Abstract
Steroid receptor coactivator 1 (SRC-1) interacts with nuclear receptors and other transcription factors (TFs) to initiate transcriptional networks and regulate downstream genes which enable the cancer cell to evade therapy and metastasise. Here we took a top–down discovery approach to map out the SRC-1 transcriptional network in endocrine resistant breast cancer. First, rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) was employed to uncover new SRC-1 TF partners. Next, RNA sequencing (RNAseq) was undertaken to investigate SRC-1 TF target genes. Molecular and patient-derived xenograft studies confirmed STAT1 as a new SRC-1 TF partner, important in the regulation of a cadre of four SRC-1 transcription targets, NFIA, SMAD2, E2F7 and ASCL1. Extended network analysis identified a downstream 79 gene network, the clinical relevance of which was investigated in RNAseq studies from matched primary and local-recurrence tumours from endocrine resistant patients. We propose that SRC-1 can partner with STAT1 independently of the estrogen receptor to initiate a transcriptional cascade and control regulation of key endocrine resistant genes.
Introduction
Ligand bound estrogen receptor (ER) along with its coactivator proteins is a key driver of endocrine sensitive breast cancer [1]. Treatment induced alterations in the steroid environment however can alter the steroid receptor coactivator dynamic. Enhanced growth factor signalling, following prolonged treatment with tamoxifen and aromatase inhibitors (AIs), can lead to an initial ligand independent activity of ER and a subsequent complete loss of ER function in the advanced setting [2]. Conversely, gain of HER2 has been reported at both protein and transcript levels, particularly in metastatic brain tumours [3]. This phenotype discordance between the primary and metastatic tumour in a subset of advanced cancers has been associated with worse post recurrence and overall survival [4].
The coactivator protein SRC-1 (NCOA1) is a master regulator with a distinct role in endocrine therapy resistance and the development of metastatic disease [5]. SRC-1 can interact with p300/CBP, CARM1 and PRMT1 [6–9] and co-activate several nuclear receptors including ER, progesterone receptor (PR), glucocorticoid receptor, thyroid receptor, retinoid X receptor, hepatocyte nuclear factor 4 and peroxisome proliferator-activated receptor γ [10]. In addition, SRC-1 can also co-activate other TFs to execute its function. In vitro interactions between SRC-1 and AP-1, serum response factor and NF-κB have been described [11–13]. Importantly, SRC-1 interactions with ETS2, PEA3 and HOXC11 in clinical ex vivo and mouse in vivo studies have been associated with endocrine resistance and breast disease progression [14–16].
SRC-1 aberrant activity is one of many mechanisms of endocrine resistance, it mediates its role in tumour progression and metastasis through transcriptional activation of key genes [17]. SRC-1 interacts with the HER2-MAPkinase activated TF ETS2 to regulate MMP9 and cMyc, with AP-1 to regulate integrin α5 and with PEA3 to activate the epithelial mesenchymal transition (EMT) TF TWIST [18–20]. In essence SRC-1 can act as a master initiating regulator of a transcriptional network to control executor genes that enable the cancer cell to evade therapy and ultimately to metastasise to distant organs. To date important information has been reported regarding new transcriptional partners and target genes of the coactivator family. These however do not address the full potential of these regulatory proteins and their downstream effectors. Full network analysis is required to understand the complexity and power of the SRC family.
TFs are the key regulatory nodes of the cancer cell [21]. In this study we took a top down discovery approach in order to map out the SRC-1 transcriptional network pertinent to endocrine resistant breast cancer. In the first instance we used RIME to uncover new SRC-1 TF partners. Subsequently, we undertook RNAseq to investigate SRC-1 TF target genes and finally we took a bioinformatic approach to model second generation downstream executioner targets. Our data suggest that SRC-1 can partner with STAT1 to regulate TFs and chromatin remodellers independently of ER to initiate a transcriptional cascade and control key genes, including cell cycle regulators, to promote endocrine resistant disease progression.
Results
SRC-1 partners with STAT1 to activate TF/chromatin remodeller target genes
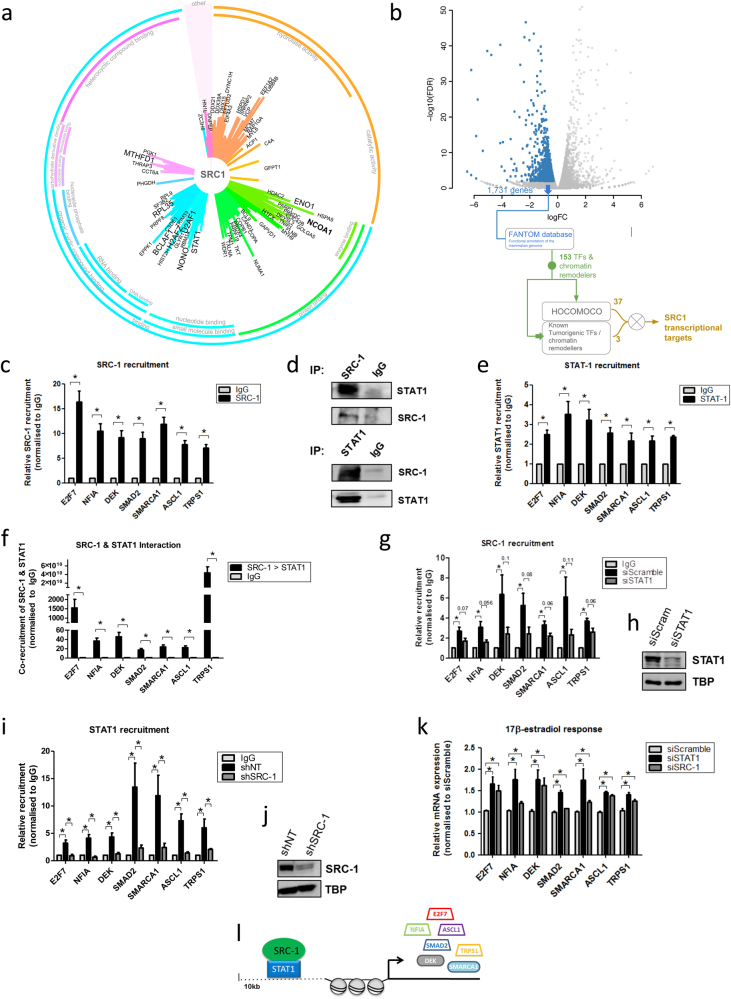
Endogenous SRC-1 was immunoprecipitated from cross linked endocrine resistant LY2 cells following 45 min of tamoxifen treatment using RIME. 148 SRC-1 associated proteins were identified in each of the three replicates (Fig. 1a), including SRC-1 (NCOA1) which was one of the most confident proteins identified. Of the 148 SRC-1 interacting proteins, 27 have been described as TF or chromatin remodellers, 9 of which have defined motifs (Supplementary Table 1). Among those identified several were known SRC-1 binding partners including PARP1, NONO and PRMT1 [22–24]. A full list of interacting proteins is provided in Extended Data 1.
Fig. 1.
Combining RIME and RNAseq analysis to discover the SRC-1 regulatory network. a RIME was conducted in three replicates of endocrine resistant breast cancer LY2 cells that were steroid depleted for 72 h and treated with tamoxifen 10−7 mol/L for 45 min to identify SRC-1 interacting proteins. Non-specific interactions (identified from two IgG control replicates) have been removed. A protein was only included if present in all three replicates with a threshold greater than two unique peptides. SRC-1-associated proteins were clustered based on molecular function with the length of the line for each protein indicating its Mascot Score in the MS-arc plot. SRC-1 and the 9 TFs with PWMs are highlighted. b Volcano plot displaying differentially expressed genes identified from RNAseq of four replicates of LY2-shSRC-1 and LY2-shNT cells that were steroid depleted for 72 h and treated with tamoxifen 10−7 mol/L for 8 h, with genes positively regulated by SRC-1 highlighted in blue. Flow diagram of the filters used to identify SRC-1 regulated TFs. c ChIP qPCR confirmation of SRC-1 recruitment to the promoter of E2F7, NFIA, DEK, SMAD2, SMARCA1, ASCL1 and TRPS1 in LY2 cells that were steroid depleted for 72 h and treated with tamoxifen 10−7 mol/L for 45 min. Results normalised to IgG and are expressed as mean ± SEM, n = 3, p < 0.05. d Immunoprecipitation (IP) experiments confirmed STAT1 as an SRC-1 interactor (n = 4). e ChIP assays demonstrated STAT1 recruitment to the promoter region of RNA-seq identified TFs in tamoxifen treated LY2 cells. Results are expressed as mean ± SEM, n = 4, p < 0.05, normalised to IgG. f ChIP re-ChIP assay confirming co-occupancy of SRC-1 and STAT1 at the promoter of TFs in LY2 cells that were steroid depleted for 72 h and treated with tamoxifen 10−7 mol/L for 45 min. Results normalised to IgG and are expressed as mean ± SEM, n = 3, p < 0.05. g Recruitment of SRC-1 to TF promoters following STAT1 knockdown in LY2 cells that had been steroid depleted for 72 h and treated with tamoxifen 10−7 mol/L for 45 min. Results normalised to IgG and are expressed as mean ± SEM, n = 4, p < 0.05. P-values for ‘n.s’ results are displayed. h Confirmation of STAT1 nuclear protein knockdown for ChIP experiments assessed by western blot (n = 3). i Occupancy of STAT1 at TF promoters following SRC-1 knockdown in LY2 cells that had been steroid depleted for 72 h and treated with tamoxifen 10−7 mol/L for 45 min. Results normalised to IgG and are expressed as mean ± SEM, n = 4, p < 0.05. j Confirmation of SRC-1 nuclear protein knockdown for ChIP experiments assessed by western blot (n = 3). k qPCR analysis of relative mRNA expression of E2F7, NFIA, DEK, SMAD2, SMARCA1, ASCL1 and TRPS1 in LY2 cells following 8 h treatment with 17β-estradiol 10−8 M following siRNA gene silencing of SRC-1 and STAT1. Results normalised to vehicle and are expressed as mean ± SEM, n = 3, p < 0.05. l Schematic representation of SRC-1 interaction with STAT1 to transcriptionally activate TFs; E2F7, NFIA, SMAD2, ASCL1, TRPS1 and chromatin remodellers; DEK and SMARCA1
RNAseq of tamoxifen treated endocrine resistant LY2 cells following SRC-1 gene silencing was used to define the SRC-1 transcriptome. 1731 genes were identified as SRC-1 positively regulated genes. The FANTOM database was used to identify TF and/or chromatin remodelers. In total 153 genes were found. To select out the known TFs from this list we used HOCOMOCO, Jaspar Core and cisBP databases to identify genes with defined position weight matrices (PWMs) (Fig. 1b) [25–28]. In total 37 TFs were found with PWMs and a further three genes were included in the analysis as they are considered tumorigenic TF/chromatin remodellers (DEK, SMARCA1 and TRPS1) [29–31]. Of the 40 putative SRC-1 TFs/chromatin remodellers identified from the RNAseq and bioinformatic analysis, 14 were taken forward for molecular validation based on their described role in tumour progression (Supplementary Table 2). In SRC-1 silencing studies 7 of 14 genes (TFs, ASCL1, E2F7, NFIA, SMAD2 and TRPS1 and the chromatin remodellers, including DEK and SMARCA1) were confirmed as SRC-1 positive regulons (Supplementary Fig. 1a). To determine if SRC-1 directly regulates these genes we used SRC-1 ChIPseq data from tamoxifen treated LY2 cells and looked at SRC-1 occupancy 10 kb upstream of the transcriptional start site (TSS). Significant recruitment of SRC-1 to the promoter region of each of these genes was observed on the UCSC Genome Browser from SRC-1 ChIP studies [32] (Supplementary Fig. 1b), which was confirmed by ChIP qPCR (Fig. 1c). Of interest, though ER recruitment to the promoter regions of the 7 target genes was also observed (Supplementary Fig. 1c), transcript levels of the genes was independent of estrogen treatment (Supplementary Fig. 1d) and of ER transcriptional activity as demonstrated by ESR1 knockdown and ICI 182, 780 microarray datasets (Supplementary Fig. 1e and f, Extended Data 2) [33].
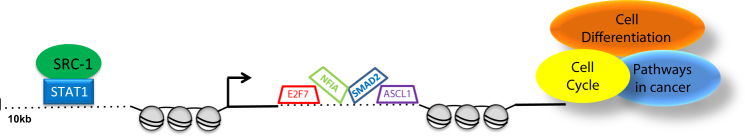
From the RIME experiment we had identified 9 potential SRC-1 TF partners with PWMs. We used the BioMART database and FIMO motif based sequence analysis to look for potential recruitment of the SRC-1 partners 10 kb upstream of the TSS of the 7 target genes. H1FX, H2AF2 and STAT1 binding regions were identified in the enhancer/promoter region of several of the target genes, however only STAT1 response elements were present upstream of all of the defined SRC-1 direct targets (Supplementary Fig. 1g) [34, 35]. SRC-1-STAT1 interactions were confirmed by immunoprecipitation (Fig. 1d) and recruitment of STAT1 alone and in combination with SRC-1 to the promoter region of each of the target genes was confirmed by ChIP and ChIP-re-ChIP qPCR respectively, in endocrine resistant LY2 cells (Fig. 1e, f). A full list of SRC-1-STAT1 target genes is given in Extended Data 3. Full recruitment of SRC-1 to the promoter of each of target gene was found to be dependent on STAT1, though this did not reach statistical significance (Fig. 1g, h). Moreover, silencing SRC-1 expression significantly impaired occupancy of STAT1 at TF promoters (Fig. 1i, j). Furthermore, silencing of both SRC-1 and STAT1 reduced transcript levels of these genes (Supplementary Fig. 1h). Of note, silencing of either SRC-1 or STAT1 induced estrogen sensitivity and enabled steroid dependent increases in transcript levels of the 7 genes (Fig. 1k). These data suggest that in the endocrine resistant setting SRC-1 can partner with STAT1, in an ER independent manner, to activate target genes ASCL1, DEK, E2F7, NFIA, SMAD2, TRPS1 and SMARCA1 (Fig. 1l). Furthermore, in the absence of SRC-1 or STAT1, the regulation of the 7 genes can be resensitised to estrogen.
The functional role of SRC-1 transcriptional targets in endocrine treatment resistance
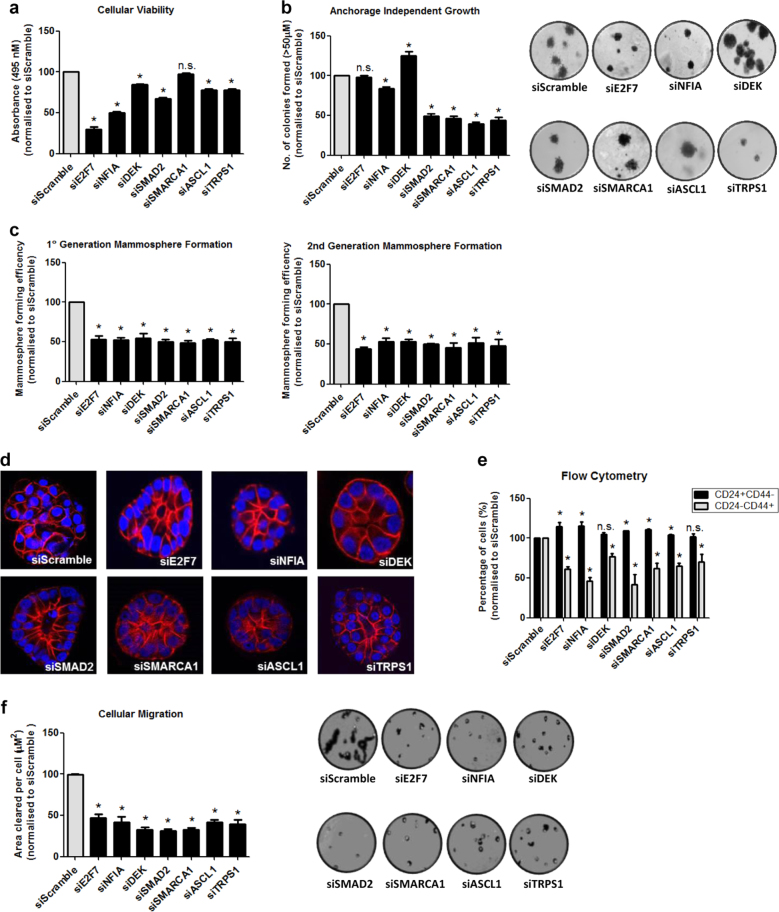
To understand the contribution of the SRC-1 transcriptional targets to tumour progression we looked at the role of each of the individual target genes in classic mechanisms of tumour aggression including cell viability, mammosphere formation, de-differentiation and migration. Expression levels of each of the 7 target genes are elevated in models of tamoxifen resistance (LY2 cells and/or TamR cells) in comparison to endocrine sensitive parent cells (MCF7 cells). Moreover, expression of E2F7, NFIA and DEK were also elevated in a model of AI resistance (LetR cells) relative to sensitive cells (Supplementary Fig. 2a).
The SRC-1 target genes have varying roles in tumour cell viability and anchorage independent growth. From transient gene silencing studies, E2F7 and NFIA were observed to play a substantial role in cell viability, whereas SMAD2, SMARCA1, ASCL1 and TRPS1 contributed to anchorage independence (Fig. 2a, b, Supplementary Fig. 2b). In contrast, each of the SRC-1 regulated genes had a significant impact on mammosphere generation, cellular depolarisation and CD24−/44+ (stem like) population (Fig. 2c, d, e) supportive of a role for this cadre of transcriptional regulators in cellular de-differentiation. Moreover each of the SRC-1 target genes was shown to significantly contribute to the migratory capacity of endocrine resistant breast cancer cells (Fig. 2f).
Fig. 2.
E2F7, NFIA, DEK, SMAD2, SMARCA1, ASCL1 and TRPS1 - roles in endocrine resistance. a Cellular viability is significantly reduced after siRNA knockdown of E2F7, NFIA, DEK, SMAD2, ASCL1 and TRPS1 in endocrine resistant LY2 cells with no significant changes observed with siSMARCA1. b Anchorage independent growth of LY2 cells was reduced following siRNA transfection with NFIA, SMAD2, SMARCA1, ASCL1 and TRPS1. Growth was increased with siDEK transfection and no significant changes were observed with siE2F7. Representative images of cells’ anchorage independent growth. c Mammosphere forming efficiency in first and second generation of mammospheres was significantly reduced following knockdown of E2F7, NFIA, DEK, SMAD2, SMARCA1, ASCL1 and TRPS1. d Representative images of acini formation from LY2 cells following siRNA transfection with E2F7, NFIA, DEK, SMAD2, SMARCA1, ASCL1 and TRPS1 showed more organised acini with superior apico-basolateral structure compared to siScramble cells. e Flow cytometry analysis of CD44 and CD24 expression in LY2 cells transfected with siRNA against E2F7, NFIA, DEK, SMAD2, SMARCA1, ASCL1 and TRPS1. The results showed a significant reduction in CD24−CD44+ (stem cell-like). f Migratory ability of the LY2 cells was significantly reduced following siRNA transfection with E2F7, NFIA, DEK, SMAD2, SMARCA1 ASCL1 and TRPS1. Representative images of cellular migration shown. All results are expressed as mean ± SEM, n = 3, p < 0.05, normalised to siScramble
Expression of SRC-1 transcriptional network in endocrine sensitive and resistant tumours
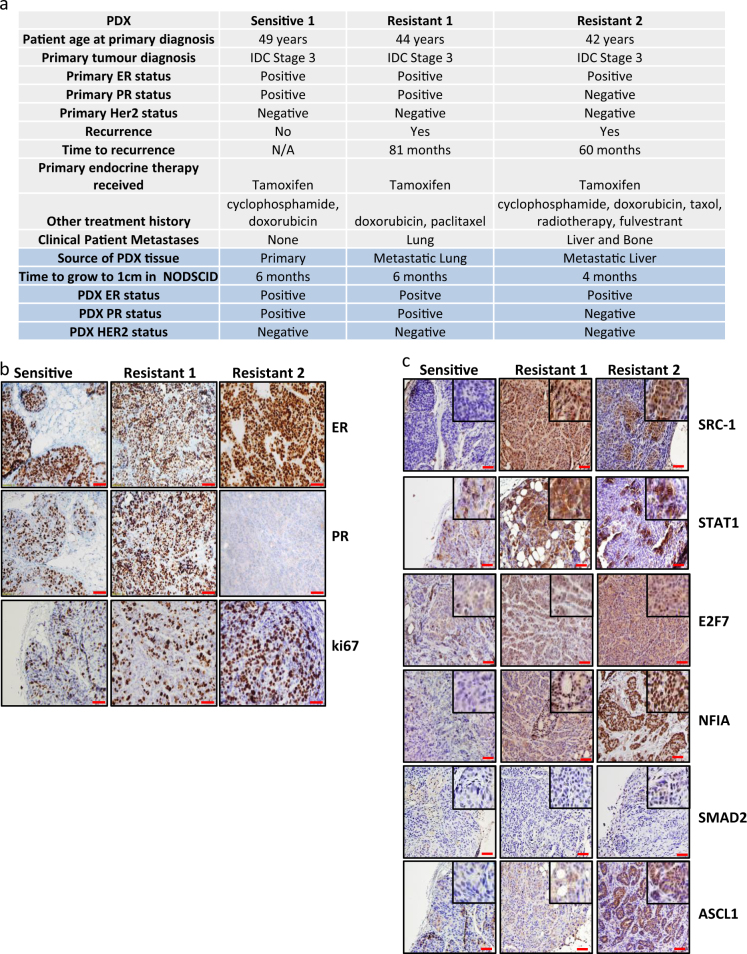
To investigate the clinical relevance of the SRC-1 transcriptional regulators we employed a patient derived xenograft (PDX) model of ER positive breast cancer. Primary and metastatic tumours from an endocrine sensitive and two endocrine resistant patients respectively were expanded in NOD SCID mice (Fig. 3a). ER expression was detected in each of the tumour models, whereas PR was expressed in the primary and lung metastatic tumours, but not the liver metastatic tissue. Elevated levels of the proliferation marker Ki67 were observed in the cancer cells of the resistant tumours in comparison to the sensitive tumour (Fig. 3b).
Fig. 3.
SRC-1 transcriptional network is expressed in endocrine resistant patient derived xenograft models compared to endocrine sensitive. a Table of characteristics and receptor status of patient breast tumours and their corresponding patient derived xenografts (PDXs). b ER, PR and ki67 expression in established PDXs showing retained biomarker status. The original clinical diagnosis for ER and PR is shown in the table. c Sections of tumour were stained by IHC for SRC-1, STAT1, E2F7, NFIA, SMAD2 and ASCL1. Representative image of 5 taken at 20X, scale bars in red correspond to 50 μm. Zoomed images at 40X
SRC-1 was found to be elevated in metastatic endocrine resistant PDX models in comparison to the primary endocrine sensitive PDX and STAT1 was observed in both the primary and metastatic tumours (Fig. 3c). Expression of SRC-1 TF targets with defined PWM (NFIA, SMAD2, E2F7 and ASCL1) (Supplementary Table 2) was also assessed. SMAD2 was upregulated in liver metastatic resistant tumour tissue whereas E2F7, NFIA and ASCL1 were elevated in both the lung and the liver metastatic resistant tumours in comparison to the sensitive primary tumour model (Fig. 3c).
Extended SRC-1 regulatory networks
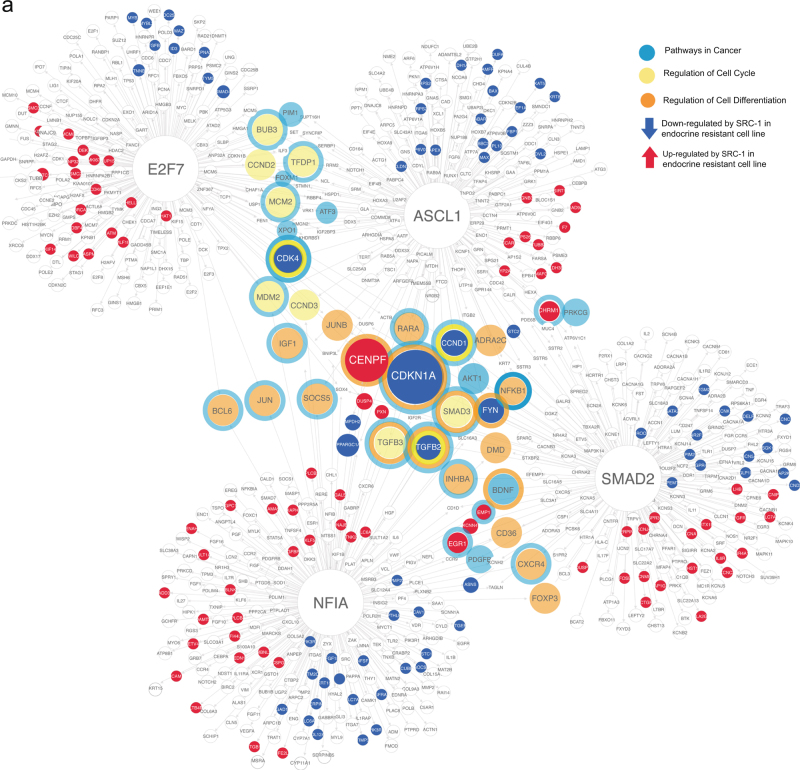
In order to understand the greater SRC-1 regulatory network we investigated the regulons of the SRC-1 TF target genes with PWM (NFIA, SMAD2, E2F7 and ASCL1). Regulons were obtained from large scale co-expression signature datasets using iRegulon [36]. 79 genes were found to be regulated by two or more of the SRC-1 TFs with PWM which includes SMAD2, E2F7, ASCL1, NFIA (Fig. 4a).
Fig. 4.
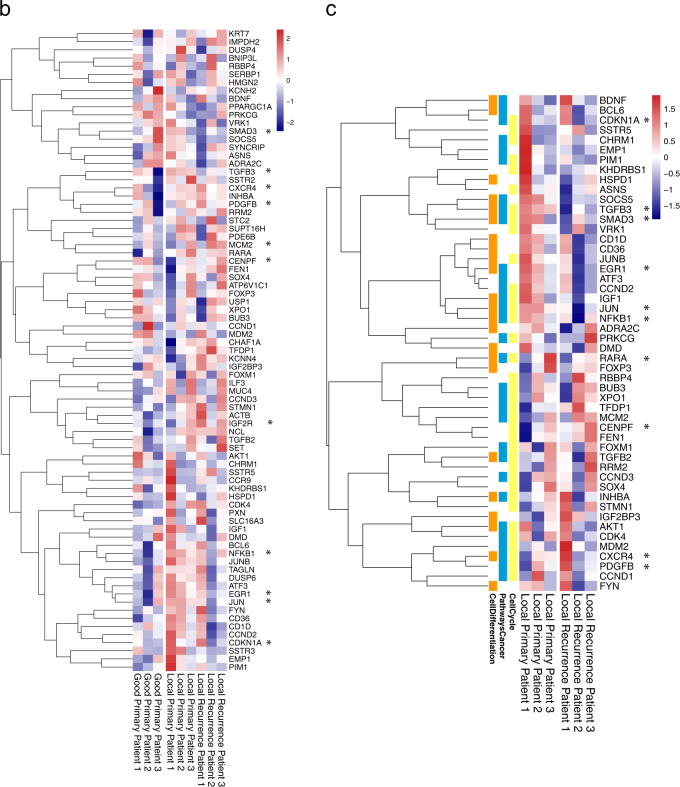
The transcriptional regulon of ASCL1, E2F7, NFIA and SMAD2. a A representative regulatory network using iRegulon in Cytoscape. The large nodes represent the genes regulated by E2F7, NFIA, SMAD2 and ASCL1. The target genes for each TF are portrayed in red if they are up regulated with SRC-1 or dark blue if they are down regulated with SRC-1 and light blue, yellow or orange for pathways in cancer, cell differentiation or cell cycle respectively. The shared targets between the 4 TFs are CDKN1A and CENPF. b From the regulatory network, 79 genes were found to be regulated by two or more of the SRC-1 regulated TFs. The expression of these genes was profiled in primary breast tumours of patients that responded well to therapy (n = 3) and in matched primary (n = 3) vs. local recurrence samples (n = 3) from patients who had disease recurrence on endocrine treatment (from left to right). c Functional annotation of a subset of these genes in primary tumours from patients that recurred on endocrine treatment and their matched local recurrence. ‘*’ denotes those genes highlighted in the text
At the core of the TF hub two key cell cycle regulators, CENPF and CDKN1A were found to be co-regulated by all four TFs (Fig. 4a). The CENPF gene which encodes centromere protein F is positively regulated by SRC-1, whereas CDKN1A which encodes the cell cycle inhibitor, p21, is negatively regulated by SRC-1 (Fig. 4a). Consistent with SRC-1’s role in endocrine resistance in breast cancer, CENPF positively associated with poor disease free survival in endocrine treated patients (p = 0.00075), whereas CDKN1A associated with good response to treatment (p = 0.046) (Supplementary Fig. 3a and b). When modelled together, (CENPF positively/CDKN1A inversely) these cell cycle regulators strongly associated with reduced disease free survival times in patients on endocrine treatment (Supplementary Fig. 3c and d).
Expression of the 79 genes of the SRC-1 network analysed in RNAseq data from ER positive primary tumours from patients with good response to endocrine therapy (n = 3) and primary tumours from those with disease recurrence on therapy and the matched local recurrence tissue (n = 3) (Supplementary Table 3). Elevations in a number of metastatic genes including, CXCR4, PDGFβ, RARA and IGF2R, were observed in the primary and matched recurrent tumours from endocrine resistant patients, in comparison to the primary tumours from endocrine sensitive patients. Whereas, a number of genes which were observed to be elevated in endocrine resistant primary tumours, were lost on local disease recurrence these include known proto-oncogenes, NF-κB, JUN and EGR1 (Fig. 4b).
Pathway analysis of the SRC-1 regulated 79 genes using ClueGo identified cell cycle regulation, cellular differentiation and pathways in cancer as key functions of the SRC-1 TF regulon in endocrine resistant breast cancer (Supplementary Fig. 4). We looked at the genes from these pathways in RNAseq data from our matched primary and recurrent tumours. Genes involved in cell differentiation were lost including TGFβ3 and SMAD3, in the endocrine resistant tumours, whereas cell cycle genes including CENPF were enhanced (Fig. 4c). An overview of the SRC-1 signalling cascade described here is illustrated in Fig. 5.
Fig. 5.
Schematic representation of SRC-1 signalling cascade. SRC-1 interacts with STAT1 at the promoter region of E2F7, NFIA, SMAD2, and ASCL1. Expression of these TFs promotes an endocrine resistant phenotype in breast cancer. Downstream network analysis revealed perturbations in several pathways involved in cancer progression including, cell differentiation, cell cycle and pathways in cancer
Discussion
The extent of molecular phenotype discordance between ER positive primary and metastatic tumours following endocrine treatment has been described in several clinical studies [4]. There is now considerable evidence to support a role for SRC-1 in coordinating the necessary changes to bring about ER independence in a significant subset of endocrine resistant tumours. Individual mechanistic studies have shown that, the SRC-1/AP1/M-CSF1 regulatory axis can enhance circulating tumour cells and lung metastasis, SRC-1 can also partner with PEA3 to regulate TWIST to drive epithelial-mesenchymal transition [37, 38]. In this study we took a high level view of SRC-1 transcriptional networks to map out SRC-1’s greater regulatory cascade which contributes to enhanced aggression of the cancer cell.
In previous work using MALDI-TOF and LC mass spectrometry screens we identified the TF HOXC11 and the chromatin remodellers, HMGB2 and Jumonji domain containing 2C as interacting partners for SRC-1 [16, 33, 39]. To specifically understand chromatin and TF/ SRC-1 complexes here we employed RIME in endocrine resistant cells. Using this technique, we identified 148 endogenous new and previously described SRC-1 complex proteins. Though this method is a powerful tool to identify new transcriptional interacting partners it is by no means comprehensive; one such caveat is that RIME of a co-activator, unlike than RIME of a TF, may fail to detect other functional co-coactivators including CBP/p300 or CARM1. Nevertheless, of the interactors identified, 27 are known TFs or chromatin remodellers, one of which is the signal transducer and activator of transcription STAT1. STAT1 is one of the 7 STAT family members which function as signal messengers and TFs participating in cellular responses to cytokines and growth factors [40]. Though interactions between SRC-1 and STAT3, STAT5, and STAT6 have previously been reported, this is the first description of a STAT1/SRC-1 interaction [41–43]. In tumour initiation processes STAT3 and STAT5 are thought to be oncogenic, whereas STAT1 has been described as a tumour suppressor [44]. In terms of endocrine resistance, both STAT3 and STAT5 have a defined role in growth factor mediated tumour progression [45]. Recently, expression and activation of STAT1 has been shown to be enhanced in endocrine resistance, leading to suggestions that STAT1 could represent a viable target in treatment resistant breast cancer [46].
SRC-1 is a metastatic oncogene with a distinct role in endocrine resistance [20]. Through interactions with key TFs, including PEA3 and MYB, it can reduce breast cancer cell differentiation and enhance tumour progression [15, 32]. To investigate first line SRC-1 mediated transcriptional regulation we combined SRC-1 RNAseq and ChIPseq, followed by bioinformatics filters which uncovered a refined list of SRC-1 regulated TFs and chromatin remodellers. Seven SRC-1 target genes, five TFs; ASCL1, E2F7, NFIA, SMAD2 and TRPS1 and two chromatin remodellers including DEK and SMARCA1 were subsequently validated in molecular studies as direct SRC-1 target genes. In this study, though each of the SRC-1 regulated TFs had varying roles in tumour cell viability and anchorage independent growth, all were found to be essential for full cellular de-differentiation and migration. These observations are consistent with the supposition that SRC-1 can manage its pro-metastatic activity though this network of TFs.
Four of the five TFs have well defined response elements, ASCL1, E2F7, NFIA, SMAD2 enabling robust modelling of extended SRC-1 regulatory networks. In PDX models of endocrine sensitivity and resistance, each of the four TFs in the hub were elevated in the endocrine resistant metastatic tumours (liver and/or lung) in comparison to the primary tumour expanded from an endocrine sensitive patient. A role for each of these TFs in endocrine cancer disease progression has previously been reported. NFIA and ASCL1 regulate FOXA1/AR and neuroendocrine differentiation, respectively in prostate cancer, whereas roles for SMAD2 and E2F7 in breast cancer disease progression and endocrine resistance have been well described [47–50].
On modelling the extended regulatory network of the clinically relevant SRC-1 TF hub, we found a preponderance of genes relevant to altered cell differentiation, pathways in cancer and cell cycle regulation. At the core of the hub key cell cycle proteins, CENPF and CDKN1A, were found to be co-regulated by all four TFs. Of interest, NFIA has previously been reported to negatively regulate cyclin-dependent kinase inhibitor p21 (CDKN1A) [51]. Either singularly or together expression of these cell cycle regulators associate with poor disease free survival in endocrine treated breast cancer patients. Of note, clinical trials targeting cell cycle have shown promise in the treatment of endocrine resistant breast cancer [52].
Differential expression of the SRC-1 network genes was observed in tumours from endocrine sensitive and resistant patients. Loss of genes pertinent to cell differentiation and gain of cell cycle genes was observed in resistant tumours relative to their matched primaries. This analysis is consistent with the observed role of the SRC-1 target TFs described here and the well-defined role of SRC-1 in cell de-differentiation and tumour progression [32].
TFs and their associated regulatory proteins respond to environmental cues to control cell fate. Due to functional redundancy and discordance between expression levels and activity, transcriptional proteins are often poor markers of cell health. Uncovering downstream networks can help us understand how these master regulators execute their function. These analyses have the potential to provide more robust prognostic markers and effective therapeutic targets to treat metastatic cancer. Here, using discovery tools, molecular and clinical studies, as well as bioinformatic modelling we describe a clinically relevant cadre of 4 TFs which execute SRC-1’s role in disease progression in a subset of endocrine resistant breast cancer. At the core of this signalling cascade are genes central to cell differentiation, pathways in cancer and cell cycle. This extended analysis has uncovered a regulatory network which provides potential new drug targets and insights into the mechanism of recent cell cycle inhibitors in the treatment of this class of breast cancer patient.
Materials and methods
Cell culture
Endocrine resistant LY2 breast cancer cells were a kind gift from Robert Clarke (Georgetown, USA). LY2 cells were derived from MCF-7 cells by selection in increasing concentrations of LY 117018 [53]. LY2 cells are also cross-resistant to tamoxifen and are ER positive with ER protein expression lower than MCF-7 cells [16, 39, 53]. LY2 cells were maintained in phenol red free Minimum Essential Medium Eagle (PRF-MEM, Sigma, Darmstadt, Germany) supplemented with 10% charcoal dextran stripped fetal calf serum (CDS-FCS, Sigma), 1% L-glutamine (LG, Sigma), 1% penicillin-streptomycin (PS, Sigma) and 4-hydroxytamoxifen (4-OHT, Sigma) 10−8 mol/L. A stable SRC-1 knock-down cell line was created, as previously described [39]. LY2 shSRC-1 and LY2 shNT cells were maintained in LY2 cell culture media containing puromycin (500 ng/mL, Sigma) to maintain stable knockdown expression. All cell lines were tested for Mycoplasma (LT07-118, Lonza, Basel, Switzerland), genotyped (Source BioSciences, Nottingham, UK) and authenticated according to ATCC guidelines.
RIME
The rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) method, mass spectrometry and data analysis were performed, as previously described [54]. LY2 cells were steroid depleted for 72 h, treated with 4-OHT 10−7 mol/L for 45 min and harvested for RIME. SRC-1 antibody (anti-human, 10 µg; SC-8995; Santa Cruz Biotechnology, Texas, USA), and IgG (10 µg; C15410206 Diagenode) as negative control, were used. Raw data were pre-processed, analysed and results visualised as per Mohammed et al. methodology [55]. RIME data will be deposited in the Gene Expression Omnibus (GEO) database.
RNA Sequencing and bioinformatics analyses
LY2 shSRC-1 and shNT cells were steroid depleted for 72-h then treated with 4-OHT 10−7 mol/L for 8 h. RNA isolation was performed using the RNeasy kit (ID-74104, Qiagen, Hilden, Germany). True-seq RNA-sequencing and library construction was performed by BGI (Hong Kong) using the Illumina HiSeq 2000 with > 40 million reads (100PE) (Extended RNASeq analysis can be found in Supplementary Information). RNASeq data are available from GEO database under series entry code GSE99649. ChIP-seq as per McCartan et al. was used to identify direct SRC-1 targets. ChIP-seq data are available from the GEO database under series entry code GSE28987 [32]. Eligible breast cancer patient cases had formalin-fixed paraffin-embedded (FFPE) tissue from primary and recurrent tumours. Informed consent was received from all eligible patients and the study was reviewed and approved by Institutional Review Boards from Royal College of Surgeons in Ireland (13/09 CTRIAL-IE Protocol 09-07). FFPE sections (5 × 10 µm) were used to extract RNA using Qiagen FFPE kit according to manufacturer’s instructions. Tru-seq RNA sequencing was carried out, as previously described [56]. Heatmaps were generated using pHeatmap in R [57].
Chromatin immunoprecipitation and ChIP-reChIP
Chromatin immunoprecipitation (ChIP) was performed, as previously described [58]. LY2 cells were steroid depleted for 72 h and treated with 4-OHT 10−7 mol/L for 45 min and lysed using a Bioruptor sonicator (B01060001, Diagenode, Belgium). SRC-1 antibody (6 µg; SC-8995; Santa Cruz Biotechnology) STAT1 (1 µg, 9172S, Cell Signalling, MA, USA), ER (6 µg, SC-543, Santa Cruz Biotechnology) and IgG (6 µg; C15410206 Diagenode), as negative control, were used. ChIP primers were designed around the promoter region of each gene of interest (Supplementary Table 5). Enrichment of the DNA to E2F7, NFIA, DEK, SMAD2, SMARCA1, ASCL1 and TRPS1 promoters was carried out using qPCR with Sybr green technology (Qiagen) on a Roche Lightcycler 2.0 (Roche, Basel, Switzerland). The ΔΔCt comparative method was used for analysis [59].
ChIP-reChIP was performed, as described above, with the exception of an additional immunoprecipitation prior to reverse cross linking.
For ChIP experiments with siRNA knockdown, cells were transfected in 15 cm2 dishes followed by 72 h steroid depletion. Cells were then treated with 4-OHT 10−7 mol/L for 45 min and ChIP protocol was carried out, as described above.
Co-Immunoprecipitation, protein extraction and western blot
LY2 cells were steroid depleted for 72 h then treated with 4-OHT 10−7 mol/L for 45 min. Cells were collected and then lysed in 80 µL of lysis buffer supplemented with protease inhibitors (Roche). In total 400 µg of total protein was immunoprecipitated with SRC-1 (6 µg, SC-8995; Santa Cruz Biotechnology), STAT1 (1 µg, 9172S, Cell Signalling) and IgG (6 µg, C15410206 Diagenode) pre-bound to protein A Dynabeads (10002D, Life Technologies, CA, USA). Nuclear protein was extracted from LY2 cells using the NE-PER extraction kit (ThermoFisher) as per manufacturer’s instructions. In total 25 µg of nuclear protein was electrophoresed and immunoblotted with antibodies against SRC-1, STAT1 (as above) and TBP (8515, Cell Signalling).
Transfection
siRNA directed against SRC-1, E2F7, NFIA, ER (Ambion, MA, USA), STAT1, DEK, SMAD2, SMARCA1, ASCL1 and TRPS1 (Dharmacon, CO, USA) were used to transiently knockdown gene expression. Transfection was performed using Lipofectamine 2000 (Invitrogen, CA, USA) according to manufacturer’s protocol siRNA. siRNA details can be found in Supplementary Table 4. For mRNA expression analysis, 24 h following transfection, cells were treated with tamoxifen 10−7 M for 8 h then analysed by qPCR. mRNA primer sequences can be found in Supplementary Table 6. For functional experiments, 24–48 h post transfection, cells were input into the relevant assays, as detailed below.
Cell viability
Cell viability was assessed using MTS Cell Proliferation Assay kit (Promega, WI, USA). Cells were seeded in triplicate into a 96 well plate at a density of 1 × 103 cells per well. On day 5, cells were incubated with MTS reagent and absorbance was read at 495 nM using a PerkinElmer (MA, USA) plate reader.
Anchorage independent growth
In total 2 × 104 cells suspended in PRF-MEM with 0.3% agarose (Promega) and 4-OHT 10−7 mol/L were seeded in duplicate into a 6 well plate containing a layer of solid 0.6% agarose in PRF-MEM and 4-OHT 10−7 mol/L. The agarose was allowed to solidify and was topped with 300 µL of PRF-MEM with 4-OHT 10−7 mol/L. Samples were incubated at 37 °C with 5% CO2 for 14 days. On day 15, colonies were stained with 400 μL of p-Iodoinitrotetrazolium chloride (1 mg/mL, Sigma Aldrich) and wrapped in aluminium foil for 24 h at 37 °C with 5% CO2, then stored in the fridge for 24 h prior to counting. Using a graticule, colonies over 100 µM in diameter were counted at 8 different points per well under a light microscope (Olympus, Tokyo, Japan).
Mammospheres
In total 5 × 103 cells were seeded in duplicate into a 6 well ultra-low adherence plate (Corning, MA, USA) containing 2 mL of mammosphere media (DMEM F12 (Sigma) supplemented with 1% PS, EGF (20 ng/mL, Sigma) and 1× B27 (Sigma)). Mammosphere culture and analysis was performed, as previously described [60].
3D acini
24 h post siRNA transfection, 5 × 103 cells were suspended in 400 µL of PRF-MEM containing 2% Matrigel (BD, Biosciences, CA, USA) and were seeded in duplicate into a Matrigel coated 8-well chamber slide (BD Biosciences). Cells were cultured for 21 days then fixed and stained as per Debnath et al. [61]. Phalloidin 594 (A12381, Life Technologies) was used to stain F-actin and 4′, 6-diamidino-2-phenylindole (DAPI, 0.5 µg/ml, Sigma) stained the nucleus. Acini were examined on a confocal microscope (Carl Zeiss LSM 710 NLO, Oberkochen, Germany).
Flow cytometry
After 48 h of siRNA transfection, 2 × 105 cells were stained with CD24 (1:20, 555428; BD Biosciences, CA, USA) and CD44 (1:6.6, 555478; BD Biosciences) before analysis on BD FACS Canto II (BD Biosciences). Results were analysed using FlowJo Software (FlowJo, OR, USA).
Migration
Cellomics Cell Motility Kit (K0800011, Thermo Scientific, MA, USA) was used to assess individual cell movement as per manufacturer’s instructions. Briefly, cells were seeded in triplicate with 500 cells per well and treated with PRF-MEM supplemented with 4OHT 10−7 mol/L. Following 24 h of culture, cells were fixed, stained and migration was analysed by examining the average track areas. A minimum of 100 tracks per sample was examined on an Olympus light microscope using DAPI and TRITC filters and CellSens Entry software.
Patient derived xenograft
All mouse experiments were performed in accordance with the European Communities Council Directive 2010/63/EU and were reviewed and approved by Research Ethics Committee under license from the Health Products Regulatory Authority of Ireland. PDX models were generated from three individual patients (T060_1, T638_1 and T1233_3) and were maintained, as previously described [62]. Sensitive and resistant PDXs were established using orthotopic implantation in Nod-SCID mice (Charles Rivers, MA, USA) supplemented with E2 pellets (Innovative Research of America) from three ER positive tamoxifen-treated patient tumours. One primary sample from an endocrine sensitive patient tumour and two metastatic samples (liver and lung) from resistant tumours were included in the study (also see Fig. 3a). Upon tumour harvesting, tumours were formalin fixed and paraffin embedded (FFPE).
Immunohistochemistry
Immunohistochemistry was performed on 5 μM sections of PDX samples using DAKO EnVison + HRP kit (Agilent, CA, USA) according to manufacturer’s instructions. Primary antibodies to detect ER (rabbit, 1:50, Ventana 790-4324), PR (rabbit, 1:50, Ventana 790-2223), ki67 (M7240, DAKO), SRC-1 (rabbit; 1:30 SC-8995; Santa Cruz Biotechnology), E2F7 (rabbit, 1:50, sc66870, SantaCruz Biotechnology), NFIA (anti-human, 1:100, 41,851, Abcam) were incubated at 25 °C for 1 h, ASCL1 (anti-human, 10 µg, 556,604, BD Biosciences) at 37 °C for 2 h and STAT1 (anti-human, 1:750, 9172 S, Cell Signalling) and pSMAD2 (anti-human, 1:250, 3108, Cell Signalling) at 4 °C overnight.
Network analysis
The iRegulon (Version: 1.3 Build ID: 1024) Cytoscape (v.3.4.0) plugin was used to query the cisTargetDB for direct downstream targets of candidate TFs (meta-regulon) [63]. The cisTargetDB integrates cancer related gene signatures from GeneSigDB(v.4), MSigDB(v3.2) and from gene modules generated across 91 cancer microarray data sets [64]. The meta-regulon is defined in iRegulon as the genes which are directly targeted by a given TF significantly enriched across the cisTargetDB gene signatures. Meta-regulons for each TF were obtained using an occurrence count threshold of 5 and number of nodes of 200 and then merged using firstly set union to remove duplicate genes and secondarily set intersection in Cytoscape to find those genes common to all TFs relevant to this study.
Statistical analysis
Statistical analyses performed using GraphPad Prism Version 5 software (La Jolla, CA, USA). Data are shown as mean ± standard error of the mean (SEM), with a minimum three replicates per experiment. Exact n numbers for each graph are reported in the Figure Legends. Significance was determined using unpaired, two-tailed, Students t-test. P values < 0.05 were considered significant and are denoted by ‘*’ while no significance is denoted n.s.
Electronic supplementary material
Acknowledgements
We kindly acknowledge the funding support from the Science Foundation Ireland (08-IN1-B1853 and 12/1A/1294), the Health Research Board of Ireland (HRB/POR/2012/101). This work was partly supported by the Irish Cancer Society Collaborative Cancer Research Centre grant, BREASTPREDICT, CCRC13GAL.
Author contributions
Conception and design: AB, SCh, LY. Development of methodology: AB, SCh, DV, AF, NC, SCo, JC, AR, LY. Acquisition of data: AB, SCh, DV, AF, NC, SCo, SP, EW, FB, LH, AR, LY, Analysis and interpretation of data: AB, SCh, DV, AF, NC, EW, AR, LY. Writing, review and/or revision of the manuscript: AB, SCh, DV, AF, NC, SCo, LY. Study supervision: AH, LY.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
The online version of this article (10.1038/s41388-017-0042-x) contains supplementary material, which is available to authorized users.
References
- 1.Carroll JS. Mechanisms of oestrogen receptor (ER) gene regulation in breast cancer. Eur J Endocrinol. 2016;175:R41–9. doi: 10.1530/EJE-16-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoefnagel LD, van de Vijver MJ, van Slooten HJ, Wesseling P, Wesseling J, Westenend PJ, et al. Receptor conversion in distant breast cancer metastases. Breast Cancer Res. 2010;12:R75. doi: 10.1186/bcr2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Priedigkeit N, Hartmaier RJ, Chen Y, Vareslija D, Basudan A, Watters RJ, et al. Intrinsic subtype switching and acquired ERBB2/HER2 amplifications and mutations in breast cancer brain metastases. JAMA Oncol. 2016;3:666–671. doi: 10.1001/jamaoncol.2016.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dieci MV, Barbieri E, Piacentini F, Ficarra G, Bettelli S, Dominici M, et al. Discordance in receptor status between primary and recurrent breast cancer has a prognostic impact: a single-institution analysis. Ann Oncol. 2013;24:101–8. doi: 10.1093/annonc/mds248. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, Yuan Y, Liao L, Kuang SQ, Tien JCY, O’Malley BW, et al. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc Natl Acad Sci Usa. 2009;106:151–6. doi: 10.1073/pnas.0808703105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao TP, Ku G, Zhou N, Scully R, Livingston DM. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci Usa. 1996;93:10626–31. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheppard HM, Harries JC, Hussain S, Bevan C, Heery DM. Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1. Mol Cell Biol. 2001;21:39–50. doi: 10.1128/MCB.21.1.39-50.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 9.Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2001;276:1089–98. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- 10.Stashi E, York B, O’Malley BW. Steroid receptor coactivators: servants and masters for control of systems metabolism. Trends Endocrinol Metab. 2014;25:337–47. doi: 10.1016/j.tem.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SK, Kim HJ, Na SY, Kim TS, Choi HS, Im SY, et al. Steroid receptor coactivator-1 coactivates activating protein-1-mediated transactivations through Interaction with the c-Jun and c-Fos subunits. J Biol Chem. 1998;273:16651–4. doi: 10.1074/jbc.273.27.16651. [DOI] [PubMed] [Google Scholar]
- 12.Kim HJ, Kim JH, Lee JW. Steroid receptor coactivator-1 interacts with serum response factor and coactivates serum response element-mediated transactivations. J Biol Chem. 1998;273:28564–7. doi: 10.1074/jbc.273.44.28564. [DOI] [PubMed] [Google Scholar]
- 13.Na SY, Lee SK, Han SJ, Choi HS, Im SY, Lee JW. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor κB-mediated transactivations. J Biol Chem. 1998;273:10831–4. doi: 10.1074/jbc.273.18.10831. [DOI] [PubMed] [Google Scholar]
- 14.Myers E, Hill ADK, Kelly G, McDermott EW, O’Higgins NJ, Buggy Y, et al. Associations and interactions between Ets-1 and Ets-2 and coregulatory proteins, SRC-1, AIB1, and NCoR in breast cancer. Clin Cancer Res. 2005;11:2111–22. doi: 10.1158/1078-0432.CCR-04-1192. [DOI] [PubMed] [Google Scholar]
- 15.Fleming FJ, Myers E, Kelly G, Crotty TB, McDermott EW, O’Higgins NJ, et al. Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1. J Clin Pathol. 2004;57:1069–74. doi: 10.1136/jcp.2004.016733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIlroy M, McCartan D, Early S, Gaora POacute, Pennington S, Hill ADK, et al. Interaction of developmental transcription factor HOXC11 with steroid receptor coactivator SRC-1 mediates resistance to endocrine therapy in breast cancer. Cancer Res. 2010;70:1585–94. doi: 10.1158/0008-5472.CAN-09-3713. [DOI] [PubMed] [Google Scholar]
- 17.Walsh CA, Qin L, Tien JC, Young LS, Xu J. The function of steroid receptor coactivator-1 in normal tissues and cancer. Int J Biol Sci. 2012;8:470–85. doi: 10.7150/ijbs.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBryan J, Theissen SM, Byrne C, Hughes E, Cocchiglia S, Sande S, et al. Metastatic progression with resistance to aromatase inhibitors is driven by the steroid receptor coactivator SRC-1. Cancer Res. 2012;72:548–59. doi: 10.1158/0008-5472.CAN-11-2073. [DOI] [PubMed] [Google Scholar]
- 19.Qin L, Chen X, Wu Y, Feng Z, He T, Wang L, et al. Steroid receptor coactivator-1 upregulates integrin alpha(5) expression to promote breast cancer cell adhesion and migration. Cancer Res. 2011;71:1742–51. doi: 10.1158/0008-5472.CAN-10-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res. 2009;69:3819–27. doi: 10.1158/0008-5472.CAN-08-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–43. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda Y, Hunter TC, Loudy DE, Dave V, Schreiber V, Whitsett JA. PARP-2 interacts with TTF-1 and regulates expression of surfactant protein-B. J Biol Chem. 2006;281:9600–6. doi: 10.1074/jbc.M510435200. [DOI] [PubMed] [Google Scholar]
- 23.Dammer EB, Leon A, Sewer MB. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3’,5’-monophosphate-dependent cytochrome P450c17 transcription rate. Mol Endocrinol. 2007;21:415–38. doi: 10.1210/me.2006-0361. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Lonard DM, O’Malley BW. The role of steroid receptor coactivators in hormone dependent cancers and their potential as therapeutic targets. Horm Cancer. 2016;7:229–35. doi: 10.1007/s12672-016-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–52. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulakovskiy IV, Medvedeva YA, Schaefer U, Kasianov AS, Vorontsov IE, Bajic VB, et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013;41:D195–D202. doi: 10.1093/nar/gks1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–7. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–43. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Privette Vinnedge LM, Benight NM, Wagh PK, Pease NA, Nashu MA, Serrano-Lopez J, et al. The DEK oncogene promotes cellular proliferation through paracrine Wnt signaling in Ron receptor-positive breast cancers. Oncogene. 2015;34:2325–36. doi: 10.1038/onc.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Y, Xiao Y, Wang W, Wang Q, Yearsley K, Wani AA, et al. Inhibition of expression of the chromatin remodeling gene, SNF2L, selectively leads to DNA damage, growth inhibition, and cancer cell death. Mol Cancer Res. 2009;7:1984–99. doi: 10.1158/1541-7786.MCR-09-0119. [DOI] [PubMed] [Google Scholar]
- 31.Wu L, Wang Y, Liu Y, Yu S, Xie H, Shi X, et al. A central role for TRPS1 in the control of cell cycle and cancer development. Oncotarget. 2014;5:7677–90. doi: 10.18632/oncotarget.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCartan D, Bolger JC, Fagan A, Byrne C, Hao Y, Qin L, et al. Global characterization of the SRC-1 transcriptome identifies ADAM22 as an ER-independent mediator of endocrine-resistant breast cancer. Cancer Res. 2012;72:220–9. doi: 10.1158/0008-5472.CAN-11-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redmond AM, Byrne C, Bane FT, Brown GD, Tibbitts P, O’Brien K, et al. Genomic interaction between ER and HMGB2 identifies DDX18 as a novel driver of endocrine resistance in breast cancer cells. Oncogene. 2015;34:3871–80. doi: 10.1038/onc.2014.323. [DOI] [PubMed] [Google Scholar]
- 34.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–91. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rs Janky, Verfaillie A, Imrichová H, Van de Sande B, Standaert L, Christiaens V, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol. 2014;10:e1003731. doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin L, Wu YL, Toneff MJ, Li D, Liao L, Gao X, et al. NCOA1 directly targets M-CSF1 expression to promote breast cancer metastasis. Cancer Res. 2014;74:3477–88. doi: 10.1158/0008-5472.CAN-13-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Hu B, Qin L, Zhao L, Wang Q, Wang Q, et al. SRC-1 and Twist1 Expression Positively Correlates with a Poor Prognosis in Human Breast Cancer. Int J Biol Sci. 2014;10:396–403. doi: 10.7150/ijbs.8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walsh CA, Bolger JC, Byrne C, Cocchiglia S, Hao Y, Fagan A, et al. Global gene repression by the steroid receptor coactivator SRC-1 promotes oncogenesis. Cancer Res. 2014;74:2533–44. doi: 10.1158/0008-5472.CAN-13-2133. [DOI] [PubMed] [Google Scholar]
- 40.Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. 2001;13:211–7. doi: 10.1016/S0955-0674(00)00199-X. [DOI] [PubMed] [Google Scholar]
- 41.Giraud S, Bienvenu F, Avril S, Gascan H, Heery DM, Coqueret O. Functional interaction of STAT3 transcription factor with the coactivator NcoA/SRC1a. J Biol Chem. 2002;277:8004–11. doi: 10.1074/jbc.M111486200. [DOI] [PubMed] [Google Scholar]
- 42.Litterst CM, Kliem S, Marilley D, Pfitzner E. NCoA-1/SRC-1 is an essential coactivator of STAT5 that binds to the FDL motif in the alpha-helical region of the STAT5 transactivation domain. J Biol Chem. 2003;278:45340–51. doi: 10.1074/jbc.M303644200. [DOI] [PubMed] [Google Scholar]
- 43.Litterst CM, Pfitzner E. Transcriptional activation by STAT6 requires the direct interaction with NCoA-1. J Biol Chem. 2001;276:45713–21. doi: 10.1074/jbc.M108132200. [DOI] [PubMed] [Google Scholar]
- 44.Clevenger CV. Roles and regulation of Stat family transcription factors in human breast cancer. Am J Pathol. 2004;165:1449–60. doi: 10.1016/S0002-9440(10)63403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113:365–71. doi: 10.1038/bjc.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meissl K, Macho-Maschler S, Muller M, Strobl B. The good and the bad faces of STAT1 in solid tumours. Cytokine. 2017;89:12–20. doi: 10.1016/j.cyto.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 47.Grabowska MM, Elliott AD, DeGraff DJ, Anderson PD, Anumanthan G, Yamashita H, et al. NFI transcription factors interact with FOXA1 to regulate prostate-specific gene expression. Mol Endocrinol. 2014;28:949–64. doi: 10.1210/me.2013-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uysal-Onganer P, Kawano Y, Caro M, Walker MM, Diez S, Darrington RS, et al. Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells. Mol Cancer. 2010;9:55. doi: 10.1186/1476-4598-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Kong J, Chang H, Kim H, Kim A. EGF induces epithelial-mesenchymal transition through phospho-Smad2/3-Snail signaling pathway in breast cancer cells. Oncotarget. 2016;7:85021–85032. doi: 10.18632/oncotarget.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu J, Zhu Y, Liu Y, Sun L, Lv X, Wu Y, et al. E2F7 overexpression leads to tamoxifen resistance in breast cancer cells by competing with E2F1 at miR-15a/16 promoter. Oncotarget. 2015;6:31944–57. doi: 10.18632/oncotarget.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glasgow SM, Laug D, Brawley VS, Zhang Z, Corder A, Yin Z, et al. The miR-223/nuclear factor I-A axis regulates glial precursor proliferation and tumorigenesis in the CNS. J Neurosci: J Soc Neurosci. 2013;33:13560–8. doi: 10.1523/JNEUROSCI.0321-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13:417–30. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 53.Bronzert DA, Greene GL, Lippman ME. Selection and characterization of a breast cancer cell line resistant to the antiestrogen LY 117018. Endocrinology. 1985;117:1409–17. doi: 10.1210/endo-117-4-1409. [DOI] [PubMed] [Google Scholar]
- 54.Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, D’Santos CS. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11:316–26. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 55.Mohammed H, D’Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–9. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McBryan J, Fagan A, McCartan D, Bane FT, Vareslija D, Cocchiglia S, et al. Transcriptomic profiling of sequential tumors from breast cancer patients provides a global view of metastatic expression changes following endocrine therapy. Clin Cancer Res: J Am Assoc Cancer Res. 2015;21:5371–9. doi: 10.1158/1078-0432.CCR-14-2155. [DOI] [PubMed] [Google Scholar]
- 57.Kolde R pheatmap: Pretty Heatmaps. 1.0.8 ed: R; 2015.
- 58.Vareslija D, McBryan J, Fagan A, Redmond AM, Hao Y, Sims AH, et al. Adaptation to AI therapy in breast cancer can induce dynamic alterations in ER activity resulting in estrogen-independent metastatic tumors. Clin Cancer Res: J Am Assoc Cancer Res. 2016;22:2765–77. doi: 10.1158/1078-0432.CCR-15-1583. [DOI] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Shaw FL, Harrison H, Spence K, Ablett MP, Simoes BM, Farnie G, et al. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J Mammary Gland Biol Neoplasia. 2012;17:111–7. doi: 10.1007/s10911-012-9255-3. [DOI] [PubMed] [Google Scholar]
- 61.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 62.Vareslija D, Cocchiglia S, Byrne C, Young L. Patient-derived xenografts of breast cancer. Methods Mol Biol. 2017;1501:327–36. doi: 10.1007/978-1-4939-6475-8_17. [DOI] [PubMed] [Google Scholar]
- 63.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verfaillie A, Imrichova H, Janky R, Aerts S. iRegulon and i-cistarget: reconstructing regulatory networks using motif and track enrichment. Curr Protoc Bioinf. 2015;52:2.16.1–39. doi: 10.1002/0471250953.bi0216s52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.