child aged 8 years from the control group.
By using UDPS, 1 (aged 1 year) of 7 children (14.3%) from the case-group harbored viruses with K103N (8.3% prevalence; mutational load: 190,567 copies/mL), a nonpolymorphic mutation causing high-level resistance to NVP and efavirenz (EFV). This infant was born from an RTI-treated mother (AZT + 3TC + NVP). Thus, Sanger sequencing and UDPS were performed also for the mother (ID-18613). UDPS revealed a virus harboring 2 major DRMs: L74 V at minority-level (2.5%), causing high- and intermediate-level resistance respectively to didanosine and to ABC; Y181C at population-level (96.7%), causing high- and intermediate-level resistance respectively to NVP and to EFV, ETR, and RPV (Table 3). No minority DRMs were found in any of all other 6 children from the case-group.
In the control-group, UDPS detected V179D at minority-level (2.9%), a polymorphic accessory mutation selected under EFV, in a child aged 6 years (Table 3).
Other variants, found even at RTI-associated drug resistance positions, were with minimal or no effect on drug susceptibility or virological response. Of note, in either group, no major DRMs to ritonavir-boosted protease inhibitors (PI/r) were found by both Sanger sequencing and UDPS.
3.4. HIV-1 co-receptor tropism in the children analyzed.
V3 loop sequencing was successful by both Sanger sequencing and UDPS for 15 of 18 children and the mother ID-18613, with an overall viral-tropism concordance of 87.5% (14/16) between Sanger sequencing and UDPS (Table 4).
Table 4.
Viral-tropism according to sequencing technologies.
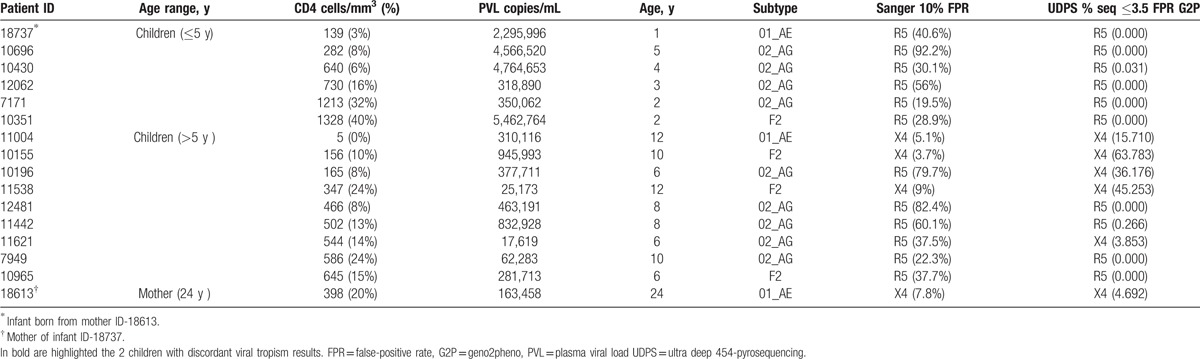
X4-tropic viruses were found in 5 of 15 (33.3%) children (including 2 cases detected only by UDPS), all aged above 5 years. Specifically, in 1 child (ID-11621) UDPS provided an added value in tropism-determination compared to Sanger sequencing. Indeed, a clinically relevant quantity of minority X4-tropic variants (frequency: 3.9%) was detected by UDPS in this child (low mutational load: 679 copies/ml). In another child (ID-10196), despite an R5-tropism (FPR = 79.7%) determined by Sanger sequencing, a discordant tropism was observed through UDPS with a high percentage of X4-tropic variants (36.2%, high mutational load: 136,641 copies/mL), because of insertions detected only at minority levels.
Of relevance, the rate of X4-tropic viruses was 0% (0/6) among children under 5 years (also as minority species at 1% the threshold), and became significantly higher as from 5 years and above (55.6% [5/9], P = .040). As expected, X4-tropic viruses were higher with CD4 ≤15% (4/9 [44.4%]) versus CD4 >15% (1/6 [16.7%], P = .580); similarly for CD4 ≤200 (3/4 [75%]) versus CD4 >200 (2/11 [18.2%] cells/mm3, P = .077). No statistical difference was found in X4-variants between the 2 PMTCT-groups: 2 of 7 (28.6%) case group versus 3 of 8 (37.5%) control group, P = 1.000.
4. Discussion
Sustaining HAART success remains challenging for children in a long term, especially in a context where adherence and drug options are limited.[2,4,5] Thus, novel strategies are required to limit the spread of preventable HIVDR and provide alternative therapeutics with utmost potency for SSA children.[29,30]
In this high CRF02_AG-infected population,[6,9,31,32] HAART-naïve children appeared with wild-type viruses at population-levels, confirming the low-level of HIVDR previously reported of this target-group.[9,33] Interestingly, a vertically transmitted minority DRM (K103N), known to be associated with resistance to NNRTIs used both for PMTCT and first-line HAART in SSA, was found in a PMTCT-exposed infant, thus suggesting NNRTI-sparing regimens for such children.[7,30,34] Discrepancy in DRMs between mother and infant would be due to sample collection later after delivery (at the moment of infant HIV diagnosis), with possible selection following prophylaxis/breastfeeding; as previously reported in similar RLS (Kyela, Tanzania).[35] This infant (aged 1 year), compared to the median age of the study population (6 years), suggests that circulating DRMs might have fade-up with increasing age.[7,33] NNRTI mutations (E138A and V179D), found in children without PMTCT-exposure, are known as polymorphisms with little or no effect on drug susceptibility or virological response.[29] The ability of NGS in excluding minority RTI-mutations (in all but one children) supports the safe use of NNRTIs/NRTIs in those without such mutations, thus sparing from inappropriate switch to PI/r- or integrase inhibitor-containing regimens.[7,8,13,17,33–35]
Coreceptor usage in these children provides a clue for clinical application. Indeed, X4-variants appeared to be associated with older ages and lower CD4 cells, suggesting limited vertical transmission by CXCR4-tropic viruses, and later appearance of X4-variants with chronicity, immunological impairment,[36,37] as well as a baseline FPR <60 as previously demonstrated.[38,39] Further investigations might help in establishing novel public health strategies for an eventual usage of maraviroc in children.[18,40] As current PMTCT-practice might not be an independent factor for viral-tropism (i.e., similar distribution in X4-variants irrespective of PMTCT-history), CCR5-antagonist (maraviroc) could be a useful therapeutic weapon for pediatric HAART.[15,18,40]
Of the two children showing discordant results between the two sequencing techniques, the added value of UDPS in detecting X4-tropic minority variants is in accordance with previous reports.[13,39] Interestingly, by detecting minority insertions associated with a complete discrepant result on Sanger sequencing, UDPS appears very useful in validating tropism determination for non-B subtypes.[41]
Therefore, UDPS might provide additional information in detecting DRMs and viral-tropism, confirming the added value of this technology for both clinical diagnostics and management of non-B HIV-infected children.[21,22,41]
In spite of this added value of UDPS, implementing NGS is more challenging in RLS (costs, technical complexity, maintenance), suggesting the need for simpler and affordable approaches integrating minority variants (point-of-care or pragmatic sequencing).[42,43]
A potential study limitation could be the relatively small sample size, which makes the study probability relatively large. Also, in the PMTCT-exposed group, only 3 of 7 were exposed to triple ART, calling for subsequent investigations with scale-up of option B+. Moreover, HIV-1 variants were investigated only in plasma compartment, suggesting the need for exploring HIV variability in several compartments (cellular reservoirs, central nervous systems, among others) and the impact on treatment and monitoring strategies in SSA.[12,13,44–46] This study therefore provides relevant data to be used as base for further/enlarged studies.
In a nutshell, NGS could help in identifying PMTCT-exposed children harboring minority NNRTI-DRMs, therefore serving for a timely switch of treatment and limiting failure rate. NGS also reveals a possible absence of X4-variants among children below 5 years, thus suggesting possible public health approaches using maraviroc. These preliminary evidences, generated on a small sample of mainly CRF02_AG-infected individuals, merit further investigations for improved pediatric-HAART strategies in RLS.
Author contributions
Conceptualization: A. Nanfack, C-F. Perno, C. Fokunang, C. Tangimpundu, D. Armenia, D. Takou, E. Temgoua, F. Ceccherini-Silberstein, G. Cappelli, J. Fokam, M-M. Santoro, P. Koki, V. Colizzi.
Data curation: D. Armenia, F. Ceccherini-Silberstein, J. Fokam, L. Carioti, M. Bellocchi, M-M. Santoro.
Formal analysis: D. Armenia, F. Ceccherini-Silberstein, J. Fokam, L. Carioti, M. Bellocchi, M-M. Santoro.
Funding acquisition: C-F. Perno, J. Fokam, V. Colizzi.
Investigation: A. Ndjolo, A. Nanfack, C-F. Perno, C. Fokunang, C. Tangimpundu, D. Takou, E. Temgoua, F. Ceccherini-Silberstein, G. Cappelli, J. Fokam, J. Torimiro, M-M. Santoro, P. Koki, V. Colizzi.
Methodology: D. Armenia, D. Takou, F. Continenza, F. Ceccherini-Silberstein, J. Fokam, L. Carioti, M. Bellocchi, M-M. Santoro.
Project administration: A. Ndjolo, C-F. Perno, C. Fokunang, F. Ceccherini-Silberstein, G. Cappelli, J. Fokam, J. Torimiro, P. Koki, V. Colizzi.
Resources: A. Ndjolo, C-F. Perno, J. Fokam, V. Colizzi.
Software: D. Armenia, D. Takou, F. Continenza.
Supervision: A. Ndjolo, C-F. Perno, C. Fokunang, F. Ceccherini-Silberstein, G. Cappelli, M-M. Santoro, P. Koki, V. Colizzi.
Validation: A. Ndjolo, C-F. Perno, C. Fokunang, C. Tangimpundu, D. Armenia, E. Temgoua, F. Continenza, F. Ceccherini-Silberstein, G. Cappelli, J. Fokam, J. Torimiro, L. Carioti, M. Bellocchi, M-M. Santoro, P. Koki, V. Colizzi.
Visualization: A. Nanfack, C-F. Perno, C. Tangimpundu, D. Takou, E. Temgoua, F. Continenza, J. Fokam, M-M. Santoro.
Writing – original draft: C-F. Perno, F. Ceccherini-Silberstein, J. Fokam, M-M. Santoro.
Writing – review & editing: A. Ndjolo, A. Nanfack, C. Fokunang, C. Tangimpundu, D. Armenia, D. Takou, E. Temgoua, F. Continenza, G. Cappelli, J. Torimiro, L. Carioti, M. Bellocchi, P. Koki, V. Colizzi.
Acknowledgements
We are appreciative to our institutional staff that participated locally in the enrolment and in sample processing. We thank Domenico Di Carlo for statistical analyses.
Footnotes
Abbreviations: 3TC = lamivudine, ABC = abacavir, AZT = zidovudine, DRMs = drug resistance mutations, EFV = efavirenz, ETR = etravirine, HAART = highly active antiretroviral therapy, HIV-1 = human immunodeficiency virus type 1, HIVDR = HIV-1 drug-resistance, NGS = next-generation sequencing, NNRTI = non-nucleoside reverse transcriptase inhibitors, NRTI = nucleoside reverse transcriptase inhibitors, NVP = nevirapine, PCR = polymerase chain reaction, PI/r = protease inhibitors boosted with ritonavir, PMTCT = prevention of mother-to-child transmission, PR = Protease, RLS = resource-limited setting, RPV = rilpivirine, RT = reverse transcriptase, RT-PCR = reverse transcriptase polymerase chain reaction, Sd-NVP = single dose nevirapine, SSA = sub-Saharan Africa, UDPS = ultra-deep 454-pyrosequencing, VF = virological failure.
Funding: This work was financially supported by the University of Rome Tor Vergata (F3-2014-0017/MIUR-FIRB-RBAP11YS7K_001-CUP: E81J11000550001), the AVIRALIA Foundation (unrestricted grant), the HIV Research Trust (HIVRT-081), and the PRD College (HEALTH-F3-2009-223581).
The authors report no conflicts of interest.
References
- [1].Joint United Nations Programme on HIV/AIDS. A progress report on the Global Plan towards the elimination of new HIV infections among children by 2015 and keeping their mothers alive. UNAIDS (2012). Available at: http://www.zero-hiv.dreamhosters.com/wp-content/uploads/2012/08/UNAIDS_ProgressReportGlobalPlan_FINAL_July17_Web.pdf. [Google Scholar]
- [2].World Health Organisation. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: Recommendations for a public health approach. Geneva: World Health Organisation (2013). Available at: http://www.who.int/hiv/pub/guidelines/arv2013/en/. [PubMed] [Google Scholar]
- [3].Joint United Nations Programme on HIV/AIDS. Access to Antiretroviral Therapy in Africa: Status Report on Progress Towards the 2015 Targets. UNAIDS (2013). Available at: http://www.unaids.org/sites/default/files/media_asset/20131219_AccessARTAfricaStatusReportProgresstowards2015Targets_en_1.pdf. [Google Scholar]
- [4].Fokam J, Billong SC, Bissek AC, et al. Declining trends in early warning indicators for HIV drug resistance in Cameroon from 2008–2010: lessons and challenges for low-resource settings. BMC Public Health 2013;13:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sigaloff KC, Hamers RL, Menke J, et al. Early warning indicators for population-based monitoring of HIV drug resistance in 6 African countries. Clin Infect Dis 2012;54(suppl 4):294–9. [DOI] [PubMed] [Google Scholar]
- [6].Ceccarelli L, Salpini R, Moudourou S, et al. Characterization of drug resistance mutations in naïve and ART-treated patients infected with HIV-1 in Yaounde, Cameroon. J Med Virol 2012;84:721–7. [DOI] [PubMed] [Google Scholar]
- [7].Chakanyuka-Musanhu CC, Penazzato M, Apollo T, et al. World Health Organization HIV drug resistance surveillance in children less than 18 months newly diagnosed with HIV in Zimbabwe. Paper presented at: The 7th International AIDS Society Conference on HIV Pathogenesis, Treatment and Prevention; 2013 June 30–July 3; Kuala Lumpur, Malaysia. Available at: http://pag.ias2013.org/Abstracts.aspx?AID=2198. [Google Scholar]
- [8].Boerma RS, Boender TS, van Hensbroek MB, et al. Sequencing paediatric antiretroviral therapy in the context of a public health approach. J Int AIDS Soc 2015;18(7 suppl 6):20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fokam J, Salpini R, Santoro MM, et al. Drug resistance among drug-naive and first-line antiretroviral treatment-failing children in Cameroon. Pediatr Infect Dis J 2011;30:1062–8. [DOI] [PubMed] [Google Scholar]
- [10].Nicot F, Sauné K, Raymond S, et al. Minority resistant HIV-1 variants and the response to first-line NNRTI therapy. J Clin Virol 2015;62:20–4. [DOI] [PubMed] [Google Scholar]
- [11].Samuel R, Paredes R, Parboosing R, et al. Minority HIV-1 drug-resistant mutations and prevention of mother-to-child transmission: perspectives for resource-limited countries. AIDS Rev 2014;16:187–98. [PubMed] [Google Scholar]
- [12].Moscona R, Ram D, Wax M, et al. Comparison between next-generation and Sanger-based sequencing for the detection of transmitted drug-resistance mutations among recently infected HIV-1 patients in Israel, 2000–2014. J Int AIDS Soc 2017;20:21846.doi: 10.7448/IAS.20.1.21846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fisher RG, Smith DM, Murrell B, et al. Next generation sequencing improves detection of drug resistance mutations in infants after PMTCT failure. J Clin Virol 2015;62:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Quiñones-Mateu ME, Avila S, Reyes-Teran G, et al. Deep sequencing: becoming a critical tool in clinical virology. J Clin Virol 2014;61:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].World Health Organisation. Meeting report on HIV/AIDS: Paediatric ARV Drug Optimization 2. World Health Organisation (2014). Available at: http://www.who.int/hiv/pub/meetingreports/paediatric-arv-optimization/en/. [Google Scholar]
- [16].PENPACT-1 (PENTA 9/PACTG 390) Study Team. First-line antiretroviral therapy with a protease inhibitor versus nonnucleoside reverse transcriptase inhibitor and switch at higher versus low viral load in HIV-infected children: An open-label, randomised phase 2/3 trial. Lancet Infect Dis 2011;11:273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].MacLeod IJ, Rowley CF, Thior I, et al. Minor resistant variants in nevirapine-exposed infants may predict virologic failure on nevirapine-containing ART. J Clin Virol 2010;48:162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gulick RM, Lalezari J, Goodrich J, et al. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med 2008;359:1429–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mbondji-Wonje C, Ragupathy V, Zhao J, et al. Genotypic prediction of tropism of highly diverse HIV-1 strains from Cameroon. PLoS One 2014;9:e112434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Church JD, Huang W, Mwatha A, et al. HIV-1 tropism and survival in vertically infected Ugandan infants. J Infect Dis 2008;197:1382–8. [DOI] [PubMed] [Google Scholar]
- [21].Lehmann C, Däumer M, Boussaad I, et al. Stable coreceptor usage of HIV in patients with ongoing treatment failure on HAART. J Clin Virol 2006;37:300–4. [DOI] [PubMed] [Google Scholar]
- [22].Surdo M, Alteri C, Puertas MC, et al. Effect of maraviroc on non-R5 tropic HIV-1: refined analysis of subjects from the phase IIb study A4001029. Clin Microbiol Infect 2015;21:103. [DOI] [PubMed] [Google Scholar]
- [23].St John EP, Simen BB, Turenchalk GS, et al. A follow-up of the multicenter collaborative study on HIV-1 drug resistance and tropism testing using 454 ultra deep pyrosequencing. PLoS One 2016;11:e0146687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fokam J, Salpini R, Santoro MM, et al. Performance evaluation of an in-house human immunodeficiency virus type-1 protease-reverse transcriptase genotyping assay in Cameroon. Arch Virol 2011;156:1235–43. [DOI] [PubMed] [Google Scholar]
- [25].Svicher V, D’Arrigo R, Alteri C, et al. Performance of genotypic tropism testing in clinical practice using the enhanced sensitivity version of Trofile as reference assay: results from the OSCAR Study Group. New Microbiol 2010;33:195–206. [PubMed] [Google Scholar]
- [26].Alteri C, Surdo M, Bellocchi MC, et al. Incomplete APOBEC3G/F Neutralization by HIV-1 Vif mutants facilitates the genetic evolution from CCR5 to CXCR4 usage. Antimicrob Agents Chemother 2015;59:4870–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Swenson LC, Mo T, Dong WW, et al. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J Infect Dis 2011;203:237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Santoro MM, Fabeni L, Armenia D, et al. Reliability and clinical relevance of the HIV-1 drug resistance test in patients with low viremia levels. Clin Infect Dis 2014;58:1156–64. [DOI] [PubMed] [Google Scholar]
- [29].World Health Organisation. HIV Drug Resistance Report 2012. World Health Organisation (2012). Available at http://www.who.int/hiv/pub/drugresistance/report2012/en/. [Google Scholar]
- [30].Paredes R, Marconi VC, Lockman S, et al. Impact of antiretroviral drugs in pregnant women and their children in Africa: HIV resistance and treatment outcomes. J Infect Dis 2013;207(suppl 2):93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Véras NM, Santoro MM, Gray RR, et al. Molecular epidemiology of HIV type 1 CRF02_AG in Cameroon and African patients living in Italy. AIDS Res Hum Retrovir 2011;27:1173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Billong SC, Fokam J, Aghokeng AF, et al. Population-based monitoring of emerging HIV-1 drug resistance on antiretroviral therapy and associated factors in a sentinel site in Cameroon: low levels of resistance but poor programmatic performance. PLoS One 2013;8:e72680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kityo C, Sigaloff KC, Boender TS, et al. HIV drug resistance among children initiating first-line antiretroviral treatment in Uganda. AIDS Res Hum Retroviruses 2016;32:628–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Palumbo P, Lindsey JC, Hughes MD, et al. Antiretroviral treatment for children with peripartum nevirapine exposure. N Engl J Med 2010;363:1510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hauser A, Sewangi J, Mbezi P, et al. Emergence of minor drug-resistant HIV-1 variants after triple antiretroviral prophylaxis for prevention of vertical HIV-1 transmission. PLoS One 2012;7:e32055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Foster C, Kaye S, Smith C, et al. HIV-1 co-receptor tropism and disease progression in children and young adults with perinatally acquired HIV-1 infection. The HICCUP Study. J Virus Erad 2015;1:173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Casper C, Navér L, Clevestig P, et al. Coreceptor change appears after immune deficiency is established in children infected with different HIV-1 subtypes. AIDS Res Hum Retroviruses 2002;18:343–52. [DOI] [PubMed] [Google Scholar]
- [38].Casper CH, Clevestig P, Carlenor E, et al. Link between the X4 phenotype in human immunodeficiency virus type 1-infected mothers and their children, despite the early presence of R5 in the child. J Infect Dis 2002;186:914–21. [DOI] [PubMed] [Google Scholar]
- [39].Svicher V, Cento V, Rozera G, et al. The genotypic false positive rate determined by V3 population sequencing can predict the burden of HIV-1 CXCR4-using species detected by pyrosequencing. PLoS One 2013;8:e53603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Palladino C, Gómez ML, Soler-Palacín P, et al. Off-label use of maraviroc in HIV-1-infected paediatric patients in clinical practice. AIDS 2015;29:2155–9. [DOI] [PubMed] [Google Scholar]
- [41].Cashin K, Gray LR, Harvey KL, et al. Reliable genotypic tropism tests for the major HIV-1 subtypes. Sci Rep 2015;5:8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Alteri C, Santoro MM, Abbate I, et al. ‘Sentinel’ mutations in standard population sequencing can predict the presence of HIV-1 reverse transcriptase major mutations detectable only by ultra-deep pyrosequencing. J Antimicrob Chemother 2011;66:2615–23. [DOI] [PubMed] [Google Scholar]
- [43].Nanfack AJ, Agyingi L, Noubiap JJ, et al. Use of amplification refractory mutation system PCR assay as a simple and effective tool to detect HIV-1 drug resistance mutations. J Clin Microbiol 2015;53:1662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zaccarelli M, Santoro MM, Armenia D, et al. Genotypic resistance test in proviral DNA can identify resistance mutations never detected in historical genotypic test in patients with low level or undetectable HIV-RNA. J Clin Virol 2016;82:94–100. [DOI] [PubMed] [Google Scholar]
- [45].Raymond S, Saliou A, Delobel P, et al. Evolution of HIV-1 quasispecies and coreceptor use in cell reservoirs of patients on suppressive antiretroviral therapy. J Antimicrob Chemother 2014;69:2527–30. [DOI] [PubMed] [Google Scholar]
- [46].Fabeni L, Berno G, Svicher V, et al. Genotypic tropism testing in HIV-1 proviral DNA can provide useful information at low-level viremia. J Clin Microbiol 2015;53:2935–41. [DOI] [PMC free article] [PubMed] [Google Scholar]


