Abstract
Plasmodium was first identified in a goat in Angola in 1923, and only recently characterized by DNA isolation from a goat blood sample in Zambia. Goats were first domesticated in the Fertile Crescent approximately 10,000 years ago, and are now globally distributed. It is not known if the Plasmodium identified in African goats originated from parasites circulating in the local ungulates, or if it co-evolved in the goat before its domestication. To address this question, we performed PCR-based surveillance using a total of 1,299 goat blood samples collected from Sudan and Kenya in Africa, Iran in west Asia, and Myanmar and Thailand in southeast Asia. Plasmodium DNA was detected from all locations, suggesting that the parasite is not limited to Africa, but widely distributed. Whole mitochondrial DNA sequences revealed that there was only one nucleotide substitution between Zambian/Kenyan samples and others, supporting the existence of a goat-specific Plasmodium species, presumably Plasmodium caprae, rather than infection of goats by local ungulate malaria parasites. We also present the first photographic images of P. caprae, from one Kenyan goat sample.
Introduction
Malaria is a mosquito-borne disease caused by intracellular protozoan parasites of the genus Plasmodium. In addition to human malaria, which remains a burden of morbidity and mortality in the world, malaria parasite species infect a wide range of hosts including non-human primate, rodent, ungulate, chiroptera, avian, and reptile1,2. Ungulate malaria parasites were first reported from antelope and grey duiker in 1913 (P. cephalophi)3, followed by a second parasite reported from grey duikers (P. brucei), and additional descriptions including goat (P. caprae), water buffalo (P. bubalis), mouse deer (P. traguli), and North American white-tailed deer (P. odocoilei)4–9. Long after these microscopic observations, in 2016, three studies independently reported the first Plasmodium DNA sequences from ungulates. Martinsen et al.8 detected Plasmodium parasites (presumptive P. odocoilei) from white-tailed deer and Anopheles mosquitoes in several locations in the United States of America10. Boundenga et al.11 reported Plasmodium sequences from duiker antelope in Africa11; and Templeton et al.12 reported Plasmodium parasites from water buffalo in southeast Asia, provisionally called P. bubalis based on an early report in India4. In addition, Templeton et al.11 described a Plasmodium sequence isolated from a goat in Zambia12. DNA sequences from these samples revealed that the ungulate malaria parasites form a monophyletic clade within the haemosporidian parasites, and branch prior to the clade containing other mammalian and avian/reptile Plasmodium parasites8. Recently, Plasmodium sequences detected from South American pampas deer were shown to be similar to Plasmodium spp. in North American white-tailed deer, consistent with a monophyletic grouping of all ungulate malaria parasites13.
Goat is a major livestock with a global distribution of 1 billion individuals14. Domestic goats are predicted to have originated from the bezoars ibex (Capra aegagrus aegagrus) approximately 10,000 years ago, in the Fertile Crescent in southwest Asia including the area of present-day Iran14. The first description of a goat malaria parasite, Laverania caprae, in Angola in 1923 was from a goat (Capra aegagrus hircus) having a submandibular abscess with bacterial infection but no anemia5. Images of the parasite were not provided, but morphologies of the ring to schizont stage parasites were described as like P. falciparum, and gametocytes were termed oval or sickle. The parasite was later renamed Plasmodium caprae, but not revisited in the literature until our molecular isolation of a Plasmodium sequence in one Zambian goat in 2016. It is not known if the identified Plasmodium sequence in the Zambian goat represents a parasite having wider host distribution among African ungulates; versus host specificity to goats, and co-evolution with the host during the process of domestication and global dissemination. Therefore, in this study, we conducted a molecular and morphological surveillance of malaria parasites in goat samples obtained from countries in southeast Asia, west Asia, and Africa, to investigate the distribution of goat malaria parasites, perform comparisons of DNA sequences, and describe intraerythrocytic morphology.
Results and Discussion
Plasmodium caprae are readily detected in goats in Asian and African countries
A total of 1,299 goat blood samples were collected in 5 countries (Thailand, Myanmar, Iran, Kenya, and Sudan) during 2014 to 2017 and examined by PCR assays targeting the cytochrome b (cytb) gene (Fig. 1). Plasmodium was detected from all 5 countries (Table 1), and all obtained PCR products (773 bp) matched the sequence of the provisionally called Plasmodium caprae reported from a Zambian goat12.
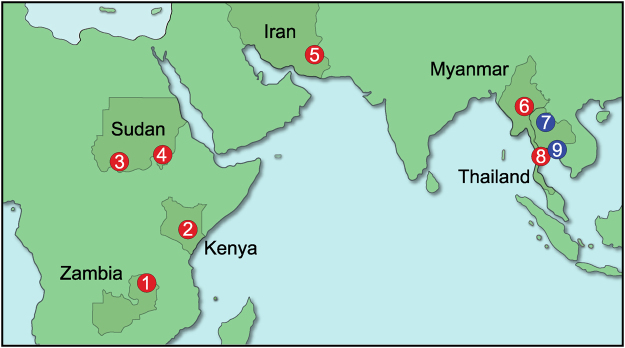
Figure 1.
Geographic locations of blood sampling sites. Goat malaria parasites in (1) Zambian samples were collected in 2010, as published12. Sampling sites in the present study are in (2) Kiambu and Kitui counties, Kenya (in 2016 and 2017, respectively); (3) West Kordufan state and (4) Blue Nile state, Sudan (2014); (5) Sistan and Baluchestan province, Iran (2016 and 2017); (6) Nay Pyi Taw, Myanmar (2016); and (7) Nan, (8) Phetchaburi, and (9) Chonburi and Rayong provinces, Thailand (2016 and 2017). Sites where Plasmodium sequences were detected are shown in red and sites where Plasmodium was not detected are shown in blue. The map was made using Adobe Illustrator CC 2017 (Adobe Systems Inc. San José, CA).
Table 1.
Prevalence of goat malaria parasites.
| Country | Location | Period | Number of samples | Positive | Positivity (%) |
|---|---|---|---|---|---|
| Thailand | Phetchaburi | May, 2016 | 126 | 5 | 4% |
| Jan, 2017 | 88 | 0 | 0% | ||
| July, 2017 | 100 | 5 | 5% | ||
| Chonburi | Jan, 2017 | 191 | 0 | 0% | |
| July, 2017 | 100 | 0 | 0% | ||
| Rayong | July, 2017 | 99 | 0 | 0% | |
| Nan | Jan, 2016 | 190 | 0 | 0% | |
| Myanmar | Nay Pyi Taw | Jun, 2016 | 10 | 4 | 40% |
| Iran | Sistan and Baluchestan | Jan, 2016 | 24 | 0 | 0% |
| Jun, 2016 | 36 | 0 | 0% | ||
| Nov, 2016 | 51 | 0 | 0% | ||
| Jul, 2017 | 89 | 28 | 31% | ||
| Sudan | West Kurdufan | Apr, 2014 | 40 | 1 | 3% |
| Blue Nile | Dec, 2014 | 76 | 6 | 8% | |
| Kenya | Kiambu county | Jun, 2016 | 13 | 0 | 0% |
| Kitui county | May, 2017 | 66 | 6 | 9% |
In Thailand, the PCR positive cases were found only in Phetchaburi province, and not in Chonburi, Rayong, or Nan provinces. Surveys in Phetchaburi were conducted in May 2016, Jan 2017, and July 2017; and 5 out of 126 (4%), 0 out of 88 (0%) and 5 out of 100 (5%) samples were positive, respectively. The prevalence of P. caprae in Thai goats was sporadic (0–5%) and much lower than that of P. bubalis in Thai water buffalo (16–45%)12. In Myanmar, a PCR-based survey was conducted in a village in Nay Pyi Taw in June 2016 and 4 out of 10 samples (40%) were positive. The rainy seasons in Thailand and Myanmar are similar, largely dominated by the monsoon, and generally spanning late May or early June to October. P. caprae positive cases were detected in May and July but not in January, which may reflect a seasonality of P. caprae infection in these areas, as is known for human malaria infections, although further long-term observations is necessary for the confirmation. This is the first description in Asian countries of goats that are infected with malaria parasites. In Iran, none of the samples collected in January, June, and November 2016 (n = 111) were positive; whereas 28 out of 89 samples (31%) collected in July 2017 were positive, corresponding to the peak season for human malaria in this region15. In Kenya, all samples (n = 13) collected in June 2016 were negative, but 6 out of 66 samples (9%) collected in May 2017 were positive. In Sudan, PCR gave positive results for 1 out of 40 samples (3%) collected in West Kordufan state in April 2014, and 6 out of 76 samples (8%) collected in Blue Nile state in December 2014. Together, our data indicate that P. caprae is widely distributed in Africa (Angola and Zambia from published reports and Sudan and Kenya in this study), west Asia (Iran), and southeast Asia (Myanmar and Thailand)5,12. Serological study would be helpful to further clarify the prevalence of the goat malaria infection.
Boundenga et al.11 detected Plasmodium sequences similar to those obtained from African duiker in Anopheles vinckei, A. obscurus and A. gabonensis11. Martinsen et al.10 reported Plasmodium sequences in A. punctipennis like those obtained from North American white-tailed deer10. Anopheles umbrosus and A. baezai (and possibly A. letifer and Mansonia crassipes) were reported as the vectors for mouse deer malaria parasite P. traguli1. These reports suggest that P. caprae is also transmitted by mosquitoes, although no arthropod vector has been reported for the goat malaria parasite. Given the wide distribution of P. caprae from Asia to Africa, a variety of local anopheline species are likely able to transmit this pathogen.
Using available data from surveys in which positive cases were detected, the associations of age, gender, or hematocrit levels of goats and P. caprae positivity were examined. We found no association between goat age and P. caprae positivity (n = 342; one out of 32 goats <1 years old, 9 out of 138 goats age between 1 and 3 years old, and 31 out of 172 goats ≥ 3 years old were positive). Additional analysis is required to see if infant goats show higher parasite positivity, because we did not collect blood from goats younger than 4 months due to the difficulty to obtain consent from local farmers. There were no associations observed between gender (45 out of 452 female and 3 out of 44 male goats were positive) or hematocrit value (n = 215, odds ratio = 0.92 (95% CI: 0.81–1.04) by logistic regression analysis) and parasite positivity. No statistical difference was detected between pregnancy and parasite positivity using 62 Kenyan female goat samples (1 out of 20 pregnant and 4 out of 42 non-pregnant goats were positive). Larger sample sizes may reveal a clinical significance, if any, of goat malaria infection.
Microscopic images of goat malaria parasite Plasmodium caprae
To evaluate the parasite loads in the infected goats, quantitative PCR (qPCR) targeting mitochondria DNA (mtDNA) was performed. Among 47 samples positive by diagnostic PCR, a Kenyan goat sample collected in 2017 (KEGoat2017–43) showed the highest copy number (~91,000 copy/µL blood equivalent) followed by a Thai sample collected in 2017 (THGoat17-448) with ~23,000 copy/µL blood equivalent (Fig. 2). Since copy numbers of mtDNA in other Plasmodium species are estimated to be approximately 30–100 per parasite16, the parasitemias of KEGoat2017-43 and THGoat17-448 were estimated to be 0.01–0.03%, and 0.002–0.008%, respectively. The other 45 samples were less than approximately 2,300 copy/µL, indicating that parasitemias in goats by P. caprae infection are usually very low and barely detectable by microscopy.
Figure 2.
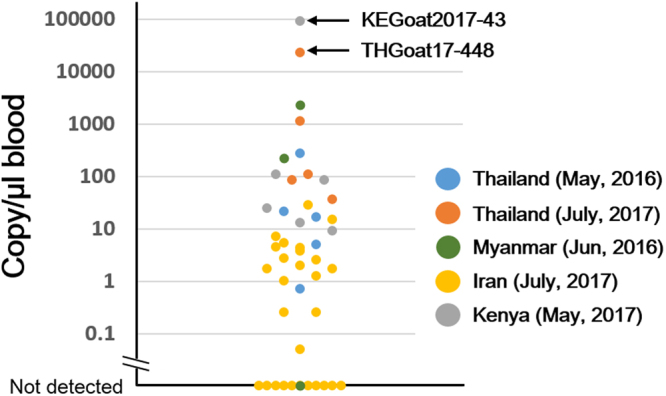
Parasite load in P. caprae infected goats as measured by qPCR. Copy numbers of mitochondrial DNA per µL of blood are shown. Blue, Thailand in May 2016; orange, Thailand in July 2017; green, Myanmar in June 2016; yellow, Iran in July 2017; and gray, Kenya in May 2017. Values for KEGoat2017-43 and THGoat17-448 samples are indicated.
Following a thorough microscopic observation of a blood smear of KEGoat2017–43 and THGoat17-448, we found Plasmodium-like organisms in the KEGoat2017-43 sample, with approximately 0.01% parasitemia by eye. Possible amoeboid late trophozoites were characterized harboring a brown pigment, likely hemozoin (Fig. 3). The first image of a putative trophozoite contained one small crystal and two vacuoles, and the size of the erythrocyte infected with this parasite was approximately 6 µm, much larger than un-infected erythrocytes (Fig. 3A). Another image contained double rod-shaped crystals, like the shape of hemozoin of other ungulate malaria parasites such as water buffalo Plasmodium bubalis4 and grey duiker Plasmodium cephalophi3. The margin of this erythrocyte infected with a potential trophozoite was not clear, thus it could not be determined if the infected erythrocytes of this pathogen are generally enlarged. Other images of putative parasites are shown in Supplementary Fig. 1. It is formally possible that the found parasites are not P. caprae, although neither Babesia nor Theileria DNA were detected from this KEGoat2017-43 sample by a PCR-based diagnostic assay.
Figure 3.
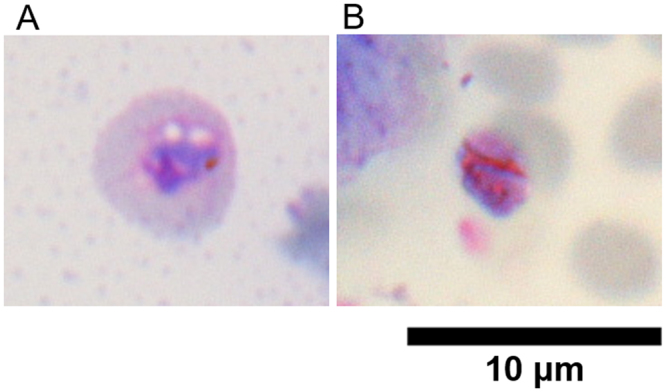
Giemsa-stained images of putative late trophozoites from KEGoat2017-43. (A) Putative trophozoite containing one small crystal and two vacuoles. (B) Putative trophozoite containing double rod-shaped crystals.
Only one nucleotide substitution was found in the whole mtDNA sequences of P. caprae among Asian and African isolates
We determined the 5,987 bp whole mitochondrial DNA sequence from the positive samples of each country. One of the sequences from a Kenyan goat (KEGoat2017-102) was identical to the published Plasmodium sequence obtained from a Zambian goat (LC090215.1)12. The other sequences (3 Thai samples, 2 Myanmar samples, 2 Iranian samples, 2 Sudanese samples [4,909 and 5,115 bp out of 5,987 bp were determined for these samples] and one Kenyan goat sample) had one synonymous substitution at nucleotide (nt) position 4421 (T to A), within the region encoding cytochrome c oxidase subunit I (coxI).
To examine the relationship of the P. caprae sequences with all other known ungulate malaria parasite sequences, including Plasmodium from African duiker11, North American white-tailed deer10, and South American pampas deer13, we used 436 bp of partial cytochrome b (cytb) sequence. P. caprae A-type and T-type sequences were monophyletic with a maximum likelihood bootstrap value (BV) of 98 and Bayesian posterior probability (BPP) of 0.91, within a clade containing all ungulate malaria parasites (BV of 100 and BPP of 1.00; Supplementary Fig. 2). When a phylogenetic tree was constructed using whole mitochondrial nucleotide sequences, the P. caprae A-type sequence was monophyletic with the T-type sequence and formed one clade with the other ungulate malaria parasites, for which whole mitochondrial nucleotide sequences were available, with BV of 100 and BPP of 1.00 (Supplementary Fig. 3), consistent with our previous report12. Together, these data confirmed that the A-type and T-type mitochondrial DNA sequences formed one clade within the ungulate malaria parasites. To evaluate the frequency distribution of this single nucleotide polymorphism (SNP) in other areas, we obtained sequences for nt 4,017–4,678 from other positive samples. We found all samples from Thailand, Iran, and Sudan were A-type, whereas both A and T types were found from Kenyan samples (Table 2).
Table 2.
Genotype at nucleotide position 4421 of mitochondrial DNA.
| Country | Nucleotide at 4421 | |
|---|---|---|
| A | T | |
| Thailand | 9/9 (100%) | 0/9 (0%) |
| Myanmar | 3/3 (100%) | 0/3 (0%) |
| Iran | 22/22 (100%) | 0/22 (0%) |
| Sudan | 6/6 (100%) | 0/6 (0%) |
| Kenya | 3/5 (60%) | 2/5 (40%) |
| Zambia* | 0/1 (0%) | 1/1 (100%) |
*Zambian data is from a published report12.
Goats were proposed to have been domesticated approximately 10,000 years ago in the region called the Fertile Crescent in southwest Asia14. From this area, goats were thought to have been introduced to southeast Asia via two routes, one through the Indian subcontinent and the other across the central Asian steppes and China14,17,18, although the age of the goat dissemination to Asia has not been clearly elucidated. Introduction to the African continent from southwest Asia is thought to have occurred roughly 7,000 years ago and spread rapidly into the central Sahara and the Ethiopian highlands around 5,000 ~ 6,500 years ago. The domestic goat did not reach southern Africa until approximately 2,000 years ago, likely due to the impact of endemic diseases such as trypanosomiasis19. Based on this history of domestic goat dissemination to southeast Asia and Africa from southwest Asia, our favored hypothesis is that the widely detected A-type mtDNA sequence is the original type in the ancestral parasite in the Fertile Crescent and spread broadly, followed by the emergence of the T-type in Africa, which was found only in Kenya and Zambia. However, it is formally possible that the Plasmodium parasites were adapted to the goat by a host-switching event after introduction to Africa, then spread to the other regions. Additional SNP information would be useful to provide further insights on this discussion. Nonetheless, the high conservation of mtDNA sequence, with only one nucleotide substitution across approximately 6 kb, suggests that the goat malaria parasite, presumably Plasmodium caprae, spread globally with its host domestic goat.
Conclusion
We performed a PCR surveillance to detect the malaria parasite P. caprae in goats in Thailand, Myanmar, Iran, Kenya, and Sudan. Putative parasite images were captured from a Giemsa-stained thin blood smear of a Kenyan goat sample. Most positive cases showed very low parasitemia by quantitative PCR and no clear clinical symptoms. The high conservation of the entire mtDNA sequence suggests that the parasite spread globally together with the goat host, rather than multiple events of host switching with local ungulate malaria parasite populations.
Methods
Sampling sites and blood collections
Blood samples were collected during the years 2014 to 2017 from indigenous goats in 5 countries; namely, Thailand, Myanmar, Iran, Sudan, and Kenya (see Fig. 1 and Table 1). In Thailand, samples were collected in Phetchaburi Province, which is along the Thai-Myanmar border; Chonburi and Rayong Provinces, in east Thailand; and Nan Province in northern Thailand. Blood was collected aseptically from the jugular vein into BD Vacutainer ACD solution A (Becton, Dickinson and Company). Samples were then divided for making thin blood smears, measuring hematocrit levels, and freezing for later extraction of DNA. Health status and pregnancy information were recorded, if available. There was no preference for blood sampling regarding gender, age, and body weight. The age of goats in Thailand was estimated by tooth numbers and were between 6 months to 8 years old, with 89% (581/655) estimated to be more than 1 year old. Eighty three percent (721/864) were female. In Kenya 13 samples were collected in Ruiru, Kiambu county (approximately 30 km north of Nairobi) and 66 samples in Mwingi, Kitui county (approximately 170 km east of Nairobi). Among these, 62 out of 79 (78%) were females, 22 goats were pregnant, 78 goats were physically healthy, and 1 was sick. In Iran, blood samples were collected in Sistan and Baluchestan province bordering Afghanistan and Pakistan. The ages of goats in Kenya and Iran were obtained from owner farmers. Kenyan and Iranian samples were collected as for the sampling in Thailand. Sudan (West Kurdufan and Blue Nile states) and Myanmar (Nay Pyi Taw) samples were archived DNA samples from the surveillance of other protozoan parasites. Among 116 Sudanese samples, 109 goats (94%) were female. Age information was not obtained for the Sudanese and Myanmar samples. Data are summarized in Table 1.
DNA extraction
Two hundred µL of blood was treated with 1 mL of 0.15% (w/v) saponin in PBS for 3–5 min and then centrifuged at 10,000 g for 5 min to remove supernatant. The pellets were washed two to three times with PBS until the supernatant turned clear. The pellet was subjected to DNA extraction using a NucleoSpin Blood QuickPure kit (Macherey-Nagel, Germany) or QIAamp DNA Mini kit (Qiagen, Germany) according to the manufacturers’ instructions. DNA was eluted with a final volume of 50 µL (Thai and Iranian samples) or 100 µL (Kenyan, Myanmar, and Sudanese samples). DNA samples were kept at ‒20 °C until use.
Parasite morphological observations
Two thin blood smears were prepared for each goat sample from Thailand and Kenya. Blood films were fixed with absolute methanol for 3 min and stained with 10% Giemsa solution. Putative P. caprae images were searched based on the description by de Mello & Paes (1923)5 and images of the other ungulate malaria parasites described by Garnham (1966)1. Images were captured using a Nikon Digital Sight system with an Eclipse E200 microscope (Nikon, Japan) and a 100× objective lens (oil, N.A. 1.40).
Parasite DNA amplification and determination of the parasite DNA sequences
DNA sequences containing a part of the cytb sequence were amplified by nested PCR with Plasmodium-specific universal primers DW2 and DW4 in the primary reaction and with the primers NCYBINF and NCYBINR in the nested PCR reaction, as described20. PCR products were subjected to ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA) treatment, and directly sequenced in both direction using BigDye v1.1 and an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). PCR products from Myanmar goats were cloned into pMD20T-vector (Mighty TA-cloning Kit, TAKARA, Japan), using E. coli DH5α competent cells. Whole mitochondrial DNA sequences were further determined for three Thai samples (THGoat16-18, THGoat17-435 and THGoat17-448), two Myanmar samples (MMGoat16-2 and MMGoat16-7), two Iranian samples (IRGoat17-15 and IRGoat17-70) and two Kenyan samples (KEGoat17-43 and KEGoat17-102) by direct sequencing of the PCR-amplified DNA fragments as described12. Two Sudanese samples (SDGoat16-5 and SDGoat16-24) were also attempted to determine the whole mitochondrial DNA sequences; however, due to sample limitations, only 5.1 kb and 4.9 kb sequences were determined, respectively (nucleotide positions corresponding to the LC326032 1–612, 741–1324, 1402–3102, 3724–3974, 4021–5987 on SDGoat16-5 and 1–1288, 1771–2848, 3006–3973, 4074–4654, 4994–5987 on SDGoat16–24). Before assembly the chromatograms of obtained DNA sequences were visually inspected to ensure that there were no ambiguous nucleotides. Co-infection with Babesia or Theileria was assayed by PCR using common primers for Babesia or Theileria and specific primers for B. ovis21 or T. ovis22. Sequence of primers used in this study are shown in Supplementary Table 1.
Phylogenetic analysis
The obtained whole mtDNA sequences were aligned with known haemosporidian mtDNA sequences using ClustalW software with manual corrections. Mitochondrial DNA sequences used in this study are listed in the Supplementary Table S1 of Templeton et al.12. Phylogenetic trees were inferred by the maximum likelihood (ML) and Bayesian inference (BI) method. Model analysis using IQ-TREE ver. 1.6.1 indicated that the GTR + I + G model was superior to other models by both Akaike and Bayesian information criterion23. Following model analysis, ML analysis was conducted using IQ-TREE 1.6.1 with 1,000 replicates of ultrafast bootstrap analysis. Bayesian posterior probabilities (BPP) were also obtained using MrBayes ver 3.2.6 with eight parallel Metropolis-coupled Markov chain Monte Carlo runs, consisting of one cold and four heated chains with a chain temperature of 0.1, for 3,000,000 generations24. Log-likelihood scores and trees with branch lengths were sampled every 1,000 generations and the first 750,000 generations were excluded as burn-in, and the remaining trees were summarized to obtain BPP. The tree was visualized by FigTree ver1.4.3. The phylogenetic tree of 436 bp of cytb sequences were also examined with the same method. In addition to the cytb sequences obtained from haemosporisdian parasites used in Supplementary Figure 3, nucleotide sequences of Plasmodium sp. in the North American white-tailed deer10, haemosporidian parasites in the African duiker11 and Plasmodium sp. in the Brazilian pampas deer13 were included in the analysis.
Quantitative measurement of parasite loads
SYBR Green-based quantitative PCR (qPCR) was performed as described12. In brief, qPCR reactions with primers TypeUnivFor and TypeUnivRevii targeting the parasite cytb gene were performed on a 7500 Real Time PCR System (Applied Biosystems) with 7500 System SDS software (Applied Biosystems). The pCR-Blunt II-TOPO plasmid containing P. bubalis cytb sequence was used to make a standard curve to obtain the copy number of the DNA in the tested samples.
Ethical statement
This study was conducted with the consent of the farm owners and was approved by the Chulalongkorn University Animal Care and Use Committee (No. 1731054). This study was approved by University of Veterinary Science, and the Ministry of Agriculture, Livestock and Irrigation, Myanmar. This study was also conducted with the consent of the farm owners in accordance with the approved guidelines in the other countries described in this study. The project was reviewed and approved by the Institutional Biosafety Committee in accordance with the faculty regulations and policies governing biosafety procedures (IBC approval No. 1631048).
The whole mitochondrial DNA sequence of THGoat16-18 was deposited in DDBJ/ENA/GenBank under the accession number LC326032.
Electronic supplementary material
Acknowledgements
We are grateful to veterinarians and veterinary students of Chulalongkorn University and students of the Faculty of Veterinary Medicine, Rajamangala University of Technology Tawan-Ok for assisting us in blood samplings. We thank Thomas Templeton for his critical reading of the manuscript. This work was conducted at the Veterinary Parasitology Research Laboratory of Chulalongkorn University and the Joint Usage/Research Center on Tropical Disease, Institute of Tropical Medicine, Nagasaki University. This study was supported in part by JSPS Grants-in-Aids for Scientific Research (B), No 16H05807 to OK and MA and No 26304035 to KK. MK was funded in part by Chulalongkorn University and the Thai Government Budget through Chulalongkorn University (grants no. GSTAR-60007310041 and GB-B-60-100-31-11). MK was a recipient of Chulalongkorn University Faculty of Veterinary Science Research Fund (RG 3/2560). ST and MK were supported in part by grant for Special Task Force for Activating Research, Chulalongkorn University (GSTAR 5900131001). EM was funded by the Ministry of Higher Education and Scientific Research, Republic of Sudan (Grant No. SRI-VS-2015-71933).
Author Contributions
M.K., M.A., and O.K. conceived and designed the study. M.K., M.T., M.A., T.S., WKae, S.T., M.C., S.C., WKat, SuT and J.P. conducted surveys in Thailand. K.K., Y.I., R.N., S.B., L.L.H. and M.M.W. conducted surveys in Myanmar. H.H. and A.S. conducted surveys in Iran. J.N.G. and D.K.M. conducted surveys in Kenya. E.M., A.A.I., A.M.I., and K.S. conducted surveys in Sudan (A.A.I. passed away in 2015). M.K., M.T., J.N.G., and M.A. prepared blood smears, extracted DNA, and performed PCR. M.T., M.A. and O.K. conducted and analyzed qPCR, mitochondrion genome sequences, and SNPs analysis. M.K., M.A., and O.K. wrote manuscript and incorporate the other author’s comments. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-018-25539-w.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24048-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/11/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
Contributor Information
Masahito Asada, Email: masahitoasada@nagasaki-u.ac.jp.
Osamu Kaneko, Email: okaneko@nagasaki-u.ac.jp.
References
- 1.Garnham, P. C. C. Malaria parasites and other Haemosporidia. Blackwell Scientific Publication, Oxford (1966).
- 2.Cox FEG. History of the discovery of the malaria parasites and their vectors. Parasites & Vectors. 2010;3:5. doi: 10.1186/1756-3305-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce D, Harvey D, Hamerton AE, Lady B. Plasmodium cephalophi sp. nov. Proc. R. Soc. B. 1913;87:45–47. doi: 10.1098/rspb.1913.0056. [DOI] [Google Scholar]
- 4.Sheather AL. A malaria parasite in the blood of a buffalo. J. Comp. Path. Ther. 1919;32:223–226. doi: 10.1016/S0368-1742(19)80026-7. [DOI] [Google Scholar]
- 5.de Mello F, Paes. S. Sur une plasmodiae du sang des chèvres. C.r. séanc. Soc. Biol. 1923;88:829–830. [Google Scholar]
- 6.Garnham PC, Edeson JF. Two new malaria parasites of the Malayan mousedeer. Rivista di malariologia. 1962;41:1–8. [PubMed] [Google Scholar]
- 7.Garnham PCC, Kuttler KL. A malaria parasite of the white-tailed deer (Odocoileus virginianus) and its relation with known species of Plasmodium in other ungulates. Proc. R. Soc. Lond. B Biol. Sci. 1980;206:395–402. doi: 10.1098/rspb.1980.0003. [DOI] [PubMed] [Google Scholar]
- 8.Templeton TJ, Martinsen E, Kaewthamasorn M, Kaneko O. The rediscovery of malaria parasites of ungulates. Parasitology. 2016;143:1501–1508. doi: 10.1017/S0031182016001141. [DOI] [PubMed] [Google Scholar]
- 9.Perkins SL, Schaer J. A modern menagerie of mammalian malaria. Trends Parasitol. 2016;32:772–782. doi: 10.1016/j.pt.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Martinsen ES, et al. Hidden in plain sight: Cryptic and endemic malaria parasites in North American white-tailed deer (Odocoileus virginianus) Sci. Adv. 2016;2:e1501486. doi: 10.1126/sciadv.1501486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boundenga L, et al. Haemosporidian parasites of antelopes and other vertebrates from Gabon, Central Africa. PLoS ONE. 2016;11:e0148958. doi: 10.1371/journal.pone.0148958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Templeton TJ, et al. Ungulate malaria parasites. Sci. Rep. 2016;6:23230. doi: 10.1038/srep23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asada M, et al. Close relationship of Plasmodium sequences detected from South American pampas deer (Ozotoceros bezoarticus) to Plasmodium spp. in North American white-tailed deer. Int. J. Parasitol. Parasites Wildl. 2018;7:44–47. doi: 10.1016/j.ijppaw.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amills M, Capote J, Tosser-Klopp G. Goat domestication and breeding: a jigsaw of historical, biological and molecular data with missing pieces. Anim. Genet. 2017;48:631–644. doi: 10.1111/age.12598. [DOI] [PubMed] [Google Scholar]
- 15.Soleimani-Ahmadi M, Vatandoost H, Zare M, Alizadeh A, Salehi M. Community knowledge and practices regarding malaria and long-lasting insecticidal nets during malaria elimination programme in an endemic area in Iran. Malar J. 2014;13:511. doi: 10.1186/1475-2875-13-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Putaporntip C, Buppan P, Jongwutiwes S. Improved performance with saliva and urine as alternative DNA sources for malaria diagnosis by mitochondrial DNA-based PCR assays. Clin. Microbiol. Infect. 2011;17:1484–1491. doi: 10.1111/j.1469-0691.2011.03507.x. [DOI] [PubMed] [Google Scholar]
- 17.Pereira F, et al. Tracing the history of goat pastoralism: new clues from mitochondrial and Y chromosome DNA in North Africa. Mol. Biol. Evol. 2009;26:2765–2773. doi: 10.1093/molbev/msp200. [DOI] [PubMed] [Google Scholar]
- 18.Pereira, F. & Amorim, A. Origin and spread of goat pastoralism. In: Encyclopedia of Life Sciences. John Wiley & Sons, Chichester (2010).
- 19.Smith AB. Origins and spread of pastoralism in Africa. Annu. Rev. Anthropol. 1992;21:125–141. doi: 10.1146/annurev.an.21.100192.001013. [DOI] [Google Scholar]
- 20.Abkallo HM, et al. DNA from pre-erythrocytic stage malaria parasites is detectable by PCR in the faeces and blood of hosts. Int. J. Parasitol. 2014;44:467–473. doi: 10.1016/j.ijpara.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Aktaş M, Altay K, Dumanli N. Development of a polymerase chain reaction method for diagnosis of Babesia ovis infection in sheep and goats. Vet. Parasitol. 2006;133:277–281. doi: 10.1016/j.vetpar.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 22.Aktas M, Altay K, Dumanli N. PCR-based detection of Theileria ovis in Rhipicephalus bursa adult ticks. Vet. Parasitol. 2006;140:259–263. doi: 10.1016/j.vetpar.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.