Abstract
Cancer metastasis causes approximately 90% of all cancer-related death and independent of the advancement of cancer therapy, a majority of late stage patients suffers from metastatic cancer. Metastasis implies cancer cell migration and invasion throughout the body. Migration requires the formation of pseudopodia in the direction of movement, but a detailed understanding of this process and accordingly strategies of prevention remain elusive. Here, we use quantitative proteomic profiling of human cancer pseudopodia to examine this mechanisms essential to metastasis formation, and identify potential candidates for pharmacological interference with the process. We demonstrate that Prohibitins (PHBs) are significantly enriched in the pseudopodia fraction derived from cancer cells, and knockdown of PHBs, as well as their chemical inhibition through Rocaglamide (Roc-A), efficiently reduces cancer cell migration.
Introduction
Cancer metastasis is responsible for over 90% of cancer-related death. In spite of numerous advances in cancer treatment, including surgical techniques, radiation therapy, as well as chemotherapy, a significant number of patients will display metastatic disease regardless of local control. Virtually all cancers are able to metastasize to tissues distant from the tumor of origin, including brain, bone, lungs and liver. The metastatic process is, however, inefficient and relies on several critical steps, of which the entry of cancer cells into the circulatory system constitutes the first step. Thus, poor prognosis is linked to lymph node involvement as well as vascular tumor emboli.
Prominent among genetic changes correlated with invasive characteristics of cancer cells is the capacity to develop specialized protrusive and adhesive cellular structures: pseudopodia, invadopodia and podosomes1–7. Invadopodia and podosomes are dynamic actin-based cytoskeleton projections into the plasma membrane and observed on the surface of cells plated on extracellular matrix. They facilitate proteolytic degradation of extracellular matrix by secretion of matrix metalloproteinase4,5. In contrast, pseudopodia are larger dynamic actin cytoskeleton-based structures formed at the cell front and promote directional migration in response to chemo-attractive stimuli3. Invasive cancer cells produce pseudopodia to penetrate constraining tissue and structures, and migrate e.g. into the lymphatic system and on to distant organs. On the molecular level, several studies report that the Arp2/3 complex, Wave3, Eps8, cortactin, α-actinin, Lim-kinase, fascin, and filamin are associated with pseudopodia formation8–11.
Based on these findings from the literature, we hypothesize that pseudopodia-specific proteins associated with regulation of pseudopodia function remain to be discovered and continued characterization of cell protrusions may lead to identification of further players in the development of metastatic disease and thus new therapeutic strategies. Accordingly, we investigated in this study metastatic cancer cell protrusions using deep quantitative proteomic profiling of cancer cell pseudopodia isolated by physical fractionation, in order to identify new target proteins that may be used for blocking cancer cell migration.
Results
Extension of Pseudopodia in Response to Serum Stimulation
To establish a model system for the study of pseudopodia in the context of cancer, MDA-MB-231 cells were starved for 16 h, followed by incubation in pseudopodia-stimulating conditions such as LPA or serum stimulation (both in DMEM) and DMEM alone as a control12–14. The resulting amount of pseudopodia extension by LPA or serum stimulation was quantified using polycarbonate transwell membrane filters14. To prevent cell bodies from penetrating the filter pores, 3-μm pore size was employed to prevent nuclear passage. To image pseudopodia and cell body, the top and bottom of the membrane filters were stained using phalloidin. Figure 1 shows that MDA-MB-231 cells extended pseudopodia through the filters in response to LPA or serum stimulation, but not when DMEM was used alone. Cultures exposed to serum resulted in superior numbers of pseudopodia as compared to LPA. Further experiments and deep quantitative proteomic profiling of pseudopodia were thus performed using 1% serum stimulation.
Figure 1.
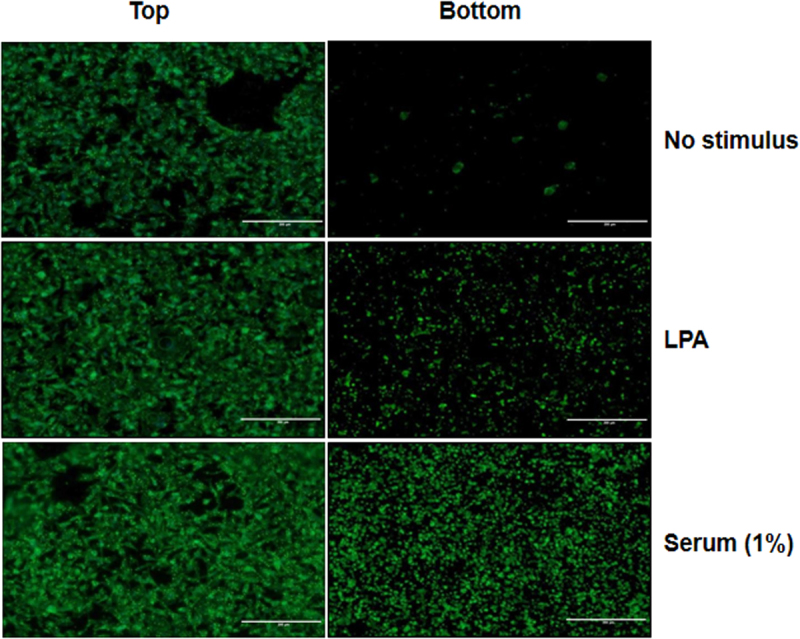
Extension of pseudopodia by MDA-MB-231 cells in response to LPA and serum. Microscope images of MDA-MB-231 cell bodies (top) or pseudopodia (bottom) separated by filters with 3 μm pore size in response to the indicated stimuli. Cells were stained for F-actin with phalloidin. Scale bar = 200 μm.
Comparative Quantitative Proteomic Analysis of Pseudopodia and Cell Body Fractions
Figure 2 shows the strategy employed for quantitative pseudopodia proteomics in MDA-MB-231 cells. Purification of pseudopodia and cell bodies, was achieved through a previously described protocol based again on microporous transwell filters to separate the structures14,15. In order to perform quantitative proteomic analysis of pseudopodia, we cultured MDA-MB-231 cells using stable isotope labeling by amino acids in cell culture16,17 for in excess of 6 cell divisions and then isolated pseudopodia and cell body fractions after serum stimulation for 2 h. To check incorporation rate of heavy labeled Arginine and Lysine, we confirmed heavy labeling peptides in cell lysates in heavy SILAC by LC/MS/MS (Supplementary Fig. 2A) and measured the ratio of heavy labeled peptides to all peptides identified. The overall incorporation rate was approximately 99%, reflecting the enrichment of the reagents used (Supplementary Fig. 2B). For SILAC-based comparative proteomic analysis between pseudopodia and cell bodies, we employed a strategy where light and heavy labeled cell body samples were combined with heavy and light labeled pseudopodia samples, respectively, in a label swapping mixing scheme.
Figure 2.
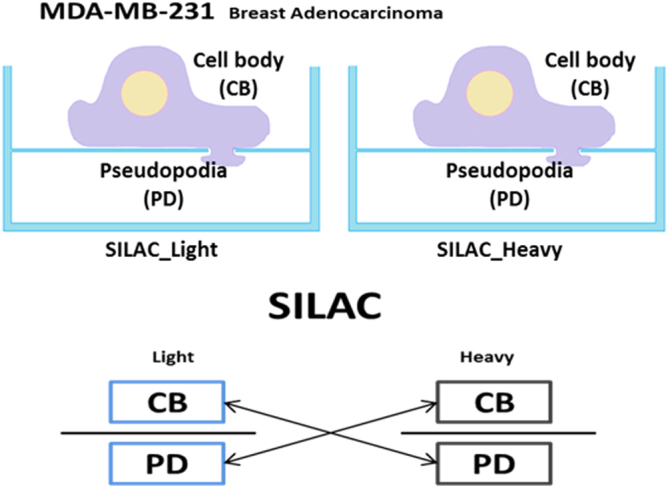
Quantitative proteomics profiling strategy for pseudopodia of MDA-MB-231 cells using SILAC. Illustration of MDA-MB-231 cell extending pseudopodia. Cells are cultured in SILAC MDEM and extend pseudopodia through 3-μm pores of membrane filter in response to serum stimulation (up). Pseudopodia are separated from cell bodies manually. The isolated cell bodies and pseudopodia are mixed 1:1 in a label swapping scheme and then profiled using quantitative proteomics (bottom).
By comparing SILAC labeling ratios between pseudopodia and cell body, we identified 578 proteins to be enriched in pseudopodia (Supplementary data), including known pseudopodial proteins such as calpain, filamin, IGQAP1, β1 integrin, and ROCK18, which validate the approach. The small number of cell body-deriving proteins, contained in the data set may stem from leaking of highly abundant proteins during the extraction procedure, which in conjunction with a comparatively low protein concentration in the pseudopodia fraction may lead to an apparent over-representation. We proceeded with annotation of pseudopodia enriched proteins using gene ontology (GO) analysis including GO Biological process, GO molecular function, GO cellular component19, and KEGG pathways20 (Supplementary data). Enriched GO categories include cytoskeleton, plasma membrane, mitochondria and cytosol. Enriched KEGG pathways include regulation of actin cytoskeleton, endocytosis, proteasome, GAP junction, and glycolysis. Groups with log2 ratios in excess of 3 (pseudopodia vs. cells bodies) were associated with GAP junction, endocytosis, tight junction, and glycolysis, while log2 ratios between 3 and 1 were mainly involved in regulation of actin cytoskeleton and the ubiquitin/proteasome system. Among the proteins identified as enriched in pseudopodia were, IQGAP1 and ARPC (Actin related protein complex). IQGAP1, a key regulator of adhesion and migration, is known to regulate an array of molecular events, such as N-Wasp induced branched actin assembly by ARPC21. Further pseudopodia-enriched proteins are ILK1, PARVA (Alpha-parvin) and RSU (Ras suppressor protein 1), which are known to cooperate in promotion of metastasis. ILK1 is a serine/threonine protein kinase which interacts with integrin subunits to regulate integrin mediated signal transduction22. It is upregulated in several cancer types23,24 and was accordingly interrogated for influence on cancer cell migration below.
To validate the quantitative proteomic analysis of pseudopodia, a subset of proteins enriched in pseudopodia or cell body fractions were examined by Western blotting (Fig. 3) and showed good correlations with the proteomics results. Histone proteins were present mostly in the cell body fraction, and IQGAP1 and ROCK1/2 were present in the pseudopodia fraction.
Figure 3.
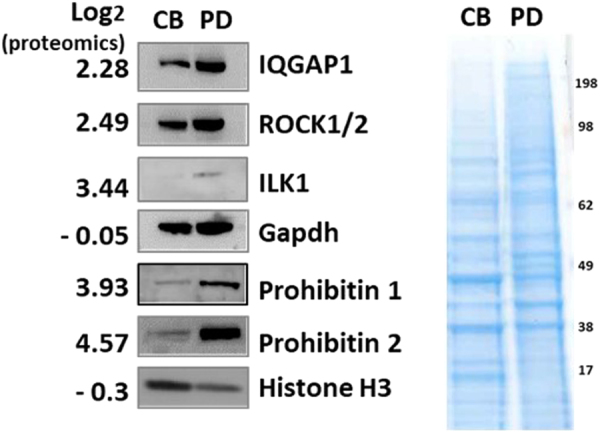
Validation of results from the quantitative proteomics screen using Western Blot. Western Blots were performed for proteins from MDA-MB-231 cell pseudopodia (PD) and cell body samples (CB; left). Equal amounts of protein samples were separated by SDS PAGE, plotted and probed with antibodies specific for the indicated proteins, as well as stained with Coomasie Blue (right).
Table 1 shows the top 50 proteins identified as highly enriched in the pseudopodia fraction. Interestingly, this includes a number of mitochondrial proteins like SLC25A, LRPPRC, GOT2, ATP5A, ACADM and PHBs (Prohibitions). PHBs proteins are evolutionarily conserved and widely expressed in diverse cellular processes such as proliferation and metabolism, are known to interact with various receptors, and are involved with activation of ERK signaling in the plasma membrane. They localize to the mitochondrial inner membrane25–27.
Table 1.
List of the top 50 proteins identified as highly enriched in pseudopodia over cell bodies.
| Gene_symbol | Protein_names | Log 2 ratio | p_values | FDR * _values |
|---|---|---|---|---|
| SLC25A6 | ADP/ATP translocase 3 | 5.40 | 1.53E-05 | 3.51E-04 |
| TBB2 | Tubulin beta-2 chain | 5.05 | 2.87E-07 | 1.93E-04 |
| GOT2 | Aspartate aminotransferase, mitochondrial | 4.97 | 2.46E-05 | 4.25E-04 |
| PHB2 | Prohibitin-2 | 4.58 | 7.21E-05 | 7.15E-04 |
| SLC25A1 | Tricarboxylate transport protein, mitochondrial | 4.50 | 1.48E-04 | 1.05E-03 |
| SLC25A24 | Calcium-binding mitochondrial carrier protein SCaMC-1 | 4.46 | 9.19E-04 | 3.49E-03 |
| ATP5A1 | ATP synthase subunit alpha, mitochondrial | 4.37 | 9.73E-04 | 3.60E-03 |
| ARHGAP1 | Rho GTPase-activating protein 1 | 4.34 | 6.93E-04 | 2.82E-03 |
| TUBB3 | Tubulin beta-3 chain | 4.13 | 1.25E-04 | 9.64E-04 |
| TUBB8 | Tubulin beta-8 chain | 4.05 | 1.12E-04 | 8.94E-04 |
| WDR1 | WD repeat-containing protein 1 | 4.02 | 7.00E-06 | 2.65E-04 |
| GDI2 | Rab GDP dissociation inhibitor beta | 3.97 | 4.86E-06 | 2.55E-04 |
| PHB | Prohibitin | 3.94 | 6.01E-04 | 2.56E-03 |
| Eif3e | Eukaryotic translation initiation factor 3 subunit E | 3.88 | 2.21E-04 | 1.32E-03 |
| CCT7 | T-complex protein 1 subunit eta | 3.84 | 3.90E-05 | 5.02E-04 |
| TUBB | Tubulin beta chain | 3.81 | 6.30E-04 | 2.64E-03 |
| TUBA3 | Tubulin alpha-3 chain;Tubulin alpha-5 chain | 3.81 | 3.38E-03 | 8.70E-03 |
| FH | Fumarate hydratase, mitochondrial | 3.80 | 1.63E-04 | 1.11E-03 |
| EHD4 | EH domain-containing protein 4 | 3.80 | 4.61E-04 | 2.15E-03 |
| ETF1 | Eukaryotic peptide chain release factor subunit 1 | 3.77 | 7.09E-05 | 7.12E-04 |
| UMPS | Uridine 5-monophosphate synthase;Orotate phosphoribosyltransferase | 3.73 | 1.29E-04 | 9.75E-04 |
| SLC25A3 | Phosphate carrier protein, mitochondrial | 3.72 | 2.01E-04 | 1.25E-03 |
| ACTR10 | Actin-related protein 10 | 3.71 | 2.07E-03 | 6.07E-03 |
| TUBB2B | Tubulin beta-2B chain | 3.66 | 2.08E-06 | 2.21E-04 |
| EPHX1 | Epoxide hydrolase 1 | 3.62 | 1.29E-04 | 9.75E-04 |
| NDA3 | Tubulin beta chain | 3.61 | 1.03E-03 | 3.74E-03 |
| SLC16A3 | Monocarboxylate transporter 4 | 3.60 | 1.87E-06 | 2.21E-04 |
| SLC7A5 | Large neutral amino acids transporter small subunit 1 | 3.60 | 1.68E-05 | 3.72E-04 |
| TUBG1 | Tubulin gamma-1 chain;Tubulin gamma-2 chain | 3.56 | 8.38E-04 | 3.25E-03 |
| ACADM | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 3.53 | 4.04E-04 | 1.99E-03 |
| PICALM | Phosphatidylinositol-binding clathrin assembly protein | 3.53 | 3.38E-03 | 8.70E-03 |
| ATP1B1 | Sodium/potassium-transporting ATPase subunit beta-1 | 3.52 | 1.24E-05 | 3.22E-04 |
| PAICS | Multifunctional protein ADE2 | 3.50 | 1.08E-03 | 3.87E-03 |
| NXN | Nucleoredoxin | 3.48 | 5.29E-05 | 6.02E-04 |
| PSMD12 | 26 S proteasome non-ATPase regulatory subunit 12 | 3.48 | 6.91E-04 | 2.82E-03 |
| SLC3A2 | 4F2 cell-surface antigen heavy chain | 3.47 | 7.52E-05 | 7.34E-04 |
| CORO1C | Coronin-1C | 3.47 | 1.78E-03 | 5.44E-03 |
| OXSR1 | Serine/threonine-protein kinase OSR1 | 3.45 | 1.37E-04 | 1.01E-03 |
| CCT | T-complex protein 1 subunit gamma | 3.45 | 1.25E-06 | 2.21E-04 |
| GPI | Glucose-6-phosphate isomerase | 3.45 | 4.87E-04 | 2.23E-03 |
| ILK1 | Integrin-linked protein kinase1 | 3.45 | 5.62E-04 | 2.44E-03 |
| PSMC3 | 26 S protease regulatory subunit 6 A | 3.44 | 3.42E-05 | 4.84E-04 |
| PNP | Purine nucleoside phosphorylase | 3.42 | 1.83E-05 | 3.84E-04 |
| HSD17B11 | Estradiol 17-beta-dehydrogenase 11 | 3.41 | 1.47E-05 | 3.43E-04 |
| EHD1 | EH domain-containing protein 1 | 3.39 | 1.94E-04 | 1.23E-03 |
| FERMT2 | Fermitin family homolog 2 | 3.38 | 2.29E-03 | 6.45E-03 |
| G6PD | Glucose-6-phosphate 1-dehydrogenase | 3.35 | 3.52E-04 | 1.84E-03 |
| SLC1A5 | Neutral amino acid transporter B | 3.33 | 1.76E-04 | 1.15E-03 |
| CS | Citrate synthase, mitochondrial | 3.33 | 1.91E-03 | 5.76E-03 |
| ASNS | Asparagine synthetase [glutamine-hydrolyzing] | 3.32 | 2.03E-04 | 1.25E-03 |
*FDR: False discovery rate (corrected p-value according to Benjamini-Hochberg1).
To exclude the possibility that recovery of mitochondrial proteins in the pseudopodia fraction was artifactual, we checked whether mitochondria localize to pseudopodia using phalloidin (green) and mitotracker (red) for staining of F-actin and mitochondria, respectively, in transwell culture. Although the majority of mitochondria were present in the cell body, they also show a clear presence in the pseudopodia (Supplementary Fig. 3). This finding is further supported by a recent report on a pseudopodia-subtype involved in myelination of Schwann cells, which also describes PHBs and mitochondria to be present in pseudopodia – with functional implications for myelination28. We conclude that mitochondria and in particular PHBs are likely components of pseudopodia in cancer cells.
PHBs are Necessary for Cancer Migration in vitro
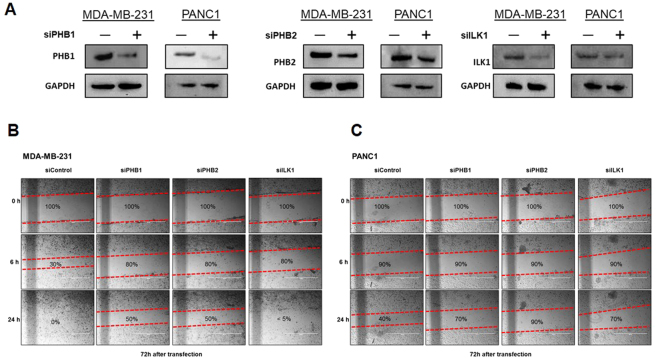
The role of pseudopodia in cell migration is well established. We thus hypothesized that disrupting a key player in pseudopodia physiology impacts migration. To explore the relevance of our candidate proteins in this context, we used two different invasive cancer cell lines. MDA-MB-231 and PANC1, models for breast and pancreatic cancer, respectively, were transfected with siRNA knocking down PHB1 and PHB2. In addition, expression of ILK1, which was also found enriched in pseudopodia above, was knocked down by siRNA in both cell lines. Reduction of protein expression by siRNA is shown for all experiments by western blot in Fig. 4A. To check the effect of knockdown PHB1, PHB2 and ILK1 on cell proliferation, we performed cell proliferation assay, observing slightly reduced proliferation in all cases (Supplementary Fig. 4A). Using a wound healing assay, we found that the PHB1 and PHB2 knockdown in both MDA-MB-231 (Fig. 4B) and PANC1 (Fig. 4C) had a reduced propensity for migration and gap closure when compared to control cells expressing a Non-targeting siRNA. Interestingly, migration in cells with ILK1 knockdown, was impacted only for PANC1, while MDA-MB-231 showed very little difference in migration pattern or rate.
Figure 4.
Wound healing assay of siRNA knockdown in MDA-MB-231 and PANC1 cells. (A) Western Blot demonstrating knockdown of PHB1, PHB2, and ILK1 in MDA-MB-231 and PANC1 cells. (B and C) siRNA knockdown of PHBs or ILK1 inhibits migration of different types of cancer cells. Each gap was created in confluent cells transfected with siRNA against PHBs or ILK1 and the evolution of the gap width was measured over time. Scale bar = 200 μm.
Rocaglamide Inhibits Directional Migration of Cancer Cells
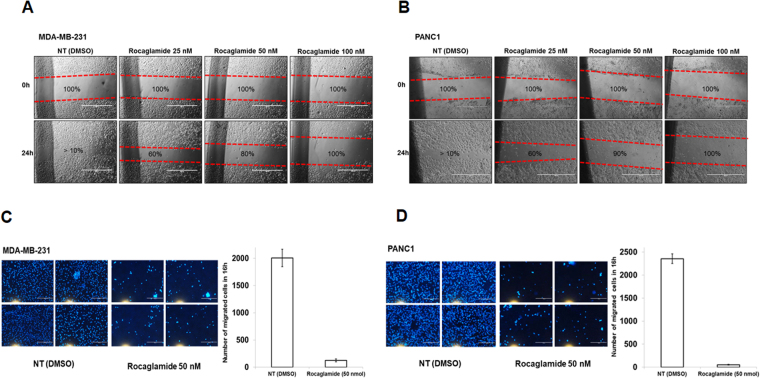
Rocaglamide (Roc-A), a naturally occurring flavagline29,30, is reported to inhibit the RAF-MEK-ERK pathway by targeting PHB1 and PHB231. Given our findings above, we therefore sought to determine its effect on cancer cell migration and repeated the wound healing assay described above in the presence of increasing doses (25 to 100 nM) of Roc-A. In alignment with the knockdown data described above, we found Roc-A to reduce migration of MDA-MB-231 (Fig. 5A) and PANC1 (Fig. 5B) cells after 24 h of treatment even at low concentrations (25 nM). Translating this finding to an alternative experimental system, we performed a directional cell migration assay. Migration of MDA-MB-231 (Fig. 5C) and PANC1 (Fig. 5D) cells into the bottom chamber of a transwell assembly was strongly stimulated by serum, while cells treated additionally with Roc-A barely moved, in spite of serum stimulation. To check the effect of Roc-A on cell proliferation, we repeated the cell proliferation assay in its presence. While we observed a dose dependent proliferative effect of Roc-A with significantly reduced proliferation at 100 nM final concentration, the effect at 25 nM and 50 nM used in the assays above was mild, supporting interpretation of our observations as a migration, rather than proliferative phenotype (Supplementary Fig. 4B).
Figure 5.
Wound healing assay and directional migration assay of Roc-A treated MDA-MB-231 and PANC1 cells. (A and B) Roc-A inhibits migration of cancer cells. MDA-MB-231 and PANC1 cells were treated without (control) or with Roc-A (25 to 100 nM) and the evolution of gap closure was measured. (C and D) Roc-A inhibits directional migration of cancer cells. MDA-MB-231 and PANC1 cells treated without (control) or with 50 nM Roc-A for 24 h were subjected to a transwell migration assay in response to serum stimulation. Scale bar = 200 μm.
Discussion
Metastasis is the major cause of mortality in cancer-affected patients and there is currently no generic treatment for patients suffering from it. Therefore, development of new anticancer drugs that efficiently tackle metastasis is crucial.
Cancer cell migration is the essential first step for invasion and metastasis formation. Using SILAC-based quantitative proteomics in combination with physical pseudopodia fractionation in MDA-MB-231 cancer cells, we devise in this study a strategy to identify proteins that may be involved with cell migration and thus serve as potential anti-metastatic targets for therapeutic purposes. Our results provide robust and reliable data for investigation of cancer metastasis research.
In the top 50 proteins most enriched in pseudopodia over cell bodies we found, as expected, many factors involved in cell adhesion, migration, and metabolism. As numerous studies suggest that cancer cell metabolism, such as the switch from oxidative metabolism to glycolysis, plays an important part in tumor cell migration, invasion, as well as growth32–37 and it was also reported that localization of mitochondria within moving cells have strong correlations with the speed of its movement as well as directional persistence during directional migration38, the inclusion of a large number of mitochondrial proteins in the enriched set may not be as surprising as it initially appears.
Pseudopodia are large dynamic actin cytoskeleton based protrusions formed at the cell front and intimately linked to directional migration. The implied high level of energy consumption may require redistribution of mitochondria to pseudopodia in moving cancer cells for directional migration and thus possibly increased invasiveness. Future studies of the mitochondrial localization may thus contribute to a better understanding of the mechanism of cancer invasion, migration and finally cancer metastasis.
The pseudopodia/mitochondria connection hypothesized is exemplarized by our finding that PHBs were significantly enriched in pseudopodia of cancer cells. Multiple studies have reported PHBs as predominantly localized to mitochondria, and playing a key role in maintenance of mitochondrial function and morphology39,40. Our follow-up experiments demonstrate that depletion of PHBs strongly reduces cancer cell migration in general and directional migration more specifically-even in the context of serum stimulation.
Our findings are supported by a similar study published recently describing PHBs as enriched in pseudopodia of Schwann cells with possible important implications for central nervous system myelination28.
Accordingly, we assume that accumulation of PHBs in pseudopodia is mechanistically linked to cell migration in general and cancer cell migration more specifically.
We proceeded with exploiting this hypothesis pharmacologically and show that Roc-A, a natural anticancer compound known to bind to PHB1 and PHB2 directly31, indeed appears to interfere with cancer cell migration in a wound healing and directional migration assay, both, even at low concentrations (25 to 100 nM). Replicating our results in cellular models deriving from different cancers (MDA-MB-231 from breast cancer and PANC1 from pancreatic cancer) hints at a general, rather than cell type-specific nature of the observed effect. We thus believe Roc-A to be an interesting candidate drug for anti-cancer metastatic activity in cancer in general.
In summary, we present a quantitative proteomics strategy for the identification of factors involved in cancer cell motility and metastasis and utilize it to identify PHBs as proteins likely involved in the process. This in turn leads us to hypothesize that localized metabolism supported by mitochondrial relocalization may be a vital factor in function of pseudopodia. We further extend our findings to describe Roc-A, a PHB directed compound, as an inhibitor of cancer cell migration, leading us to conclude that it may represent a promising drug candidate targeting metastasis reduction or prevention.
While this study focused on the effects of PHBs and a single PHB inhibitor, we believe that other proteins characterized by us as enriched in pseudopodia may provide further insight into the process of cancer cell migration, invasion and metastasis, as well as serving as targets for further pharmacological interference with those processes. For instance, our result show that ILK1 knockdown in PANC1 cells reduced migration capacity. As an agent called compound 22 (cpd 22) is an ILK1 inhibitor already described in the literature41, this target/compound pairing may warrant future exploration in at least a subset of cancers. Similarly, the vacuolar protein sorting-associated protein (VPS) family and STAT proteins may be inhibited by Bafilomycin42 and Nifuroxazide, respectively43. Our proteomic analysis of pseudopodia of cancer cells may thus offer promising avenues for the development of strategies to combat metastatic disease.
Methods
Pseudopodia Assay
MDA-MB-231 human breast cancer cells were starved for 16 h, subsequently plated into the top Boyden chamber of polycarbonate transwell inserts with 3-μm pores (Corning #3414; 1.5 × 106 cells), and incubated for 2 h at 37 °C. To stimulate formation of pseudopodia from the cell surface, the inserts were transferred to wells containing serum-free Dulbecco’s modified Eagle’s medium (DMEM) (HyClone, Thermo Scientific) (no stimulus) and DMEM with 100 ng/mL lysophosphatidic acid (LPA), or DMEM with 1% fetal bovine serum (FBS), respectively and incubated for 2 h at 37 °C. We also tested 0.1% and 0.5% of serum stimulation (Supplementary Fig. 1). Cell bodies in the top chamber and pseudopodia in the bottom chamber were stained using phalloidin (DyLight 488 nm, #21833, Thermo Scientific) after three PBS washes and fixing with 4% paraformaldehyde.
SILAC Based Cell Culture and Pseudopodia Extraction for Quantitative Proteomics
MDA-MB-231 cells were grown in SILAC DMEM (#88425, Thermo Scientific) containing natural lysine and arginine in the ‘light’ or 13C615N2-lysine (Lys8, #CNLM-291, (13C6, 99%; 15N2, 99%), Cambridge Isotope laboratory Inc) and 13C615N4-arginine (Arg10, #CNLM539, (13C6, 99%; 15N4, 99%), Cambridge Isotope laboratory Inc) in the ‘heavy’ condition, along with antibiotics and dialyzed FBS. For complete labeling, cells were cultured in excess of 6 doublings in SILAC DMEM and an incorporation test was performed to measure the ratio of heavy labeled peptides to all peptides identified (heavy and light). In order to prepare a pseudopodia fraction, we used the specific enrichment method described in Klemke et al.15. Protein yield was measured using a BCA assay. Twenty micrograms of cell body sample from the top and pseudopodia sample from the bottom chamber were prepared, respectively, and for comparative proteome profiling mixed 1:1 in a label swapping scheme.
Proteomics Analysis
For LC/MS/MS analysis, a tryptic digest of each sample was injected using a nano-flow chromatographic system (Easy nLC-II, Thermo Fisher Scientific, Bremen, Germany) and separated on in-house packed column emitters using a 20 cm fused silica capillary (i.d. 75 μm, o.d. 360 μm) filled with C18 (particle size 3 μm, 100 Å) resin (Dr. Maisch GmbH, Ammerbuch-Entrigen, Germany). Separation of peptides was performed by a 120 min gradient from buffer A (0.5% acetic acid in water) to buffer B (80% acetonitrile, 0.5% acetic acid). Eluted peptides were electrospray ionized and introduced into a Q Exactive mass spectrometer. Precursor ion scans were acquired at a resolution of 70,000 (m/z 300). A data dependent analysis was used, where the 10 most intense ions detected in the full scan are isolated (3 Th isolation width) and fragmented using higher energy collisional dissociation (HCD, collision energy 25) in the context of dynamic exclusion for 25 s. Unassigned charge states and singly charged ions were also excluded.
Protein identification and quantification were performed using the MaxQuant suite of algorithms (version 1.4.1.2)44 and a Homo sapiens UniprotKB data base (downloaded on 04.06.2015;109,411 protein entries). The search parameters employed are as follows: first search mass accuracy tolerance 20 ppm, main search mass accuracy tolerance 4.5 ppm, FTMS MS/MS tolerance 20 ppm, minimum peptide length of 7 amino acids, peptide spectrum match false discovery rate (FDR) and protein FDR both set to 0.01 as calculated by the reverse database approach45,46. Carbamidomethylation of cysteine was searched as a fixed modification, and N-terminal acethylation and oxidation of methionine were searched as variable modifications. Heavy labeled arginine (Arg10) and lysine (Lys8) were specified for quantitative analysis.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE47 partner repository with the dataset identifier PXD004658.
Western Blotting for Candidate Validation
To validate proteomics results, western blot analysis was performed. After the transfer of target proteins from SDS-PAGE, blocked membranes were incubated with Millipore anti-ROCK2 (Rho-associated protein kinase 2) (#07-1458), anti- IQGAP1 (Ras GTPase-activating-like protein 1) (#abt186), anti-PHB1 (#abn293), anti-PHB2 (#mabc953); Cell Signaling anti-GAPDH (#2118), anti-ILK1 (Integrin-linked protein kinase 1) (#3862), and Anti-Histone H3 (#4499) overnight at 4 °C. The membrane was rinsed with 1x TBS buffer prior to incubation with secondary antibody for 1 h. Blots were developed using ECL buffer (GE Healthcare) and the Geliance 600 imaging system (PerkinElmer).
Bioinformatical and Statistical Analyses
All statistical analysis of MaxQuant results was performed by autonomics, an in-house suite for the statistical analysis of omics data (A.M. Bhagwat, R.J. Cotton, L. Cougnaud, and J. Graumann, manuscript in preparation; https://bitbucket.org/graumannlabtools/autonomics). The experimental design consisted of 4 replicates for both pseudopodia as well as cell body. MaxQuant was used to transform the raw spectra into a matrix of protein group ration (row = protein group, column = sample). From this matrix, we retained proteins which were not tagged by MaxQuant as contaminants or reverse hits, and which had at least two ratios associated. We subsequently log2 transformed the ratios to obtain approximately symmetric distributions. Log2 protein ratios were quantile normalized prior to statistical analysis48. For statistical analysis, the model “protein ratio ~ 1” was fitted in parallel for each protein using the R49 package limma48, For each protein group, this model tests whether the log2 ratio is significantly different from zero, using limma’s built-in moderated t-test. Benjamini-Hochberg FDR values were calculated using the p.adjust function in R50,51.
siRNA Knockdown of Protein Expression and Wound Healing Assay
The MDA-MB-231 human breast cancer cell line and PANC1 human pancreatic cancer cell line were transiently transfected using siRNA against PHB (siGENOME Human PHB (5254) siRNA – SMART pool; Dharmacom), PHB2 (siGENOME Human Phb2 (11331) siRNA – SMART pool; Dharmacon), ILK1 (siGENOME Human Ilk (3611) siRNA – SMART pool; Dharmacon), and control (siGENOME Non-Targeting siRNA Pool; Dharmacon) using DharmaFECT1 transfection reagent (#T-2001-02; Dharmacon) for 72 h. For validation of the transfection efficiency, western blot analysis was employed. In a wound healing assay, cells were plated in 12-well plates (#3043, Becton Dickinson) and cultured. After transfection, a narrow gap was created with a pipette tip in the confluent cell monolayer and monitored for 24 h for recolonization.
Treatment with Rocaglamides and Wound Healing/Migration Assay
MDA-MB-231 and PANC1 cells were cultured in DMEM with 10% FBS until 90–100% confluence in 12-well plates (#3043, Becton Dickinson) and subsequently treated with DMSO (control), 25 nM, 50 nM, and 100 nM of Roc-A (Rocaglamides, #84573-16-0 ≥96% pure, Sigma Aldrich) dissolved in DMSO. Wound healing was assayed for 24 h as described above.
For the migration assay, polycarbonate transwell inserts with 8-μm pores (Corning #3422) were used. MDA-MB-231 and PANC1 cells were cultured in DMEM with 10% FBS and subsequently starved for 16 h. Trypsinized cells were plated into the top Boyden chamber inserts and incubated for 2 h at 37 °C. The inserts were transferred to a plate containing DMEM with 1% FBS and incubated overnight at 37 °C. Subsequently cell presence in top and bottom chambers was evaluated using DAPI (#p36935, molecular probes by Life Technologies).
Cellular Proliferation Assay
For cellular proliferation assay, Cell Proliferation ELISA, BrdU (colorimetric) Version 14.0 (#11647229001, Roche Applied Science, Mannheim, Germany) was used. Cells were labeled with BrdU labeling solution for 2 h at 37 °C. After fixing the labeled cells, cells were labeled with Anti-BrdU-POD working solution for 90 min. Cells were washed with Washing solution and then incubated with Substrate solution for 15 min. BrdU assay was performed by measuring the absorbance at 450 nm. All tests were conducted in triplicates and repeated twice.
Electronic supplementary material
Supplementary Dataset Pseudopodia protein ID
Acknowledgements
The authors thank Arash Rafii Tabrizi (Weill Cornell Medicine – Qatar) for critical reading of the manuscript. JG and the Proteomics & Imaging Cores at Weill Cornell Medicine – Qatar are supported by “Biomedical Research Program” funds at WCM-Q, a program funded by Qatar Foundation. The work was supported by Qatar National Research Fund’s National Priority Research Program grant no. 7-082-1-014. The statements made herein are solely the responsibility of the authors.
Author Contributions
S.C. carried out the study, which S.C. and J.G. designed and J.G. directed. A.M.B. analyzed proteomic data and performed bioinformatics analysis. R.A.M., N.G. & H.B.H. provided technical and infrastructural support. L.S. operated microscopy. S.C. and J.G. drafted the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24256-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Havrylov S, Park M. MS/MS-based strategies for proteomic profiling of invasive cell structures. Proteomics. 2015;15:272–286. doi: 10.1002/pmic.201400220. [DOI] [PubMed] [Google Scholar]
- 2.Linder S, Wiesner C, Himmel M. Degrading devices: invadosomes in proteolytic cell invasion. Annual review of cell and developmental biology. 2011;27:185–211. doi: 10.1146/annurev-cellbio-092910-154216. [DOI] [PubMed] [Google Scholar]
- 3.Klemke RL. Trespassing cancer cells: ‘fingerprinting’ invasive protrusions reveals metastatic culprits. Current opinion in cell biology. 2012;24:662–669. doi: 10.1016/j.ceb.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nature reviews. Molecular cell biology. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoshino D, Branch KM, Weaver AM. Signaling inputs to invadopodia and podosomes. Journal of cell science. 2013;126:2979–2989. doi: 10.1242/jcs.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi H. Pathological roles of invadopodia in cancer invasion and metastasis. European journal of cell biology. 2012;91:902–907. doi: 10.1016/j.ejcb.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 7.McNiven MA. Breaking away: matrix remodeling from the leading edge. Trends in cell biology. 2013;23:16–21. doi: 10.1016/j.tcb.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwaya K, et al. Correlation between liver metastasis of the colocalization of actin-related protein 2 and 3 complex and WAVE2 in colorectal carcinoma. Cancer science. 2007;98:992–999. doi: 10.1111/j.1349-7006.2007.00488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwaya K, Norio K, Mukai K. Coexpression of Arp2 and WAVE2 predicts poor outcome in invasive breast carcinoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2007;20:339–343. doi: 10.1038/modpathol.3800741. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochimica et biophysica acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiological reviews. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 12.De Saedeleer CJ, et al. Glucose deprivation increases monocarboxylate transporter 1 (MCT1) expression and MCT1-dependent tumor cell migration. Oncogene. 2014;33:4060–4068. doi: 10.1038/onc.2013.454. [DOI] [PubMed] [Google Scholar]
- 13.Zhen C, et al. Gankyrin promotes breast cancer cell metastasis by regulating Rac1 activity. Oncogene. 2013;32:3452–3460. doi: 10.1038/onc.2012.356. [DOI] [PubMed] [Google Scholar]
- 14.Cho SY, Klemke RL. Purification of pseudopodia from polarized cells reveals redistribution and activation of Rac through assembly of a CAS/Crk scaffold. The Journal of cell biology. 2002;156:725–736. doi: 10.1083/jcb.200111032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemke RL, Jiang X, Choi S, Kelber JA. Proteomic and biochemical methods to study the cytoskeletome. Methods in molecular biology. 2013;1046:203–218. doi: 10.1007/978-1-62703-538-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibarrola N, Kalume DE, Gronborg M, Iwahori A, Pandey A. A proteomic approach for quantitation of phosphorylation using stable isotope labeling in cell culture. Analytical chemistry. 2003;75:6043–6049. doi: 10.1021/ac034931f. [DOI] [PubMed] [Google Scholar]
- 17.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & cellular proteomics: MCP. 2002;1:376–386. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Jia Z, et al. Tumor cell pseudopodial protrusions. Localized signaling domains coordinating cytoskeleton remodeling, cell adhesion, glycolysis, RNA translocation, and protein translation. The Journal of biological chemistry. 2005;280:30564–30573. doi: 10.1074/jbc.M501754200. [DOI] [PubMed] [Google Scholar]
- 19.Gene Ontology, C. The Gene Ontology project in 2008. Nucleic acids research. 2008;36:D440–444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nature reviews. Molecular cell biology. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 22.Hannigan GE, et al. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- 23.Dai DL, et al. Increased expression of integrin-linked kinase is correlated with melanoma progression and poor patient survival. Clinical cancer research: an official journal of the American Association for Cancer Research. 2003;9:4409–4414. [PubMed] [Google Scholar]
- 24.Marotta A, et al. Dysregulation of integrin-linked kinase (ILK) signaling in colonic polyposis. Oncogene. 2001;20:6250–6257. doi: 10.1038/sj.onc.1204791. [DOI] [PubMed] [Google Scholar]
- 25.Rajalingam K, et al. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nature cell biology. 2005;7:837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 26.Ikonen E, Fiedler K, Parton RG, Simons K. Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS letters. 1995;358:273–277. doi: 10.1016/0014-5793(94)01444-6. [DOI] [PubMed] [Google Scholar]
- 27.McClung JK, Jupe ER, Liu XT, Dell’Orco RT. Prohibitin: potential role in senescence, development, and tumor suppression. Experimental gerontology. 1995;30:99–124. doi: 10.1016/0531-5565(94)00069-7. [DOI] [PubMed] [Google Scholar]
- 28.Poitelon Y, et al. Spatial mapping of juxtacrine axo-glial interactions identifies novel molecules in peripheral myelination. Nature communications. 2015;6:8303. doi: 10.1038/ncomms9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebada SS, Lajkiewicz N, Porco JA, Jr, Li-Weber M, Proksch P. Chemistry and biology of rocaglamides (=flavaglines) and related derivatives from aglaia species (meliaceae) Progress in the chemistry of organic natural products. 2011;94:1–58. doi: 10.1007/978-3-7091-0748-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li-Weber M. Molecular mechanisms and anti-cancer aspects of the medicinal phytochemicals rocaglamides (=flavaglines) International journal of cancer. 2015;137:1791–1799. doi: 10.1002/ijc.29013. [DOI] [PubMed] [Google Scholar]
- 31.Polier G, et al. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chemistry & biology. 2012;19:1093–1104. doi: 10.1016/j.chembiol.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Han T, et al. How does cancer cell metabolism affect tumor migration and invasion? Cell adhesion & migration. 2013;7:395–403. doi: 10.4161/cam.26345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enns L, Ladiges W. Mitochondrial redox signaling and cancer invasiveness. Journal of bioenergetics and biomembranes. 2012;44:635–638. doi: 10.1007/s10863-012-9467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goetze K, Walenta S, Ksiazkiewicz M, Kunz-Schughart LA, Mueller-Klieser W. Lactate enhances motility of tumor cells and inhibits monocyte migration and cytokine release. International journal of oncology. 2011;39:453–463. doi: 10.3892/ijo.2011.1055. [DOI] [PubMed] [Google Scholar]
- 35.Gatenby RA, Gawlinski ET. The glycolytic phenotype in carcinogenesis and tumor invasion: insights through mathematical models. Cancer research. 2003;63:3847–3854. [PubMed] [Google Scholar]
- 36.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nature reviews. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 37.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. The Journal of general physiology. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desai SP, Bhatia SN, Toner M, Irimia D. Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophysical journal. 2013;104:2077–2088. doi: 10.1016/j.bpj.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Artal-Sanz M, Tavernarakis N. Prohibitin and mitochondrial biology. Trends in endocrinology and metabolism: TEM. 2009;20:394–401. doi: 10.1016/j.tem.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Theiss AL, et al. Prohibitin protects against oxidative stress in intestinal epithelial cells. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21:197–206. doi: 10.1096/fj.06-6801com. [DOI] [PubMed] [Google Scholar]
- 41.Lee SL, et al. Identification and characterization of a novel integrin-linked kinase inhibitor. Journal of medicinal chemistry. 2011;54:6364–6374. doi: 10.1021/jm2007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. The Journal of biological chemistry. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 43.Nelson EA, et al. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood. 2008;112:5095–5102. doi: 10.1182/blood-2007-12-129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 45.Billing AM, et al. Comprehensive transcriptomic and proteomic characterization of human mesenchymal stem cells reveals source specific cellular markers. Scientific reports. 2016;6:21507. doi: 10.1038/srep21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods in molecular biology. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vizcaino JA, et al. Update of the PRIDE database and its related tools. Nucleic acids research. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun, J. & Murdoch, D. J. A first course in statistical programming with R. Second edition. edn.
- 50.Klipper-Aurbach Y, et al. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Medical hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 51.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in medicine. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset Pseudopodia protein ID