Abstract
Purpose
Mosaicism probably represents an underreported cause of genetic disorders due to detection challenges during routine molecular diagnostics. The purpose of this study was to evaluate the frequency of mosaicism detected by next-generation sequencing in genes associated with epilepsy-related neurodevelopmental disorders.
Methods
We conducted a retrospective analysis of 893 probands with epilepsy who had a multigene epilepsy panel or whole-exome sequencing performed in a clinical diagnostic laboratory and were positive for a pathogenic or likely pathogenic variant in one of nine genes (CDKL5, GABRA1, GABRG2, GRIN2B, KCNQ2, MECP2, PCDH19, SCN1A, or SCN2A). Parental results were available for 395 of these probands.
Results
Mosaicism was most common in the CDKL5, PCDH19, SCN2A, and SCN1A genes. Mosaicism was observed in GABRA1, GABRG2, and GRIN2B, which previously have not been reported to have mosaicism, and also in KCNQ2 and MECP2. Parental mosaicism was observed for pathogenic variants in multiple genes including KCNQ2, MECP2, SCN1A, and SCN2A.
Conclusion
Mosaic pathogenic variants were identified frequently in nine genes associated with various neurological conditions. Given the potential clinical ramifications, our findings suggest that next-generation sequencing diagnostic methods may be utilized when testing these genes in a diagnostic laboratory.
Keywords: epilepsy, genetic testing, mosaicism, next-generation sequencing, neurodevelopmental disorders
Introduction
The development and adoption of next-generation sequencing (NGS) based diagnostics have revolutionized the ability to simultaneously analyze multiple genes in an accurate and efficient manner. Compared with traditional sequencing methods, the digital nature of NGS technology allows quantification of sequence variants such that a high read depth can enable the detection of low-level mosaic variants. As diagnostic NGS is increasingly used for sequence-based variant detection, there is mounting evidence that somatic and gonosomal mosaicism are more common and play a greater role in human genetic disorders than previously recognized. In affected patients, enhanced detection of somatic mosaicism can provide a definitive molecular diagnosis to patients who may have received negative results from less sensitive testing and can also directly affect disease management.1 Additionally, the detection and reporting of mosaicism is relevant to the families of probands for accurate estimates of recurrence risk. When mosaicism is detected in apparently healthy parents of an affected child, it is essential for assigning a more accurate recurrence risk, where that risk would increase from negligible for a true de novo variant to up to the published theoretical risk of 33%.2
Mosaicism can be classified into three types depending on the affected stage of development and the affected tissue(s): germ-line mosaicism, somatic mosaicism, and gonosomal mosaicism (a combination of germ-line and somatic mosaicism).3 Owing to limitations in tissue sampling, it is difficult to precisely identify the type of mosaicism in all situations. Several genetic disorders often involve mosaicism, for example, neurofibromatosis type 1, Proteus syndrome, linear nevus sebaceous syndrome, McCune–Albright syndrome, Beckwith–Wiedemann syndrome, and tuberous sclerosis complex. Based on case reports, mosaicism also plays a role in other types of genetic disorders such as neurodevelopmental and epilepsy-related disorders.4, 5, 6, 7, 8, 9, 10, 11
To date, the frequency of mosaicism in neurodevelopmental disorders has been evaluated mostly in research studies of individuals with disorders such as autism and intellectual disability.12, 13 A systematic study of the role of mosaicism in epilepsy or other neurodevelopmental disorders, especially in a clinical diagnostic setting, is currently lacking. Although it has been hypothesized that the frequency of mosaicism in certain epilepsy-related genes may be higher than previously appreciated, no information is available regarding the overall frequency of mosaic pathogenic variants in specific genes or disorders associated with epilepsy.4, 5, 6, 7, 8, 9, 10, 11 Additionally, parental mosaicism has been reported in some epilepsy-related genes. For example, a high incidence (at least 7%) of parental mosaicism has been detected by direct sequencing in families with inherited SCN1A-related epilepsies. In these families, the parents included individuals who were asymptomatic, mildly affected, and severely affected, though this type of clinical information for other genes may not be available.14
In this study, we report for the first time the extent and level of mosaicism detectable by multigene or whole-exome NGS for patients with epilepsy undergoing testing in a clinical diagnostic laboratory.
Materials and methods
Patient population
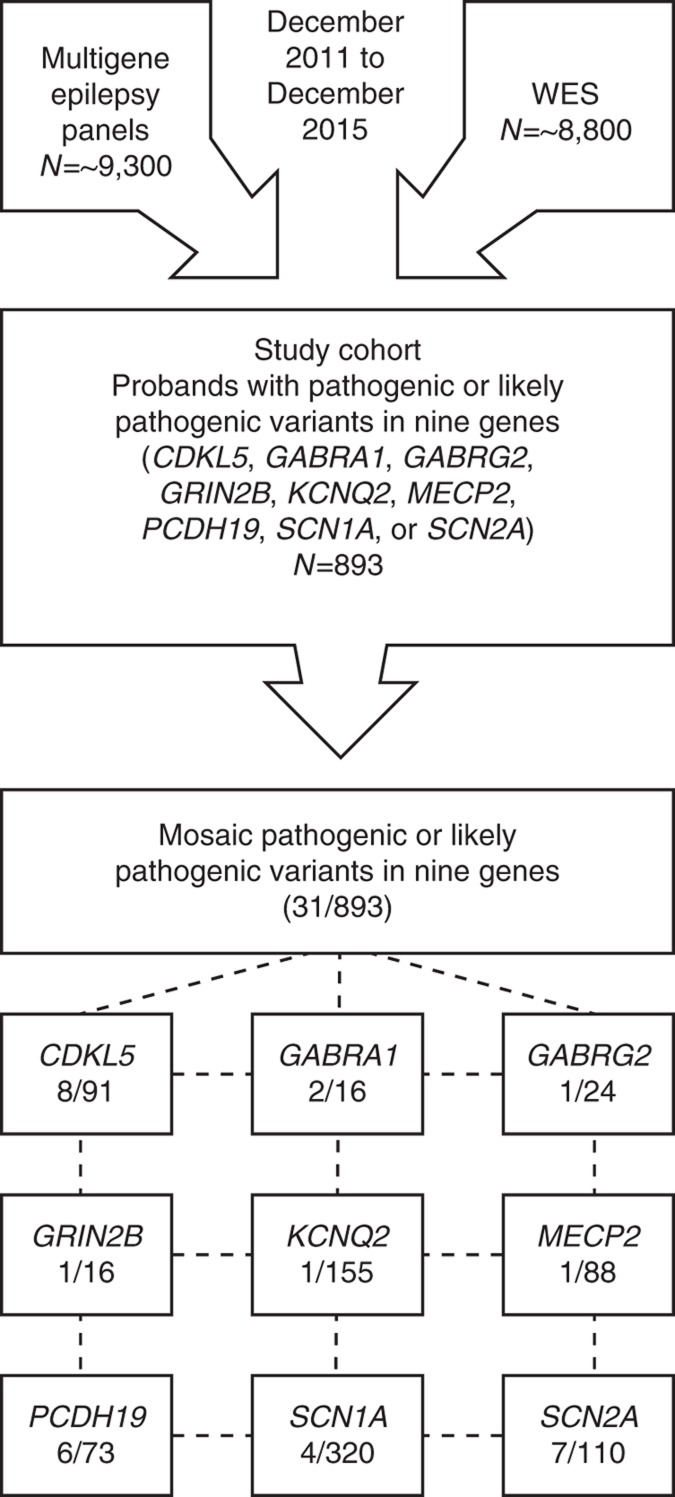
The cohort included 893 probands, referred from December 2011 to December 2015, who had undergone NGS testing for either a panel of up to 70 epilepsy-related genes or whole-exome sequencing (WES) and were found to be positive for a pathogenic or likely pathogenic variant in one of the following genes: CDKL5, GABRA1, GABRG2, GRIN2B, KCNQ2, MECP2, PCDH19, SCN1A, or SCN2A (Figure 1). These nine genes were selected because mosaic pathogenic and likely pathogenic variants were observed in one or more probands with epilepsy by NGS and frequencies of mosaicism for these genes have not been previously reported, to our knowledge. In addition to epilepsy, clinical indications for testing often included other neurodevelopmental features such as developmental delay, intellectual disability, learning disability, developmental regression, autism spectrum disorder, behavioral/psychiatric abnormalities, hypotonia, microcephaly/macrocephaly, and movement disorders. Clinical information, family history, and ethnicity were provided using a clinical checklist and/or clinic notes submitted by the referring clinicians, who were primarily neurologists or geneticists working in private practice, private hospitals, or university-based medical centers. The majority of patients resided in the United States or Canada. In a subset of probands with heterozygous or hemizygous pathogenic or likely pathogenic variants, parental mosaicism was evaluated either by NGS or targeted Sanger sequencing. Informed consent for clinical testing was obtained by the referring clinicians.
Figure 1.
Flow chart of testing. Schematic overview of the breakdown of testing for the study cohort. WES, whole-exome sequencing.
Next-generation sequencing
A targeted capture panel (ranging from 18 to 70 genes) or WES was used to investigate genes associated with epilepsy-related disorders. For all panels, targeted regions were enriched from genomic DNA extracted from blood using either multiplex polymerase chain reaction (PCR) by Raindance Technologies (Raindance Technologies, Lexington, MA) or RNA base hybridization capture using a custom designed library and SureSelect kit (Agilent Technologies, Santa Clara, CA) following the manufacturer’s instructions. Specifically, for libraries generated with Raindance Technologies (Billerica, MA), DNA was sheared on a Covaris instrument (Woburn, MA) to produce 5-kb fragments that were purified using the Qiagen QIAQuick (Hilden, Germany) mini-elute kit. DNA fragments were merged with PCR primers in microdroplets and transferred to PCR tubes for PCR amplification. The amplified products were enzymatically fragmented and tagged with adaptors using the standard Nextera protocol (Illumina, San Diego, CA). A final PCR enrichment amplification was performed at 65°C for 15 cycles. Products were purified using the Qiagen QIAQuick mini-elute kit. For Agilent capture processing, DNA was sheared into approximately 400-bp fragments, which were then repaired, ligated to adaptors, and purified for subsequent PCR amplification. Amplified products were then captured by biotinylated RNA library baits in solution following the manufacturer’s instructions. Bound DNA was isolated with streptavidin-coated beads and reamplified.
The final library products for both methods were sequenced using the Illumina HiSeq 2000 or 2500 sequencing system with either 50-bp single-end or 100-bp paired-end reads, respectively (Illumina, San Diego, CA), following manufacturer’s instructions and described previously.15 All targeted gene regions were created as custom contigs based on RefSeq gene sequences. For Raindance, reads were mapped to only the custom contig targets, while for capture, to avoid misalignment of any genomic carryover, read alignment used the curated gene regions plus a masked version of hg19 to filter any genomic carryover. Local realignment around insertion/deletion sites was performed using the Genome Analysis Toolkit v1.6.16 Regions of interest were defined as the exon and flanking noncoding sequences. All samples were sequenced to an average coverage of at least 1200 ×. Positions within the region of interest covered at less than 15 × were compensated for by conventional Sanger DNA sequencing using an ABI 3730 (Life Technologies, Carlsbad, CA) and standard protocols. All variants called within the region of interest by any of these programs were analyzed, and all novel variants that appeared real based on manual inspection of NGS alignments and all previously classified, nonbenign variants were confirmed by conventional Sanger DNA sequence analysis with a new DNA preparation.
For WES, genomic DNA was extracted from whole blood of affected probands and, when available, their parents and other similarly affected relatives. Next-generation sequencing was performed on exon targets captured using the Agilent SureSelect Human All Exon V4 (SS V4) or Clinical Research Exome kit (Agilent Technologies) to an average target depth of more than 100 ×. The sequencing methodology, data processing pipeline, and variant filtering and interpretation protocol have been described previously.17 All clinically relevant variants were confirmed by conventional Sanger DNA sequence analysis with a new DNA preparation.
The read depth varied based on the specific NGS method, Raindance, custom capture, or whole exome sequencing (Supplementary Table S1 online and Supplementary Figure S1). Supplementary Figure S1 shows that the per-target coverage is more consistent for capture-based methods. The capture panel provides the best coverage for consistent mosaic variant calling with nearly all targets consistently covered to >100 × and most targets consistently covered to >200 ×. While the Raindance panel provides higher coverage for some targets, its coverage uniformity is poor.
Variant analysis and interpretation
Mosaicism was hypothesized when variants in autosomal genes and X-linked genes in females were observed at a lower-than-expected ratio of variant to wild-type NGS reads and exhibited unequal amplification by Sanger sequencing. For males, mosaicism was suspected for X-linked genes when both variant and wild-type alleles were observed and when there was no indication of a sex chromosome aneuploidy based on either concurrent exon-level array comparative genomic hybridization data or prior chromosome analysis by karyotype.
The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (http://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/). Variants were categorized using the five-tier classification system recommended by the American College of Medical Genetics and Genomics. Clinical variant interpretation was conducted in the context of the proband’s phenotype when available. Likely pathogenic variants have a high probability of being pathogenic. Pathogenic and likely pathogenic variants were used for the analysis of this manuscript. For the sake of simplicity, all of the qualifying variants are referred to as “pathogenic” in this manuscript, although many are classified in ClinVar as “likely pathogenic.”
Results
Mosaicism in probands
Molecular diagnostic testing for epilepsy-related disorders utilizing NGS yielded positive diagnostic results in 893 probands, including pathogenic variants in nine genes analyzed (CDKL5, GABRA1, GABRG2, GRIN2B, KCNQ2, MECP2, PCDH19, SCN1A, and SCN2A) (Figure 1). For the multigene NGS panels, more than 9,300 probands were tested in the specified time period, and these nine genes accounted for approximately 57% of the overall positive cases. Among all positive cases identified through the multigene NGS panels, missense variants accounted for the majority (52%) of pathogenic variants detected; frameshift accounted for 23%, nonsense 14%, splice 10%, and insertion/deletion 1%. Among probands with positive results in these nine genes (n = 893, Supplementary Figure S1), 31 demonstrated a lower-than-expected ratio of variant to wild-type NGS reads, which is indicative of mosaicism in the nine selected genes. The mean read depth at the site of pathogenic changes was 1,299 (range: 42–3,444 reads) (Table 1). The level of observed mosaicism ranged from 9 to 40% of sequencing reads in probands with autosomal dominant disorders and females with X-linked disorders. Males with an X-linked disorder had mosaic variants in 8–87% of sequencing reads (Table 1). In this study, the breakdown of the types of mosaic variants in these nine genes were missense (68%), nonsense (16%), frameshift (10%), and splice site (6%) variants (Table 1). The mean read depth across the nine genes with mosaic pathogenic variants is provided in Supplementary Table S1 for each of the different testing methods. Density plots of per-sample, per-target coverage for each of the four methods are illustrated in Supplementary Figure S1.
Table 1. Cases with mosaic pathogenic variant(s).
| Gene | Gender | Variant (hg19 position; coding DNA; protein) | Type of variant | Testing method | % of variant reads | Coverage (read depth at variant position) |
|---|---|---|---|---|---|---|
| Autosomal dominant inheritance | ||||||
| GABRA1 | Male | chr5:161317988 T>C; c.788 T>C; p.Met263Thr | Missense | Raindance | 11.7% | 803 |
| GABRA1 | Male | chr5:161317999 C>A; c.799 C>A; p.Leu267Ile | Missense | Raindance | 18.6% | 483 |
| GABRG2 | Male | chr5:161522522 C>A; c.281 C>A; p.Thr94Lys | Missense | Raindance | 14.4% | 667 |
| GRIN2B | Male | chr12:13720105 A>C; c.2452 A>C; p.Met818Leu | Missense | Custom capture | 24.4% | 2,574 |
| KCNQ2 | Female | chr20:62044866 T>A; c.1700 T>A; p.Val567Asp | Missense | Custom capture | 30.2% | 463 |
| SCN1A | Male | chr2:166852531 C>T; c.4573 C>T; p.Arg1525Ter | Nonsense | Raindance | 9.2% | 1,021 |
| SCN1A | Male | chr2:166915131 T>A; c.332 T>A; p.Leu111Ter | Nonsense | Custom capture | 20.2% | 3,406 |
| SCN1A | Female | chr2:166903453 T>C; c.1204 T>C; p.Phe402Leu | Missense | Custom capture | 24.7% | 3,107 |
| SCN1A | Female | chr2:166929992 A>-; c.140delA; p.Asn47MetfsTer45 | Frameshift | Raindance | 26.0% | 412 |
| SCN2A | Male | chr2:166210777 G>A; c.2995 G>A; p.Glu999Lys | Missense | Raindance | 11.6% | 627 |
| SCN2A | Male | chr2:166201203 C>G; c.2701 C>G; p.Gln901Glu | Missense | Custom capture | 13.0% | 3,444 |
| SCN2A | Male | chr2:166198881 G>A; c.2464 G>A; p.Gly822Ser | Missense | WES | 14.7% | 95 |
| SCN2A | Female | chr2:166237619 T>A; c.4463 T>A; p.Ile1488Asn | Missense | Raindance | 16.8% | 1,413 |
| SCN2A | Female | chr2:166201153 T>A; c.2651 T>A; p.Leu884His | Missense | Raindance | 20.3% | 488 |
| SCN2A | Female | chr2:166165888 G>A; c.632 G>A; p.Gly211Asp | Missense | Custom capture | 22.1% | 961 |
| SCN2Aa | Male | chr2:166231223 T>C, T>A c.4001 T>C, c.4001 T>A; p.Ile1334Thr, p.Ile1334Asn | Two missense at same nucleotide position | Custom capture | 27.1% 39.5% | 3,320 3,320 |
| X-linked inheritance | ||||||
| CDKL5 | Female | chrX:18613489 C>T; c.766 C>T; p.Gln256Ter | Nonsense | Custom capture | 10.1% | 2,353 |
| CDKL5 | Male | chrX:18602452 G>A; c.533 G>A; p.Arg178Gln | Missense | WES | 14.3% | 42 |
| CDKL5 | Female | chrX:18600056 A>G; c.449 A>G; p.Lys150Arg | Missense | Raindance | 16.0% | 562 |
| CDKL5 | Male | chrX:18627690 G>A; c.2152 G>A; p.Val718Met | Missense | Raindance | 25.3% | 2,961 |
| CDKL5 | Male | chrX:18598085 C>T; c.400 C>T; p.Arg134Ter | Nonsense | Raindance | 31.1% | 380 |
| CDKL5 | Male | chrX:18622719 C>T; c.1675 C>T; p.Arg559Ter | Nonsense | WES | 36.1% | 180 |
| CDKL5 | Male | chrX:18528948 C>A; c.73 G>A; p.Gly25Arg | Missense | Raindance | 41.2% | 284 |
| CDKL5 | Male | chrX:18627686-18627687 CA>-; c.2148_2149delCA; p.Tyr716Ter | Frameshift | Raindance | 56.3% | 895 |
| MECP2 | Male | chrX:153297671 G>A; c.364 G>A; p.Val122Met | Missense | Custom capture | 31.4% | 1,797 |
| PCDH19 | Male | chrX:99661637-99661640 CTCT>-; c.1956_1959delCTCT; p.Ser653ProfsTer6 | Frameshift | Custom capture | 8.1% | 1,551 |
| PCDH19 | Female | chrX:99662962 G>C; c.634 G>C; p.Asp212His | Missense | Raindance | 12.5% | 2,556 |
| PCDH19 | Male | chrX:99605642 T>C; c.2534+2 T>C | Splice | Raindance | 22.2% | 189 |
| PCDH19 | Male | chrX:99661447 T>C; c.2147+2 T>C | Splice | Custom capture | 43.6% | 1,338 |
| PCDH19 | Male | chrX:99661723 A>G; c.1873 A>G; p.Arg625Gly | Missense | Raindance | 83.1% | 688 |
| PCDH19 | Male | chrX:99663226 G>C; c.370 G>C; p.Asp124His | Missense | Raindance | 87.2% | 1,219 |
WES, whole-exome sequencing.
Single patient is mosaic for two different nucleotide substitutions at same codon, known as double mosaicism.
The highest frequency of mosaicism among positive probands was observed for two X-linked genes, 8.8% for CDKL5 (0.088; 95% CI, 0.039–0.166) and 8.2% for PCDH19 (0.082; 95% CI, 0.031–0.170) (Table 2). Of the individuals with pathogenic variants in CDKL5, 21% (19/91) were male. Additionally, 75% (6/8) of mosaic pathogenic variants identified in CDKL5 were in affected males noted to have epilepsy and/or developmental delay, and 25% (2/8) were in affected females with epilepsy and developmental delay. Although PCDH19-related epilepsy is an X-linked dominant disorder almost exclusively affecting females, five males were found to be mosaic for a pathogenic variant in the PCDH19 gene and presented with epilepsy and/or developmental delay, similar to females with PCDH19 pathogenic variants.
Table 2. Frequency of mosaicism for each reported gene considering probands who were mosaic for a pathogenic or likely pathogenic variant in the corresponding gene.
| Gene | Disease/phenotype | Primary transcript | Percent of probands with mosaic variants | No. of mosaic variants/total no. of pathogenic and likely pathogenic variants | 95% CI | Percent of de novo variants detected by multigene panels when both parents tested by targeted dideoxy sequencing (no. of de novo variants/no. of cases tested) | Percent of de novo variants for positive cases when tested via trio-based WES |
|---|---|---|---|---|---|---|---|
| CDKL5 | Atypical RS, EIEE | NM_003159.2 | 8.8% | 8/91 | 0.039–0.166 | 100% (26/26) | 100% (6/6) |
| PCDH19 | EIEE (EFMR) | NM_001184880.1 | 8.2% | 6/73 | 0.031–0.170 | 75.9% (22/29) | 40% (2/5) |
| SCN2A | EIEE, BFNIS, GEFS+, IS, ICE | NM_021007.2 | 6.4% | 7/110 | 0.026–0.128 | 89.8% (44/49) | 95.2% (20/21) |
| SCN1A | GEFS+, ICE-GTCS, EIEE, SMEI, DS | NM_001165963.1 | 1.3% | 4/320 | 0.003–0.032 | 75.5% (74/98) | 73.7% (14/19) |
| GABRA1 | OS, DS, IS, GEFS+, JME, CAE | NM_000806.5 | 12.5% | 2/16 | 0.016–0.384 | 100% (8/8) | 100% (2/2) |
| GRIN2B | EIEE, ID | NM_000834.3 | 6.3% | 1/16 | 0.002–0.302 | 50% (1/2) | 100% (12/12) |
| GABRG2 | FS, FS with CAE, GEFS+ | NM_000816.3 | 4.2% | 1/24 | 0.001–0.211 | 73.3% (11/15) | 50% (1/2) |
| MECP2 | RS, atypical RS | NM_004992.3 | 1.1% | 1/88 | 0.0003–0.062 | 77.8% (7/9) | 81% (17/21) |
| KCNQ2 | BFNS, EIEE | NM_172107.2 | 0.6% | 1/155 | 0.001–0.035 | 81.5% (44/54) | 88.2% (15/17) |
| Total | 3.5% | 31/893 | 0.024–0.049 | 81.7% (237/290) | 84.8% (89/105) |
BFNIS, benign familial neonatal–infantile seizures; BFNS, benign familial neonatal seizures; CAE, childhood absence epilepsy; DS, Dravet syndrome; EFMR, epilepsy and mental retardation limited to females; EIEE, early infantile epileptic encephalopathy; FS, febrile seizures; GEFS+, genetic (generalized) epilepsy with febrile seizures plus; ICE, intractable childhood epilepsy; ICE-GTCS, intractable childhood epilepsy with generalized tonic–clonic seizures; ID, intellectual disability; IS, infantile spasms; JME, juvenile myoclonic epilepsy; OS, Ohtahara syndrome; RS, Rett syndrome; SMEI, severe myoclonic epilepsy of infancy; WES, whole-exome sequencing.
95% confidence interval (CI) is provided.
Mosaic pathogenic variants were observed in 6.4% of patients with pathogenic variants in SCN2A (0.064; 95% CI, 0.026–0.128) (Table 2). All patients with mosaic SCN2A pathogenic variants had epilepsy and many (5/7) were also noted to have developmental delay based on limited clinical information provided. Additionally, an unusual finding of double mosaicism, previously referred to as triple mosaicism,18 was identified in one patient who was mosaic for two different pathogenic variants at the same nucleotide position in the SCN2A gene, with each sequence change having different variant allele frequencies (Table 1). The frequency of mosaicism in SCN1A (0.013; 95% CI, 0.003–0.032) was significantly lower than in SCN2A (P < 0.01; two-tailed Fisher’s exact test) (Table 2).
Of note, we also detected mosaic pathogenic variants in several additional epilepsy-related genes including some of the GABA receptor genes, GABRA1 (0.125; 95% CI, 0.016–0.384) and GABRG2 (0.042; 95% CI, 0.001–0.211); GRIN2B (0.063; 95% CI, 0.002–0.302), which encodes the glutamate-binding NR2B subunit of the N-methyl-d-aspartate receptor; the potassium channel gene KCNQ2 (0.006; 95% CI, 0.001–0.035); and MECP2 (0.011; 95% CI, 0.0003–0.062) in a male with atypical Rett syndrome (Table 2).
These data indicate that the overall frequency of mosaic pathogenic variants in the two genes, CDKL5 and PCDH19 (0.085; 95% CI, 0.048–0.139), was significantly higher than the overall frequency of mosaic pathogenic variants in the other seven genes combined, GABRA1, GABRG2, GRIN2B, KCNQ2, MECP2, SCN1A, and SCN2A (0.023; 95% CI, 0.014–0.037), (P < 0.01; two-tailed Fisher’s exact test).
Mosaicism in parents
Parental testing was performed for probands undergoing trio-based WES and recommended for all probands with positive results identified via panel testing. For probands tested by multigene NGS panels and identified to have pathogenic variants in CDKL5, GABRA1, GABRG2, GRIN2B, KCNQ2, MECP2, PCDH19, SCN1A, or SCN2A, 290 sets of parents underwent targeted testing for the variant by Sanger sequencing. Of these, 237 (81.7%) were determined to be de novo and 53 (18.3%) were inherited (Table 2). Similarly, for probands tested by WES trio with a pathogenic variant in one of these nine genes, the overall frequency of de novo variants was 84.8% (89/105) (Table 2). For parents tested either by targeted Sanger sequencing after the proband had a positive result by multigene NGS panel or by WES trio testing, we observed that 12 parents were suspected to have mosaic variants in either SCN1A (6), SCN2A (3), KCNQ2 (2), or MECP2 (1) (Table 3). Clinical information was not provided for one parent, but it was provided for the other 11 parents. Based on the clinical information provided, 27% (3/11) of these parents were clinically affected with clinical features consistent with the disorders associated with these genes (Table 3). However, two of the three affected parents were reported to have milder or later-onset symptoms compared with their children. The remaining 73% (8/11) of the parents who carried a mosaic pathogenic variant and for whom clinical information was provided were reportedly unaffected (Table 3). In addition, we observed one case of suspected germ-line mosaicism for an SCN1A pathogenic variant in a family with two affected siblings (Table 3). This SCN1A variant was undetectable via targeted Sanger sequencing of DNA obtained from blood from both parents.
Table 3. Clinical information for cases of parental mosaicism.
| Gene | Variant (hg19 position; coding DNA; protein) | Type of variant | Percent of variant reads | Coverage (read depth at variant position) | Parent: clinical information | Parent: onset of seizures | Proband: onset of seizures |
|---|---|---|---|---|---|---|---|
| SCN1A | chr2:166929897 G>A; c.235 G>A; p.Asp79Asn | Missense | 12.6% | 495 | Unknown | Unknown | Unknown |
| KCNQ2 | chr20:62076020 C>T; c.682 C>T; p.His228Tyr | Missense | 27.1% | 266 | Unaffected | N/A | Neonatal |
| MECP2 | chrX:153296354 C>T; c.925 C>T; p.Arg309Trp | Missense | 19.9% | 540 | Unaffected | N/A | Infancy |
| SCN1A | chr2: 166909392 C>T; c.664 C>T; p.Arg222Ter | Nonsense | 6.0% | 847 | Unaffected | N/A | Infancy |
| SCN1A | chr2:166852557 C>A; c.4547 C>A; p.Ser1516Ter | Nonsense | 14.2% | 690 | Unaffected | N/A | Unknown |
| SCN1A | chr2:166929950 T>C; c.182 T>C; p.Leu61Pro | Missense | 20.2% | 1,011 | Unaffected | N/A | Infancy |
| SCN1A | chr2:166850727 C>A; c.4781 C>A; p.Ser1594Tyr | Missense | 15.8% | 38 | Unaffected | N/A | Early childhood |
| SCN1Aa | chr2:166894519 G>A; c.2713 G>A; p.Ala905Thr | Missense | N/A | N/A | Unaffected | N/A | Unknown |
| SCN2A | chr2: 166237633-166237635 GAA>-c.4477_4479delGAA; p.Glu1493del | In-frame deletion | 9.0% | 885 | Unaffected | N/A | Infancy |
| SCN2A | chr2:166245211 G>A; c.4895 G>A; p.Arg1632Lys | Missense | 16.4% | 764 | Unaffected | N/A | Neonatal |
| KCNQ2 | chr20:62073785 T>A; c.790 T>A; p.Tyr264Asn | Missense | 14.0% | 172 | Affected (mild compared with proband) | Infancy | Neonatal |
| SCN1A | chr2:166848302 T>C; c.5483 T>C; p.Leu1828Ser | Missense | 18.1% | 533 | Affected (more severe compared with proband) | Neonatal | Unknown (asymptomatic in infancy) |
| SCN2A | chr2:166245224 C>G; c.4908 C>G; p.Ile1636Met | Missense | 7.6% | 885 | Affected (mild compared with proband) | Infancy | Neonatal |
N/A, not applicable.
Suspected germ-line mosaicism: both parents were negative by targeted dideoxy sequencing and two affected offspring were heterozygous for variant.
Discussion
Mosaicism has been reported in the literature in genes associated with epilepsy and neurodevelopmental disorders. However, for most genes, information has been limited to case reports. While these publications provide evidence that mosaicism can occur in genes causing epilepsy, the frequency of mosaic findings is not known for most genes. Based on our analyses, mosaic pathogenic variants were identified at an overall frequency of 3.5% (0.035; 95% CI, 0.024–0.049) in nine genes associated with epilepsy-related disorders (Table 2). Additionally, we report on the observed occurrence of mosaic pathogenic variants in several epilepsy-related genes. The CDKL5 gene encodes a serine/threonine protein kinase that is highly expressed in the brain, mediates phosphorylation of MECP2, and is associated with variable clinical phenotypes.19, 20, 21 CDKL5-related disorders are X-linked dominant and often result in male lethality or, less commonly, in a severe phenotype with early-onset epileptic encephalopathy and intellectual disability. Mosaic pathogenic variants have been reported occasionally in affected females and males, but may be more prevalent than previously understood.4, 9, 10, 20, 22, 23 In our cohort, mosaic variants in CDKL5 account for nearly 9% of all CDKL5-positive cases. Additionally, of the males who were positive for a pathogenic variant in CDKL5, 32% were found to be mosaic.
The PCDH19 gene encodes protocadherin-19, a calcium-dependent cell–cell adhesion molecule primarily expressed in the brain. Pathogenic variants in PCDH19 cause a type of early infantile epileptic encephalopathy that is sometimes referred to as epilepsy and mental retardation limited to females. This condition follows an unusual X-linked inheritance pattern in which females are affected while males who are hemizygous for a pathogenic variant are unaffected carriers. Although the mechanism of disease is not completely understood, cellular interference has been speculated.7, 24 Mosaic pathogenic variants have been reported in affected males and females.7, 8, 11, 24, 25, 26 We identified mosaic pathogenic variants in PCDH19 in over 8% of patients with a PCDH19-related disorder. As expected based on the hypothesized dominant-negative etiology for PCDH19-related disorders, all males with a mosaic pathogenic PCDH19 variant were found to be clinically affected. Additionally, two affected males in our cohort were hemizygous for pathogenic variants in the PCDH19 gene (one frameshift and one missense); these patients had no evidence of wild-type allele in the DNA sample extracted from blood. It is possible that the phenotype in these male patients was not caused by the pathogenic PCDH19 variants identified and the patients were coincidentally found to be carriers of the variants. Another possibility is that these male patients are mosaic, but we were unable to detect the wild-type allele in the provided samples extracted from blood. A prior publication included a report of a male who appeared hemizygous for a pathogenic PCDH19 variant with no evidence of mosaicism in one tissue (blood), while a sample from another tissue (skin) revealed the presence of mosaicism.7 In the patients reported here, alternate tissue types were not submitted, limiting certain conclusions.
The SCN2A gene encodes the alpha subunit of a neuronal sodium channel, which mediates normal neuronal firing.27 Pathogenic variants in SCN2A cause benign familial neonatal–infantile seizures, neonatal epileptic encephalopathy, and other types of epilepsy.27, 28 Mosaic pathogenic variants in SCN2A have only been reported in the literature in two cases to date.29, 30 Paternal germ-line mosaicism for an SCN2A pathogenic variant that was transmitted to two affected half-siblings has also been reported.31 Our data demonstrate that more than 6% of patients with a pathogenic SCN2A variant were mosaic.
The SCN1A gene encodes the alpha subunit of a neuronal voltage-gated sodium channel that regulates the excitability of neurons.32 Pathogenic variants in SCN1A cause a variety of epilepsy phenotypes, ranging from simple febrile seizures to severe infantile epileptic encephalopathies. The frequency of mosaic pathogenic variants detected in SCN1A in probands was 1.3% the overall frequency of mosaicism in SCN1A that we observed for families with pathogenic variants in the SCN1A gene was 3.1% (10/320). In addition to the four probands found to harbor mosaic pathogenic variants in SCN1A, there were six instances of parental mosaicism for SCN1A pathogenic variants. The majority of parents with mosaic SCN1A pathogenic variants were reportedly unaffected. These data support previous reports that parental mosaicism in SCN1A may be more common than previously thought.33 In addition, other investigators have indicated that parental mosaicism was not detected in their testing cohort by targeted Sanger sequencing, and that more sensitive tools for detecting low-level mosaicism in parents could allow for more accurate diagnosis.33, 34, 35
Mosaic pathogenic variants were also observed in the GABRA1, GRIN2B, GABRG2, MECP2, and KCNQ2 genes. Although previous case studies have included reports of mosaic pathogenic variants in KCNQ2 and MECP2 in probands and/or their parents, ours is, to our knowledge, the first report of mosaic variants in the GABRA1, GABRG2, and GRIN2B genes. Although these genes had a relatively high frequency of mosaicism, limited data were available due to the small number of positive cases with pathogenic variants in these genes (Table 2).
We report parental mosaicism for pathogenic variants in the SCN1A, SCN2A, KCNQ2, and MECP2 genes. Parental mosaicism in these genes has been reported in unaffected, mildly affected, and more severely affected parents; therefore, the likelihood of parental mosaicism appears to be difficult to predict based on parental phenotype.5, 34, 36, 37, 38, 39 In our data set, of the parents who were identified as mosaic carriers of a pathogenic variant, and for whom we received clinical information, 73% (8/11) were reported to have no related neurological features. The parents identified as mosaic carriers of a pathogenic variant who were also reported to have a neurodevelopmental phenotype typically had features that were less severe than the features reported for their affected child. Taken together, our NGS-based data suggest that parental mosaicism may be a relatively frequent and underappreciated occurrence for pathogenic variants in genes causing epilepsy, and that the possibility of parental mosaicism should be considered when providing counseling about reproductive risks to parents of a child with a pathogenic variant in an epilepsy-related gene. Targeted parental testing is routinely performed in clinical laboratories utilizing Sanger sequencing, which is a qualitative assay and therefore has limited sensitivity in detecting mosaicism. NGS with high read depth should be considered for parental samples when testing for variants in genes with a high frequency of mosaic pathogenic variants for better sensitivity of low-level mosaicism.
In summary, individuals with epilepsy who previously tested negative for pathogenic variants in these genes by Sanger sequencing may benefit from a repeat analysis using NGS, which is more sensitive in the detection of mosaic variants. For any proband with a pathogenic variant in these genes, targeted testing of both parents is indicated and should be performed by NGS or another quantitative assay to better evaluate for possible parental mosaicism and more accurately estimate the recurrence risk. A careful and systematic review of laboratory diagnostic data may reveal that the frequency of mosaicism in genes associated with other disorders is higher than currently reported. To determine this, the methodology and threshold of variant calls would need to be optimized for consistent detection of low-level mosaicism and multiple tissue types would need to be screened. This endeavor could possibly be accomplished by multicenter collaborations and/or clinic–research collaborations.
Acknowledgments
We gratefully acknowledge all the past and present members of the neurogenetics, laboratory, and bioinformatics teams at GeneDx.
Footnotes
Supplementary material is linked to the online version of the paper at http://www.nature.com/gim
This cohort was tested at GeneDx and reviewed by M.B.S., A.S.L., E.B., K.R., G.R., and D.A.M., all of whom are employees of GeneDx, a wholly owned subsidiary of OPKO Health.
Supplementary Material
References
- Qin L, Wang J, Tian X et al. Detection and quantification of mosaic mutations in disease genes by next-generation sequencing. J Mol Diagn 2016;18:446–453. [DOI] [PubMed] [Google Scholar]
- Campbell M, Stewart JR, James RA et al. Parent of origin, mosaicism, and recurrence risk: probabilistic modeling explains the broken symmetry of transmission genetics. Am J Hum Genet 2014;95:345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Spinner NB. A genomic view of mosaicism and human disease. Nat Rev Genet 2013;14:307–320. [DOI] [PubMed] [Google Scholar]
- Bartnik M, Derwińska K, Gos M et al. Early-onset seizures due to mosaic exonic deletions of CDKL5 in a male and two females. Genet Med 2011;13:447–452. [DOI] [PubMed] [Google Scholar]
- Christodoulou J, Ho G.MECP2-related disorders. In: Pagon RA, Adam MP, Ardinger HH et al (eds). GeneReviews. University of Washington: Seattle:, WA, 28 June 2012. https://www.ncbi.nlm.nih.gov/books/NBK1497/.16 February, 2016. [Google Scholar]
- Clayton-Smith J, Watson P, Ramsden S, Black GC. Somatic mutation in MECP2 as a non-fatal neurodevelopmental disorder in males. Lancet 2000;356:830–832. [DOI] [PubMed] [Google Scholar]
- Depienne C, Bouteiller D, Keren B et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet 2009;5:e1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, Kneen R, Bayly MA et al. Recurrence risk of epilepsy and mental retardation in females due to parental mosaicism of PCDH19 mutations. Neurology 2011;76:1514–1519. [DOI] [PubMed] [Google Scholar]
- Masliah-Plachon J, Auvin S, Nectoux J, Fichou Y, Chelly J, Bienvenu T. Somatic mosaicism for a CDKL5 mutation as an epileptic encephalopathy in males. Am J Med Genet 2010;152A:2110–2111. [DOI] [PubMed] [Google Scholar]
- Mei D, Darra F, Barba C et al. Optimizing the molecular diagnosis of CDKL5 gene-related epileptic encephalopathy in boys. Epilepsia 2014;55:1748–1753. [DOI] [PubMed] [Google Scholar]
- Terracciano A, Trivisano M, Cusmai R et al. PCDH19-related epilepsy in two mosaic male patients. Epilepsia 2016;57:e51–55. [DOI] [PubMed] [Google Scholar]
- Freed D, Pevsner J. The contribution of mosaic variants to autism spectrum disorder. PLoS Genet 2016;12:e1006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna-Hidalgo R, Bo T, Kwint MP et al. Post-zygotic point mutations are an underrecognized source of de novo genomic variation. Am J Hum Genet 2015;97:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Trouillard O, Gourfinkel-An I et al. Mechanisms for variable expressivity of inherited SCN1A mutations causing Dravet syndrome. J Med Genet 2010;47:404–410. [DOI] [PubMed] [Google Scholar]
- Susswein LR, Marshall ML, Nusbaum R et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med 2016;18:823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retterer K, Juusola J, Cho MT et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med 2016;18:696–704. [DOI] [PubMed] [Google Scholar]
- Draaken M, Giesen CA, Kesselheim AL et al. Maternal de novo triple mosaicism for two single OCRL nucleotide substitutions (c.1736A>T, c.1736A>G) in a Lowe syndrome family. Hum Genet 2011;129:513–519. [DOI] [PubMed] [Google Scholar]
- Artuso R, Mencarelli MA, Polli R et al. Early-onset seizure variant of Rett syndrome: definition of the clinical diagnostic criteria. Brain Dev 2010;32:17–24. [DOI] [PubMed] [Google Scholar]
- Russo S, Marchi M, Cogliati F et al. Novel mutations in the CDKL5 gene, predicted effects and associated phenotypes. Neurogenetics 2009;10:241–250. [DOI] [PubMed] [Google Scholar]
- Bahi-Buisson N, Nectoux J, Rosas-Vargas H et al. Key clinical features to identify girls with CDKL5 mutations. Brain 2008;131:2647–2661. [DOI] [PubMed] [Google Scholar]
- Boutry-Kryza N, Ville D, Labalme A et al. Complex mosaic CDKL5 deletion with two distinct mutant alleles in a 4-year-old girl. Am J Med Genet 2014;164A:2025–2028. [DOI] [PubMed] [Google Scholar]
- Kato T, Morisada N, Nagase H et al. Somatic mosaicism of a CDKL5 mutation identified by next-generation sequencing. Brain Dev 2015;37:911–915. [DOI] [PubMed] [Google Scholar]
- Depienne C, LeGuern E. PCDH19-related infantile epileptic encephalopathy: an unusual X-linked inheritance disorder. Hum Mutat 2012;33:627–634. [DOI] [PubMed] [Google Scholar]
- Terracciano A, Specchio N, Darra F et al. Somatic mosaicism of PCDH19 mutation in a family with low-penetrance EFMR. Neurogenetics 2012;13:341–345. [DOI] [PubMed] [Google Scholar]
- Thiffault I, Farrow E, Smith L et al. PCDH19-related epileptic encephalopathy in a male mosaic for a truncating variant. Am J Med Genet 2016;170:1585–1589. [DOI] [PubMed] [Google Scholar]
- Shi X, Yasumoto S, Kurahashi H et al. Clinical spectrum of SCN2A mutations. Brain Dev 2012;34:541–545. [DOI] [PubMed] [Google Scholar]
- Howell KB, McMahon JM, Carvill GL et al. SCN2A encephalopathy: a major cause of epilepsy of infancy with migrating focal seizures. Neurology 2015;85:958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kato M, Osaka H et al. Clinical spectrum of SCN2A mutations expanding to Ohtahara syndrome. Neurology 2013;81:992–998. [DOI] [PubMed] [Google Scholar]
- Thiels C, Hoffjan S, Köhler C, Wolff M, Lücke T. SCN2A a sequence variant in mosaic state in a patient with infantile epileptic encephalopathy. Neuropediatrics 2016;47:PS01–PS11. [Google Scholar]
- Zerem A, Lev D, Goldberg-Stern H et al. Paternal germline mosaicism of a SCN2A mutation results in Ohtahara syndrome in half siblings. Eur J Paediatr Neurol 2014;18:567–571. [DOI] [PubMed] [Google Scholar]
- Miller IO, Sotero de Menezes MA.SCN1A-related seizure disorders. In: Pagon RA, Adam MP, Ardinger HH et al (eds). GeneReviews. University of Washington: : Seattle, WA, 15 May 2014. https://www.ncbi.nlm.nih.gov/books/NBK1318/.16 February, 2016. [Google Scholar]
- Xu X, Yang X, Wu Q et al. Amplicon resequencing identified parental mosaicism for approximately 10% of “de novo” SCN1A mutations in children with Dravet syndrome. Hum Mutat 2015;36:861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Arzimanoglou A, Trouillard O et al. Parental mosaicism can cause recurrent transmission of SCN1A mutations associated with severe myoclonic epilepsy of infancy. Hum Mutat 2006;27:389. [DOI] [PubMed] [Google Scholar]
- Campbell IM, Yuan B, Robberecht C et al. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am J Hum Genet 2014;95:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Yamagata T, Kubota M et al. Clinical spectrum of early onset epileptic encephalopathies caused by KCNQ2 mutation. Epilepsia 2013;54:1282–1287. [DOI] [PubMed] [Google Scholar]
- Marini C, Mei D, Helen Cross J, Guerrini R. Mosaic SCN1A mutation in familial severe myoclonic epilepsy of infancy. Epilepsia 2006;47:1737–1740. [DOI] [PubMed] [Google Scholar]
- Miceli F, Soldovieri MV, Joshi N, Weckhuysen S, Cooper E, Taglialatela M.KCNQ2-related disorders. In: Pagon RA, Adam MP, Ardinger HH et al, (eds). GeneReviews. University of Washington: : Seattle, WA, 31 March 2016. https://www.ncbi.nlm.nih.gov/books/NBK32534/.16 February, 2016. [Google Scholar]
- Milh M, Lacoste C, Cacciagli P et al. Variable clinical expression in patients with mosaicism for KCNQ2 mutations. Am J Med Genet 2015;167A:2314–2318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.