Abstract
Psychotic disorders including schizophrenia are commonly accompanied by cognitive deficits. Recent studies have reported negative genetic correlations between schizophrenia and indicators of cognitive ability such as general intelligence and processing speed. Here we compare the effect of polygenetic risk for schizophrenia (PRSSCZ) on measures that differ in their relationships with psychosis onset: a measure of current cognitive abilities (the Brief Assessment of Cognition in Schizophrenia, BACS) that is greatly reduced in psychotic disorder patients, a measure of premorbid intelligence that is minimally affected by psychosis onset (the Wide-Range Achievement Test, WRAT); and educational attainment (EY), which covaries with both BACS and WRAT. Using genome-wide single nucleotide polymorphism (SNP) data from 314 psychotic and 423 healthy research participants in the Bipolar-Schizophrenia Network for Intermediate Phenotypes (B-SNIP) Consortium, we investigated the association of PRSSCZ with BACS, WRAT, and EY. Among apparently healthy individuals, greater genetic risk for schizophrenia (PRSSCZ) was significantly associated with lower BACS scores (r = −0.17, p = 6.6 × 10−4 at PT = 1 × 10−4), but not with WRAT or EY. Among individuals with psychosis, PRSSCZ did not associate with variations in any of these three phenotypes. We further investigated the association between PRSSCZ and WRAT in more than 4500 healthy subjects from the Philadelphia Neurodevelopmental Cohort. The association was again null (p > 0.3, N = 4511), suggesting that different cognitive phenotypes vary in their etiologic relationship with schizophrenia.
Introduction
Schizophrenia is a debilitating psychiatric disorder that commonly involves severe cognitive deficits that compromise functional ability1,2. Underperformance in general intelligence tasks as well as tasks designed to be specific to cognitive domains such as memory, executive function, and motor function have been noted in psychosis patients3.
Many schizophrenia-associated cognitive deficits are present many years prior to the onset of the illness4,5. A meta-analysis of 4396 schizophrenia cases and 745,000 controls showed that every point decrease in premorbid IQ associated with a 3.7% increase in schizophrenia risk6. In a nationwide cohort of over 900,000 Swedish individuals, children with the lowest grades showed a 4-fold increased risk of developing schizophrenia and schizoaffective disorder and a 3-fold increased risk of developing other psychotic illnesses7. Additionally, studies of clinically high-risk (CHR) groups have shown that people with attenuated psychotic symptoms were cognitively impaired compared to healthy controls (HC) and that, within the CHR group, those that converted to a chronic psychotic disorder within one or 2 years of ascertainment displayed lower cognitive performance compared to those that did not convert8–11. Together these results indicate that cognitive deficits are significantly associated with risk of developing a psychotic illness.
Both cognitive performance and psychotic disorders such as schizophrenia are heritable12–19, and significant genetic overlap has been consistently reported between schizophrenia and some indicators of cognitive ability, such as general intelligence or processing speed20–26. However, it is still unclear how the genetic differences associated with schizophrenia influence cognitive function, and which domains of cognitive function are most associated with schizophrenia risk.
Motivated by these earlier findings, we investigated the relationship between polygenic risk for schizophrenia—as defined by large constellations of common variants that associate with schizophrenia risk (PRSSCZ)—and three cognitive phenotypes in the Bipolar-Schizophrenia Network for Intermediate Phenotypes27,28 (B-SNIP) cohort: (1) the Brief Assessment of Cognition in Schizophrenia (BACS)29, which provides a composite score of current general cognitive function; (2) the Wide-Range Achievement Test (WRAT)30–32 reading score, a measure of premorbid intellectual potential; and (3) educational attainment (as measured by years of education, EY). These phenotypes are correlated but differentially associated with psychosis-spectrum case status. Compared to BACS or general cognition, WRAT scores are minimally affected by psychosis onset8, and are commonly used as a measure for premorbid intelligence in people with psychotic disorders30–32; educational attainment is phenotypically associated with WRAT and BACS and also strongly genetically overlaps with cognition33,34. A companion analysis was conducted in the large Philadelphia Neurodevelopmental Cohort (PNC, N = 4511)35-37 investigating the relationship between WRAT and PRSSCZ since WRAT measures were also available in the PNC.
As an additional validation analysis we investigated the relationship between the polygenic score of educational attainment (PRSEDUC) and these three cognitive phenotypes because of the significant genetic overlap between educational attainment and cognition25,34.
Methods
Study design and participants
Demographic information about the B-SNIP and the PNC cohorts can be found in Table 1. The B-SNIP analysis included 737 Caucasians from the Bipolar-Schizophrenia Network for Intermediate Phenotypes (B-SNIP)27,28, which is a five-site consortium (Maryland Psychiatric Research Center, University of Chicago/University of Illinois at Chicago, University of Texas-Southwestern, Wayne State University/Harvard University, and the Institute of Living/Yale University) organized to address questions about diagnostic boundaries and familiality of intermediate phenotypes. Previous work using this cohort reported BACS performance to be consistent with a dimensional model of psychosis27,38; Hill et al. (2013) showed that cognitive performance declined progressively as affective symptoms became less prominent and psychotic features became more pronounced and pervasive. Due to these findings, we combined all psychotic probands to form the PSYCH group (N = 314) consisting of schizophrenia (N = 100), psychotic bipolar disorder (N = 143), and schizoaffective disorder patients (N = 71). The NPSYCH group consisted of unrelated nonpsychotic individuals combining samples collected as controls (HC, N = 180) and first-degree relatives of probands with no history of psychosis (NPFAM, N = 243) and without elevated axis II traits27 (cluster A or cluster B). While the NPFAM members of the NPSYCH group were related to probands in the PSYCH group, none of the analyses included related individuals (e.g., group differences were calculated between HC and PSYCH or between NPFAM and HC; correlation analyses with PRSSCZ, PRSEDUC or between the three cognitive phenotypes were conducted within the PSYCH and the NPSYCH groups separately). All participants provided written informed consent. Institutional review boards at each site approved the study and all sites used identical diagnostic, clinical, and recruitment techniques28.
Table 1.
Demographic information for the B-SNIP and PNC cohorts
| B-SNIP | PNC | |||
|---|---|---|---|---|
| NPSYCH | PSYCH | Controls | ||
| HC | NPFAM | |||
| N | 180 | 243 | 314 | 4511 |
| Age (years) | 38.7 (12.8) | 46.5 (14.5) | 34.9 (1.3) | 13.8 (3.7) |
| Sex (%F) | 51.6 | 73.7 | 45.9 | 50.0 |
| Years of education | 15.2 (2.5) | 14.9 (2.5) | 13.9 (2.3) | N/A |
B-SNIP Bipolar-Schizophrenia Network for Intermediate Phenotypes, PNC Philadelphia Neurodevelopmental Cohort, NPSYCH B-SNIP nonpsychotic group consisting of healthy controls (HC) and nonpsychotic relatives (NPFAM), PSYCH B-SNIP psychotic proband group consisting of schizophrenia (N = 100), psychotic bipolar (N = 143), and schizoaffective disorder (N = 71) patients. Mean values are shown with standard deviations in parentheses. Years of Education was not an applicable measure for the young PNC cohort. Only samples with European ancestry were used in this study.
The Philadelphia Neurodevelopmental Cohort (PNC) is a sample from the greater Philadelphia area, including over 9000 individuals aged 8–21 years who received medical care at the Children’s Hospital at Philadelphia network35–37. The overall inclusion criteria for the cohort included: (1) Ability to provide signed informed consent (parental consent was required for participants under age 18), (2) English language proficiency, and (3) Physical and cognitive ability to participate in computerized cognitive testing. Only unrelated participants (pi-hat <0.2) of European ancestry were used in this work. Individuals with significant medical conditions that can impact brain function, as well as those with either an invalid or incomplete neurocognitive battery were excluded. After genetic quality control (described below and in Supplementary Material) the final sample for this study consisted of 4511 unrelated individuals (mean age 13.76 years, S.D. 3.66 years). All analyses in the PNC cohort were done in this entire sample.
Cognitive measures
Three cognitive measures were available in the B-SNIP cohort: BACS, WRAT, and educational attainment. General cognitive function in the B-SNIP was measured by the BACS, which is a 30 min test of global neuropsychological function29. Premorbid intellectual potential was measured using the reading score of the Wide-Range Achievement Test (WRAT IV), which has a phenotypic correlation of ~0.4 with full-scale intelligent quotient30,39. Self-reported years of education completed at the time of recruitment was used as a measure of EY. WRAT was similarly assessed in the PNC sample. A BACS equivalent was not available in the PNC and due to the young age of the subjects (8–21 years) EY would be largely redundant to age itself.
Genetic analyses
Genetic data for the B-SNIP project were collected for 2053 subjects (multi-ethnic sample) using the Illumina Infinium PsychArray BeadChip™ platform. Genotypes underwent quality control using PLINK 1.940,41 based on a standardized protocol42 (Supplementary Material). After initial quality control, and removal of individuals with missing cognitive phenotypes, 1528 samples remained of whom 927 were self-reported Caucasians (SRC). To avoid population stratification, only SRC samples were used in all analyses. The ancestries of these SRC samples were verified by principal component analysis combining the B-SNIP genotype data with the 1000 Genomes phase 1 data43. Samples that were more than four standard deviations away from the SRC group mean along the first ten principal components were excluded resulting in a final sample size of 737 (Figure S1). Imputation of the B-SNIP genetic data was performed using HAPI-UR for pre-phasing44 and IMPUTE2 for imputation45,46 using a multi-ethnic (the 1000 Genomes phase 1 reference panel43) reference panel47. Poorly imputed single nucleotide polymorphisms (SNPs) were filtered post-imputation (SNPs with information score <0.548 were removed) resulting in 22.5 million imputed SNPs.
Genotype data for 8211 multi-ethnic PNC samples were downloaded from dbGAP. These data were distributed across five different Illumina genotyping chips (as described in the Supplementary Material). Quality control was performed with the programs PLINK41 and GCTA49. After principal component analysis of the PNC data combined with the HapMap reference panel50, only samples with European ancestry were retained by visual inspection (overlapping with CEU and TSI, Figure S2). Following these steps 4733 samples and 204,597 markers were retained for imputation. The Michigan Imputation Server51 was used for genetic imputation of the PNC data (Minimac351 for imputation and HAPI-UR44 for phasing) with the 1000 genome phase 3 data52 as reference panel resulting in a total of 18 million imputed markers. The imputed variants were filtered for info score ≥0.6 (7.9 million markers) for polygenic score calculation with PLINK. Filtering samples for medical criteria and missing cognitive phenotypes (see Study Design and Participants) resulted in a final PNC sample of 4511 unrelated healthy individuals.
Schizophrenia polygenic profile scores (PRSSCZ) and educational attainment polygenic scores (PRSEDUC) were calculated using the schizophrenia GWAS summary statistics of the Psychiatric Genome Consortium (PGC)19 (https://www.med.unc.edu/pgc/results-and-downloads) and the summary statistics from Okbay et al.34, respectively. Score calculation was done using custom scripts in the B-SNIP and using PLINK in the PNC. Of the 120,636 PGC schizophrenia polygenic score training SNPs, 101,927 overlapped with the imputed B-SNIP data and 85,598 overlapped with the imputed PNC data. Of the 626,000 educational attainment GWAS markers (clumped using the 1000 Genome43 European Ancestry group; r2 < 0.1 within a 500 kb window of a more significantly associated SNP), 530,894 and 210,501 SNPs were in common with the imputed data in B-SNIP and the PNC, respectively. Polygenic scores were calculated for seven p-value thresholds of significance of association: P ≤ 10−4, 0.001, 0.01, 0.05, 0.1, 0.5, and 1.0. The first 10 principal components from ancestry analyses of B-SNIP and PNC were used as covariates for correlation analyses in the respective cohorts.
Statistical analyses
All statistical analyses in B-SNIP were performed using Matlab (version 2012b). Correlations between BACS, WRAT, and EY and the polygenic scores were calculated within the PSYCH group and the NPSYCH group (HC + NPFAM) separately using the Spearman Rank method, which is a nonparametric measure of correlation (deviation from normal distribution was noted in WRAT, EY, and PRSSCZ in specific groups). Age, sex, data collection site, the first 10 principal components from the genetic ancestry analysis, and DSM diagnosis (schizophrenia/bipolar disorder/schizoaffective disorder status for members of the PSYCH group and respective relative’s diagnosis for member’s of the NPFAM group) were regressed out for correlation analyses within each group. As an additional precaution, the samples’ HC/NPFAM status was used as a covariate for all analyses within the NPSYCH group. Differences in BACS, WRAT, and EY (Figure S3) between the HC, PSYCH, and NPFAM groups were calculated using the Kruskal–Wallis test (a nonparametric method for testing whether samples originate from the same distribution, which was used due to unequal variances in BACS between groups) after regressing out the effects of age, sex, data collection site, and the first ten principal components from the genetic ancestry analysis. These group differences were calculated between HC/PSYCH and HC/NPFAM instead of NPSYCH/PSYCH so that only unrelated individuals were compared. This was not a concern for correlation analyses within the NPSYCH group since the HC and the NPFAM subgroups were unrelated. Group differences in PRSSCZ and PRSEDUC were calculated between the HC and PSYCH groups (Fig. 1, Table S1, Kruskal–Wallis test was used due to unequal variance between groups for PRSEDUC) after regressing out the effects of data collection site and the first ten ancestry principal components. To correct for multiple hypotheses testing in analyses of the B-SNIP cohort a false discovery rate (FDR) approach53 was used following the example of recent studies that used polygenic risk scores25,54. For analyses with polygenic scores in B-SNIP the combined PFDR-PRS was 0.0064 at α = 0.05. Analysis specific FDR p-values are reported with each result.
Fig. 1. Mean Polygenic scores of schizophrenia (PRSSCZ) in the psychotic (PSYCH, N = 314) and the healthy controls (HC, N = 180) in B-SNIP.
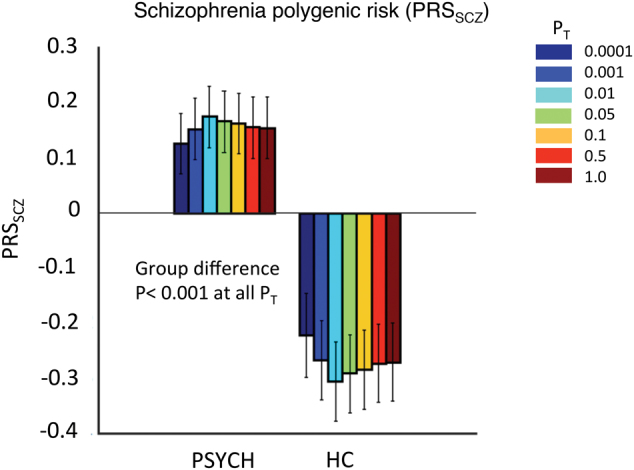
The vertical black lines show the standard errors of the mean (SEM). Scores were calculated at seven p-value thresholds (PT): 0.0001, 0.001, 0.01, 0.05, 0.1, 0.5, and 1.0 (shown in different colors). All scores were z-transformed before mean and SEM calculation. PRSSCZ was significantly higher (p ≤ PFDR = 2.6 × 10−4, Kruskal–Wallis test) in the PSYCH group compared to the HC group at all PT. Table S1 shows the p-values for this analysis. NPFAM (nonpsychotic family members of PSYCH group probands) were excluded from this case-control analysis so that only unrelated individuals were compared
All analyses in the PNC were done using RStudio55 (Version 1.0.44). Since individuals in the PNC sample were controls and unrelated, correlations between polygenic scores and WRAT were calculated within the entire sample controlling for effects of age, sex, and the first 10 ancestry principal components using the Spearman Rank method. The FDR-corrected53 p-value threshold for PNC was PFDR-PNC = 3.1 × 10−12 at α = 0.05 for all analyses using PRSSCZ and PRSEDUC.
Results
Genetic risk for schizophrenia was higher among individuals with psychosis in the mixed diagnostic group in B-SNIP
An individual’s polygenic risk of schizophrenia, PRSSCZ, estimates genome-wide common genetic influences on the risk of developing schizophrenia. Compared to the HC (Fig. 1), individuals with psychosis from 3 diagnosis groups in the B-SNIP sample (schizophrenia, psychotic bipolar, schizoaffective disorder) showed significantly higher PRSSCZ (p ≤ PFDR = 2.6 × 10−4, Table S1) at all PT. Among the psychosis probands schizophrenia patients had highest PRSSCZ (Figure S4). In our sample PRSEDUC did not differ significantly between the PSYCH and the HC groups (Table S1). Figure S4 shows the distributions of PRSSCZ and PRSEDUC for the different DSM diagnosis groups.
Psychosis did not alter the correlations between EY, BACS, and WRAT in B-SNIP
An individual’s educational attainment, cognitive functioning and intellectual potential are interdependent traits56. We examined these relationships within the PSYCH and the NPSYCH groups separately in the B-SNIP sample and found that the presence of psychosis did not alter the extent to which the phenotypes are independent (Fig. 2). Although BACS, WRAT, and EY were significantly lower in the PSYCH group compared to the HC group (Figure S3), the effect size of deficit in BACS (Cohen’s d = 1.24, p = 8.1 × 10−32) was more than three times greater than that of EY or WRAT. Additionally, partial correlation analyses between pairs of these three phenotypes controlling for the third phenotype revealed that, (1) EY and WRAT shared a positive correlation that could not be accounted for by BACS; (2) WRAT and BACS shared a positive correlation that could not be accounted for by EY; and (3) although EY and BACS were weakly positively correlated, this correlation was mediated via factors that could be captured by WRAT (Table S2).
Fig. 2. Relationship between the Brief Assessment of Cognition in Schizophrenia score (BACS), educational attainment (EY) and premorbid intellectual potential (WRAT) in B-SNIP.
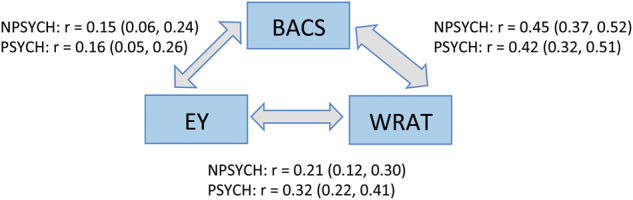
Correlation coefficients (Spearman’s Rank method) and 95% confidence intervals are shown. The three phenotypes were positively correlated in both the PSYCH (N = 314) and the NPSYCH (N = 423) groups and the magnitudes of the correlations were not significantly different between the groups. More details can be found in Table S2
Higher polygenetic risk for schizophrenia was significantly associated with lower BACS scores, but not WRAT or EY in nonpsychotic individuals in B-SNIP
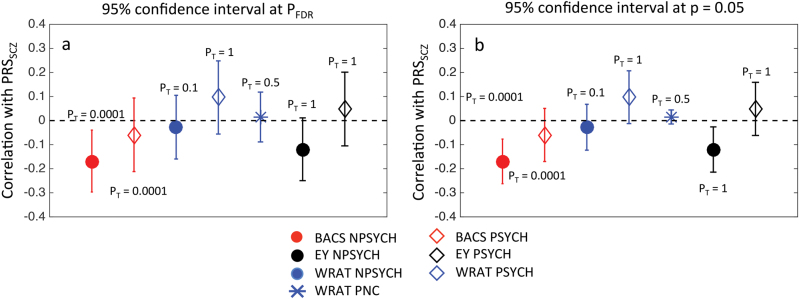
To evaluate whether the genetic risk for schizophrenia associates with variations in BACS, EY, and WRAT, correlations of PRSSCZ with these measures were calculated within the PSYCH and the NPSYCH groups in the B-SNIP sample separately. Figure 3 shows the strongest correlations for each phenotype. The numerical values for the correlation coefficients and the p-values for both groups at all PT can be found in Table S3. BACS showed significant negative association with PRSSCZ in the NPSYCH group (Fig. 3a, r = −0.17 and p = 6.6 × 10−4 at PT = 1 × 10−4), but not in the PSYCH group. This association remained significant when variability due to EY and WRAT were accounted for by additionally controlling for those two phenotypes (Table S3). Nominally significant (p < 0.05) negative association was seen between PRSSCZ and EY in the NPSYCH group, but not in the PSYCH group (Fig. 3b). WRAT was not significantly or nominally associated with PRSSCZ in either group.
Fig. 3. Correlations of the polygenic risk of schizophrenia (PRSSCZ) with the Brief Assessment of Cognition in Schizophrenia score (BACS), premorbid intellectual potential (WRAT) and educational attainment (EY).
For B-SNIP: N = 314 (PSYCH), N = 423 (NPSYCH) and for the PNC: N = 4511. All markers other than the blue star, which represents PNC, show results for B-SNIP. Correlation coefficients are shown with 95% confidence intervals for FDR-corrected p-value threshold (PFDR-B-SNIP = 0.0064, PFDR-PNC = 3.1 × 10−12) (a) and p = 0.05 (b). Only the strongest correlation (Spearman’s Rank method) for each phenotype is shown with the corresponding PT labeled. Correlation coefficients and corresponding p-values for all PT can be found in Tables S3 and S6
The correlation between BACS and PRSSCZ within the schizophrenia proband group only (SZP, N = 100) was also not significant, similar to the results of the entire PSYCH group. Adding illness duration, number of hospitalization, chlorpromazine dose equivalent, number of psychotropic drugs, and social-functional scale score as covariates in the correlation analysis between BACS and PRSSCZ did not alter the lack of significant results in the PSYCH group.
Since the NPSYCH group consisted of nonpsychotic individuals recruited as HC as well as the nonpsychotic family members of the psychosis probands (NPFAM, all subjects within this group were unrelated), the significance of the association of PRSSCZ with BACS was additionally investigated within the HC and the NPFAM groups individually (Supplementary Material). At the subgroup level, statistically significant correlation (Table S4) between BACS and PRSSCZ was seen at PT = 10−4 in the HC group (r = −0.25, p = 1.9 × 10−3), which remained significant when EY and WRAT were regressed out. In the NPFAM subgroup, significant negative correlation was detected at PT = 0.01 (r = −0.19, p = 6.4 × 10−3) when EY and WRAT were regressed out (Table S4).
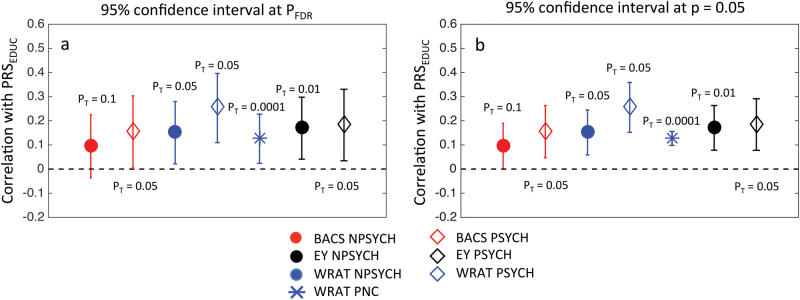
The polygenic score of educational attainment, PRSEDUC, showed significant positive correlations with EY in both the PSYCH group (Fig. 4, strongest correlation of r = 0.19, p = 0.0016 at PT = 0.05) and the NPSYCH group (Fig. 4, strongest correlation of r = 0.17, p = 7 × 10−4 at PT = 0.01). Significant positive correlations were observed between PRSEDUC and WRAT also in both the PSYCH group and the NPSYCH group (Fig. 4) at several PT (strongest correlation of r = 0.26, p = 1.1 × 10−5 at PT = 0.05 in PSYCH and strongest correlation of r = 0.15, p = 2.4 × 10−3 at PT = 0.05 in NPSYCH). No significant correlation was found between PRSEDUC and BACS in either group. The numerical values for all the correlation coefficients and p-values can be found in Table S5.
Fig. 4. Correlations of the polygenic score of educational attainment (PRSEDUC) with the Brief Assessment of Cognition in Schizophrenia score (BACS), premorbid intellectual potential (WRAT), and educational attainment (EY).
For B-SNIP: N = 314 (PSYCH), N = 423 (NPSYCH) and for PNC: N = 4511. All markers other than the blue star, which represents PNC, show results for B-SNIP. Correlation coefficients are shown with 95% confidence intervals for FDR-corrected threshold (PFDR-B-SNIP = 0.0064, PFDR-PNC = 3.1 × 10−12) (a) and p = 0.05 (b). Only the strongest correlation (Spearman’s Rank method) for each phenotype is shown with the corresponding PT labeled. Correlation coefficients and corresponding p-values for all PT can be found in Tables S5 and S6
Polygenic risk for schizophrenia and WRAT were unrelated in the PNC
We investigated the relationship between PRSSCZ, PRSEDUC, and WRAT in unrelated healthy individuals from the Philadelphia Neurocognitive Cohort (N = 4511). Our analyses showed lack of significant association between PRSSCZ and WRAT at all PT (Table S6, Fig. 3, p > 0.3 at all PT) and significant positive association between PRSEDUC and WRAT at all PT (Fig. 4, Table S6, p ≤ PFDR-PNC = 3.1 × 10−12 at all PT; maximum correlation of r = 0.13 and p < 2.2 × 10−16 at PT = 1 × 10−4). These results were consistent with the findings in the B-SNIP cohort.
Discussion
Cognitive deficits are widespread in psychotic disorder patients, especially in schizophrenia. Recent molecular genetics studies have shown that schizophrenia is genetically negatively correlated with multiple measures of cognition25,57. Our work focused on the relationship between the common polygenic risk of schizophrenia and three cognitive measures that are phenotypically correlated (Fig. 2), but differentially associated (Fig S3) with psychosis-spectrum case status. Our main findings were: (1) BACS, a measure of general cognitive function (most strongly affected in the patient group) was negatively associated with the polygenic risk of schizophrenia in apparently healthy individuals, (2) WRAT, often used as a measure of premorbid intelligence in psychosis-spectrum patients, was not associated with the common genetic risk of schizophrenia in healthy or psychotic individuals, and (3) the negative association between BACS and the polygenic risk of schizophrenia did not appear to hold in the psychotic patient group.
The first finding is consistent with other recent reports of genetic overlap between general cognitive function and schizophrenia24,25,57. For example, Trampush et al.57, reported a genetic correlation of −0.17 between schizophrenia and general intelligence. Our B-SNIP sample was not large enough for applying the recently developed methods of LD Score Regression58 or GCTA49 to calculate genetic correlation between traits. However, PRS correlations to phenotype are mathematically translatable to genetic correlations between two traits59. In other words, a positive PRS association would translate to a positive genetic correlation of estimable magnitude. Hence, the negative association between PRSSCZ and BACS is consistent with the above-mentioned negative genetic correlations between cognitive measures and schizophrenia25,37. Such negative associations have been shown in young cohorts also—Riglin et al.60 recently showed that lower performance intelligence quotient is associated with higher common genetic risk of schizophrenia in 14,701 samples of the ALSPAC cohort (age 7–9 years). Also, within the PNC cohort, it has been shown previously that the common genetic risk of schizophrenia negatively influences speed of verbal reasoning and emotion identification39.
The second and third findings were intriguing and warrant further investigation. In spite of sharing a phenotypic correlation of 0.4 with BACS, WRAT did not associate even nominally with the genetic risk of schizophrenia in either of our cohorts, including the large PNC. Although both BACS and WRAT measure cognitive function, BACS measures an individual’s ability to use cognitive resources to solve problems and WRAT is more of a measure of crystallized verbal knowledge. These results indicate the possibility that cognitive domains measured by BACS—rather than other brain phenotypes that shape premorbid intelligence—may be more direct targets of the genetic risk factors of schizophrenia.
Though we observed a significant negative correlation between PRSSCZ and BACS at multiple PT in the nonpsychotic group, we did not observe such a correlation among psychosis patients. Cognitive deficits in the patient group may thus reflect morbid factors such as, disease progression, protective effects of supportive care, and the effects of medications, medical, and psychiatric comorbidity and substance use, that are not predicted by PRSSCZ. However, recently, a similar result was reported in a study of Autism Spectrum Disorder (ASD) in which the polygenic risk of ASD did not predict IQ in the ASD probands (despite a strong positive correlation in the general population61), although the polygenic scores of educational attainment and schizophrenia did62. This too could be due to factors other than the genetic risk of developing the disease playing a significant role in determining the pathologic trajectory of cognitive function in the ASD patients.
Due to the relatively small sample size of the B-SNIP cohort, one might be concerned about statistical power of these analyses. In the B-SNIP cohort 80% statistical power corresponded to a correlation of magnitude 0.2 in the PSYCH group (N = 314) and of 0.17 in the NPSYCH group at PFDR = 0.0064. The PRSSCZ used here now explains ~20% or more of case-control variation in schizophrenia risk19, but it is difficult to estimate the expected relationship between that PRS and the specific domains of cognition considered here. It is particularly difficult because domains of cognition likely differ in their relationships with schizophrenia25,39. The genetic associations to the BACS should be replicated in a larger sample.
The polygenic score for educational attainment (PRSEDUC) showed significant positive association with years of education in both the psychotic and the nonpsychotic groups in B-SNIP, and also showed significant association with WRAT in both B-SNIP and PNC. However, the association between PRSEDUC and BACS was not significant in the nonpsychotic group and was nominally significant (p < 0.05, Table S5) in the psychotic group in B-SNIP. While this lack of significant association with BACS in our work could be due to the relatively small B-SNIP sample size (BACS was not available in PNC), it is also possible that different cognitive domains have varying degrees of genetic overlap with educational attainment, and that cognitive phenotypes that assess verbal abilities are more closely genetically linked to educational performance. For example, in the UK Biobank data Hagenaars et al.25 reported strong genetic correlation between educational attainment and verbal-numerical reasoning (rg ~ 0.72), but the genetic correlation of educational attainment with memory and reaction time were not significant.
Our results indicate the need to further explore the relationship between cognitive performance and the genetic risk factors of psychiatric disorders in larger patient groups. These findings also suggest that specific domains of cognition may be more closely etiologically linked to schizophrenia than other domains are, creating an opportunity for longitudinal studies to identify the domains that best predict illness onset.
Electronic supplementary material
Acknowledgements
The B-SNIP study is in part supported by the National Institute of Mental Health (grants MH077852, MH077945, MH078113, MH077851, and MH077862). E.B.R. and D.J.W. were funded by National Institute of Mental Health grant 1K01MH099286-01A1 and Brain Behavior Research Foundation (NARSAD) Young Investigator grant 22379. Additional support was provided by the Stanley Center for Psychiatric Research at the Broad Institute of MIT and Harvard.
Conflict of interest
R.S.E.K. receives royalties for the BACS. J.A.S. has been a consultant for Takeda Pharmaceuticals. C.A.T. is on the board of IntraCellular Technology and an ad hoc consultant for Takeda, Pierre Fabre, and Atiphony.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41398-018-0124-8).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kahn RS, Keefe RS. Schizophrenia is a cognitive illness: time for a change in focus. JAMA Psychiatry. 2013;70:1107–1112. doi: 10.1001/jamapsychiatry.2013.155. [DOI] [PubMed] [Google Scholar]
- 2.Kraepelin E. Psychiatrie: Ein Lehrbuch für Studierende und Ärtze. Leipzig, Germany: Verlag von Johann Ambrosius Barth; 1893. [Google Scholar]
- 3.Bilder RM, et al. Neuropsychology of first-episode schizophrenia: initial characterization and clinical correlates. Am. J. Psychiatry. 2000;157:549–559. doi: 10.1176/appi.ajp.157.4.549. [DOI] [PubMed] [Google Scholar]
- 4.Reichenberg A, et al. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: a 30-year study. Am. J. Psychiatry. 2010;167:160–169. doi: 10.1176/appi.ajp.2009.09040574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu CH, Keshavan MS, Tronick E, Seidman LJ. Perinatal risks and childhood premorbid indicators of later psychosis: next steps for early psychosocial interventions. Schizophr. Bull. 2015;41:801–816. doi: 10.1093/schbul/sbv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khandaker GM, Barnett JH, White IR, Jones PB. A quantitative meta-analysis of population-based studies of premorbid intelligence and schizophrenia. Schizophr. Res. 2011;132:220–227. doi: 10.1016/j.schres.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCabe JH, et al. Scholastic achievement at age 16 and risk of schizophrenia and other psychoses: a national cohort study. Psychol. Med. 2008;38:1133–1140. doi: 10.1017/S0033291707002048. [DOI] [PubMed] [Google Scholar]
- 8.Seidman LJ, et al. Association of neurocognition with transition to psychosis: baseline functioning in second phase of the North American Prodome Longitudinal Study. JAMA Psychiatry. 2016;73:1239–1248. doi: 10.1001/jamapsychiatry.2016.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seidman LJ, et al. Neuropsychology of the prodrome to psychosis in the NAPLS consortium: relationship to family history and conversion to psychosis. Arch. Gen. Psychiatry. 2010;67:578–588. doi: 10.1001/archgenpsychiatry.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fusar-Poli P, et al. Cognitive functioning in prodromal psychosis: a meta-analysis. Arch. Gen. Psychiatry. 2012;69:562–571. doi: 10.1001/archgenpsychiatry.2011.1592. [DOI] [PubMed] [Google Scholar]
- 11.Simon AE, et al. Cognitive functioning in the schizophrenia prodrome. Schizophr. Bull. 2007;33:761–777. doi: 10.1093/schbul/sbm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 13.Bouchard TJ., Jr. Genes, evolution and intelligence. Behav. Genet. 2014;44:549–577. doi: 10.1007/s10519-014-9646-x. [DOI] [PubMed] [Google Scholar]
- 14.Haworth CM, et al. The heritability of general cognitive ability increases linearly from childhood to young adulthood. Mol. Psychiatry. 2015;15:1112–1120. doi: 10.1038/mp.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deary IJ, Penke L, Johnson W. The neuroscience of human intelligence differences. Nat. Rev. Neurosci. 2010;11:201–211. doi: 10.1038/nrn2793. [DOI] [PubMed] [Google Scholar]
- 16.Plomin R, Deary IJ. Genetics and intelligence differences: five special findings. Mol. Psychiatry. 2010;20:98–108. doi: 10.1038/mp.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies G, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N = 53949) Mol. Psychiatry. 2015;20:183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies G, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol. Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schizophrenia working group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toulopoulou T, et al. Substantial genetic overlap between neurocognition and schizophrenia: genetic modeling in twin samples. Arch. Gen. Psychiatry. 2007;64:1348–1355. doi: 10.1001/archpsyc.64.12.1348. [DOI] [PubMed] [Google Scholar]
- 21.Toulopoulou T, et al. Impaired intellect and memory: a missing link between genetic risk and schizophrenia? Arch. Gen. Psychiatry. 2010;67:905–913. doi: 10.1001/archgenpsychiatry.2010.99. [DOI] [PubMed] [Google Scholar]
- 22.Fowler T, Zammit S, Owen MJ, Rasmussen F. A population-based study of shared genetic variation between premorbid IQ and psychosis among male twin pairs and sibling pairs from Sweden. Arch. Gen. Psychiatry. 2012;69:460–466. doi: 10.1001/archgenpsychiatry.2011.1370. [DOI] [PubMed] [Google Scholar]
- 23.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hubbard L, et al. Evidence of common genetic overlap between schizophrenia and cognition. Schizophr. Bull. 2016;42:832–842. doi: 10.1093/schbul/sbv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagenaars SP, et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N = 112151) and 24 GWAS consortia. Mol. Psychiatry. 2016;21:1624–1632. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lencz T, et al. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics consortium (COGENT) Mol. Psychiatry. 2014;19:168–174. doi: 10.1038/mp.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill SK, et al. Neuropsychological impairments in schizophrenia and psychotic bipolar disorder: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) Am. J. Psychiatry. 2013;170:1275–1284. doi: 10.1176/appi.ajp.2013.12101298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamminga CA, et al. Bipolar and schizophrenia network for intermediate phenotypes: outcomes across the psychosis continuum. Schizophr. Bull. 2014;40:S131–S137. doi: 10.1093/schbul/sbt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keefe RS, et al. The brief assessment of cognition in schizophrenia: reliability, sensitivity and comparison with a standard neurocognitive battery. Schizophr. Res. 2014;68:283–297. doi: 10.1016/j.schres.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Kareken DA, Gur RC, Saykin AJ. Reading on the wide range achievement test-revised and parental education as predictors of iq: comparison with the Barona formula. Arch. Clin. Neuropsychol. 1995;10:147–157. doi: 10.1093/arclin/10.2.147. [DOI] [PubMed] [Google Scholar]
- 31.Bright P, Jaldow E, Kopelman MD. The national adult reading test as a measure of premorbid intelligence: a comparison with estimates derived from demographic variables. J. Int. Neuropsychol. Soc. 2002;8:847–854. doi: 10.1017/S1355617702860131. [DOI] [PubMed] [Google Scholar]
- 32.Gladsjo JA, Heaton RK, Palmer BW, Taylor MJ, Jeste DV. Use of oral reading to estimate premorbid intellectual and neuropsychological functioning. J. Int. Neuropsychol. Soc. 1999;5:247–254. doi: 10.1017/S1355617799533079. [DOI] [PubMed] [Google Scholar]
- 33.Davies G, et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N = 112151) Mol. Psychiatry. 2016;21:758–767. doi: 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okbay A, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gur RC, et al. Age group and sex differences in performance on a computerized neurocognitive battery in children age 8–21. Neuropsychology. 2012;26:251. doi: 10.1037/a0026712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson EB, et al. The genetic architecture of pediatric cognitive abilities in the Philadelphia Neurodevelopmental Cohort. Mol. Psychiatry. 2015;20:454–458. doi: 10.1038/mp.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calkins ME, et al. The Philadelphia neurodevelopmental cohort: constructing a deep phenotyping collaborative. J. Child. Psychol. Psychiatry. 2015;56:1356–1369. doi: 10.1111/jcpp.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clementz BA, et al. Identification of distinct psychosis biotypes using brain-based biomarkers. Am. J. Psychiatry. 2016;173:373–384. doi: 10.1176/appi.ajp.2015.14091200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Germine L, et al. Association between polygenic risk for schizophrenia, neurocognition, and social cognition across development. Transl. Psychiatry. 2016;6:e924. doi: 10.1038/tp.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson CA, et al. Data quality control in genetic case-control association studies. Nat. Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.1000 Genomes Project Consortium. et al. An Integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams AL, Patterson N, Glessner J, Hakonarson H, Reich D. Phasing of many thousands of genotyped samples. Am. J. Hum. Genet. 2012;91:238–251. doi: 10.1016/j.ajhg.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2011;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 49.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das S, et al. Next-generation genotype imputation service and methods. Nat. Gen. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.The 1000 Genomes project consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 54.Deary V, et al. Genetic contributions to self-reported tiredness. Mol. Psychiatry. 2017;23:609–620. doi: 10.1038/mp.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.RStudio Team. RStudio: Integrated Development for R. (RStudio, Inc. Boston, MA, 2015.).
- 56.Tomlinson-Keasey C, Little TD. Predicting educational attainment, occupational achievement, intellectual skill, and personal adjustment among gifted men and women. J. Educ. Psychol. 1990;82:442–455. doi: 10.1037/0022-0663.82.3.442. [DOI] [Google Scholar]
- 57.Trampush JW, et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol. Psychiatry. 2017;22:336–345. doi: 10.1038/mp.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J. Schizophrenia working group of the psychiatric genomics consortium et al. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riglin L, et al. Schizophrenia risk alleles and neurodevelopmental outcomes in childhood: a population-based cohort study. Lancet Psychiatry. 2017;4:57–62. doi: 10.1016/S2215-0366(16)30406-0. [DOI] [PubMed] [Google Scholar]
- 61.Clarke TK, et al. Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Mol. Psychiatry. 2016;21:419–425. doi: 10.1038/mp.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiner DJ, et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet. 2017;49:978–985. doi: 10.1038/ng.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.