Abstract
As a bacterial resistance strategy, serine β-lactamases have evolved from cell wall synthesizing enzymes known as penicillin-binding proteins (PBP), by not only covalently binding β-lactam antibiotics but, also acquiring mechanisms of deacylating these antibiotics. This critical deacylation step leads to release of hydrolyzed and inactivated β-lactams, thereby providing resistance for the bacteria against these antibiotics targeting the cell wall. To combat β-lactamase-mediated antibiotic resistance, numerous β-lactamase inhibitors were developed that utilize various strategies to inactivate the β-lactamase. Most of these compounds are “mechanism-based” inhibitors that in some manner mimic the β-lactam substrate, having a carbonyl moiety and a negatively charged carboxyl or sulfate group. These compounds form a covalent adduct with the catalytic serine via an initial acylation step. To increase the life-time of the inhibitory covalent adduct intermediates, a remarkable array of different strategies was employed to improve inhibition potency. Such approaches include post-acylation intra- and intermolecular chemical rearrangements as well as affecting the deacylation water. These approaches transform the inhibitor design process from a 3-dimensional problem (i.e., XYZ coordinates) to one with additional dimensions of complexity as the reaction coordinate and time spent at each chemical state need to be taken into consideration. This review highlights the mechanistic intricacies of the design efforts of the β-lactamase inhibitors which so far have resulted in the development of “two generations” and 5 clinically available inhibitors.
Keywords: beta-lactamase, structural biology, enzyme inhibitors, transition state, antibacterial agents
Resistance against β-lactam antibiotics is in large part mediated by β-lactamases. The expression of β-lactamases protects the intended targets of these antibiotics, the penicillin binding proteins (PBPs), transpeptidase and carboxypeptidase enzymes critical in the synthesis of peptidoglycan and the bacterial cell wall (Nikolaidis et al., 2014). β-lactamases provide this protection as they have evolved from PBPs to recognize β-lactams, yet have also acquired a deacylation machinery to inactivate/hydrolyze β-lactam antibiotics (Fisher and Mobashery, 2009). There are four classes of β-lactamases, A–D, with Classes A, C, and D being serine β-lactamases that have PBPs as a shared common ancestor; Class B is reserved for the structurally unrelated metallo β-lactamases (Bush, 2013).
The serine β-lactamases contain key motifs or features to recognize and facilitate the β-lactam for hydrolysis: (1) a polar pocket optimized to attract the carboxyl moieties of β-lactams; (2) an oxyanion hole to attract and stabilize the carbonyl oxygen of the β-lactam ring; (3) a catalytic serine hydroxyl moiety that attacks the carbonyl carbon atom which leads to breakage of the carbonyl carbon nitrogen bond in the β-lactam; (4) conserved residues involved in a deacylation step not present in PBPs (Class A β-lactamases utilize for example a deacylation water that is primed by E166/N170 in the omega loop, a structural motif not present in PBPs). These features are complemented by an intricate hydrogen bonding network involving conserved Lys and Ser/Tyr hydroxyl moieties, in addition to a likely substrate-assisted hydrogen donation step that aids in the catalytic mechanism of some of these enzymes (e.g., Class C β-lactamases; Bulychev et al., 1997; Patera et al., 2000). These above steps have been investigated for different β-lactamases (Strynadka et al., 1992; Bulychev et al., 1997; Chen et al., 2006; Docquier and Mangani, 2016) as well as analyzed with QM/MM calculations [(Meroueh et al., 2005; Li et al., 2011; Sgrignani et al., 2014, 2016; Tripathi and Nair, 2016; Lizana and Delgado, 2017)]. To combat the β-lactamase-mediated resistance against β-lactam antibiotics, many different β-lactamase inhibitors (BLIs) were developed often using novel strategies to overcome the deacylation machinery of β-lactamases (Papp-Wallace and Bonomo, 2016). This review summarizes the remarkable breadth of inhibitor development strategies often involving additional chemical bond rearrangements post-acylation. These chemical and mechanistic strategies might also be useful for targeting other enzymes. Overall, the successful efforts in this arena have led to five β-lactamase inhibitors being approved for clinical use and others that are still in preclinical development.
Clavulanic acid, sulbactam, and tazobactam; the “first generation”
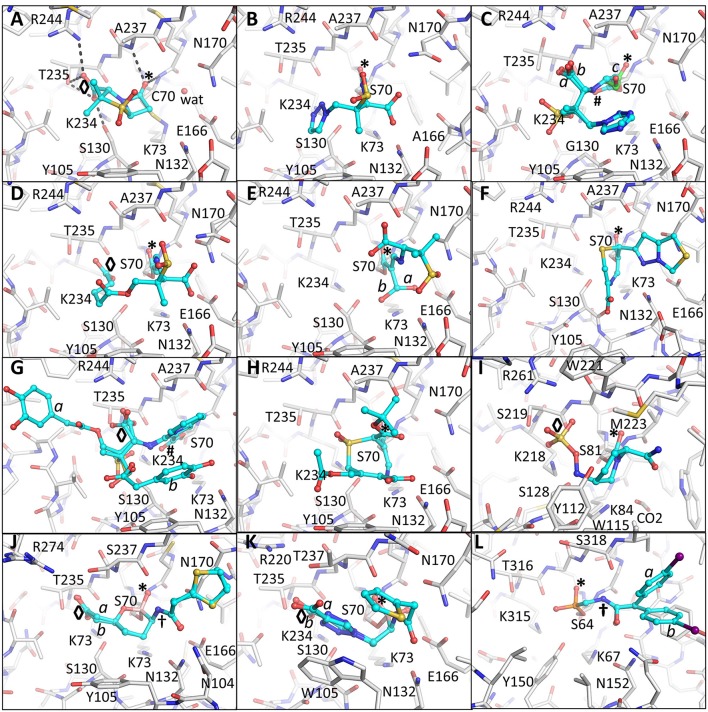
The first BLIs that were approved by the FDA were clavulanic acid, sulbactam, and tazobactam (Figure 1) (Page, 2000; Drawz and Bonomo, 2010). Each of these BLIs was paired with a β-lactam (amoxicillin/clavulanic acid, ticarcillin/clavulanic acid, ampicillin/sulbactam, cefoperazone/sulbactam, and piperacillin/tazobactam). These three inhibitors shared several features with β-lactamase substrates, such as penicillin, including a β-lactam ring fused to a 5-membered ring containing a carboxylate moiety. Sulbactam and tazobactam are penicillanic acid sulfones and differ in the C2 substituent which is a methyl group for sulbactam and a triazolyl containing moiety for tazobactam (Figure 1). In contrast, clavulanic acid is a clavam with sulfone replaced by an oxygen. The latter inhibitor differs from the other two inhibitors at the C2 position. The serine β-lactamases recognize the inhibitors by positioning the carboxyl moiety and carbonyl moieties in conserved regions in the active site as was observed in the pre-acylation Michaelis-Menten complex of sulbactam bound to the S70C mutant of SHV-1 β-lactamase (Figure 2A; Rodkey et al., 2012). The carboxyl moiety is in a pocket with hydrogen bond donors T235, S130, and within electrostatic interaction distances of R244 and K234 (Figure 2A). The carbonyl oxygen is positioned in the oxyanion hole formed by backbone nitrogens of residues 70 and 237 thereby priming the carbonyl carbon for nucleophilic attack by the hydroxyl moiety of the catalytic S70 as well as to stabilize the transition state. Finally, the hydrophobic part of the ring systems of sulbactam form hydrophobic interactions with the aromatic face of the side chain of Y105. Overall, this binding mode is similar to how these enzymes recognize β-lactam substrates (Beadle et al., 2002).
Figure 1.
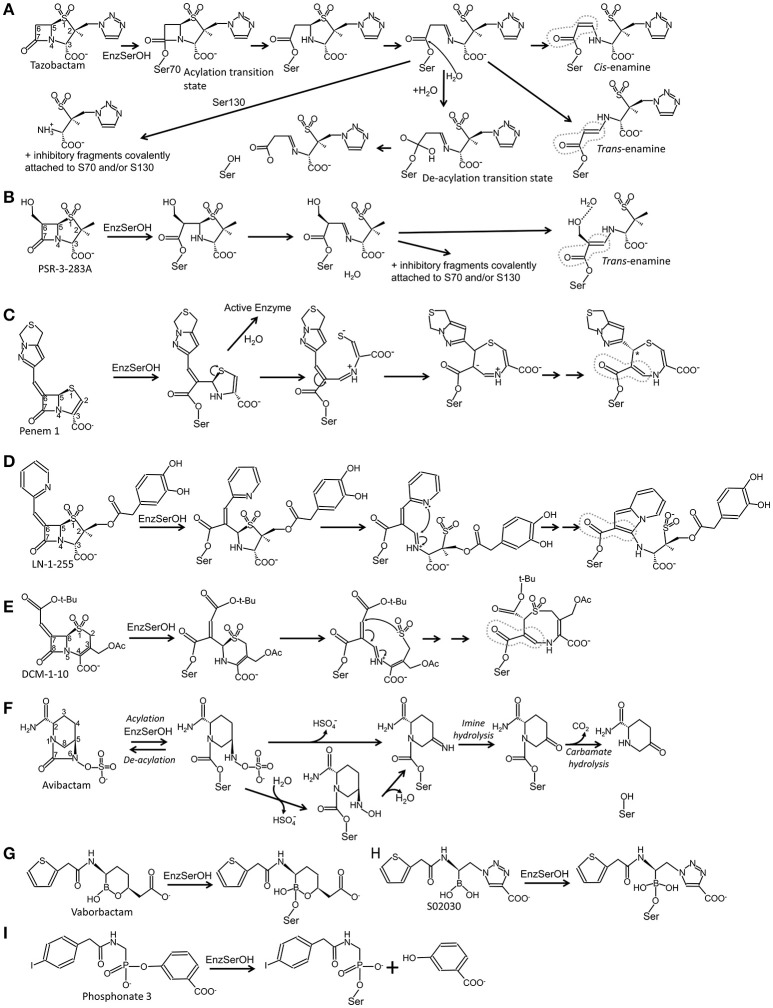
Schematic diagrams of different approaches of mechanism-based inhibition of serine β-lactamases. (A) Inhibition by tazobactam; (B) inhibition by PSR-3-283A; (C) inhibition by penem 1; (D) inhibition by LN-1-255; (E) inhibition by DCM-1-10; (F) inhibition by avibactam; (G) inhibition by vaborbactam; (H) inhibition by S02030; (I) inhibition by phosphonate 3. Instances where there is conjugation with the double bond of the carbonyl moiety are highlighted by a dashed gray line.
Figure 2.
Crystallographically determined binding modes of β-lactamase inhibitors. (A) Sulbactam bound in a pre-acylation/Michaelis-Menten binding mode in the S70C mutant of SHV-1 β-lactamase. The S70C mutations changes the reactivity of the catalytic S70 nucleophile; the C70 residue forms a covalent sulfonamide bond with the conserved K73 allowing capture of the pre-acylation complex. Hydrogen bonds between the carboxyl and carbonyl oxygens are depicted as dashed black lines. The occupied carboxyl pocket and oxyanion hole are labeled “◇” and “*”, respectively. These labels are used through subsequent panels of this figure where applicable. The deacylation water is shown as a solid red sphere labeled “wat;” (B) tazobactam, in the trans-enamine conformation, bound to the deacylation deficient E166A mutant of SHV-1; (C) tazobactam, in the cis-enamine conformation, bound to the inhibitor-resistant S130G mutant of SHV-1. Tazobactam adopts three conformations two of which are cis-enamine (0.33 occupancy with cyan carbon atoms each labeled “a” and “b”) and a fragmented species with green carbon atoms labeled “c” (also 0.33 occupancy). These labels for alternate conformations are used when needed in subsequent panels of this Figure. The cis-enamine and fragmented species have their carbonyl oxygens positioned outside (labeled “#”) and inside the oxyanion hole (labeled “*”), respectively; (D) SA2-13 complexed to SHV-1; (E) PSR-2-283A complexed to SHV-1. The hydroxymethyl moiety was observed to be in two conformations (labeled “a” and “b”). The major conformation hydrogen bonds with the deacylation water (not shown) and the second conformation does not. Residue S130 is also in two conformations; (F) penem 1 bound to SHV-1; (G) LN-1-255 complexed to SHV-1. Two conformations of the tail of LN-1-255 are observed (“a” and “b”); (H) DCM-1-10 bound to SHV-1; (I) Avibactam bound to Class D OXA-24 β-lactamase; (J) Vaborbactam complexed to CTX-M-15. Two conformations for vaborbactam were observed. The amide moiety of vaborbactam (labeled “†”) makes hydrogen bonds across the active site groove; (K) S02030 bound to KPC-2. Two conformations were observed for the carboxyl-triazole moiety (labeled “a” and “b”); (L) Phosphonate 3 complexed to P99 β-lactamase. The iodobenzene ring was present in two conformations. Like in vaborbactam, the amide moiety of phosphonate 3 makes hydrogen bonds across the active site.
All three “first generation” inhibitors are mechanism-based compounds and inhibit serine β-lactamases by limiting deacylation via additional post-acylation reaction pathways that can promote semi-stable intermediates including trans-enamine (Figure 2B) and cis-enamine (Figure 2C) inhibitory species (Figure 1A; Padayatti et al., 2004, 2005; Totir et al., 2006). The enamine species both yield a double bond in the vicinity of, and thereby a conjugated system with, the carbonyl bond (Figure 1A). This conjugation is thought to decrease the carbonyl carbon's susceptibility to nucleophilic attack by the deacylation water thus preventing deacylation.
A second inhibitory mechanism for these inhibitors likely entails fashioning an eventual irreversible inhibitory species that appears after several turn-over events; this inhibitory species is postulated to involve fragmentation of the inhibitor yielding covalent modifications on either S130 and/or the catalytic S70 residue (Figures 1A, 2C; Kuzin et al., 2001; Sun et al., 2004).
Defining the importance of a long-lived intermediate: SA2-13
Efforts to improve the longevity of the trans-enamine intermediate have yielded the inhibitor SA2-13 which increases the lifetime of this intermediate by 10-fold over tazobactam as observed in the measured kobs, react (Padayatti et al., 2006). This improvement was accomplished by changing the C2 substituent to a carboxyl linker such that the latter moiety occupies the carboxyl binding pocket thereby forming a U-shaped covalent adduct that stabilizes the trans-enamine intermediate (Padayatti et al., 2006; Sampson et al., 2011; Ke et al., 2012c; Rodkey et al., 2014; Figure 2D). SA2-13 thus positions its two carboxyl moieties in the carboxyl binding pocket during two separate steps in its reaction with the β-lactamase. The position of the original carboxyl moiety in the SA2-13 complex leads to a minor steric and electrostatic repulsion with residues in the omega-loop such that binding of SA2-13 to Extended-Spectrum-β-Lactamase (ESBL) mutants of SHV-1 causes complete disorder of the omega loop; this disorder further enhancing SA2-13's inhibitory efficacy toward ESBLs as this loop harbors residues needed for deacylation (Sampson et al., 2011).
Slowing the deacylation rate: 6β-hydroxymethyl containing inhibitors
The addition of a 6β-hydroxy-methyl moiety to a penicillanic acid sulfone improved certain inhibitory characteristics specifically slowing down the deacylation rate (Bitha et al., 1999a,b; Papp-Wallace et al., 2012; Che et al., 2015). The basis of this effect is attributed to the presence of the hydroxyl-methyl moiety interacting with the deacylation water when in the trans-enamine or imine intermediate state of the inhibitor (Figures 1B, 2E). This interaction can thereby either (1) sterically prevent the deacylation water from being able to nucleophilically attack the carbonyl carbon; and/or (2) negatively alter the nucleophilic properties of the deacylation water (Che et al., 2015). These inhibitors were also observed to undergo fragmentation yielding inhibitory adducts to Class A and C β-lactamases (Papp-Wallace et al., 2012). A 6-α-hydroxymethyl penicillanate variant yielded a similar inhibitory binding mode when bound to TEM-1 with the hydroxymethyl moiety also interacting with the deacylation water (Maveyraud et al., 1996).
Penem; a strategic design that enhances the acyl intermediate
To take advantage of and to increase the longevity of the semi-stable acyl-enzyme imine intermediate, reactive groups were added at the C6 position such as the alkylidene group in penems (Figure 1C; Nukaga et al., 2003; Venkatesan et al., 2004a,b, 2008; Mansour et al., 2007; Ke et al., 2012b). The design was to, when in the imine intermediate, allow the nucleophilic sulfur to react with the carbon in the alkylidene group via a 7-endo-trig rearrangement. This reaction would form a new 7-membered ring that via additional rearrangements can lead to an enamine species that conjugates with the carbonyl carbon bond (Figure 1C). This in turn limits deacylation by decreasing the susceptibility of the carbonyl carbon to nucleophilic attack of the deacylation water. This 7-membered ring enamine intermediate was crystallographically observed for penem 1 with the carbonyl oxygen situated in the oxyanion hole with deacylation likely being diminished due to this conjugation (Figures 1C, 2F; Ke et al., 2012b).
The alkylidene moiety at the C6 position can have different aromatic 1-, 2-, or 3-ring systems as substituents of which penem 1 contains a 2-ring heterocycle substitution (Bulychev et al., 1995; Ke et al., 2012b). When combined with piperacillin, penem 1 lowered MIC values from 64–2,048 to 4–8 μg/ml for Escherichia coli expressing SHV-1, SHV-2, and the inhibitor-resistant R244S variant (Ke et al., 2012b).
6-alkylidene-2′β-substituted penam sulfones: LN-1-255 and novel chemistry
Like the penems above, alkylidene group containing reactive groups were added at the C6 position of penam sulfones (Chen et al., 1987; Buynak et al., 1999; Phillips et al., 2005; Kalp et al., 2007; Che et al., 2012). In particular, the pyridylemethylidene moiety in LN-1-255 is potent since, when in the imine intermediate, the nitrogen of the pyridyl group reacts with the carbon atom of the imine bond to form a bicyclic ring (Figure 1D; Buynak et al., 1999; Pattanaik et al., 2009). The carbonyl carbon is now conjugated with the newly formed bicyclic ring; to maintain its conjugation and thus planarity with this bulky ring, the carbonyl oxygen “flips out” of the oxyanion hole (labeled “#” in Figure 2G). This oxygen movement and the resulting conjugation renders the carbonyl bond very resistant to deacylation making the inhibitor even more efficient with a lower turn-over number compared to tazobactam (Pattanaik et al., 2009).
Remarkably, LN-1-255 and other 6-alkylidene-2′β-substituted penam sulfones are also potent Class D β-lactamase inhibitors and have a similar mechanism of enzyme inhibition (Bou et al., 2010). An additional improvement for LN-1-255 included adding a dihydroxy-phenyl catechol moiety at the C2 position of the penam sulfone. This moiety is a siderophore and could allow improved uptake of LN-1-255 via bacterial iron-acquisition siderophore channels (Pattanaik et al., 2009). Presently, LN-1-255 is undergoing preclinical studies to establish it efficacy in treating infections.
7-alkylidenecephalosporin sulfones
Like in LN-1-255 and penem 1, the alkylidene moiety can also be incorporated on the equivalent position in cephalosporin sulfones, at the 7 position (Buynak et al., 2000). Such a 7-alkylidenecaphalosporin sulfone is DCM-1-10 (Figure 1E). DCM-1-10 undergoes a similar acyl-forming inhibitory mechanism, yet deviates from penem 1 in that it is the sulfone that reacts with the carbon of the alkylidene moiety thus forming an 8 atom cyclic intermediate (Figures 1E, 2H). The carbonyl oxygen remains in the oxyanion hole yet the intermediate is likely protected from deacylation by the stabilizing effect on the carbonyl bond by being conjugated with a neighboring double bond (Figure 1E; Rodkey et al., 2013). DCM-1-10 has only modest potency as its IC50 is 4- and 27-fold higher for clavulanic acid and tazobactam, respectively. Nevertheless, the turnover numbers for DCM-1-10 are similar to tazobactam and the kobs, react is significantly slower compared to tazobactam and clavulanic acid indicating that DCM-1-10 can form a relatively stable inhibitory complex (Rodkey et al., 2013).
Diazabicyclooctane inhibitors; the “second generation”
Avibactam (NXL104) is a diazabicyclooctane (DBO) (Coleman, 2011) and is the 4th β-lactamase inhibitor that was FDA approved as part of the formulation ceftazidime/avibactam (in 2015). Unlike the above described inhibitors, avibactam inhibition of serine β-lactamases is mostly reversible (Figure 1F; Ehmann et al., 2012, 2013). Avibactam is chemically distinct from the other inhibitors in that its rings are arranged differently with the strained 4-atom β-lactam ring being absent. Nevertheless, avibactam contains a carbonyl bond adjacent to a ring nitrogen. The carboxyl moiety, present in all previously discussed inhibitors, is replaced by a negatively charged sulfate moiety. For proper recognition in the active site, the same distance between the carbonyl oxygen and the negatively charged oxygens of the sulfate group is maintained relative to its equivalent atoms in the above β-lactam containing inhibitors: the negatively charged oxygens (in either the carboxyl or sulfate moiety) and the carbonyl oxygen are separated by 4 atoms in both classes of inhibitors (Figure 1).
Avibactam forms an acyl-enzyme complex with the serine β-lactamase upon breakage of the C-N bond and concomitant opening of the 5-membered ring (Figure 1F). Interestingly, avibactam can be removed from the enzyme via deacylation and ring closure resulting in an intact avibactam molecule being liberated. A number of crystal structures have been determined of avibactam complexes with representatives from all three serine β-lactamase classes (Xu et al., 2012; Lahiri et al., 2013, 2014, 2015; King et al., 2015; Krishnan et al., 2015; Calvopiña et al., 2017; Jin et al., 2017; Lohans et al., 2017).
Despite the mostly reversible mode of inhibition (a property not evident with the other BLIs listed above), some β-lactamases are capable of slowly desulfating avibactam once bound to the enzyme resulting in inactivation of avibactam upon carbamate hydrolysis (Ehmann et al., 2012, 2013; Figure 1F). Avibactam forms similar acyl-enzyme complexes in Class A, C, and D β-lactamases (Xu et al., 2012; Lahiri et al., 2013, 2015; King et al., 2015; Krishnan et al., 2015) in which the sulfate moiety is occupying the carboxyl binding pocket and the carbonyl oxygen is situated in the oxyanion hole (Figure 2I). One possible explanation that the acyl-enzyme is likely resistant to attack by the deacylation water could be due to having a nitrogen atom bonded to the carbonyl carbon atom (Figure 1F) thereby likely altering this bond as well as its local environment. The chirality of this tertiary amine, when bound to β-lactamase, can vary from S, R, or planar (Krishnan et al., 2015). Additional DBO β-lactamase inhibitors are currently being developed with improved efficacies with some having dual action potential by also inhibiting PBPs (Ambrose et al., 2017; Durand-Réville et al., 2017; Moya et al., 2017a,b; Shapiro et al., 2017; Zhanel et al., 2018). The DBOs in preclinical development are listed in Table 1; additional DBO analogs in earlier stages of development can be found here (King et al., 2016; Wang et al., 2016; Durand-Réville et al., 2017).
Table 1.
Promising DBO inhibitors in pre-clinical development or FDA approved.
| DBO name | Characteristics | References |
|---|---|---|
| Avibactam (NXL104) | Currently only FDA approved DBO β-lactamase inhibitor. Partnered with ceftazidime | Reviewed in Coleman, 2011 |
| WCK 4234 | Active against Pseudomonas and Acinetobacter Class A, C, and D β-lactamases. Partnered with imipenem or meropenem | Mushtaq et al., 2017 |
| WCK 5107 (Zidebactam) | Active against A. baumannii, Enterobacteriaceae, and Pseudomonas aeruginosa. Dual target inhibitor (P.a. PBP2). Partnered with cefepime or sulbactam | Livermore et al., 2017; Moya et al., 2017a,b; Sader et al., 2017a,b |
| WCK 5153 | Active against A. baumannii and P. aeruginosa. Dual target inhibitor (P.a. PBP2). Partnered with cefepime or sulbactam | Moya et al., 2017a,b |
| ETX2514 | Active against Gram-negative bacteria including A. baumannii and P. aeruginosa. Dual target inhibitor (A. b. PBP2). Partnered with sulbactam | Durand-Réville et al., 2017; McLeod et al., 2017; Shapiro et al., 2017 |
| Relebactam (MK-7655) | Active against Enterobacteriaceae, Klebsiella pneumoniae, and Pseudomonas. Partnered with imipenem | Livermore et al., 2013; Blizzard et al., 2014; Lapuebla et al., 2015; Haidar et al., 2017; Lob et al., 2017 |
| Nacubactam (OP0595) | Active against Enterobacteriaceae, P. aeruginosa, and K. pneumoniae. Dual target activity (inhibits PBP2) and has “enhancer”-activity. Partnered with cefepime, piperacillin, or meropenem | Livermore et al., 2015; Morinaka et al., 2015, 2017 |
Boronic acid and phosphonate transition state analogs
Elucidation of the reaction scheme of mechanism-based inhibition BLIs (Figure 1A) suggested that transitions states can be mimicked to obtain potent inhibitors. The reaction scheme indicates that a transition state exists for both the acylation and the deacylation component of the reaction; exploiting these transitions state for developing new BLIs will be discussed next.
The cyclic boronic acid inhibitor vaborbactam (RPX7009; Hecker et al., 2015; Lomovskaya et al., 2017) was recently FDA approved (meropenem/vaborbactam) and its complex with CTX-M-15 and P99 β-lactamases was crystallographically determined (Hecker et al., 2015; Figures 1G, 2J). Vaborbactam mimics β-lactamase inhibitors/substrates (Figure 2J) by having (1) a boron atom, like the carbonyl carbon, that can be the recipient of nucleophilic attack by the catalytic serine; (2) a carboxyl moiety that occupies the carboxyl binding pocket; (3) a hydroxyl moiety mimicking the carbonyl oxygen; (4) a ring system that makes hydrophobic interactions with the Y/W side chain often found in β-lactamase active sites; and (5) an amide moiety, found in the penicillin substrate, that can interact with the different atoms across the active site cleft (with a backbone oxygen of T237 on one side of the cleft and the amide moieties of both Asn132 and Asn102 on the other end). When bound to the active site, vaborbactam adopts an acylation transition state binding mode with its exocyclic boron oxygen in the oxyanion hole (Figure 2J). Like DBOs that can reversibly acylate and deacylated, vaborbactam can be a reversible β-lactamase inhibitor (Hecker et al., 2015; Lomovskaya et al., 2017). Cyclic boronate inhibitors can have broad spectrum efficacy as some are capable of inhibiting all 4 classes of β-lactamases including metallo β-lactamases (Cahill et al., 2017). Furthermore, these inhibitors have potential beyond inhibiting β-lactamases as a cyclic boronate inhibitor was shown to inhibit PBP5 (Brem et al., 2016).
A different boronic acid inhibitor is S02030 which when complexed to KPC-2 binds in a deacylation transition state mode (Figures 1H, 2K; Nguyen et al., 2016). This is in sharp contrast to vaborbactam. S02030 possesses two boron hydroxyl groups: one of them occupies the oxyanion hole whereas the second hydroxyl occupies the pocket normally harboring the deacylation water, but this water is now displaced (Figure 2K); these hydroxyl interactions were also observed in a KPC-2 complex with a small boronic acid fragment molecule (Ke et al., 2012a). S02030 is very potent at inhibiting β-lactamases observed in Klebsiella pneumoniae and E. coli species. Developing boronic acid β-lactamase inhibitors is a promising approach as has been shown for a number of different β-lactamases (Tondi et al., 2010, 2014; Powers et al., 2014; Bouza et al., 2017; Werner et al., 2017; Caselli et al., 2018).
In addition to boronic acid analogs, phosphonates also behave as transition state BLIs. Phosphonates are unique mechanism-based BLIs; the nucleophilic attack of the catalytic serine leads to bond breakage and release of part of the molecule adjacent to the phosphonate (Figure 1I). The structure of phosphonate 3 bound to P99 β-lactamase reveals the phosphorous atom covalently bonded to the catalytic serine (Figure 2L; Lobkovsky et al., 1994). Furthermore, one of the phosphonate oxygen atoms is in the oxyanion hole and the amide moiety makes hydrogen bond interactions across the active site cleft like vaborbactam (Figures 2J, 2L). Like with boronic acids, phosphonates have also been used to probe binding modes of transition states of β-lactams such as the phosphonate transition state analog of a cephalosporin bound to a Class C β-lactamase (Nukaga et al., 2004).
Additional inhibitor design approaches
In addition to these mechanism-based β-lactamase inhibitors, some groups have targeted developing non-covalent β-lactamase inhibitors (Eidam et al., 2012; Nichols et al., 2014). This approach is often initiated by starting from small fragments and exploiting hydrogen bond, electrostatic, and van der Waals interactions similar those observed in the mechanism-based inhibitor complexes. Despite not having a covalent bond with the catalytic serine, this approach can yield nano-molar affinity β-lactamase inhibitors (Eidam et al., 2012; Nichols et al., 2014). An important challenge here is the need to demonstrate “broad class” inhibition as was seen with DBOs. Alternatively, “narrow spectrum” inhibitors should not be discounted for therapeutic purposes as they will likely cause less damage to the patients beneficial microbiome (Boucher et al., 2017). Another approach is to utilize naturally observed protein inhibitors of β-lactamases from Streptomyces, termed β-lactamase inhibitor proteins (BLIPs), which can be altered to modulate β-lactamase specificity (Brown et al., 2013; Chow et al., 2016; Adamski and Palzkill, 2017a,b); peptides derived from BLIPs have been shown to have antimicrobial activity (Alaybeyoglu et al., 2015).
Conclusion
BLI development has made tremendous progress during the last decades and exploited numerous different chemical and/or mechanistic strategies. This includes unusual (post-acylation) reactions that can involve both intra- and/or inter-molecular rearrangements. Different areas of the reaction coordinate space have been exploited to arrive at novel and promising compounds. Despite all the progress resulting in now 5 inhibitors clinically available, resistance against these β-lactamase inhibitors has occurred including against avibactam (Wright et al., 2017); resistance against vaborbactam has not been observed yet, as the inhibitor has only recently been FDA approved (Zhanel et al., 2018). Therefore, continued efforts in this field are needed to develop BLIs with novel properties such as the dual action DBO inhibitors that are in preclinical development. Also, developing BLIs that target both metallo-β-lactamases (Class B) and serine based enzymes (Classes A, C, and D) remains a goal of the future.
Author contributions
FvdA has written the initial draft manuscript and prepared the figures. FvdA and RB have contributed to editing the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R01AI100560, R01AI063517, R21AI114508, and R01AI072219 to RB. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to RB from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 to RB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
References
- Adamski C. J., Palzkill T. (2017a). Systematic substitutions at BLIP position 50 result in changes in binding specificity for class A beta-lactamases. BMC Biochem. 18:2 10.1186/s12858-017-0077-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamski C. J., Palzkill T. (2017b). BLIP-II employs differential hotspot residues to bind structurally similar staphylococcus aureus PBP2a and class A beta-lactamases. Biochemistry 56, 1075–1084. 10.1021/acs.biochem.6b00978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaybeyoglu B., Akbulut B. S., Ozkirimli E. (2015). A novel chimeric peptide with antimicrobial activity. J. Pept. Sci. 21, 294–301. 10.1002/psc.2739 [DOI] [PubMed] [Google Scholar]
- Ambrose P. G., VanScoy B. D., Trang M., McCauley-Miller J., Conde H., Bhavnani S. M., et al. (2017). Pharmacokinetics-pharmacodynamics of CB-618 in combination with cefepime, ceftazidime, ceftolozane, or meropenem: the pharmacological basis for a stand-alone beta-lactamase inhibitor. Antimicrob. Agents Chemother. 61:e00630–17. 10.1128/AAC.00630-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle B. M., Trehan I., Focia P. J., Shoichet B. K. (2002). Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC β-lactamase. Structure 10, 413–424. 10.1016/S0969-2126(02)00725-6 [DOI] [PubMed] [Google Scholar]
- Bitha P., Li Z., Francisco G. D., Rasmussen B. A., Lin Y. I. (1999a). 6-(1-Hydroxyalkyl)penam sulfone derivatives as inhibitors of class A and class C beta-lactamases I. Bioorg. Med. Chem. Lett. 9, 991–996. [DOI] [PubMed] [Google Scholar]
- Bitha P., Li Z., Francisco G. D., Yang Y., Petersen P. J., Lenoy E., et al. (1999b). 6-(1-Hydroxyalkyl))penam sulfone derivatives as inhibitors of class A and class C beta-lactamases II. Bioorg. Med. Chem. Lett. 9, 997–1002. [DOI] [PubMed] [Google Scholar]
- Blizzard T. A., Chen H., Kim S., Wu J., Bodner R., Gude C., et al. (2014). Discovery of MK-7655, a beta-lactamase inhibitor for combination with Primaxin(R). Bioorg. Med. Chem. Lett. 24, 780–785. 10.1016/j.bmcl.2013.12.101 [DOI] [PubMed] [Google Scholar]
- Bou G., Santillana E., Sheri A., Beceiro A., Sampson J. M., Kalp M., et al. (2010). Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase. J. Am. Chem. Soc. 132, 13320–13331. 10.1021/ja104092z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Ambrose P. G., Chambers H. F., Ebright R. H., Jezek A., Murray B. E., et al. (2017). White paper: developing antimicrobial drugs for resistant pathogens, narrow-spectrum indications, and unmet needs. J. Infect. Dis. 216, 228–236. 10.1093/infdis/jix211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouza A. A., Swanson H. C., Smolen K. A., VanDine A. L., Taracila M. A., Romagnoli C., et al. (2017). Structure-based analysis of boronic acids as inhibitors of acinetobacter-derived cephalosporinase-7, a unique class c beta-lactamase. ACS Infect. Dis. 4, 325–336. 10.1021/acsinfecdis.7b00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem J., Cain R., Cahill S., McDonough M. A., Clifton I. J., Jimenez-Castellanos J. C., et al. (2016). Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates. Nat. Commun. 7:12406. 10.1038/ncomms12406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. G., Chow D. C., Palzkill T. (2013). BLIP-II is a highly potent inhibitor of Klebsiella pneumoniae carbapenemase (KPC-2). Antimicrob. Agents Chemother. 57, 3398–3401. 10.1128/AAC.00215-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulychev A., Massova I., Lerner S. A., Mobashery S. (1995). Penem BRL 42715: an effective inactivator of beta-lactamases. J. Am. Chem. Soc. 117 4797–4800. 10.1021/ja00122a009 [DOI] [Google Scholar]
- Bulychev A., Massova I., Miyashita K., Mobashery S. (1997). Nuances of mechanisms and their implications for evolution of the versatile beta-lactamase activity: from biosynthetic enzymes to drug resistance factors. J. Am. Chem. Soc. 119, 7619–7625. [Google Scholar]
- Bush K. (2013). The ABCD's of beta-lactamase nomenclature. J. Infect. Chemother. 19, 549–559. 10.1007/s10156-013-0640-7 [DOI] [PubMed] [Google Scholar]
- Buynak J. D., Doppalapudi V. R., Adam G. (2000). The synthesis and evaluation of 3-substituted-7-(alkylidene)cephalosporin sulfones as beta-lactamase inhibitors. Bioorg. Med. Chem. Lett. 10, 853–857. 10.1016/S0960-894X(00)00098-6 [DOI] [PubMed] [Google Scholar]
- Buynak J. D., Rao A. S., Doppalapudi V. R., Adam G., Petersen P. J., Nidamarthy S. D. (1999). The synthesis and evaluation of 6-alkylidene-2'beta-substituted penam sulfones as beta-lactamase inhibitors. Bioorg. Med. Chem. Lett. 9, 1997–2002. 10.1016/S0960-894X(99)00325-X [DOI] [PubMed] [Google Scholar]
- Cahill S. T., Cain R., Wang D. Y., Lohans C. T., Wareham D. W., Oswin H. P., et al. (2017). Cyclic boronates inhibit all classes of beta-lactamases. Antimicrob. Agents Chemother. 61:e02260–16. 10.1128/AAC.02260-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvopiña K., Hinchliffe P., Brem J., Heesom K. J., Johnson S., Cain R., et al. (2017). Structural/mechanistic insights into the efficacy of nonclassical beta-lactamase inhibitors against extensively drug resistant Stenotrophomonas maltophilia clinical isolates. Mol. Microbiol. 106, 492–504. 10.1111/mmi.13831 [DOI] [PubMed] [Google Scholar]
- Caselli E., Romagnoli C., Powers R. A., Taracila M. A., Bouza A. A., Swanson H. C. et al. (2018). Inhibition of acinetobacter-derived cephalosporinase: exploring the carboxylate recognition site using novel beta-lactamase inhibitors. ACS Infect. Dis. 4, 337–348. 10.1021/acsinfecdis.7b00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che T., Bonomo R. A., Shanmugam S., Bethel C. R., Pusztai-Carey M., Buynak J. D., et al. (2012). Carboxylation and decarboxylation of active site lys 84 controls the activity of OXA-24 beta-lactamase of Acinetobacter baumannii: raman crystallographic and solution evidence. J. Am. Chem. Soc. 134, 11206–11215. 10.1021/ja303168n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che T., Rodkey E. A., Bethel C. R., Shanmugam S., Ding Z., Pusztai-Carey M., et al. (2015). Detecting a quasi-stable imine species on the reaction pathway of SHV-1 beta-lactamase and 6beta-(Hydroxymethyl)penicillanic acid sulfone. Biochemistry 54, 734–743. 10.1021/bi501197t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. L., Chang C. W., Hedberg K., Guarino K., Welch W. M., Kiessling L., et al. (1987). Structure-activity relationships of 6-(heterocyclyl)-methylene penam sulfones; a new class of beta-lactamase inhibitors. J. Antibiot. 40, 803–822. [DOI] [PubMed] [Google Scholar]
- Chen Y., Minasov G., Roth T. A., Prati F., Shoichet B. K. (2006). The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution. J. Am. Chem. Soc. 128, 2970–2976. 10.1021/ja056806m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow D. C., Rice K., Huang W., Atmar R. L., Palzkill T. (2016). Engineering specificity from broad to narrow: design of a beta-lactamase inhibitory protein (BLIP) variant that exclusively binds and detects KPC beta-lactamase. ACS Infect. Dis. 2, 969–979. 10.1021/acsinfecdis.6b00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman K. (2011). Diazabicyclooctanes (DBOs): a potent new class of non-beta-lactam beta-lactamase inhibitors. Curr. Opin. Microbiol. 14, 550–555. 10.1016/j.mib.2011.07.026 [DOI] [PubMed] [Google Scholar]
- Docquier J. D., Mangani S. (2016). Structure-function relationships of class D carbapenemases. Curr. Drug Targets 17, 1061–1071. 10.2174/1389450116666150825115824 [DOI] [PubMed] [Google Scholar]
- Drawz S. M., Bonomo R. A. (2010). Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201. 10.1128/CMR.00037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Réville T. F., Guler S., Comita-Prevoir J., Chen B., Bifulco N., Huynh H., et al. (2017). ETX2514 is a broad-spectrum beta-lactamase inhibitor for the treatment of drug-resistant Gram-negative bacteria including Acinetobacter baumannii. Nat. Microbiol. 2:17104. 10.1038/nmicrobiol.2017.104 [DOI] [PubMed] [Google Scholar]
- Ehmann D. E., Jahic H., Ross P. L., Gu R. F., Hu J., Durand-Reville T. F., et al. (2013). Kinetics of avibactam inhibition against Class, A., C, and D beta-lactamases. J. Biol. Chem. 288, 27960–27971. 10.1074/jbc.M113.485979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann D. E., Jahic H., Ross P. L., Gu R. F., Hu J., Kern G., et al. (2012). Avibactam is a covalent, reversible, non-beta-lactam beta-lactamase inhibitor. Proc. Natl. Acad. Sci. U.S.A. 109, 11663–11668. 10.1073/pnas.1205073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam O., Romagnoli C., Dalmasso G., Barelier S., Caselli E., Bonnet R., et al. (2012). Fragment-guided design of subnanomolar beta-lactamase inhibitors active in vivo. Proc. Natl. Acad. Sci. U.S.A. 109, 17448–17453. 10.1073/pnas.1208337109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J. F., Mobashery S. (2009). Three decades of the class A beta-lactamase acyl-enzyme. Curr. Protein Pept. Sci. 10, 401–407. 10.2174/138920309789351967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar G., Clancy C. J., Chen L., Samanta P., Shields R. K., Kreiswirth B. N., et al. (2017). Identifying spectra of activity and therapeutic niches for ceftazidime-avibactam and imipenem-relebactam against carbapenem-resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 61, e00642–e00617. 10.1128/AAC.00642-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker S. J., Reddy K. R., Totrov M., Hirst G. C., Lomovskaya O., Griffith D. C., et al. (2015). Discovery of a cyclic boronic acid beta-lactamase inhibitor (RPX7009) with utility vs class A serine carbapenemases. J. Med. Chem. 58, 3682–3692. 10.1021/acs.jmedchem.5b00127 [DOI] [PubMed] [Google Scholar]
- Jin W., Wachino J. I., Yamaguchi Y., Kimura K., Kumar A., Yamada M., et al. (2017). Structural insights into the TLA-3 extended-spectrum beta-lactamase and its inhibition by avibactam and OP0595. Antimicrob. Agents Chemother. 61:e00501–17. 10.1128/AAC.00501-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalp M., Sheri A., Buynak J. D., Bethel C. R., Bonomo R. A., Carey P. R. (2007). Efficient inhibition of class A and class D β-lactamases by Michaelis complexes. J. Biol. Chem. 282, 21588–21591. 10.1074/jbc.C700080200 [DOI] [PubMed] [Google Scholar]
- Ke W., Bethel C. R., Papp-Wallace K. M., Pagadala S. R., Nottingham M., Fernandez D., et al. (2012a). Crystal structures of KPC-2 β-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226. Antimicrob.Agents Chemother. 56, 2713–2718. 10.1128/AAC.06099-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke W., Pattanaik P., Bethel C. R., Sheri A., Buynak J. D., Bonomo R. A., et al. (2012b). Structures of SHV-1 β-lactamase with penem and penam sulfone inhibitors that form cyclic intermediates stabilized by carbonyl conjugation. PLoS ONE 7:e49035. 10.1371/journal.pone.0049035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke W., Rodkey E. A., Sampson J. M., Skalweit M. J., Sheri A., Pagadala S. R., et al. (2012c). The importance of the trans-enamine intermediate as a β-lactamase inhibition strategy probed in inhibitor-resistant SHV β-lactamase variants. Chem. Med. Chem. 7, 1002–1008. 10.1002/cmdc.201200006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M., King D. T., French S., Brouillette E., Asli A., Alexander J. A., et al. (2016). Structural and kinetic characterization of diazabicyclooctanes as dual inhibitors of both serine-beta-lactamases and penicillin-binding proteins. ACS Chem. Biol. 11, 864–868. 10.1021/acschembio.5b00944 [DOI] [PubMed] [Google Scholar]
- King D. T., King A. M., Lal S. M., Wright G. D., Strynadka N. C. (2015). Molecular mechanism of avibactam mediated beta-lactamase inhibition. ACS Infect. Dis. 1, 175–184. 10.1021/acsinfecdis.5b00007 [DOI] [PubMed] [Google Scholar]
- Krishnan N. P., Nguyen N. Q., Papp-Wallace K. M., Bonomo R. A., van den Akker F. (2015). Inhibition of klebsiella beta-lactamases (SHV-1 and KPC-2) by avibactam: a structural study. PLoS ONE 10:e0136813. 10.1371/journal.pone.0136813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin A. P., Nukaga M., Nukaga Y., Hujer A., Bonomo R. A., Knox J. R. (2001). Inhibition of the SHV-1 β-lactamase by sulfones: crystallographic observation of two reaction intermediates with tazobactam. Biochemistry 40, 1861–1866. 10.1021/bi0022745 [DOI] [PubMed] [Google Scholar]
- Lahiri S. D., Johnstone M. R., Ross P. L., McLaughlin R. E., Olivier N. B., Alm R. A. (2014). Avibactam and class C beta-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob. Agents Chemother. 58, 5704–5713. 10.1128/AAC.03057-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S. D., Mangani S., Durand-Reville T., Benvenuti M., De Luca F., Sanyal G., et al. (2013). Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC beta-lactamases. Antimicrob. Agents Chemother. 57, 2496–2505. 10.1128/AAC.02247-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S. D., Mangani S., Jahic H., Benvenuti M., Durand-Reville T. F., De Luca F., et al. (2015). Molecular basis of selective inhibition and slow reversibility of avibactam against class D carbapenemases: a structure-guided study of OXA-24 and OXA-48. ACS Chem. Biol. 10, 591–600. 10.1021/cb500703p [DOI] [PubMed] [Google Scholar]
- Lapuebla A., Abdallah M., Olafisoye O., Cortes C., Urban C., Landman D., et al. (2015). Activity of imipenem with relebactam against gram-negative pathogens from New York city. Antimicrob. Agents Chemother. 59, 5029–5031. 10.1128/AAC.00830-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liao J. M., Gu C. R., Wang Y. T., Chen C. L. (2011). Theoretical investigation on reaction of sulbactam with wild-type SHV-1 beta-lactamase: acylation, tautomerization, and deacylation. J. Phys. Chem. B 115, 10298–10310. 10.1021/jp111572v [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Mushtaq S., Warner M., Vickers A., Woodford N. (2017). In vitro activity of cefepime/zidebactam (WCK 5222) against Gram-negative bacteria. J. Antimicrob. Chemother. 72, 1373–1385. 10.1093/jac/dkw593 [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Mushtaq S., Warner M., Woodford N. (2015). Activity of OP0595/beta-lactam combinations against Gram-negative bacteria with extended-spectrum, AmpC and carbapenem-hydrolysing beta-lactamases. J. Antimicrob. Chemother. 70, 3032–3041. 10.1093/jac/dkv239 [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Warner M., Mushtaq S. (2013). Activity of MK-7655 combined with imipenem against Enterobacteriaceae and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 68, 2286–2290. 10.1093/jac/dkt178 [DOI] [PubMed] [Google Scholar]
- Lizana I., Delgado E. J. (2017). A QM/MM study on the enzymatic inactivation of cefotaxime. J. Mol. Model. 23:209 10.1007/s00894-017-3379-8 [DOI] [PubMed] [Google Scholar]
- Lob S. H., Hackel M. A., Kazmierczak K. M., Young K., Motyl M. R., Karlowsky J. A., et al. (2017). In vitro activity of imipenem-relebactam against gram-negative ESKAPE pathogens isolated by clinical laboratories in the United States in 2015 (Results from the SMART Global Surveillance Program). Antimicrob. Agents Chemother. 61:e02209-16 10.1128/AAC.02209-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobkovsky E., Billings E. M., Moews P. C., Rahil J., Pratt R. F., Knox J. R. (1994). Crystallographic structure of a phosphonate derivative of the Enterobacter cloacae P99 cephalosporinase: mechanistic interpretation of a beta-lactamase transition-state analog. Biochemistry 33, 6762–6772. 10.1021/bi00188a004 [DOI] [PubMed] [Google Scholar]
- Lohans C. T., Wang D. Y., Jorgensen C., Cahill S. T., Clifton I. J., McDonough M. A., et al. (2017). (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions. Organ. Biomol. Chem. 15 6024–6032. 10.1039/C7OB01514C [DOI] [PubMed] [Google Scholar]
- Lomovskaya O., Sun D., Rubio-Aparicio D., Nelson K., Tsivkovski R., Griffith D. C., et al. (2017). Vaborbactam: spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob. Agents Chemother. 61, e01443–e01417. 10.1128/AAC.01443-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour T. S., Agarwal A., Venkatesan A., Abe T., Mihira A., Takasaki T., et al. (2007). On the absolute configuration in 1,4-dihydrothiazepine covalent complexes derived from inhibition of class A and C beta-lactamases with 6-methylidene penems. Chem. Med. Chem. 2, 1713–1716. 10.1002/cmdc.200700144 [DOI] [PubMed] [Google Scholar]
- Maveyraud L., Massova I., Birck I., Miyashita K., Samama J. P., Mobashery S. (1996). Crystal structure of 6alpha-(hydroxymethyl)penicillanate complexed to the TEM-1 beta-lactamase from Escherichia coli: evidence on the mechanism of action of a novel inhibitor designed by a computer-aided process. J. Am. Chem. Soc. 118, 7435–7440. [Google Scholar]
- McLeod S. M., Shapiro A. B., Moussa S. H., Johnstone M., McLaughlin R. E., de Jonge B. L. M., et al. (2017). Frequency and mechanism of spontaneous resistance to sulbactam combined with the novel beta-lactamase inhibitor ETX2514 in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 62, e01576–e01517. 10.1128/AAC.01576-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroueh S. O., Fisher J. F., Schlegel H. B., Mobashery S. (2005). Ab initio QM/MM study of class A beta-lactamase acylation: dual participation of Glu166 and Lys73 in a concerted base promotion of Ser70. J. Am. Chem. Soc. 127, 15397–15407. 10.1021/ja051592u [DOI] [PubMed] [Google Scholar]
- Morinaka A., Tsutsumi Y., Yamada K., Takayama Y., Sakakibara S., Takata T., et al. (2017). In vitro and in vivo activities of the diazabicyclooctane OP0595 against AmpC-derepressed Pseudomonas aeruginosa. J. Antibiot. 70, 246–250. 10.1038/ja.2016.150 [DOI] [PubMed] [Google Scholar]
- Morinaka A., Tsutsumi Y., Yamada M., Suzuki K., Watanabe T., Abe T., et al. (2015). OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam ‘enhancer’. J. Antimicrob. Chemother. 70, 2779–2786. 10.1093/jac/dkv166 [DOI] [PubMed] [Google Scholar]
- Moya B., Barcelo I. M., Bhagwat S., Patel M., Bou G., Papp-Wallace K. M., et al. (2017a). Potent beta-lactam enhancer activity of zidebactam and WCK 5153 against Acinetobacter baumannii, including carbapenemase-producing clinical isolates. Antimicrob. Agents Chemother. 61:01238–17. 10.1128/AAC.01238-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya B., Barcelo I. M., Bhagwat S., Patel M., Bou G., Papp-Wallace K. M., et al. (2017b). WCK 5107 (Zidebactam) and WCK 5153 are novel inhibitors of PBP2 showing potent “beta-lactam enhancer” activity against pseudomonas aeruginosa, including multidrug-resistant metallo-beta-lactamase-producing high-risk clones. Antimicrob. Agents Chemother. 61:e02529–6. 10.1128/AAC.02529-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushtaq S., Vickers A., Woodford N., Livermore D. M. (2017). WCK 4234, a novel diazabicyclooctane potentiating carbapenems against Enterobacteriaceae, Pseudomonas and Acinetobacter with class A, C and D beta-lactamases. J. Antimicrob. Chemother. 72, 1688–1695. 10.1093/jac/dkx035 [DOI] [PubMed] [Google Scholar]
- Nguyen N. Q., Krishnan N. P., Rojas L. J., Prati F., Caselli E., Romagnoli C., et al. (2016). Crystal structures of KPC-2 and SHV-1 beta-lactamases in complex with the boronic acid transition state analog S02030. Antimicrob. Agents Chemother. 60, 1760–1766. 10.1128/AAC.02643-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols D. A., Renslo A. R., Chen Y. (2014). Fragment-based inhibitor discovery against beta-lactamase. Future Med. Chem. 6, 413–427. 10.4155/fmc.14.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis I., Favini-Stabile S., Dessen A. (2014). Resistance to antibiotics targeted to the bacterial cell wall. Protein Sci. 23, 243–259. 10.1002/pro.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukaga M., Abe T., Venkatesan A. M., Mansour T. S., Bonomo R. A., Knox J. R. (2003). Inhibition of class A and class C β-lactamases by penems: crystallographic structures of a novel 1,4-thiazepine intermediate. Biochemistry 42, 13152–13159. 10.1021/bi034986b [DOI] [PubMed] [Google Scholar]
- Nukaga M., Kumar S., Nukaga K., Pratt R. F., Knox J. R. (2004). Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes. J. Biol. Chem. 279, 9344–9352. 10.1074/jbc.M312356200 [DOI] [PubMed] [Google Scholar]
- Padayatti P. S., Helfand M. S., Totir M. A., Carey M. P., Carey P. R., Bonomo R. A., et al. (2005). High resolution crystal structures of the trans-enamine intermediates formed by sulbactam and clavulanic acid and E166A SHV-1 β-lactamase. J. Biol. Chem. 280, 34900–34907. 10.1074/jbc.M505333200 [DOI] [PubMed] [Google Scholar]
- Padayatti P. S., Helfand M. S., Totir M. A., Carey M. P., Hujer H. M., Carey P. R., et al. (2004). Tazobactam forms a stoichiometric trans-enamine intermediate in the E166A variant of SHV-1 β-lactamase: 1.63 Å crystal structure. Biochemistry 43, 843–848. 10.1021/bi035985m [DOI] [PubMed] [Google Scholar]
- Padayatti P. S., Sheri A., Totir M. A., Helfand M. S., Carey M. P., Anderson V. E., et al. (2006). Rational design of a β-lactamase inhibitor achieved via stabilization of the trans-enamine intermediate: 1.28Å crystal structure of wt SHV-1 complex with a penam sulfone. J. Am. Chem. Soc. 128, 13235–13242. 10.1021/ja063715w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M. G. P. (2000). β-Lactamase inhibitors. Drug Resist. Updat. 3, 109–125. 10.1054/drup.2000.0137 [DOI] [PubMed] [Google Scholar]
- Papp-Wallace K. M., Bethel C. R., Gootz T. D., Shang W., Stroh J., Lau W., et al. (2012). Inactivation of a class A and a class C β-lactamase by 6β-(hydroxymethyl)penicillanic acid sulfone. Biochem. Pharmacol. 83, 462–471. 10.1016/j.bcp.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace K. M., Bonomo R. A. (2016). New beta-lactamase inhibitors in the clinic. Infect. Dis. Clin. North Am. 30, 441–464. 10.1016/j.idc.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patera A., Blasczcak L. C., Shoichet B. (2000). Crystal structures of substrate and inhibitor complexes with AmpC beta-lactamase: possible implications for substrate-assisted catalysis. J. Am. Chem. Soc. 122, 10504–10512. 10.1021/ja001676x [DOI] [Google Scholar]
- Pattanaik P., Bethel C. R., Hujer A. M., Hujer K. M., Distler A. M., Taracila M., et al. (2009). Strategic design of an effective β-lactamase inhibitor: LN-1-255, a 6-alkylidene-2'-substituted penicillin sulfone. J. Biol.Chem. 284, 945–953. 10.1074/jbc.M806833200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips O. A., Reddy A. V., Setti E. L., Spevak P., Czajkowski D. P., Atwal H., et al. (2005). Synthesis and biological evaluation of penam sulfones as inhibitors of beta-lactamases. Bioorg. Med. Chem. 13, 2847–2858. 10.1016/j.bmc.2005.02.020 [DOI] [PubMed] [Google Scholar]
- Powers R. A., Swanson H. C., Taracila M. A., Florek N. W., Romagnoli C., Caselli E., et al. (2014). Biochemical and structural analysis of inhibitors targeting the ADC-7 cephalosporinase of Acinetobacter baumannii. Biochemistry 53, 7670–7679. 10.1021/bi500887n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodkey E. A., Drawz S. M., Sampson J. M., Bethel C. R., Bonomo R. A. van den Akker, F. (2012). Crystal structure of a pre-acylation complex of the b-lactamase inhibitor sulbactam bound to a sulfenamide bond-containing thiol-beta-lactamase. J. Am. Chem. Soc. 134, 16798–16804. 10.1021/ja3073676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodkey E. A., McLeod D. C., Bethel C. R., Smith K. M., Xu Y., Chai W., et al. (2013). beta-Lactamase inhibition by 7-alkylidenecephalosporin sulfones: allylic transposition and formation of an unprecedented stabilized acyl-enzyme. J. Am. Chem. Soc. 135, 18358–18369. 10.1021/ja403598g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodkey E. A., Winkler M. L., Bethel C. R., Pagadala S. R., Buynak J. D., Bonomo R. A., et al. (2014). Penam sulfones and beta-lactamase inhibition: SA2-13 and the importance of the C2 side chain length and composition. PLoS ONE 9:e85892. 10.1371/journal.pone.0085892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Huband M., Jones R. N., Flamm R. K. (2017a). WCK 5222 (Cefepime-Zidebactam) antimicrobial activity against clinical isolates of gram-negative bacteria collected worldwide in 2015. Antimicrob. Agents Chemother. 61:e00072–17. 10.1128/AAC.00072-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Rhomberg P. R., Flamm R. K., Jones R. N., Castanheira M. (2017b). WCK 5222 (cefepime/zidebactam) antimicrobial activity tested against Gram-negative organisms producing clinically relevant beta-lactamases. J. Antimicrob. Chemother. 72, 1696–1703. 10.1093/jac/dkx050 [DOI] [PubMed] [Google Scholar]
- Sampson J. M., Ke W., Bethel C. R., Pagadala S. R., Nottingham M. D., Bonomo R. A., et al. (2011). Ligand-dependent disorder of the Ω-loop observed in extended-spectrum SHV-type β-lactamase. Antimicrob. Agents Chemother. 55, 2303–2309. 10.1128/AAC.01360-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgrignani J., Grazioso G., De Amici M. (2016). Insight into the mechanism of hydrolysis of meropenem by OXA-23 serine-beta-lactamase gained by quantum mechanics/molecular mechanics calculations. Biochemistry 55, 5191–5200. 10.1021/acs.biochem.6b00461 [DOI] [PubMed] [Google Scholar]
- Sgrignani J., Grazioso G., De Amici M., Colombo G. (2014). Inactivation of TEM-1 by avibactam (NXL-104): insights from quantum mechanics/molecular mechanics metadynamics simulations. Biochemistry 53, 5174–5185. 10.1021/bi500589x [DOI] [PubMed] [Google Scholar]
- Shapiro A. B., Gao N., Jahic H., Carter N. M., Chen A., Miller A. A. (2017). Reversibility of covalent, broad-spectrum serine beta-lactamase inhibition by the diazabicyclooctenone ETX2514. ACS Infect. Dis. 3, 833–844. 10.1021/acsinfecdis.7b00113 [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., Adachi H., Jensen S. E., Johns K., Sielecki A., Betzel C., et al. (1992). Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 A resolution. Nature 359, 700–705. [DOI] [PubMed] [Google Scholar]
- Sun T., Bethel C. R., Bonomo R. A., Knox J. R. (2004). Inhibitor-resistant class A β-lactamases: consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant. Biochemistry 43, 14111–14117. 10.1021/bi0487903 [DOI] [PubMed] [Google Scholar]
- Tondi D., Calo S., Shoichet B. K., Costi M. P. (2010). Structural study of phenyl boronic acid derivatives as AmpC beta-lactamase inhibitors. Bioorg. Med. Chem. Lett. 20, 3416–3419. 10.1016/j.bmcl.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondi D., Venturelli A., Bonnet R., Pozzi C., Shoichet B. K., Costi M. P. (2014). Targeting class A and C serine beta-lactamases with a broad-spectrum boronic acid derivative. J. Med. Chem. 57, 5449–5458. 10.1021/jm5006572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totir M. A., Padayatti P. S., Helfand M. S., Carey M. P., Bonomo R. A., Carey P. R., et al. (2006). Effect of the inhibitor-resistant M69V substitution on the structures and populations of trans-enamine beta-lactamase intermediates. Biochemistry 45, 11895–11904. 10.1021/bi060990m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi R., Nair N. N. (2016). Deacylation mechanism and kinetics of acyl-enzyme complex of class C beta-lactamase and cephalothin. J. Phys. Chem. B 120, 2681–2690. 10.1021/acs.jpcb.5b11623 [DOI] [PubMed] [Google Scholar]
- Venkatesan A. M., Agarwal A., Abe T., Ushirogochi H., Ado M., Tsuyoshi T., et al. (2008). 5,5,6-Fused tricycles bearing imidazole and pyrazole 6-methylidene penems as broad-spectrum inhibitors of beta-lactamases. Bioorg. Med. Chem. 16, 1890–1902. 10.1016/j.bmc.2007.11.006 [DOI] [PubMed] [Google Scholar]
- Venkatesan A. M., Agarwal A., Abe T., Ushirogochi H., Yamamura I., Kumagai T., et al. (2004a). Novel imidazole substituted 6-methylidene-penems as broad-spectrum beta-lactamase inhibitors. Bioorg. Med. Chem. 12, 5807–5817. 10.1016/j.bmc.2004.08.039 [DOI] [PubMed] [Google Scholar]
- Venkatesan A. M., Gu Y., Dos S. O., Abe T., Agarwal A., Yang Y., et al. (2004b). Structure-activity relationship of 6-methylidene penems bearing tricyclic heterocycles as broad-spectrum beta-lactamase inhibitors: crystallographic structures show unexpected binding of 1,4-thiazepine intermediates. J. Med. Chem. 47, 6556–6568. 10.1021/jm049680x [DOI] [PubMed] [Google Scholar]
- Wang D. Y., Abboud M. I., Markoulides M. S., Brem J., Schofield C. J. (2016). The road to avibactam: the first clinically useful non-beta-lactam working somewhat like a beta-lactam. Future Med. Chem. 8, 1063–1084. 10.4155/fmc-2016-0078 [DOI] [PubMed] [Google Scholar]
- Werner J. P., Mitchell J. M., Taracila M. A., Bonomo R. A., Powers R. A. (2017). Exploring the potential of boronic acids as inhibitors of OXA-24/40 beta-lactamase. Protein Sci. 26, 515–526. 10.1002/pro.3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright H., Bonomo R. A., Paterson D. L. (2017). New agents for the treatment of infections with Gram-negative bacteria: restoring the miracle or false dawn? Clin. Microbiol. Infect. 23, 704–712. 10.1016/j.cmi.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Xu H., Hazra S., Blanchard J. S. (2012). NXL104 irreversibly inhibits the beta-lactamase from Mycobacterium tuberculosis. Biochemistry 51, 4551–4557. 10.1021/bi300508r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel G. G., Lawrence C. K., Adam H., Schweizer F., Zelenitsky S., Zhanel M., et al. (2018). Imipenem-relebactam and meropenem-vaborbactam: two novel carbapenem-beta-lactamase inhibitor combinations. Drugs 78, 65–98. 10.1007/s40265-017-0851-9 [DOI] [PubMed] [Google Scholar]