Abstract
Colorectal cancer (CRC) accounts for ~9% of all cancers in the Veteran population, a fact which has focused a great deal of the attention of the VA’s research and development efforts. A field-based meeting of CRC experts was convened to discuss both challenges and opportunities in precision medicine for CRC. This group, designated as the VA Colorectal Cancer Cell-genomics Consortium (VA4C), discussed advances in CRC biology, biomarkers, and imaging for early detection and prevention. There was also a discussion of precision treatment involving fluorescence-guided surgery, targeted chemotherapies and immunotherapies, and personalized cancer treatment approaches. The overarching goal was to identify modalities that might ultimately lead to personalized cancer diagnosis and treatment. This review summarizes the findings of this VA field-based meeting, in which much of the current knowledge on CRC prescreening and treatment was discussed. It was concluded that there is a need and an opportunity to identify new targets for both the prevention of CRC and the development of effective therapies for advanced disease. Also, developing methods integrating genomic testing with tumoroid-based clinical drug response might lead to more accurate diagnosis and prognostication and more effective personalized treatment of CRC.
Keywords: Biomarkers, Cancer stem cells, Clinical drug response, Colorectal cancer, FiSS, Genomic testing, Next generation sequencing, Precision Oncology Program (POP), Tumoroids
Introduction
Colorectal cancer (CRC) is the second most common cause of cancer-related deaths in the USA with an estimated 5% lifetime risk [1]. Another 3% of individuals diagnosed with CRC have a high-risk hereditary cancer syndrome such as Lynch syndrome. In recent years, a major emphasis has been placed on screening of conventional adenomatous and serrated polyps with the intention of removing them prior to their progression to CRC. The serrated polyp has been recognized as a progenitor of 15–30% of all CRCs. Technological advancements in CRC screening procedures and modalities, such as fecal immunochemical test (FIT), air contrast barium enema, and colonoscopy, have led to a decrease in mortality from CRC since the mid-1980s [1]. Despite these screening tools, only 40% of CRCs are diagnosed at an early stage (I or II) [2]. This is due in part to a lack of patient compliance, limited access to screening colonoscopy, and the low sensitivity/specificity of more commonly used tests such as FIT [2]. Furthermore, a “one drug fits all” cancer treatment approach has been unsuccessful in effectively reducing cancer deaths. For these reasons, there is an urgent need to identify new strategies for the prevention of CRC as well as the development of effective therapeutics for advanced disease.
Specifically, the US Veteran population represents 3% of all cancer patients in the USA and CRC accounts for ~9% of all cancers among Veterans [4]. This is because a large proportion of Veterans receiving care under Veterans Health Administration (VHA) have poor health, multiple comorbidities, and lower income [3]. Further, the incidence of cancer among Veterans is likely to increase with the aging population, an increase in military personnel, and an increase in Veterans seeking health care in VHA [4]. VHA has supported a Precision Oncology Program (POP) that uses multi-gene, next-generation sequencing (NGS) panels for patients diagnosed with Stage III or IV CRC, with an intent to provide precision cancer treatment (PCTx). These panels screen for ~ 200 genes, cost about $1500 per panel, have a turnaround time of about 2 weeks, and are not validated for PCTx use. Also, since genomic testing only provides a predictive approach, several tumor spheroid culture systems have been examined for their potential use in PCTx, but this technology also has several critical barriers. Thus, there is a need to integrate genomics and cell-based approaches, referred to as “cell-genomics,” to accurately diagnose, prognosticate, and treat gastrointestinal malignancies including pancreatic and CRCs. To this end, in a VA R&D supported field-based meeting, a panel of CRC experts met to review research advances on the biology, biomarkers, and treatment of CRC. The panel discussed the state-of-the-art science, challenges, and opportunities and pragmatically established a “VA Colorectal Cancer Cell-genomics Consortium” (VA4C) with the goal of fostering multi-site collaborative studies aimed at early detection, prevention, and diagnosis as well as novel treatments for CRC. This review summarizes these discussions on the latest developments in biology and biomarkers to individualize diagnostics and prognostication and precision oncology approaches, and charts future collaborative research directions.
Precision Diagnostics and Prognostics for CRC
Advances in Biology, Biomarkers, and Imaging for Early Detection of CRCs
To date the principal modalities for identifying early-stage CRCs have been through use of screening colonoscopy or fecal-based methods such as FIT. The rationale for this approach is to identify either polyps (precancerous) or early-stage CRCs before they develop into lethal tumors. Early-stage CRC is curable with surgical treatment, whereas advanced or metastatic disease may only be treated to prolong survival without curative intent [5]. The traditional paradigm is that sporadic CRC progresses in a stepwise fashion from normal epithelium, to aberrant crypt foci, to low-grade then high-grade adenomas and ultimately to adenocarcinoma which may then metastasize to other organs. This pattern of tumor progression was first identified by Vogelstein and colleagues some 30 years ago [6]. It is now known that cancer cell heterogeneity and other factors can modulate this progression. For example, recent studies suggest that CRCs originating from different locations may have different metastatic targets, for example, colon cancer spreads primarily to liver, whereas rectal cancer spreads primarily to lung [7]. Moreover, early in tumorigenesis, some cells within the primary tumor already possess the ability to invade and seed distant organs [8]. Such cells have acquired mesenchymal traits through the epithelial-to-mesenchymal transition (EMT). EMT is defined by a variety of genetic and morphological changes, which allow cells to invade normal colon epithelium, vascular endothelium, and eventually the parenchyma of target organs. Recent work indicates that the genetic signatures identifying EMT may also be cell context-dependent [9]. Metastatic cell deposits may grow slowly or lie dormant until circulating and stroma-dependent factors stimulate their arousal and growth.
An important anatomical factor very specific to CRC is the recently identified “serrated polyp-neoplasia” pathway [10–12]. Sessile serrated adenoma/polyps (SSA/Ps), established precursors of CRC, exhibit microsatellite instability (MSI) and account for approximately 15–30% of sporadic CRCs [13]. SSA/Ps are predominantly located in the right/proximal colon and observed in “interval cancers” occurring within a short time after a screening procedure (2–6% of all CRCs) [14, 15]. Serrated polyps are characterized by a variety of histologic features including irregular distribution of crypts, dilatation of crypt bases, serration at crypt bases, branched crypts, horizontal extension of crypt bases, dysmaturation of crypts, and herniation of crypts through the muscularis mucosae [16]. According to the American Gastroenterology Association criteria, even one crypt showing any of the above characteristics is sufficient to diagnosis SSA/P. Approximately 17.6% of SSA/Ps (> 2 cm) show residual adenomatous tissues when reexamined [17]. Unlike other adenomatous polyps, SSA/Ps are characterized by mutations in the BRAF proto-oncogene that causes the development of polyps. Hyper-methylation of CpG islands in the promoter regions of tumor suppressor genes, including MLH1, leads to a decrease in tumor suppression and an increase in sporadic MSI [10, 18–21]. Because benign hyperplastic polyps (HPs) and SSA/Ps have a different risk of developing into malignant disease, their accurate classification is critical for making clinical decisions (e.g., surveillance intervals, intervention). Understanding these complex processes and testing all prognostic and therapeutic measures in animal models before proceeding to clinical trials is challenging. It is noteworthy that although colonoscopic screening and polypectomy have led to a decline in advanced CRC, its utility for differentiating SSA/Ps from HPs has not yet been realized.
Genetic and Epigenetic CRC Biomarkers
The multistep process of CRC development is associated with the accumulation of genetic and epigenetic alterations in epithelial cells via distinct molecular pathways, including the common “adenoma-carcinoma” pathway as well as the recently identified “serrated polyp-neoplasia” pathway [6–8]. Hence, identifying these CRC molecular markers, i.e., biomarkers, is key to early detection, staging, and assessment of therapeutic efficacy, as reviewed recently [22]. Whereas current guidelines recommend surgery alone for Stage I and adjuvant chemotherapy for Stage III cancers, the appropriate treatment for Stage II cancers, which have breached the colon wall but not invaded lymph nodes or other organs, is uncertain. Although it is proposed that molecular biomarker panels, including genetic, epigenetic, protein and carbohydrate-based markers, may identify Stage II cancers that warrant aggressive treatment after surgical resection [23], none have been validated prospectively.
Circulating tumor DNA and RNA-based biomarkers offer high specificity and are ideal as predictive biomarkers for monitoring the response to chemotherapy as well as tumor progression, as reviewed recently [24]. However, due to low DNA yields and low mutational burden, they are inadequate for diagnosis. Cell-free microRNAs (miRNAs) and small non-coding RNAs (ncRNAs) are now emerging as noninvasive and highly sensitive biomarkers, but specificity remains a concern. For these reasons, large-scale prospective clinical studies aimed at carefully evaluating the sensitivity and specificity of these biomarkers are needed before their adoption to clinical practice.
Hereditary cancer syndromes have provided powerful insights into our understanding of somatic mutations present in sporadic cancers, as well as implicated cell signaling pathways [20–24]. One clear example is the identification of germline, inactivating mutations in the APC gene, which encodes a 300-kD wnt pathway adaptor protein [23]. Although germline mutations in APC are responsible for familial adenomatous polyposis (FAP), a rare condition affecting about 1 in 7000 individuals in the USA, somatic mutations in the APC gene are present in more than 70% of colonic adenomatous polyps and carcinomas [24]. Hereditary non-polyposis colon cancer (HNPCC), or Lynch syndrome, involves a germline mutation in a DNA mismatch repair (MMR) genes, leading to defective DNA repair. Recent studies identifying inactivation of TGF-β signaling through loss of the Smad3 adaptor ß-2 spectrin could potentially provide new insights into CRC development from stem-like tumor-initiating cells (STICs) for targeted chemoprevention.
Further, genome-wide association studies (GWAS) have identified numerous genetic loci that are associated with CRC, but hereditability remains unknown. More than 40 low-penetrant polymorphic variants are reported [25]. More recently in a Finnish cohort, a single-nucleotide polymorphism (SNP) rs992157 at chromosome 2q35 intronic to PNKD and TMBIMI was found to be associated with CRC [26]. Genome-wide alterations in DNA methylation influence gene expression resulting in altered aberrant crypt foci, an early precursor of CRC [27]. Recently, single-cell RNA sequencing (e.g., GemCode technology 10 × Genomics, San Francisco, CA) has permitted an assessment of transcriptional events that may be important in the development of cancers and could potentially predict therapeutics. In addition to genomic profiling approaches, depending upon the molecule of interest, several additional “omics” approaches are being utilized, including proteomics, metabolomics, lipidomics (chloroform/methanol), glycomics (glycoprotein isolation), and peptidomics.
TGF-β Receptor and SMAD Gene Mutations in Gastrointestinal/CRC
In normal and premalignant cells, TGF-β enforces homeostasis and suppresses tumor progression through tumor-suppressive effects on the stroma. Current data strongly support TGF-β signaling as a suppressor of early CRCs [28, 29]. In advanced disease, metastatic CRCs escape the tumor suppressor effects of TGF-β signaling by becoming resistant to TGF-β-induced growth inhibition [30]. Inactivating mutations in the TGFBR2 gene occur in most human CRC and gastric carcinomas that demonstrate MSI [31]. The TGF-β signaling network is also disrupted in cancer by mutations in Smad4, which occur in more than 30% of CRCs [32, 33]. Smad-deficient mice display phenotypes that suggest a tumor suppressor role for the Smads [34]. When mice with one mutated APC allele are crossed with heterozygous Smad4 mice, the compound heterozygotes develop larger polyps that can progress into malignant adenocarcinomas [35]. Although fewer Smad3 mutations have been found in human cancers, mice with a Smad3 homozygous deletion develop aggressive CRC at an early age in a manner that seems to be highly dependent on the genetic background of the mice.
Cross talk between CEA, TGF-β signaling, and STAT3
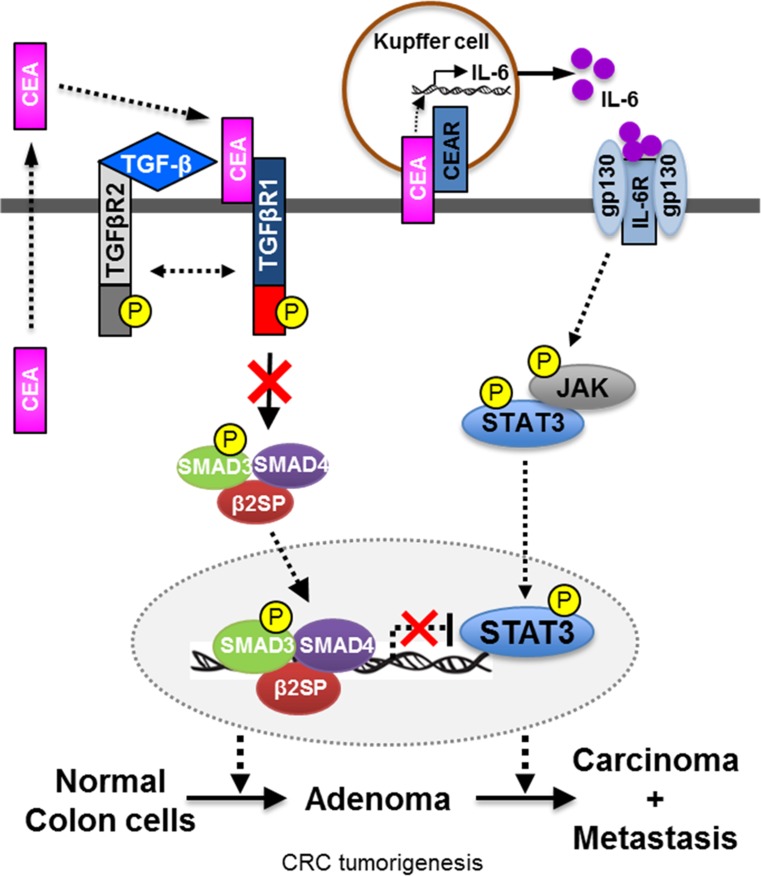
Figure 1 depicts cross talk between carcinoembryonic antigen (CEA), TGF-β signaling, and STAT3. Advanced CRC is associated with high levels of circulating CEA and a dismal survival; the elevated levels of CEA are especially associated with Stage IV cancers. Consequently, there is a need to identify new targets for both the prevention of CRC as well as the development of novel therapies for advanced cancer. CEA (also known as CEACAM5) is one of the few biomarkers approved by the U.S. Food and Drug Administration (FDA) for CRC [36]. TCGA studies have validated that elevated CEA levels and disruption of TGF-β signaling are observed in over 80% of right-sided CRCs [37]. Activation of STAT3 is also observed in this subgroup of patients. TGF-β induces the secretion of CEA in a dose-dependent manner [38]. Given the importance of CEA, TGF-β, and STAT3 signaling pathways in CRC tumorigenesis, cross talk between these pathways is a focus of investigation. CEA and CEACAM6 are known Smad3 target genes. An important direction for the future is designing studies that delineate the molecular mechanisms underlying the cross talk between CEA and the TGF-β and STAT3 signaling pathways during CRC development.
Fig. 1.
Schematics of cross talk between CEA, TGF-β signaling, and IL6-STAT3 pathway
Mucins as Biomarkers for CRC Early Detection and Prognosis
Several studies showed moderate to high inter-observer disagreement between pathologists in the histologic differentiation of SSA/Ps (highly malignant) and HPs (benign) [21, 39–46]. Considering this variability, studies have focused on identifying molecular markers for distinguishing between these two entities. For example, MUC5AC, TFF1, and Annexin A10 have emerged as potential markers for differentiating benign polyps from SSA/P [47, 48]. Aberrant changes in the expression, localization, and glycosylation of colonic mucin have been observed in very early stages of colon cancer. SSA/Ps exhibit an elevated expression of transmembrane mucin MUC17 and secreted mucin MUC5AC in comparison to HP [49]. Notably, in conjunction with these expressional changes, distinct differences were also observed among the studied polyp groups in MUC5AC and MUC4 localization. This signifies the ability of these markers to discriminate benign HP from SSA/P polyp subtypes. In multivariate regression models in conjunction with ROC curve analyses, the combination of MUC17/MUC5AC effectively discriminated SSA/Ps from HP [49]. The trio (CA 19-9, MUC17, and MUC5AC) emerged as a combined panel for discriminating polyp subtypes and thus accurately classifying colorectal polyps. Based on the lack of clear tools to identify SSA/Ps, histologic markers can aid in improving the differentiation of SSA/Ps from benign polyp subtypes. Due to difficulties in their diagnosis by endoscopists, their inter-observer variability among pathologists, and their highly malignant nature, a means of identifying SSA/Ps is central to the improved detection and timely treatment of CRC. There is a broad consensus that future studies should focus on identifying alternative molecular markers that can more successfully achieve these goals.
Biomarkers for Colon Cancer Dissemination
One novel approach to stratify the malignant potential of CRC is to identify cancer cells with distinct invasive potential at the tumor “front,” namely the interface between tumor and normal tissue. Nuclear accumulation of β-catenin distinguishes the cells at the “front” from those closer to the tumor core [50]. This is consistent with observations that cells tend to invade in clusters, not as individuals [9]; these cells likely share attributes that predispose them to behave as an aggressive unit. Cell clusters at the tumor front are more likely to co-overexpress M3 muscarinic receptors (M3R) and matrix metalloproteinase-1 (MMP1) within cytoplasmic vesicles [51]. This is potentially important since M3R activation robustly induces expression of MMP1, a secreted collagenase that promotes colon cancer invasion [52–54]. Validating the use of such histochemical tests in formalin-fixed paraffin-embedded tissues may yield an efficient and economical way to identify those Stage II cancers that require more aggressive therapy.
Histopathology with Neuroendocrine Tumor (NET) Stains Identifies Neuroendocrine Features of CRC
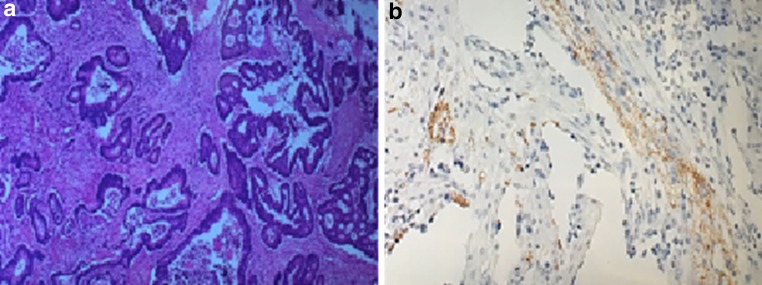
A subset of adenocarcinomas and adenomas possess neuroendocrine features [55]. Neuroendocrine cells possess bioactive amines and peptides that may have an important role in promoting the growth of transformed cells such as those within adenocarcinomas, as has been demonstrated with peptides such as PACAP [56]. The expression of receptors on CRCs can differentially couple to intracellular signaling molecules that promote proliferation [57]. A recent study identified neuroendocrine features in up to one-third of 40 patients with CRC (unpublished data); assays of chromogranin A and neuron-specific enolase were performed (Fig. 2). The presence of neuroendocrine features may suggest a stem cell component or portend a more aggressive tumor state. Neuroendocrine features may also suggest different chemotherapeutic or biologic strategies in advanced-stage CRC.
Fig. 2.
a NET features by H&E staining in colorectal adenocarcinoma specimen. b NET features by chromogranin A immunohistochemistry staining in colorectal adenocarcinoma specimen. Unpublished data
Role of Microbiome in CRCs
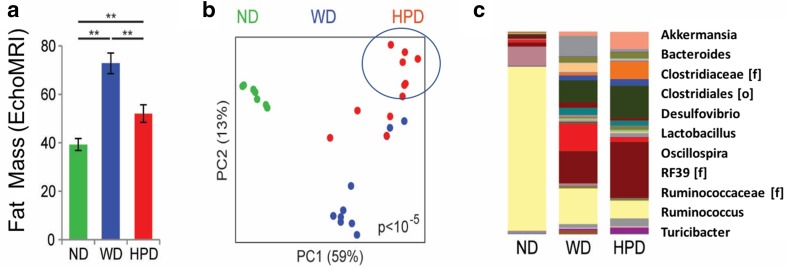
Well-established risk factors for colorectal adenomas and cancer include: a personal or family history of CRC or polyps; an inherited condition that predisposes to colon polyps and cancer; inflammatory bowel disease (e.g., Crohn’s disease or ulcerative colitis); obesity; type 2 diabetes; heavy alcohol use; a diet high in red meat and processed meats; smoking; and physical inactivity. Additional risk factors that have been described include fatty liver [58], hepatitis C [59], and Agent Orange exposure [60]; the latter is very relevant to Vietnam Veterans. Modifying known dietary and lifestyle risk factors and chemoprevention with aspirin and nonsteroidal anti-inflammatory drug (NSAID) use are associated with reduced risk for colon polyps and CRC [61]. In relation to the role of the diet and intestinal microbiome in CRC pathogenesis, the microbiome of subjects consuming a Western diet (WD) was compared to that of subjects consuming a high-protein diet (HPD) [62] (Fig. 3a, b). There was a highly significant difference in composition by weighted UniFrac analysis (p < 10−5) (Fig. 3b). At the genus level, the HPD microbiome was characterized by expansion of Akkermansia, Ruminococcus, and Bacteroides and depletion of Lactobacillus and Turicibacter (Fig. 3c). The HPD group had increased abundance of 114 operational taxonomic units (OTUs) selected at 97% similarity (corresponding approximately to species) at a threshold of q < 0.05 (representing significance adjusted for multiple hypothesis testing).
Fig. 3.
A high-protein diet induces fat loss and an altered intestinal microbiome in rats with Western diet-induced obesity. a Body fat mass of rats kept on a WD or switched to a HPD. Age-matched control rats on a ND are shown for comparison. **p < 0.01 b Principal coordinates analysis (PCoA) of the cecal luminal microbiome colored by diet. P value across groups calculated using Adonis. c Mean abundance of common genera. Some reads were identified only at the family (f) or order (o) level
Of these, only Akkermansia mucinophila (q = 0.008) and an unclassified Clostridiales (q = 0.04) had a statistically significant inverse correlation with fat mass after adjustment for diet [62]. Fusobacterium enrichment is associated with specific molecular subsets of CRCs, suggesting that there is a potential pathogenic role in the development of CRC [63]. Furthermore, it has also been shown that environmental pollutants may affect the microbiome, such as was observed with ultrafine particles [64]. Whether ultrafine particles affect the steps involved in colon cancer pathogenesis remains to be investigated.
Precision Imaging for Prevention of CRCs
Screening colonoscopy has emerged as perhaps the most effective lifesaving intervention against CRC to date. Pooled analysis of numerous large observational studies indicates that systematic removal of all visualized polyps including adenomatous and serrated polyps [43, 65] at colonoscopy decreases proximal and distal CRC incidence and mortality by > 60% [2]. Currently, a VA clinical trial (50,000-subject, prospective randomized controlled study) is underway (Cooperative Studies Program #577) to assess screening colonoscopy versus FIT for reducing CRC deaths. While cancer death rates in persons older than 50 years are now declining, likely due to the adoption of screening programs, recent data indicate that age-specific CRC incidence and mortality are actually rising in those < 50 years of age. Colonoscopy has not been fully protective due to several uncontrollable factors including missed polyps, operator factors [66–69], and an inability to detect atypical [70] serrated polyps and poorly visualized flat neoplasms [71]. Thus, colonoscopy can benefit from assistive technologies that improve the neoplastic polyp detection rate as a primary performance benchmark [72]. To this end, standard colonoscopy has undergone recent enhancements to improve detection of more subtle lesions and has been supplemented with other technologies that provide “real-time histology” to enable selective removal [73–75]. Clinical trials, however, have shown that both high definition colonoscopy and dye spray chromoendoscopy only marginally improve neoplastic polyp detection [76, 77]. Using retrograde and ultra-wide angle camera systems that reveal surface area between folds can improve neoplastic polyp detection by 23–70% [78, 79]. Selective wavelength imaging and transparent cap-assisted colonoscopy have had mixed results [80, 81]. Thus, despite some improvements there remains a major unmet need in the ability to detect and remove atypical and serrated polyps.
CRC: Advances in Precision Surgery and Treatment
Although surgical resection is the mainstay of treatment in early-stage CRC, incomplete removal of tumors leads to local or widely metastatic disease which is resistant to conventional chemotherapeutics. For patients whose disease has progressed to metastatic CRC, molecular profiling may shed light on why various treatments fail. The discussions in this section focus first on advancing CRC precision treatment through increased precision in CRC surgery. Secondarily it reviews the state of the art in targeted chemotherapies.
Fluorescence-Guided Surgery for CRC
Surgical resection for CRC has the greatest potential for cure. Since the application of complete mesocolic excision for colon cancer surgery, local 5-year recurrence rates have decreased [82]. Furthermore, achieving negative microscopic margins and complete resection of metastatic tumor (R0 resection) can significantly improve 5-year survival rates [83–87]. Despite achieving R0 resection rates in patients with CRC, local and distant recurrence has still been reported to be as high as 34% [84, 88, 89]. Furthermore, the prognosis of patients with local recurrence is poor [90]. Real-time, reliable imaging to detect positive surgical margins at the time of surgery would improve outcomes. Emerging techniques are revolutionizing the performance of cancer surgery [91]. Intraoperative fluorescence imaging, or fluorescence-guided surgery (FGS), can provide high fidelity tumor visualization for localization, resection, and margin confirmation [92]. Thus, targeted fluorescent labeling of cancer cells may change the way we find and treat cancer [93, 94].
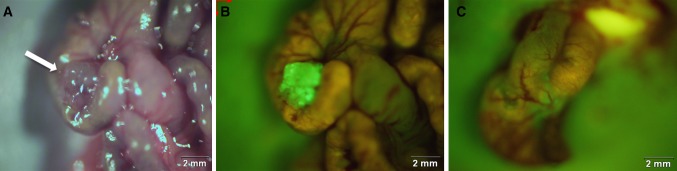
FGS combines advanced imaging platforms with targeted fluorescent agents to improve their intraoperative detection [95]. One technique is to covalently bind organic dye to a monoclonal antibody which labels a known tumor-specific antigen [91]. One such tumor biomarker is anti-CEA antibodies which label human CRC in murine models [92, 96–98] (Fig. 4). Mouse and chimeric (mouse/human) CEA antibodies, conjugated to fluorescent dyes, are capable of enhancing visualization of submillimeter tumor deposits [92] and aiding with FGS [99–101]. In an effort to improve therapeutic efficacy in the clinic, Yazaki et al. [102] humanized the murine anti-CEA T84.66 antibody with structurally similar “human” segments through a technique known as complementary determining region (CDR) grafting. This humanized anti-CEA antibody is currently used in clinical trials for PET imaging of CRC. In Europe, this technique was recently applied in clinical trials to localize CRC using SGM-101, an antibody-dye conjugate in which the fluorochrome BM104 is coupled to a chimeric monoclonal antibody against CEA [103]. Other fluorophore-conjugated antibodies to tumor-specific antigens such as insulin-like growth factor-1 receptor have been used in preclinical FGS for colon cancer [104]. In addition, viral vectors such as adenovirus and herpes virus can be used to deliver fluorescent proteins to cancer cells [105]. Activatable cell-penetrating peptides have been engineered to highlight tumors based on enzymatic cleavage of peptidases [106]. Indocyanine green (ICG), used clinically to define liver tumor margins, tissue perfusion, and biliary anatomy, has been shown to reduce leak rates after low anterior resection for CRC [107]. Thus, the use of FGS as a surgical guide has the potential to increase both survival and quality of life for patients [108]. FGS has great potential for a broad range of clinical applications, including in CRC.
Fig. 4.
Labeling of orthotopic human colon tumors in nude mice with fluorophore-conjugated chimeric anti-CEA antibodies. Panel A shows bright-field image of an orthotopic colon tumor (white arrow) and panel B shows the bright fluorescence of the tumor 48 h after labeling the fluorophore chimeric anti-CEA antibodies. The absence of any fluorescence signal after fluorescence-guided surgery in panel C indicates a complete resection
Targeted Chemotherapies
Targeted therapies have recently been developed to join traditional therapeutic approaches to CRC including surgery, chemotherapy, and radiotherapy [109]. Targeted chemotherapy may be used for neoadjuvant treatment or adjuvant treatment. For advanced cancers that have spread to other organs, such as the liver, chemo can also be used to help shrink tumors and relieve symptoms. Although it is not likely to cure the cancer, it often prolongs life. Common drugs used for CRC include: 5-fluorouracil (5-FU), often given with leucovorin (also called folinic acid) or levo-leucovorin; capecitabine (Xeloda tablets) metabolized to 5-FU in tumors [110]; irinotecan (Camptosar); and a combination tablet containing oxaliplatin (Eloxatin) and trifluridine and tipiracil (Lonsurf).
About 85% of all sporadic CRCs occur due to chromosomal instability, and ~ 15% of all CRCs are caused by various deficiencies in the DNA mismatch repair system. In the order of decreasing frequencies, TP53, KRAS, PIK3CA, BRAF, and PTEN are among the most commonly altered genes in CRC [111]. TP53 was the most commonly mutated gene, affecting 17/24 cell lines; hyperactivating KRAS mutations were found in 15 cancer cell lines of which five were homozygous, and BRAF mutations were found in another five cell lines.
Several cutting-edge biological modalities are under investigation. Mannosylated liposomes have been used to improve cell uptake and tumor inhibition rate [112]. This was shown without increase in toxicity. Thus, Patras et al. [113] have shown the effects of liposomal paclitaxel encapsulated in long-circulating liposomes together with liposomal prednisolone phosphate to be superior in anti-tumor activity mainly related to anti-angiogenic and anti-inflammatory effects. Attempts have been made to detail the functional pathways microRNA array, and considerable knowledge has been accumulated [114]. Yao et al. [115] have shown the importance of microRNA-215 as a tumor suppressor in colon cancer, which paradoxically acts as oncogene in breast cancer, and have proposed this to be a novel therapeutic target. However, knowledge of several microRNAs is very limited in terms of their targeted genes [113, 114]. Zheng et al. studied the effects of caudal type homeobox (CDX2) and human telomerase reverse transcriptase (hTERT) [115]. hTERT and loss of CDX2 have been associated with colon cancer in humans. In their elegant experiment, they showed over-expression of CDX2 is associated with suppression of colon cancer progression [116]. Finally, as stated before, TGF-β has also been implicated in treatment of metastatic colon cancer [117].
Immunotherapies for CRC
Immune surveillance is a key mechanism to protect against malignancy. Successful CRC immune surveillance relies on the effective function of antigen-presenting cells and T cells. It is now evident that this inadequate function of host immune system is due to suppressive factors like myeloid-derived suppressor cells (MDSCs). It is one of the major mechanisms of tumor escape from immune control as well as an important factor limiting the success of cancer immunotherapy. MDSCs play an important role in tumor non-responsiveness by suppressing antigen-specific T cell responses [118–121]. These cells take up antigen delivered by vaccination, present it to activated T cells, and thereby inhibit the same antigen-specific T cells that the vaccination strategy is aiming to activate. Cancer patients or tumor-bearing mice have a considerable number of MDSCs, which cause even the most effective antigen-delivery strategies to fail [120, 121]. CRC therapy will only be successful if strategies involve either the differentiation or elimination of suppressive cells. This strategy will be more effective than direct cancer therapy that attempts to eliminate tumor cells. Consideration of immune suppressive parameters will provide better stratification of CRC patient treatment.
Currently there are about 46 active (recruiting) immunotherapy trials against CRC (Clin Trials. Gov accessed on July 23, 2017). Peptide vaccines targeting HER-2/Neu, P-53, SART-3, and CEA in CRC have been studied. These vaccines are generally well tolerated but are far from a successful treatment. Most of the current CRC immunotherapies can be grouped as checkpoint blockade inhibitors like anti-CTLA4 [122], PDL-L1 [123], PD-1 [124], LAG-3; approved FDA targeted antibodies like bevacizumab, cetuximab, ramucirumab, and panitumumab; ACT (adoptive T cell therapy) and virus-mediated therapies. These therapies are tried either as adjuvants or as combinations. Current CRC immunotherapy targets the highly expressed tumor antigens like CEA, APC, MAGE, MUC-1, and survivin [125], or common mutations in the WNT, MAPK, TGF, P53, or apoptosis pathway [126–128]. A recent paper targeting mutant KRAS in CRC patients using ACT observed objective progressive regression [129]. ACT targeting various known CRC mutations in combination with checkpoint inhibitors and modulating the persistent immune suppressive factors would lead to effective therapies. Data from a phase 2 trial of pembrolizumab for the treatment of tumors with and without mismatch repair deficiency support the hypothesis that mismatch repair-deficient tumors are more responsive to PD-1 blockade than are mismatch repair-proficient tumors [130].
Emerging Approaches to Personalizing Cancer Treatment
Genomic Testing
Personalizing cancer treatment that is tailored to an individual’s genomic profile is a cornerstone of precision oncology [131]. NGS of DNA from tumor tissue can identify gene variants that could be targets of specific cancer therapies. These variants can be inherited (germline) or acquired (somatic) [132]. For example, poly(ADP-ribose) polymerase (PARP) inhibitors are effective in killing cancer cells that are deficient in repair of double-stranded DNA breaks. This is due to a pathogenic variant in the BRCA1 or BRCA2 gene [133] and other germline variants, which cause hereditary breast-ovarian cancer syndrome [134]. Tumors deficient in DNA mismatch repair (MMR) due to a pathogenic variant in the MSH2, MLH1, MSH6, PMS2, or EPCAM genes are responsive to antibodies or ligands that block the programmed death 1 (PD1) pathway [130]. These germline variants cause Lynch syndrome, the most common cause of hereditary CRC [135].
The VHA has undertaken feasibility studies on the use of NGS testing of tumor samples [136]. Tumors tested with multi-gene NGS sequencing through 1 of 2 contracted vendors were identified from POP records, and cancer characteristics were extracted from POP and medical records. Drug use data were obtained from the VA Corporate Data Warehouse. NGS testing results and annotations were extracted from POP records. Of a total of 1292 tumor samples sent for NGS testing since program inception in 2015, the most common diagnoses have been lung (561: adeno 418, squamous 143), gastrointestinal [109], lymph node [75], liver [56], head and neck [52], and prostate [43]. The rate of sample test requests increased rapidly after national dissemination in July 2016 (from a mean of 23 samples/month prior to launch to a mean of 126 samples/month 3 months later), as did the number of participating facilities (from a mean of 8/month to a mean of 27/month). Sequencing success rate increased from 68 to 71% over the same interval, while mean turnaround time remained similar at 19.7 and 19.1 days, respectively. NGS was requested for 61 patients with CRC, with almost all requested after dissemination of POP across the USA. Only one case failed to generate NGS results (98% success rate). Twenty-two patients had mutations targetable with an off-label FDA-approved drug, but no patient received NGS-directed treatment. These results suggest that tumor NGS testing as part of POP in the VA healthcare system is feasible with a high rate of testing success in CRC, but effective implementation strategies are needed to ensure uptake of treatments targeting tumor sequencing results.
The Spheroid and Organoid Culture Approaches to Assess Clinical Drug Response
Animal models of human CRC do not recapitulate human disease and frequently yield overly optimistic results [137, 138]. In the absence of accurate murine models, robust cellular models of CRC are considered critical. With over 150 CRC cell lines having been investigated, there is an abundance of cell line resources to investigate [139]. A number of different colon two-dimensional (2D) cell techniques, also referred to as “flat” cell cultures, have been described including the flow cell used for organ-on-chip models [140]. Despite progress made, these flat cultures are significantly different from in vivo tumor cells biochemically, physiologically, and genetically. Another approach that has been developed is in vitro multi-cell tumor-like cultures referred to as spheroids, tumoroids, or organoid culture systems [141]. Whether they are scaffold-free or scaffold-based 3D platforms, they fail to fully recapitulate in vivo tumors [141]. More importantly, they are cumbersome, costly, and not adaptable to perfusion-driven culture. Though very few of these have progressed to clinical testing realm the spheroid technologies are under intense investigation for their use in clinical drug response analyses.
Notably, several studies reported success in predicting CRC clinical drug response using organoid culture, also referred to as histoculture technique. Thus, an evaluation of 86 chemotherapy regimens using the 3D histoculture drug response assay (HDRA) for CRCs showed that the correlation of HDRA to the clinical response to chemotherapy was 66% (sensitivity 73%, specificity 55%) [142]. In another study, ATP-CRA-guided chemotherapy group showed better treatment response (48.4 vs. 21.9%, p = 0.027) and a higher rate of resection of hepatic lesions (35.5 vs. 12.5%, p = 0 [143]. Also, the extreme drug response assay (EDRA), developed as an exclusion test to identify drugs unlikely to elicit a response [144], was used to analyze the therapeutic efficacy of treatment for CRC patients. Thus, in the clinical correlation of 25 Duke’s D patients with EDRA, the sensitivity and specificity of the assay were 100 and 95%, respectively, suggesting that EDRA obtained at initial diagnosis may be useful for the selection of therapeutic regimens for metastatic disease [145, 146]. While these studies indicate the value of ex vivo organoid cultures, these results need to be replicated in multicenter clinical trials.
Tumoroid Technology for Clinical Drug Response
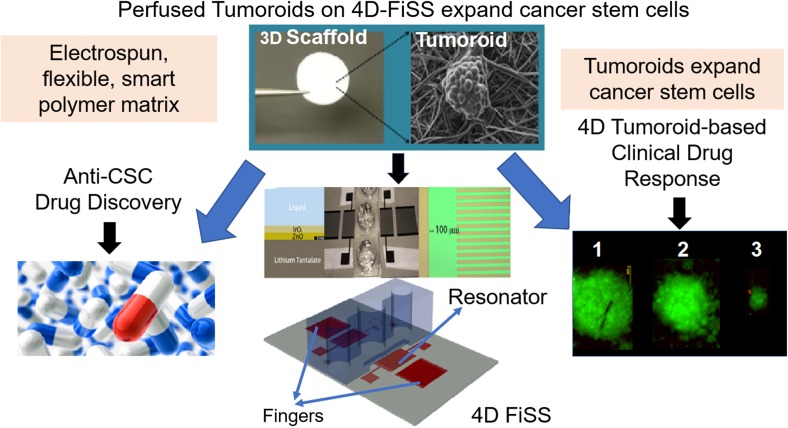
A fiber-inspired smart scaffold (FiSS) tumoroid culture platform has been developed that allows the formation of tumor-like organoids with molecular signatures of tumors. These cancer cells obtained from ex vivo tumor biopsies mimic patient tumors [141, 147, 148] (Fig. 5). The novelty of this platform lies in its simplicity, reproducibility, and similarity to in vivo tumors in terms of its drug responsiveness. Recent pilot proof-of-concept studies showed that cells of tumors from cancer patients, when cultured on a tumoroid platform, formed tumoroids that exhibited differential drug response [141, 147, 148]. Tumoroids were also found to expand cancer stem cells (CSCs) up to 30-fold, with cancer cells originating from several cancer cell lines including cells from breast, lung, and colon tumor xenografts.
Fig. 5.
Schematics of tumoroid technology platform for anti-CSC drug discovery and tumoroid-based clinical drug response assay
In the post-genomic era the development of microarray technology has allowed use of genomic data to help define which patients would benefit most from a specific therapy [149]. For example, gene signatures have been developed and validated against large retrospective databases to risk stratify patients to selectively receive adjuvant treatments for breast cancer [150]. However, genomic/genetic tests are based on pathway analysis of several genes with mutations out of more than 1000 genes that play a role in cancer pathogenesis. They may reveal potential adverse responses to drugs. However, the results do not indicate one or two potential drugs for effective treatments but rather suggest excluding therapies that may cause adverse effects.
One novel approach to PCT involves use of the patient-derived perfused tumoroid (PPT) platform, which mimics patient's tumor. PPT provides a simple, rapid, scalable, and inexpensive in vitro tumor model that better replicates the structure, physiology, and function of tissues. It also recreates the in vivo morphology and arrangement of individual cells and the concentration gradients of both signaling molecules and therapeutic agents [151]. This platform can be used to culture tumoroids utilizing human tumor cells (Fig. 5) with or without stromal cells. These are derived from biopsy or surgery discard tumor samples from patients. The PPT also promotes tumoroids in a manner that can be adapted for cell viability measurements (e.g., Celltiter-Glo or PrestoBlue). Preliminary data suggest that PCT tests will fully recapitulate in vivo tumors and will provide evidence for the best drug to treat each patient’s tumor. Further, expanding PCT to include first genetic/genomic testing that predicts potential therapies, followed by tumoroid-based clinical drug response assay, is expected to significantly improve the standard of personalized therapy. Adding evidence-based, simple-to-analyze, and ready-to-prescribe therapies will significantly improve personalized cancer treatment. Thus, a cell-genomic approach that combines NGS-based genomic testing with a PPT-based clinical drug response assay is expected to provide an exciting approach for PCT and may aid in discovery of novel anticancer agents.
Cancer Stem Cell Biology and Drug Discovery
Pivotal to successful CRC treatments are CSCs, referred to as the “seeds” of tumors, which play a central role in processes such as tumor initiation, drug resistance, invasion, and metastasis. While chemo-, radio- and immune-therapies are more effective at killing bulk tumor cells, the quiescent CSCs manage to escape and seed new tumor growth. CSC phenotype in human colon cancer is associated with poor prognosis [152, 153]. Hence, a therapeutic approach has been to target CSCs through either inhibition of self-renewal or induction of differentiation [154, 155]. A major barrier to CSC research, however, is the small proportion of these cells in tumors. With the possibility to expand CSCs by tumoroid cultures has opened new avenues for anti-CSC drug discovery. An NCI library of FDA-approved drugs was screened using FiSS tumoroid platform, which revealed that mithramycin A, idarubicin, and daunorubicin were the most potent inhibitors of HT-29 cell viability; and cisplatin, 6-thioguanine, and cytarabine were the least potent inhibitors. Further studies have revealed that mithramycin A treatment can reduce CSC self-renewal as well as expression of stemness genes. These results suggest that mithramycin A may be used as a single agent or in combination with other chemo- or immuno-therapy to most effectively treat or possibly ‘cure’ CRCs.
In addition to these efforts, two major pathways have been exploited to screen for anti-CSC drugs. First, application of HTS on a library of small molecules targeting EMT in breast cancer cells led to identification of four candidates: salinomycin, a polyketide synthase-derived natural product, and probes ML239, ML243, and ML245, which are chemically synthesizable, small hydrophobic molecules [156–160]. A second target signaling pathway for CSCs involves interaction of glycosaminoglycans (GAGs) with factors involved in growth and/or differentiation signaling that regulate CSCs, including growth factors (e.g., fibroblast growth factors, TGF-β, bone morphogenetic proteins), cytokines (e.g., interleukins-6 and -8), or morphogens (e.g., Hedgehog, Notch). These in turn influence many transcription factors including Nanog, Oct4, and SOX2 and c-MYC, which play critical roles in deciding the fate of CSCs [161–163]. In sum, the identification of only four CSC targeting molecules so far using CSC-pathway screening approach for breast CSCs highlights the difficulty of targeting CSCs. For the anti-CSC paradigm to translate into therapeutic success, a more fundamental and generalizable approach is needed.
Concluding Remarks and Opportunities
Our review has identified key research directions in the field of CRC, as summarized below. a) Current genomic testing includes primarily somatic mutations only, and not genetic testing for germline mutations. Adding the latter might allow for more accurate CRC risk assessment, surveillance and prevention. Also, marker(s) such as specific mucins could serve as a powerful adjunct to colonoscopy by enabling efficient detection of SSA/Ps. b) Several emerging genetic biomarkers as well as cell-free miRNAs and small non-coding RNAs have been suggested as noninvasive and highly sensitive candidate markers for CRCs. They will require large clinical trial-based validation. c) In relation to precision treatment approaches, developments in ex vivo cell-genomics technologies and the potential to combine genomic/genetic testing with tumoroid-based drug response testing might provide more accurate clinical efficacy of certain drugs in individual patients. d) Integrating targeted tumor imaging preoperatively and delineating tumor margins intraoperatively with fluorescent labeling can improve the precision of surgical resection. e) Given the pivotal role of cancer-initiating stem cells in CRC progression, the stem cell niches discovered thus far, such as CEACAM/TGF-β and M3R, need further investigation. Other stem cell niches, such as telomerase, CTCF, CEA, E-cadherin, and β-catenin, may be equally important in developing targeted, preventive therapies against CRC. f) CRC tumoroid/organoid technology using polymeric nanofiber scaffolds to expand CSCs ex vivo is of considerable interest. Specifically, tumoroid technology can be used to expand CSCs of hard-to-detect SSA/P neoplasms, which can be characterized and investigated for CRC stem cell niches and used for evaluating existing chemo- and immunotherapies.
Acknowledgments
This VA Colorectal Cancer Cell-genomics Consortium (VA4C) meeting was supported by a Department of Veterans Affairs Biomedical Laboratory Research and Development Service field-based meeting award to Dr. Shyam Mohapatra and Dr. Subhra Mohapatra. The contents of any report, written material or manuscripts emanating from this meeting do not represent the views of the Department of Veterans Affairs or the United States Government. The VA4C would like to acknowledge the support of Ms. Christen Bouchard in preparation of this manuscript.
Compliance with ethical standards
Conflict of interest
All authors except the following have indicated no financial conflicts of interests in relation to writing of this review. Even though not relevant to this review, Dr. Hemant K Roy has declared that he is a co-founder and shareholder in startup company Nanocytomics and American BioOptics LLC. Additionally, Subhra Mohapatra (co-founder and scientific advisor), Shyam Mohapatra (co-founder and scientific advisor) and the University of South Florida have equity interest in Transgenex Nanobiotech Inc. The terms of the conflict management have been reviewed and approved by the University of South Florida, Tampa, in accordance with its conflict of interest policies.
Contributor Information
Shyam S. Mohapatra, Email: shyam.mohapatra@va.gov, Email: smohapat@health.usf.edu
Surinder K. Batra, Email: sbatra@unmc.edu
Michael Bouvet, Email: mbouvet@ucsd.edu.
Bard Cosman, Email: Bard.Cosman@va.gov.
Ajay Goel, Email: Ajay.Goel@BSWHealth.org.
Michael J. Kelley, Email: kelleym@duke.edu
Lopa Mishra, lmishra@email.gwu.edu.
Subhra Mohapatra, Email: smohapa2@health.usf.edu.
Bhaumik Patel, Email: Bhaumik.Patel@va.gov.
Joseph R. Pisegna, Email: Joseph.Pisegna@va.gov
Jean-Pierre Raufman, Email: jraufman@som.umaryland.edu.
Hemant Roy, Email: hkroy@bu.edu.
Maren Scheuner, Email: Maren.Scheuner@va.gov.
Satish Singh, Email: Satish.Singh@va.gov.
Gitanjali Vidyarthi, Email: Gitanjali.Vidyarthi@va.gov.
References
- 1.Zauber AG, Winawer SJ, O’Brien MJ, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687–696. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan J, Xin L, Ma YF, et al. Colonoscopy reduces colorectal cancer incidence and mortality in patients with non-malignant findings: a meta-analysis. Am J Gastroenterol. 2016;111:355–365. doi: 10.1038/ajg.2015.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson KM, Starkebaum GA, Reiber GE. Veterans using and uninsured veterans not using Veterans Affairs (VA) health care. Public Health Rep. 2007;122:93–100. doi: 10.1177/003335490712200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zullig LL, Williams CD, Fortune-Britt AG. Lung and colorectal cancer treatment and outcomes in the Veterans Affairs health care system. Cancer Manag Res. 2015;7:19–35. doi: 10.2147/CMAR.S75463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fakih MG. Metastatic colorectal cancer: current state and future directions. J Clin Oncol. 2015;33:1809–1824. doi: 10.1200/JCO.2014.59.7633. [DOI] [PubMed] [Google Scholar]
- 6.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 7.Qiu M, Hu J, Yang D, et al. Pattern of distant metastases in colorectal cancer: a SEER based study. Oncotarget. 2015;6:38658–38666. doi: 10.18632/oncotarget.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enquist IB, Good Z, Jubb AM, et al. Lymph node-independent liver metastasis in a model of metastatic colorectal cancer. Nat Commun. 2014;5:3530. doi: 10.1038/ncomms4530. [DOI] [PubMed] [Google Scholar]
- 9.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149:1177e3–1190e3. doi: 10.1053/j.gastro.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149:1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pancione M, Remo A, Colantuoni V. Genetic and epigenetic events generate multiple pathways in colorectal cancer progression. Patholog Res Int. 2012;2012:509348. doi: 10.1155/2012/509348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buda A, De Bona M, Dotti I, et al. Prevalence of different subtypes of serrated polyps and risk of synchronous advanced colorectal neoplasia in average-risk population undergoing first-time colonoscopy. Clin Transl Gastroenterol. 2012;3:e6. doi: 10.1038/ctg.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crockett SD, Snover DC, Ahnen DJ, et al. Sessile serrated adenomas: an evidence-based guide to management. Clin Gastroenterol Hepatol. 2015;13:11–26.e1. doi: 10.1016/j.cgh.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 15.IJSPEERT JE, Rana SA, Atkinson NS, et al. Clinical risk factors of colorectal cancer in patients with serrated polyposis syndrome: a multicentre cohort analysis. Gut. 2017;66:278–284. doi: 10.1136/gutjnl-2015-310630. [DOI] [PubMed] [Google Scholar]
- 16.East JE, Atkin WS, Bateman AC, et al. British Society of Gastroenterology position statement on serrated polyps in the colon and rectum. Gut. 2017;66:1181–1196. doi: 10.1136/gutjnl-2017-314005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman DA, Rex DK, Winawer SJ, et al. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2012;143:844–857. doi: 10.1053/j.gastro.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Freeman HJ. Heterogeneity of colorectal adenomas, the serrated adenoma, and implications for screening and surveillance. World J Gastroenterol. 2008;14:3461–3463. doi: 10.3748/wjg.14.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S, Farraye FA, Mack C, et al. BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol. 2004;28:1452–1459. doi: 10.1097/01.pas.0000141404.56839.6a. [DOI] [PubMed] [Google Scholar]
- 21.Huang CS, O’Brien MJ, Yang S, et al. Hyperplastic polyps, serrated adenomas, and the serrated polyp neoplasia pathway. Am J Gastroenterol. 2004;99:2242–2255. doi: 10.1111/j.1572-0241.2004.40131.x. [DOI] [PubMed] [Google Scholar]
- 22.Das V, Kalita J, Pal M. Predictive and prognostic biomarkers in colorectal cancer: a systematic review of recent advances and challenges. Biomed Pharmacother. 2017;87:8–19. doi: 10.1016/j.biopha.2016.12.064. [DOI] [PubMed] [Google Scholar]
- 23.Felton J, Raufman JP. Is the ColDx assay a valid prognostic mrker for stage II colon cancer? Transl Cancer Res. 2017;5:S1157–S1159. doi: 10.21037/tcr.2016.11.64. [DOI] [Google Scholar]
- 24.Singh MP, Rai S, Suyal S, et al. Genetic and epigenetic markers in colorectal cancer screening: recent advances. Expert Rev Mol Diagn. 2017;17:665–685. doi: 10.1080/14737159.2017.1337511. [DOI] [PubMed] [Google Scholar]
- 25.Barontini J, Antinucci M, Tofanelli S, et al. Association between polymorphisms of TAS2R16 and susceptibility to colorectal cancer. BMC Gastroenterol. 2017;17:104. doi: 10.1186/s12876-017-0659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanskanen T, van den Berg L, Valimaki N, et al. Genome-wide association study and meta-analysis in Northern European populations replicate multiple colorectal cancer risk loci. Int J Cancer. 2018;142:540–546. doi: 10.1002/ijc.31076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanley MP, Hahn MA, Li AX, et al. Genome-wide DNA methylation profiling reveals cancer-associated changes within early colonic neoplasia. Oncogene. 2017;36:5035–5044. doi: 10.1038/onc.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Cao H, Jiao Z, et al. Carcinoembryonic antigen interacts with TGF-{beta} receptor and inhibits TGF-{beta} signaling in colorectal cancers. Cancer Res. 2010;70:8159–8168. doi: 10.1158/0008-5472.CAN-10-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–128. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 30.Hoosein NM, McKnight MK, Levine AE, et al. Differential sensitivity of subclasses of human colon carcinoma cell lines to the growth inhibitory effects of transforming growth factor-beta 1. Exp Cell Res. 1989;181:442–453. doi: 10.1016/0014-4827(89)90101-8. [DOI] [PubMed] [Google Scholar]
- 31.Yashiro M, Hirakawa K, Boland CR. Mutations in TGFbeta-RII and BAX mediate tumor progression in the later stages of colorectal cancer with microsatellite instability. BMC Cancer. 2010;10:303. doi: 10.1186/1471-2407-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takagi Y, Kohmura H, Futamura M, et al. Somatic alterations of the DPC4 gene in human colorectal cancers in vivo. Gastroenterology. 1996;111:1369–1372. doi: 10.1053/gast.1996.v111.pm8898652. [DOI] [PubMed] [Google Scholar]
- 33.Takagi Y, Koumura H, Futamura M, et al. Somatic alterations of the SMAD-2 gene in human colorectal cancers. Br J Cancer. 1998;78:1152–1155. doi: 10.1038/bjc.1998.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 35.Takaku K, Oshima M, Miyoshi H, et al. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/S0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 36.Tiernan JP, Perry SL, Verghese ET, et al. Carcinoembryonic antigen is the preferred biomarker for in vivo colorectal cancer targeting. Br J Cancer. 2013;108:662–667. doi: 10.1038/bjc.2012.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakrabarty S, Tobon A, Varani J, et al. Induction of carcinoembryonic antigen secretion and modulation of protein secretion/expression and fibronectin/laminin expression in human colon carcinoma cells by transforming growth factor-beta. Cancer Res. 1988;48:4059–4064. [PubMed] [Google Scholar]
- 39.Michalopoulos G, Tzathas C. Serrated polyps of right colon: guilty or innocent? Ann Gastroenterol. 2013;26:212–219. [PMC free article] [PubMed] [Google Scholar]
- 40.Sandmeier D, Seelentag W, Bouzourene H. Serrated polyps of the colorectum: is sessile serrated adenoma distinguishable from hyperplastic polyp in a daily practice? Virchows Arch. 2007;450:613–618. doi: 10.1007/s00428-007-0413-8. [DOI] [PubMed] [Google Scholar]
- 41.Yang HM, Mitchell JM, Sepulveda JL, et al. Molecular and histologic considerations in the assessment of serrated polyps. Arch Pathol Lab Med. 2015;139:730–741. doi: 10.5858/arpa.2014-0424-RA. [DOI] [PubMed] [Google Scholar]
- 42.Schreiner MA, Weiss DG, Lieberman DA. Proximal and large hyperplastic and nondysplastic serrated polyps detected by colonoscopy are associated with neoplasia. Gastroenterology. 2010;139:1497–1502. doi: 10.1053/j.gastro.2010.06.074. [DOI] [PubMed] [Google Scholar]
- 43.Rex DK, Ahnen DJ, Baron JA, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107:1315–1329. doi: 10.1038/ajg.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Brien MJ. Hyperplastic and serrated polyps of the colorectum. Gastroenterol Clin North Am. 2007;36:947–968. doi: 10.1016/j.gtc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 45.IJspeert JE, Vermeulen L, Meijer GA, et al. Serrated neoplasia-role in colorectal carcinogenesis and clinical implications. Nat Rev Gastroenterol Hepatol. 2015;12:401–409. doi: 10.1038/nrgastro.2015.73. [DOI] [PubMed] [Google Scholar]
- 46.Hetzel JT, Huang CS, Coukos JA, et al. Variation in the detection of serrated polyps in an average risk colorectal cancer screening cohort. Am J Gastroenterol. 2010;105:2656–2664. doi: 10.1038/ajg.2010.315. [DOI] [PubMed] [Google Scholar]
- 47.Khaidakov M, Lai KK, Roudachevski D, et al. Gastric proteins MUC5AC and TFF1 as potential diagnostic markers of colonic sessile serrated adenomas/polyps. Am J Clin Pathol. 2016;146:530–537. doi: 10.1093/ajcp/aqw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalo DH, Lai KK, Shadrach B, et al. Gene expression profiling of serrated polyps identifies annexin A10 as a marker of a sessile serrated adenoma/polyp. J Pathol. 2013;230:420–429. doi: 10.1002/path.4200. [DOI] [PubMed] [Google Scholar]
- 49.Krishn SR, Kaur S, Sheinin YM, et al. Mucins and associated O-glycans based immunoprofile for stratification of colorectal polyps: clinical implication for improved colon surveillance. Oncotarget. 2017;8:7025–7038. doi: 10.18632/oncotarget.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Cheng K, Shang AC, Drachenberg CB, et al. Differential expression of M3 muscarinic receptors in progressive colon neoplasia and metastasis. Oncotarget. 2017;8:21106–21114. doi: 10.18632/oncotarget.15500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Said AH, Hu S, Abutaleb A, et al. Interacting post-muscarinic receptor signaling pathways potentiate matrix metalloproteinase-1 expression and invasion of human colon cancer cells. Biochem J. 2017;474:647–665. doi: 10.1042/BCJ20160704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raufman JP, Cheng K, Saxena N, et al. Muscarinic receptor agonists stimulate matrix metalloproteinase 1-dependent invasion of human colon cancer cells. Biochem Biophys Res Commun. 2011;415:319–324. doi: 10.1016/j.bbrc.2011.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lew EA, Lewin KJ, Zarchy T, et al. Adenocarcinoma of the colon with neuroendocrine features and secretory diarrhea. Am J Gastroenterol. 1999;94:1692–1694. doi: 10.1111/j.1572-0241.1999.01167.x. [DOI] [PubMed] [Google Scholar]
- 56.Le SV, Yamaguchi DJ, McArdle CA, et al. PAC1 and PACAP expression, signaling, and effect on the growth of HCT8, human colonic tumor cells. Regul Pept. 2002;109:115–125. doi: 10.1016/S0167-0115(02)00194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Germano PM, Le SV, Oh DS, et al. Differential coupling of the PAC1 SV1 splice variant on human colonic tumors to the activation of intracellular cAMP but not intracellular Ca2 + does not activate tumor proliferation. J Mol Neurosci. 2004;22:83–92. doi: 10.1385/JMN:22:1-2:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ze EY, Kim BJ, Jun DH, et al. The fatty liver index: a simple and accurate predictor of colorectal adenoma in an average-risk population. Dis Colon Rectum. 2018;61:36–42. doi: 10.1097/DCR.0000000000000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rustagi T, Zarookian EI, Qasba O, et al. Chronic hepatitis C as a risk factor for colorectal adenoma. Int J Colorectal Dis. 2014;29:75–80. doi: 10.1007/s00384-013-1763-0. [DOI] [PubMed] [Google Scholar]
- 60.Yi SW, Ohrr H, Hong JS, et al. Agent orange exposure and prevalence of self-reported diseases in korean vietnam veterans. J Prev Med Public Health. 2013;46:213–225. doi: 10.3961/jpmph.2013.46.5.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw E, Warkentin MT, McGregor SE, et al. Intake of dietary fibre and lifetime non-steroidal anti-inflammatory drug (NSAID) use and the incidence of colorectal polyps in a population screened for colorectal cancer. J Epidemiol Community Health. 2017;71:961–969. doi: 10.1136/jech-2016-208606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Jacobs JP, Lagishetty V, et al. High-protein diet promotes sensitivity to cholecystokinin and shifts the cecal microbiome without altering brain inflammation in diet-induced obesity in rats. Am J Physiol Regul Integr Comput Physiol. 2017 doi: 10.1152/ajpregu.00105.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang LJJ, Lagishetty V, Yuan PQ, Wu SV, Million M, Reeve JR, Jr, Pisegna JR, Tache Y. Am J Physiol Regul Integr Comput Physiol. 2017;313:R473–R486. doi: 10.1152/ajpregu.00105.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li R, Yang J, Saffari A, et al. Ambient ultrafine particle ingestion alters gut microbiota in association with increased atherogenic lipid metabolites. Sci Rep. 2017;7:42906. doi: 10.1038/srep42906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Brien MJ, Gibbons D. The adenoma-carcinoma sequence in colorectal neoplasia. Surg Oncol Clin N Am. 1996;5:513–530. [PubMed] [Google Scholar]
- 66.Lakoff J, Paszat LF, Saskin R, et al. Risk of developing proximal versus distal colorectal cancer after a negative colonoscopy: a population-based study. Clin Gastroenterol Hepatol. 2008;6:1117–1121. doi: 10.1016/j.cgh.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 67.Kaminski MF, Regula J, Kraszewska E, et al. Quality indicators for colonoscopy and the risk of interval cancer. N Engl J Med. 2010;362:1795–1803. doi: 10.1056/NEJMoa0907667. [DOI] [PubMed] [Google Scholar]
- 68.Pickhardt PJ, Nugent PA, Mysliwiec PA, et al. Location of adenomas missed by optical colonoscopy. Ann Intern Med. 2004;141:352–359. doi: 10.7326/0003-4819-141-5-200409070-00009. [DOI] [PubMed] [Google Scholar]
- 69.Rex DK. Maximizing detection of adenomas and cancers during colonoscopy. Am J Gastroenterol. 2006;101:2866–2877. doi: 10.1111/j.1572-0241.2006.00905.x. [DOI] [PubMed] [Google Scholar]
- 70.Matsumoto T, Iida M, Kuwano Y, et al. Small nonpolypoid neoplastic lesions of the colon: endoscopic features with emphasis on their progression. Gastrointest Endosc. 1995;41:135–140. doi: 10.1016/S0016-5107(05)80595-4. [DOI] [PubMed] [Google Scholar]
- 71.Brooker JC, Saunders BP, Shah SG, et al. Total colonic dye-spray increases the detection of diminutive adenomas during routine colonoscopy: a randomized controlled trial. Gastrointest Endosc. 2002;56:333–338. doi: 10.1016/S0016-5107(02)70034-5. [DOI] [PubMed] [Google Scholar]
- 72.Anderson JC, Butterly LF. Colonoscopy: quality indicators. Clin Transl Gastroenterol. 2015;6:e77. doi: 10.1038/ctg.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rex DK, Kahi C, O’Brien M, et al. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and incorporation of valuable endoscopic innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2011;73:419–422. doi: 10.1016/j.gie.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 74.Hassan C, Pickhardt PJ, Rex DK. A resect and discard strategy would improve cost-effectiveness of colorectal cancer screening. Clin Gastroenterol Hepatol. 2010;8:865–869. doi: 10.1016/j.cgh.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 75.Ignjatovic A, East JE, Suzuki N, et al. Optical diagnosis of small colorectal polyps at routine colonoscopy (Detect InSpect ChAracterise Resect and Discard; DISCARD trial): a prospective cohort study. Lancet Oncol. 2009;10:1171–1178. doi: 10.1016/S1470-2045(09)70329-8. [DOI] [PubMed] [Google Scholar]
- 76.Subramanian V, Mannath J, Hawkey CJ, et al. High definition colonoscopy vs. standard video endoscopy for the detection of colonic polyps: a meta-analysis. Endoscopy. 2011;43:499–505. doi: 10.1055/s-0030-1256207. [DOI] [PubMed] [Google Scholar]
- 77.Kahi CJ, Anderson JC, Waxman I, et al. High-definition chromocolonoscopy vs. high-definition white light colonoscopy for average-risk colorectal cancer screening. Am J Gastroenterol. 2010;105:1301–1307. doi: 10.1038/ajg.2010.51. [DOI] [PubMed] [Google Scholar]
- 78.Leufkens AM, DeMarco DC, Rastogi A, et al. Effect of a retrograde-viewing device on adenoma detection rate during colonoscopy: the TERRACE study. Gastrointest Endosc. 2011;73:480–489. doi: 10.1016/j.gie.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 79.Gralnek IM, Siersema PD, Halpern Z, et al. Standard forward-viewing colonoscopy versus full-spectrum endoscopy: an international, multicentre, randomised, tandem colonoscopy trial. Lancet Oncol. 2014;15:353–360. doi: 10.1016/S1470-2045(14)70020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Floer M, Biecker E, Fitzlaff R, et al. Higher adenoma detection rates with endocuff-assisted colonoscopy—a randomized controlled multicenter trial. PLoS ONE. 2014;9:e114267. doi: 10.1371/journal.pone.0114267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dik VK, Gralnek IM, Segol O, et al. Multicenter, randomized, tandem evaluation of EndoRings colonoscopy–results of the CLEVER study. Endoscopy. 2015;47:1151–1158. doi: 10.1055/s-0034-1392421. [DOI] [PubMed] [Google Scholar]
- 82.Hohenberger W, Weber K, Matzel K, et al. Standardized surgery for colonic cancer: complete mesocolic excision and central ligation–technical notes and outcome. Colorectal Dis. 2009;11:354–364. doi: 10.1111/j.1463-1318.2008.01735.x. [DOI] [PubMed] [Google Scholar]
- 83.Andreoni B, Chiappa A, Bertani E, et al. Surgical outcomes for colon and rectal cancer over a decade: results from a consecutive monocentric experience in 902 unselected patients. World J Surg Oncol. 2007;5:73. doi: 10.1186/1477-7819-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campos FG, Calijuri-Hamra MC, Imperiale AR, et al. Locally advanced colorectal cancer: results of surgical treatment and prognostic factors. Arq Gastroenterol. 2011;48:270–275. doi: 10.1590/S0004-28032011000400010. [DOI] [PubMed] [Google Scholar]
- 85.Mayo SC, Pulitano C, Marques H, et al. Surgical management of patients with synchronous colorectal liver metastasis: a multicenter international analysis. J Am Coll Surg. 2013;216:707–716. doi: 10.1016/j.jamcollsurg.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ruo L, Guillem JG. Surgical management of primary colorectal cancer. Surg Oncol. 1998;7:153–163. doi: 10.1016/S0960-7404(99)00033-X. [DOI] [PubMed] [Google Scholar]
- 87.Yedibela S, Klein P, Feuchter K, et al. Surgical management of pulmonary metastases from colorectal cancer in 153 patients. Ann Surg Oncol. 2006;13:1538–1544. doi: 10.1245/s10434-006-9100-2. [DOI] [PubMed] [Google Scholar]
- 88.Manfredi S, Benhamiche AM, Meny B, et al. Population-based study of factors influencing occurrence and prognosis of local recurrence after surgery for rectal cancer. Br J Surg. 2001;88:1221–1227. doi: 10.1046/j.0007-1323.2001.01861.x. [DOI] [PubMed] [Google Scholar]
- 89.Manfredi S, Bouvier AM, Lepage C, et al. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br J Surg. 2006;93:1115–1122. doi: 10.1002/bjs.5349. [DOI] [PubMed] [Google Scholar]
- 90.Abulafi AM, Williams NS. Local recurrence of colorectal cancer: the problem, mechanisms, management and adjuvant therapy. Br J Surg. 1994;81:7–19. doi: 10.1002/bjs.1800810106. [DOI] [PubMed] [Google Scholar]
- 91.Bouvet M, Hoffman RM. Glowing tumors make for better detection and resection. Sci Transl Med. 2011;3:110fs10. doi: 10.1126/scitranslmed.3003375. [DOI] [PubMed] [Google Scholar]
- 92.Metildi CA, Kaushal S, Luiken GA, et al. Fluorescently labeled chimeric anti-CEA antibody improves detection and resection of human colon cancer in a patient-derived orthotopic xenograft (PDOX) nude mouse model. J Surg Oncol. 2014;109:451–458. doi: 10.1002/jso.23507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nguyen QT, Tsien RY. Fluorescence-guided surgery with live molecular navigation–a new cutting edge. Nat Rev Cancer. 2013;13:653–662. doi: 10.1038/nrc3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vahrmeijer AL, Hutteman M, van der Vorst JR, et al. Image-guided cancer surgery using near-infrared fluorescence. Nat Rev Clin Oncol. 2013;10:507–518. doi: 10.1038/nrclinonc.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.DeLong JC, Hoffman RM, Bouvet M. Current status and future perspectives of fluorescence-guided surgery for cancer. Expert Rev Anticancer Ther. 2016;16:71–81. doi: 10.1586/14737140.2016.1121109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hiroshima Y, Lwin TM, Murakami T, et al. Effective fluorescence-guided surgery of liver metastasis using a fluorescent anti-CEA antibody. J Surg Oncol. 2016;114:951–958. doi: 10.1002/jso.24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hiroshima Y, Maawy A, Metildi CA, et al. Successful fluorescence-guided surgery on human colon cancer patient-derived orthotopic xenograft mouse models using a fluorophore-conjugated anti-CEA antibody and a portable imaging system. J Laparoendosc Adv Surg Tech A. 2014;24:241–247. doi: 10.1089/lap.2013.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaushal S, McElroy MK, Luiken GA, et al. Fluorophore-conjugated anti-CEA antibody for the intraoperative imaging of pancreatic and colorectal cancer. J Gastrointest Surg. 2008;12:1938–1950. doi: 10.1007/s11605-008-0581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Metildi CA, Kaushal S, Pu M, et al. Fluorescence-guided surgery with a fluorophore-conjugated antibody to carcinoembryonic antigen (CEA), that highlights the tumor, improves surgical resection and increases survival in orthotopic mouse models of human pancreatic cancer. Ann Surg Oncol. 2014;21:1405–1411. doi: 10.1245/s10434-014-3495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Metildi CA, Kaushal S, Snyder CS, et al. Fluorescence-guided surgery of human colon cancer increases complete resection resulting in cures in an orthotopic nude mouse model. J Surg Res. 2013;179:87–93. doi: 10.1016/j.jss.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hiroshima Y, Lwin TM, Murakami T, et al. Effective fluorescence-guided surgery of liver metastasis using a fluorescent anti-CEA antibody. J Surg Oncol. 2016;114:951–958. doi: 10.1002/jso.24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yazaki PJ, Sherman MA, Shively JE, et al. Humanization of the anti-CEA T84.66 antibody based on crystal structure data. Protein Eng Des Sel. 2004;17:481–489. doi: 10.1093/protein/gzh056. [DOI] [PubMed] [Google Scholar]
- 103.Gutowski M, Framery B, Boonstra MC, et al. SGM-101: An innovative near-infrared dye-antibody conjugate that targets CEA for fluorescence-guided surgery. Surg Oncol. 2017;26:153–162. doi: 10.1016/j.suronc.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 104.Park JY, Murakami T, Lee JY, et al. Fluorescent-antibody targeting of insulin-like growth factor-1 receptor visualizes metastatic human colon cancer in orthotopic mouse models. PLoS ONE. 2016;11:e0146504. doi: 10.1371/journal.pone.0146504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yano S, Takehara K, Miwa S, et al. Improved resection and outcome of colon-cancer liver metastasis with fluorescence-guided surgery using in situ GFP labeling with a telomerase-dependent adenovirus in an orthotopic mouse model. PLoS ONE. 2016;11:e0148760. doi: 10.1371/journal.pone.0148760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Metildi CA, Felsen CN, Savariar EN, et al. Ratiometric activatable cell-penetrating peptides label pancreatic cancer, enabling fluorescence-guided surgery, which reduces metastases and recurrence in orthotopic mouse models. Ann Surg Oncol. 2015;22:2082–2087. doi: 10.1245/s10434-014-4144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jafari MD, Wexner SD, Martz JE, et al. Perfusion assessment in laparoscopic left-sided/anterior resection (PILLAR II): a multi-institutional study. J Am Coll Surg. 2015;220(82–92):e1. doi: 10.1016/j.jamcollsurg.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 108.Tipirneni KE, Warram JM, Moore LS, et al. Oncologic procedures amenable to fluorescence-guided surgery. Ann Surg. 2017;266:36–47. doi: 10.1097/SLA.0000000000002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Inoue Y, Kusunoki M. Advances and directions in chemotherapy using implantable port systems for colorectal cancer: a historical review. Surg Today. 2014;44:1406–1414. doi: 10.1007/s00595-013-0672-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ahmed S, Johnson K, Ahmed O, et al. Advances in the management of colorectal cancer: from biology to treatment. Int J Colorectal Dis. 2014;29:1031–1042. doi: 10.1007/s00384-014-1928-5. [DOI] [PubMed] [Google Scholar]
- 111.Ciombor KK, Wu C, Goldberg RM. Recent therapeutic advances in the treatment of colorectal cancer. Annu Rev Med. 2015;66:83–95. doi: 10.1146/annurev-med-051513-102539. [DOI] [PubMed] [Google Scholar]
- 112.Xiong M, Lei Q, You X, et al. Mannosylated liposomes improve therapeutic effects of paclitaxel in colon cancer models. J Microencapsul. 2017;34:513–521. doi: 10.1080/02652048.2017.1339739. [DOI] [PubMed] [Google Scholar]
- 113.Patras L, Sylvester B, Luput L, et al. Liposomal prednisolone phosphate potentiates the antitumor activity of liposomal 5-fluorouracil in C26 murine colon carcinoma in vivo. Cancer Biol Ther. 2017;18:616–626. doi: 10.1080/15384047.2017.1345392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Slattery ML, Herrick JS, Stevens JR, et al. An assessment of database-validated microRNA target genes in normal colonic mucosa: implications for pathway analysis. Cancer Inform. 2017;16:1176935117716405. doi: 10.1177/1176935117716405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yao J, Zhang P, Li J, et al. MicroRNA-215 acts as a tumor suppressor in breast cancer by targeting AKT serine/threonine kinase 1. Oncol Lett. 2017;14:1097–1104. doi: 10.3892/ol.2017.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zheng J, He S, Qi J, et al. Targeted CDX2 expression inhibits aggressive phenotypes of colon cancer cells in vitro and in vivo. Int J Oncol. 2017;51:478–488. doi: 10.3892/ijo.2017.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Villalba M, Evans SR, Vidal-Vanaclocha F, et al. Role of TGF-beta in metastatic colon cancer: it is finally time for targeted therapy. Cell Tissue Res. 2017;370:29–39. doi: 10.1007/s00441-017-2633-9. [DOI] [PubMed] [Google Scholar]
- 118.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nagaraj S, Gabrilovich DI. Myeloid-derived suppressor cells. Adv Exp Med Biol. 2007;601:213–223. doi: 10.1007/978-0-387-72005-0_22. [DOI] [PubMed] [Google Scholar]
- 120.Nagaraj S, Gabrilovich DI. Myeloid-derived suppressor cells in human cancer. Cancer J. 2010;16:348–353. doi: 10.1097/PPO.0b013e3181eb3358. [DOI] [PubMed] [Google Scholar]
- 121.Nagaraj S, Gupta K, Pisarev V, et al. Altered recognition of antigen is a novel mechanism of CD8 + T cell tolerance in cancer. Nat Med. 2007;13:828–835. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chung KY, Gore I, Fong L, et al. Phase II study of the anti-cytotoxic T-lymphocyte-associated antigen 4 monoclonal antibody, tremelimumab, in patients with refractory metastatic colorectal cancer. J Clin Oncol. 2010;28:3485–3490. doi: 10.1200/JCO.2010.28.3994. [DOI] [PubMed] [Google Scholar]
- 123.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nagaraj S, Pisarev V, Kinarsky L, et al. Dendritic cell-based full-length survivin vaccine in treatment of experimental tumors. J Immunother. 2007;30:169–179. doi: 10.1097/01.cji.0000211329.83890.ba. [DOI] [PubMed] [Google Scholar]
- 126.Koido S, Ohkusa T, Homma S, et al. Immunotherapy for colorectal cancer. World J Gastroenterol. 2013;19:8531–8542. doi: 10.3748/wjg.v19.i46.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xiang B, Snook AE, Magee MS, et al. Colorectal cancer immunotherapy. Discov Med. 2013;15:301–308. [PMC free article] [PubMed] [Google Scholar]
- 128.Signorini L, Delbue S, Ferrante P, et al. Review on the immunotherapy strategies against metastatic colorectal carcinoma. Immunotherapy. 2016;8:1245–1261. doi: 10.2217/imt-2016-0045. [DOI] [PubMed] [Google Scholar]
- 129.Tran E, Robbins PF, Lu YC, et al. T-Cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Millner LM, Strotman LN. The future of precision medicine in oncology. Clin Lab Med. 2016;36:557–573. doi: 10.1016/j.cll.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 132.Duzkale H, Shen J, McLaughlin H, et al. A systematic approach to assessing the clinical significance of genetic variants. Clin Genet. 2013;84:453–463. doi: 10.1111/cge.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meehan RS, Chen AP. New treatment option for ovarian cancer: PARP inhibitors. Gynecol Oncol Res Pract. 2016;3:3. doi: 10.1186/s40661-016-0024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Petrucelli N, Daly MB, Pal T, et al. et al. BRCA1- and BRCA2-associated hereditary breast and ovarian cancer. In: Pagon RA, Adam MP, Ardinger HH, et al.et al., editors. GeneReviews(R) Seattle, WA: University of Washington; 1993. [PubMed] [Google Scholar]
- 135.Kohlmann W, Gruber SB, et al. et al. BRCA1- and BRCA2-associated hereditary breast and ovarian cancer. In: Pagon RA, Adam MP, Ardinger HH, et al.et al., editors. GeneReviews(R) Seattle, WA: University of Washington; 1993. [PubMed] [Google Scholar]
- 136.Dong L, Wang W, Li A, et al. Clinical next generation sequencing for precision medicine in cancer. Curr Genomics. 2015;16:253–263. doi: 10.2174/1389202915666150511205313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6:114–118. [PMC free article] [PubMed] [Google Scholar]
- 138.Day CP, Merlino G, Van Dyke T. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell. 2015;163:39–53. doi: 10.1016/j.cell.2015.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pereira JF, Awatade NT, Loureiro CA, et al. The third dimension: new developments in cell culture models for colorectal research. Cell Mol Life Sci. 2016;73:3971–3989. doi: 10.1007/s00018-016-2258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Huh D, Kim HJ, Fraser JP, et al. Microfabrication of human organs-on-chips. Nat Protoc. 2013;8:2135–2157. doi: 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- 141.Nair R, Padhee S, Das T, Green R, Howell M, Mohapatra SS, Mohapatra S. Three- and four-dimensional spheroid and fiss tumoroid cultures: platforms for drug discovery and development, and translational research. Crit Rev Ther Drug Carr Syst. 2017;34(3):185–208. doi: 10.1615/CritRevTherDrugCarrierSyst.2017018042. [DOI] [PubMed] [Google Scholar]
- 142.Yoon YS, Kim JC. Recent applications of chemosensitivity tests for colorectal cancer treatment. World J Gastroenterol. 2014;20:16398–16408. doi: 10.3748/wjg.v20.i44.16398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hur H, Kim NK, Kim HG, et al. Adenosine triphosphate-based chemotherapy response assay-guided chemotherapy in unresectable colorectal liver metastasis. Br J Cancer. 2012;106:53–60. doi: 10.1038/bjc.2011.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kern DH, Weisenthal LM. Highly specific prediction of antineoplastic drug resistance with an in vitro assay using suprapharmacologic drug exposures. J Natl Cancer Inst. 1990;82:582–588. doi: 10.1093/jnci/82.7.582. [DOI] [PubMed] [Google Scholar]
- 145.Mechetner E, Brunner N, Parker RJ. In vitro drug responses in primary and metastatic colorectal cancers. Scand J Gastroenterol. 2011;46:70–78. doi: 10.3109/00365521.2010.510573. [DOI] [PubMed] [Google Scholar]
- 146.Fan CW, Fan HA, Hsu SH, et al. An in vitro short time-high dose drug exposure assay for predicting 5FU-resistance of colorectal cancer. Cancer Lett. 2004;214:181–188. doi: 10.1016/j.canlet.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 147.Das T, Nair RR, Green R, et al. Actinomycin D down-regulates SOX2 expression and induces death in breast cancer stem cells. Anticancer Res. 2017;37:1655–1663. doi: 10.21873/anticanres.11496. [DOI] [PubMed] [Google Scholar]
- 148.Girard YK, Wang C, Ravi S, et al. A 3D fibrous scaffold inducing tumoroids: a platform for anticancer drug development. PLoS ONE. 2013;8:e75345. doi: 10.1371/journal.pone.0075345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gluck S, de Snoo F, Peeters J, et al. Molecular subtyping of early-stage breast cancer identifies a group of patients who do not benefit from neoadjuvant chemotherapy. Breast Cancer Res Treat. 2013;139:759–767. doi: 10.1007/s10549-013-2572-4. [DOI] [PubMed] [Google Scholar]
- 150.Gluck S, McKenna EF, Jr, Royce M. XeNA: capecitabine plus docetaxel, with or without trastuzumab, as preoperative therapy for early breast cancer. Int J Med Sci. 2008;5:341–346. doi: 10.7150/ijms.5.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stachelscheid H, Wulf-Goldenberg A, Eckert K, et al. Teratoma formation of human embryonic stem cells in three-dimensional perfusion culture bioreactors. J Tissue Eng Regen Med. 2013;7:729–741. doi: 10.1002/term.1467. [DOI] [PubMed] [Google Scholar]
- 152.Chen J, Xia Q, Jiang B, et al. Prognostic value of cancer stem cell marker ALDH1 expression in colorectal cancer: a systematic review and meta-analysis. PLoS ONE. 2015;10:e0145164. doi: 10.1371/journal.pone.0145164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iinuma H, Watanabe T, Mimori K, et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J Clin Oncol. 2011;29:1547–1555. doi: 10.1200/JCO.2010.30.5151. [DOI] [PubMed] [Google Scholar]
- 154.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 155.Scatena R, Bottoni P, Pontoglio A, et al. Cancer stem cells: the development of new cancer therapeutics. Expert Opin Biol Ther. 2011;11:875–892. doi: 10.1517/14712598.2011.573780. [DOI] [PubMed] [Google Scholar]
- 156.Carmody L, Germain A, Morgan B, et al. Identification of a selective small-molecule inhibitor of breast cancer stem cells—probe 3. In: Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD); 2010.
- 157.Carmody L, Germain A, Morgan B, et al. Identification of a selective small-molecule inhibitor of breast cancer stem cells—probe 1. In: Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD; 2010.
- 158.Carmody LC, Germain A, Morgan B, et al. Identification of a Selective Small-Molecule Inhibitor of Breast Cancer Stem Cells—Probe 2. In: Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD; 2010.
- 159.Carmody LC, Germain AR, VerPlank L, et al. Phenotypic high-throughput screening elucidates target pathway in breast cancer stem cell-like cells. J Biomol Screen. 2012;17:1204–1210. doi: 10.1177/1087057112458317. [DOI] [PubMed] [Google Scholar]
- 160.Germain AR, Carmody LC, Nag PP, et al. Cinnamides as selective small-molecule inhibitors of a cellular model of breast cancer stem cells. Bioorg Med Chem Lett. 2013;23:1834–1838. doi: 10.1016/j.bmcl.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 161.Hirano K, Sasaki N, Ichimiya T, et al. 3-O-sulfated heparan sulfate recognized by the antibody HS4C3 contributes [corrected] to the differentiation of mouse embryonic stem cells via fas signaling. PLoS ONE. 2012;7:e43440. doi: 10.1371/journal.pone.0043440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Johnson CE, Crawford BE, Stavridis M, et al. Essential alterations of heparan sulfate during the differentiation of embryonic stem cells to Sox1-enhanced green fluorescent protein-expressing neural progenitor cells. Stem Cells. 2007;25:1913–1923. doi: 10.1634/stemcells.2006-0445. [DOI] [PubMed] [Google Scholar]
- 163.Nairn AV, Kinoshita-Toyoda A, Toyoda H, et al. Glycomics of proteoglycan biosynthesis in murine embryonic stem cell differentiation. J Proteome Res. 2007;6:4374–4387. doi: 10.1021/pr070446f. [DOI] [PMC free article] [PubMed] [Google Scholar]