Abstract
Huntington disease (HD) is an autosomal dominant neurodegenerative disorder caused by a mutation in the huntingtin (HTT) gene which results in progressive neurodegeneration in the striatum, cortex, and eventually most brain areas. Despite being a monogenic disorder, environmental factors influence HD characteristics. Both human and mouse studies suggest that mutant HTT (mHTT) leads to gene expression changes that harbor potential to be modulated by the environment. Yet, the underlying mechanisms integrating environmental cues into the gene regulatory program have remained largely unclear. To better understand gene-environment interactions in the context of mHTT, we employed RNA-seq to examine effects of maternal separation (MS) and environmental enrichment (EE) on striatal gene expression during development of BACHD rats. We integrated our results with striatal consensus modules defined on HTT-CAG length and age-dependent co-expression gene networks to relate the environmental factors with disease progression. While mHTT was the main determinant of expression changes, both MS and EE were capable of modulating these disturbances, resulting in distinctive and in several cases opposing effects of MS and EE on consensus modules. This bivalent response to maternal separation and environmental enrichment may aid in explaining their distinct effects observed on disease phenotypes in animal models of HD and related neurodegenerative disorders.
Introduction
Huntington disease (HD) is an autosomal dominantly inherited neurodegenerative disorder caused by a polyglutamine expansion in exon 1 of the huntingtin gene (HTT)1. The mutation results in progressive neurodegeneration throughout the brain with strongest damages in the striatum which shows marked atrophy and loss of medium spiny neurons2–4. As a consequence, HD patients are burdened with motor deficits, cognitive impairments, and neuropsychiatric symptoms1.
The pathomechanical trajectory between mutant HTT (mHTT) and brain damage seems highly complex and different cellular functions have been implicated in mediating the toxicity. Previous studies in cell and animal models as well as human brains show that accumulation of mHTT leads to disturbed expression of genes associated with different neuronal functions5,6. Recently, these disturbances have been further explored in different HD mouse models carrying either fragment or full-length mHTT7–10, with respect to age and CAG length11, and also in BACHD (bacterial artificial chromosome HD) rats12.
Adding to the complexity, HD and related neurodegenerative disorders including Alzheimer disease, Parkinson’s disease, and Amyotrophic lateral sclerosis have been associated with environmental factors that bivalently correlate with disease risk and characteristics13–16. Previous studies in animal models showed positive effects on HD-like phenotypes through enriched housing, cognitive enrichment, and exercise. Specifically, enriched environments caused expression changes of distinct genes17, influenced markers of brain pathology9,18–21, altered behavioural19,22–25 as well as other parameters such as survival rate and body weight26. On the other hand, worsening of disease phenotypes in mouse models of HD was observed for various stressors. While acute stress led to increased depressive-like behaviour19 and impaired memory acquisition25, chronic stress affected olfactory sensitivity27 and accelerated memory deficits28. Despite the increasing body of evidence that environmental effects influence HD phenotypes in animal models, little is known about how these environmental cues are integrated into the gene regulatory program.
Towards a better understanding of these mechanisms, we examined interactions between environment and genotype in BACHD rats expressing full-length human mHTT and exhibiting motor impairments, cognitive deficits, emotional changes, and characteristic neuropathology of HD29–32. Given the greater sensitivity of the brain to environmental changes and challenges during development with known consequences for later stages in life33–35, we manipulated the rearing environment of BACHD rats in early life through maternal separation and environmental enrichment, respectively. While one group of rat pups was repeatedly separated from their mothers during the first 14 days of life to implement the stress paradigm, the other group was housed in the enriched environment from weaning till adulthood. As both environmental conditions are known to affect brain function and induce gene expression changes in wildtype animals33–36, subsequent profiling of the striatal transcriptome of wildtype and BACHD rats using RNA-seq enabled comprehensive analyses of gene expression changes in the context of mHTT and their modulations through the respective environmental paradigm.
Results
Full-length mutant HTT disturbed striatal gene expression depending on the environment
In order to assess the impact of environmental conditions on gene expression in the context of mHTT, we used a BAC transgenic rat model that expresses the full-length human mHTT gene with 97 polyQ repeats (BACHD)29 and exposed groups of animals (n=6 each) to either a standard environment (SE), enriched environment (EE), or maternal separation (MS) before profiling their striatal transcriptome using deep-sequencing of polyA-enriched RNA.
The enriched environment was set-up as a combination of larger cages with a cohort of eight animals supplied with nesting and bedding material as well as repeatedly rearranged toys for a total period of six weeks after weaning (Supplementary Fig. S1). In a second cohort, pups were separated daily for 4 h from their mothers from day 1 to 14 after birth. Afterwards, till the age of nine weeks, they were housed in the standard environment like the control group with four animals per standard cage (Supplementary Fig. S1). Body weight for animals in all experimental groups increased significantly over time with no observable difference between WT and BACHD (TG) animals (Supplementary Fig. S2) as shown before29. While rats in the MS group had a lower weight relative to the SE groups (Supplementary Fig. S2A), there were no significant body weight differences between the EE and SE groups (Supplementary Fig. S2B). Comparing MS to SE, animals showed an age effect (F(5,168)=1190; p<0.0001) as well as a treatment effect (F(1,168)=27.48; p<0.0001), but no genotype differences. Additionally, there were no significant interactions indicating that treatment effects were unspecific for age or genotype. Comparing EE to SE, animals showed an effect of age (F(5,168)=996.9; p<0.0001), but not of treatment, genotype, or their interaction.
Using an experimental design that describes all six animal groups with respect to genotype (wildtype, WT; transgenic, TG) and environmental condition (SE, EE, and MS) and modelling gene expression as a function of genotype, environment, and their interaction allowed determining differentially expressed genes across all pair-wise comparisons (Fig. 1a).
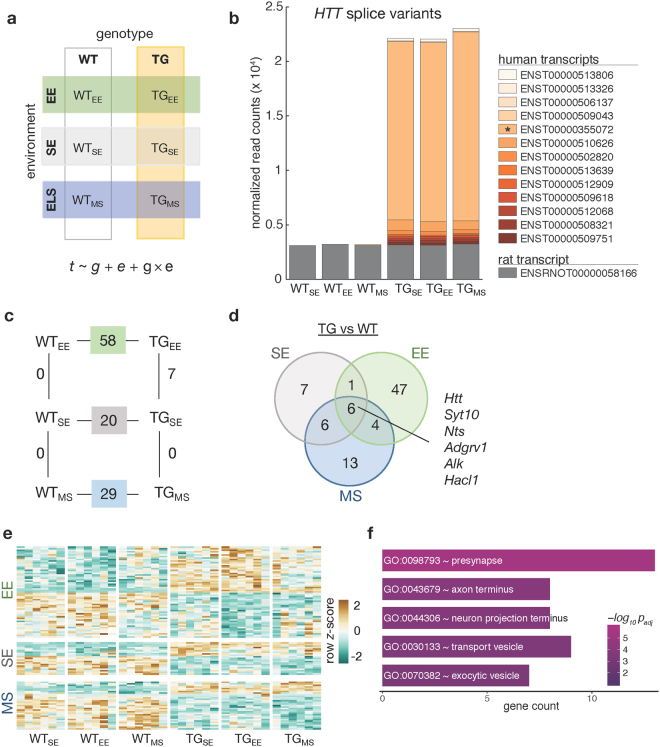
Figure 1.
Full-length mHTT disturbed the striatal transcriptome. (a) Schematic diagram of six experimental groups in a 2 × 3 factorial design based on two genotypes (WT, TG) and three environmental conditions (SE, EE, MS) that was used to reveal effects of environmental enrichment and maternal separation on the striatal transcriptome of 2-month-old WT and BACHD rats. Gene expression (t) was modelled as a function of genotype (g), environment (e), and their interaction (g × e). (b) Composition and expression level of rat and human HTT splice variants in the striatum of 2-month-old rats with respect to genotype and environmental condition. (c) Diagram showing number of differentially expressed genes between main comparisons in the factorial design. (d) Venn diagram showing overlaps of differentially expressed genes between BACHD and WT animals in each of the tested environmental conditions. (e) Heatmap of hierarchically clustered z-scores across all experimental groups for differentially expressed genes that were identified in comparing TGSE/WTSE, TGEE/WTEE, and TGMS/WTMS. (f) Overrepresented Gene Ontology terms of merged differentially expressed genes in TGSE/WTSE, TGEE/WTEE, and TGMS/WTMS.
Specifically, examining levels of endogenous as well as mHTT firstly, we observed no change in the endogenous rat transcript but an intriguing addition of human transcripts in transgenic animals irrespective of the environmental condition (Fig. 1b). The human protein-coding isoform spanning all 67 exons represented the largest fraction. In total, a ~7-fold increase of HTT expression was observed, in contrast to the 4.5-fold increase previously reported29 which likely results from the more sensitive RNA-seq as well as the alignment against the mHTT construct-extended rat genome compared to the previous microarray-based interrogation of the transcriptome.
In addition, overexpression of mHTT led to 20 differentially expressed genes (DEGs) in the striatum in the standard environment (TGSE/WTSE), 58 DEGs under environmental enrichment (TGEE/WTEE), and 29 DEGs after maternal separation (TGMS/WTMS) (Fig. 1c). In these three comparisons, six common DEGs were consistently identified irrespective of the environmental condition (Fig. 1d). Besides HTT, Alk was up- and Syt10, Nts, Hacl1, and Adgrv1 downregulated (Supplementary Fig. S3). For the latter, we identified a transcript isoform-specific downregulation (Supplementary Fig. S3b).
While the relatively small overlap of only six genes suggested an environment-dependent modulation of the mHTT-induced disturbances, most genes showed similar trends of expression changes across the environmental conditions (Fig. 1e). Therefore, merging the DEGs of all three TG/WT comparisons allowed investigating the general transgenic effect on cellular functions resulting in overrepresented Gene Ontology terms including presynapse, axon terminus, neuron projection terminus, transport vesicle, and exocytic vesicle (Fig. 1f) that were in line with previous findings6.
Early changes in striatal gene expression agree with dysregulation in later stages
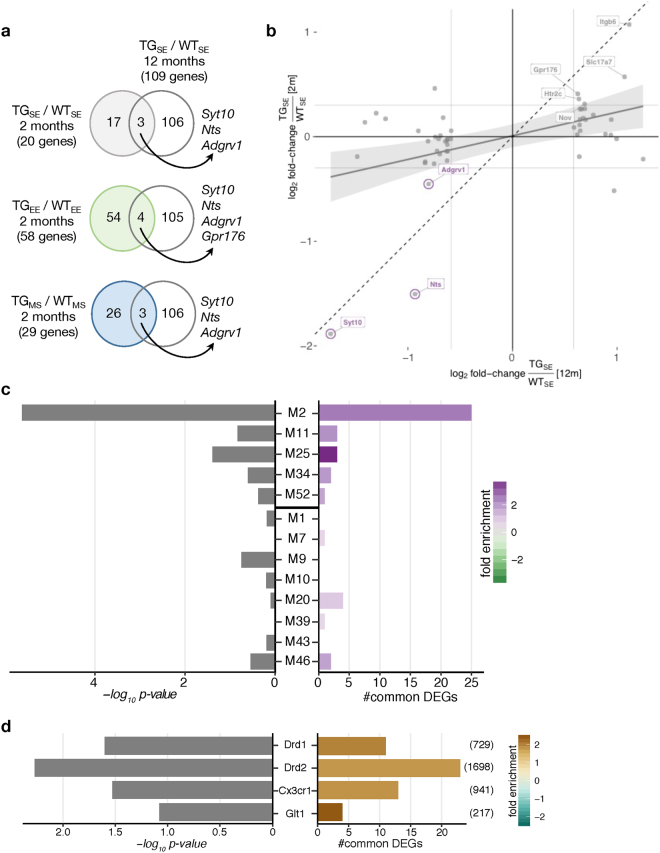
To relate these DEGs, identified at an early age, to later changes in the context of HD, we compared them with striatal DEGs of 12-month-old BACHD rats12. When comparing gene expression changes under SE conditions between 2 and 12 months of age for the 109 DEGs identified at the later time point, similarities of dysregulation were observed (Fig. 2b). The strongest correlation was found for Syt10, Nts, and Adgrv1 that showed differential expression across all three environmental treatments (Fig. 2a), suggesting these genes to represent characteristic markers for early mHTT disease unfolding irrespective of environmental condition.
Figure 2.
Later-state dysregulation of striatal gene expression induced by mHTT surfaced early on. (a) Venn diagram comparing differentially expressed genes in 2-month-old BACHD rats housed in one of the three tested environmental conditions with 12-month-old BACHD rats housed in the standard environment12. (b) Scatter plot of gene expression changes between WT and BACHD rats at 2 months (x-axis) and 12 months of age (y-axis). Striatal DEGs in 12-month-old BACHD rats12 plotted in grey. Striatal DEGs also identified in 2-month-old rats labelled in purple. Linear regression line with standard error shown in grey. Dashed line represents ordinary diagonal. Grey labels indicate DEGs that reached significance in 12-month-old but not in 2-month-old rats. (c) Enrichment of DEGs in 2-month-old rats (TG/WT for all three environments) for genes in HD consensus modules11. Genes in the upper five modules are downregulated and in the lower eight modules upregulated in a CAG length-dependent manner. Bars show p-values of two-sided Fisher’s exact test and number of common genes. (d) Enrichment of DEGs in 2-month-old rats (TG/WT for all three environments) for distinct striatal cell types characterized by sorting Drd1-, Drd2-, Glt1-, and Cx3cr1-positive striatal cells in mice. Number of cell type-specific genes (padj < 0.1) indicated in brackets on right. Bars show p-values of two-sided Fisher’s exact test and number of common genes.
To further explore these age-dependent effects, we integrated our data with available data from striatal consensus modules based on HTT-CAG length and age-dependent gene co-expression networks11. The most significant enrichment of DEGs in these modules was observed for module M2, M11, and M25, which contain genes that become increasingly downregulated with CAG length and age (Fig. 2c). Module M2 contains genes involved in cAMP signalling, while modules M11 and M25 are enriched for glia-related genes11.
The link to glia-related disturbances was further supported by comparing DEGs in BACHD rats to genes attributed to distinct striatal cell types in mouse. Differential expression in BACHD rats correlated significantly with the expression of their orthologous genes in Glt1-, and Cx3cr1-positive striatal cells in mice (Fig. 2d). In these cell type-specific comparisons, the most significant enrichment was found between BACHD DEGs and genes attributed to striatal medium spiny neurons sorted by Drd2 (Fig. 2d), in line with the critical role of this cell type in the context of mHTT 37.
Genotype-dependent activation of genes in response to environmental enrichment
Having integrated early striatal gene expression changes in BACHD rats with age-dependent aspects of HD, we next explored the impact of environmental changes in wildtype and transgenic rats. Intriguingly, neither MS nor EE led to any differentially expressed genes in the striatum of WT rats (Fig. 3a). Genes such as Oprk1, Nrxn1, and Nfia38,39 that are known to be responsive to early life stress showed trends of differential expression (Supplementary Fig. S4). Other markers such as Bdnf and Il1b showed very low expression in the striatum making conclusive results difficult to derive (Supplementary Fig S4). In TG animals, no DEGs were identified upon MS, however, seven genes (Ankrd13b, Clcf1, Dnajc30, LOC681282, Nr1d1, Slc22a3, and Syndig1l) were significantly upregulated upon EE (Fig. 3a,b). We confirmed this genotype-specific response with qPCR on Nr1d1 as a representative of their activation pattern (Fig. 3c).
Figure 3.
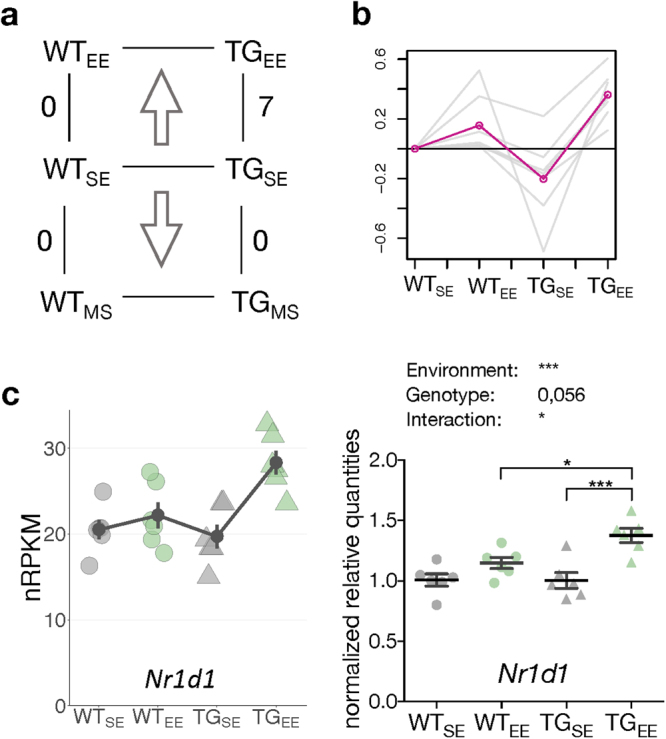
Environmental enrichment activated genes specifically in transgenic animals. (a) DEGs comparing WT and TG rats in EE and MS. (b) Gene expression profiles of seven DEGs in TGEE/TGSE relative to WTSE across four experimental groups. Mean profile in pink. (c) Left panel shows expression level for Nr1d1 plotted as individual data points with mean ± SEM. Right panel shows quantitative PCR data for Nr1d1 as normalized quantities relative to WTSE (n = 6 mice per group) plotted as individual data points with mean ± SEM. Two-way ANOVA was performed followed by Tukey’s multiple comparisons test. *p < 0.05, ***p < 0.001.
Environmental enrichment modulated several genes against their later-stage trend of dysregulation
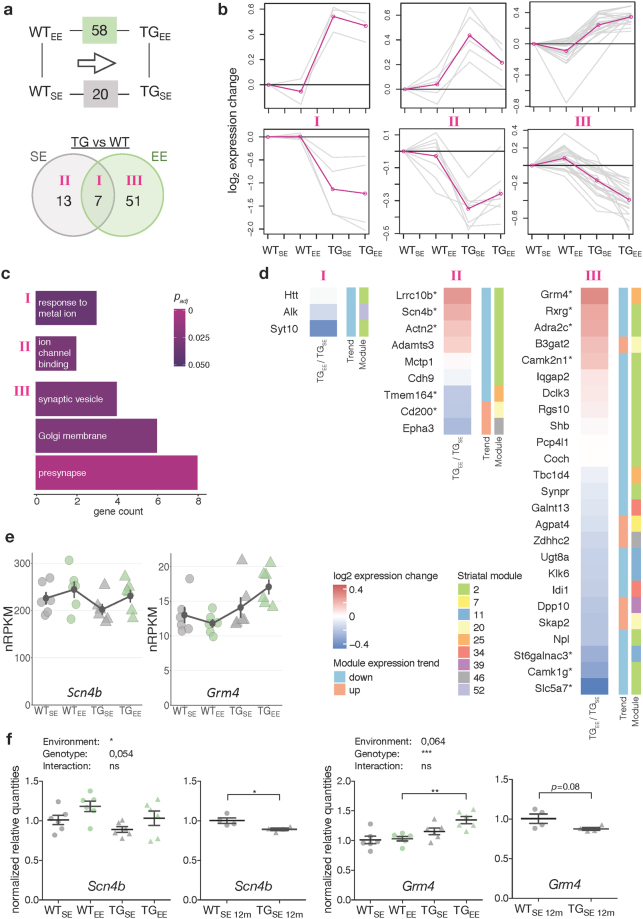
After having explored the transgenic and environmental impact on the striatal transcriptome individually, we studied their combined influence in order to assess environment-dependent modulation of gene expression changes in the context of mHTT. Focusing on the 58 DEGs in TGEE/WTEE and comparing them to the 20 DEGs found in TGSE/WTSE, seven overlapped (class I), while 13 were exclusively identified in SE (class II), and 51 DEGs were exclusively found in EE (class III) (Fig. 4a). To better understand the characteristics of the genes in these classes, their expression profiles were clustered (Fig. 4b). Class I was enriched for the GO term response to metal ion, and class II for ion channel binding (Fig. 4b,c). The class II expression profile reflects the intuitive amelioration, yet no complete prevention, of disturbed gene expression through the EE. Class III contained genes overrepresented for the GO terms synaptic vesicle, Golgi membrane, and presynapse (Fig. 4b,c) and stronger differential expression in EE than in SE, counter-intuitive to a protective influence at first glance.
Figure 4.
Environmental enrichment modulated striatal expression of disturbed genes. (a) Schematic and Venn diagram comparing number of DEGs between TGSE/WTSE and TGEE/WTEE. Roman numbers indicate sets analysed individually in (b). (b) Cluster analysis of the subsets of DEGs in (a) using k-means on gene expression ratios relative to WTSE across four experimental groups. Grey lines indicate expression changes per gene, pink lines cluster centroids. (c) Gene count and significance of overrepresented Gene Ontology terms for subsets of DEGs in (a). (d) Subsets of DEGs in (a) plotted with respect to their expression change between TGEE/TGSE, consensus module association and trend11. DEGs with no module information omitted. Rows sorted by mean expression change. Asterisks indicate DEGs with p-value < 0.05 when comparing TGEE/TGSE. (e) Expression levels for Scn4b and Grm4 plotted as individual data points with mean ± SEM. (f) Quantitative PCR data for Scn4b and Grm4 shown as normalized quantities relative to WTSE (n = 6 and n = 4 rats per group in 2-month-old and 12-month-old cohorts, respectively) plotted as individual data points with mean ± SEM. Significance determined by two-way ANOVA followed by Tukey’s multiple comparisons test for the 2-month-old cohort, and unpaired two-tailed t-test for the 12-month-old cohort. *p < 0.05, **p < 0.01, ***p < 0.001.
To interpret these modulations through the EE, we compared the DEGs in these classes with their associated consensus module characteristics11. For most DEGs in class II the modulating effect of the EE between TGEE/TGSE was indeed against their associated module trend, further supporting the intuitive ameliorating effect (Fig. 4d). Intriguingly, the modulation of expression changes through the EE in class III, in particular on upregulated genes, was against the predicted changes according to the consensus modules, too (Fig. 4d). This result resolves the contradiction of alleged aggravated disturbances and renders the EE-induced modulations into ameliorations against the age trend.
To validate these results, gene expression changes were measured for selected candidates in 2- and 12-month-old rats using quantitative PCR. In line with the computationally predicted expression changes, Scn4b in class II and Grm4 in class III were downregulated at 12 months (Fig. 4e,f), and showed less downregulation at 2 months of age in EE conditions (Fig. 4e,f).
Maternal separation aggravated expression changes of some genes towards their later-stage level of dysregulation.
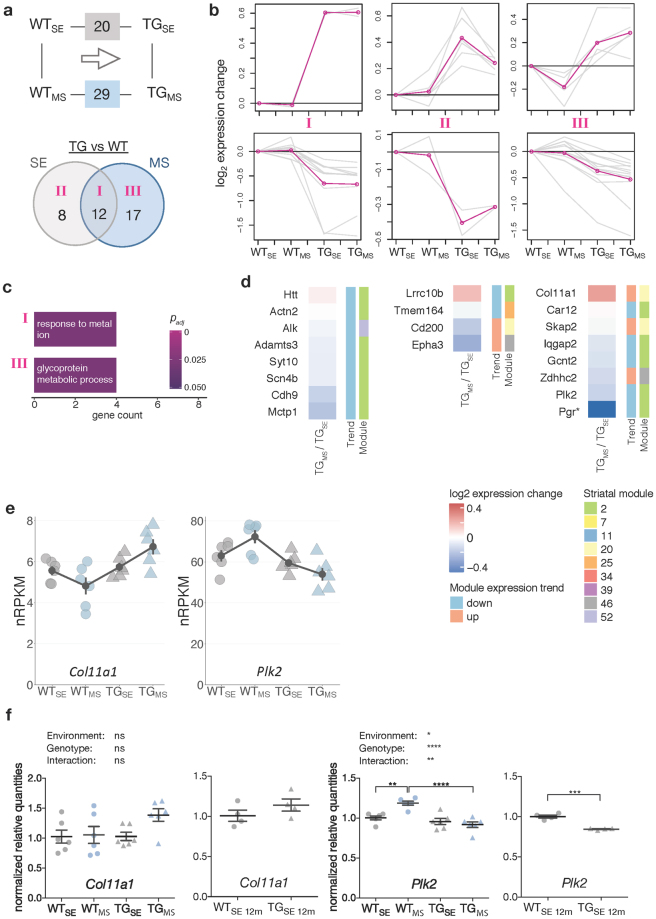
After exploring EE-induced modulations, we next focused on the impact of MS on mHTT-induced striatal gene expression disturbances. Compared to 20 DEGs in TGSE/WTSE, 29 DEGs were identified in TGMS/WTMS (Fig. 5a). Similar to the approach described for the EE, we grouped the three subsets of genes (class I, II, III) based on overlaps between both comparisons (Fig. 5b). Class I was enriched for the GO term response to metal ion (Fig. 5c). Class II, containing genes whose transgene-induced disturbance in SE was ameliorated after MS, was not enriched for any GO term. Interestingly, some of these genes responded similar under EE and MS conditions (Supplementary Fig. S5). Class III contained DEGs with stronger gene expression changes after MS, which were enriched for the GO term glycoprotein metabolic process (Fig. 5c).
Figure 5.
Maternal separation modulated striatal expression of disturbed genes. (a) Schematic and Venn diagram comparing number of DEGs between TGSE/WTSE and TGMS/WTMS. Roman numbers indicate subsets analysed individually in (b). (b) Cluster analysis of the DEG subsets in (a) using k-means on gene expression ratios relative to WTSE across four experimental groups. Grey lines indicate expression changes per gene, pink lines cluster centroids. (c) Gene count and significance of overrepresented Gene Ontology terms for subsets of DEGs in (a). (d) Subsets of DEGs from (a) plotted with respect to their expression change between TGMS/TGSE, module association, and trend11. DEGs with no module information omitted. Rows sorted by mean expression change. Asterisks indicate DEGs with p-value < 0.05 when comparing TGMS/TGSE. (e) Expression levels of Col11a1 and Plk2 plotted as individual data points with mean ± SEM. (f) Quantitative PCR data for Col11a1 and Plk2 shown as normalized quantities relative to WTSE (n = 6 and n = 4 rats per group in 2-month-old and 12-month-old cohort, respectively) plotted as individual data points with mean ± SEM. Significance determined by two-way ANOVA followed by Tukey’s multiple comparisons test for the 2-month-old cohort, and unpaired two-tailed t-test for the 12-month-old cohort. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Computationally associating genes in these three classes with consensus module information, an ameliorating effect for most class II genes against the module trend was apparent similar to effect observed in EE (Fig. 5d, Supplementary Fig. S5). In contrast, for several genes in class III the stronger expression changes after MS pointed in the same direction as the predicted changes according to the consensus modules, thereby representing a reinforcement of the transgenic disturbance expected with age by MS (Fig. 5d). These computational associations were confirmed for Col11a1 and Plk2 using quantitative PCR. Col11a1 indeed showed upregulated levels with age and after MS already in young animals (Fig. 5e,f). Plk2 was downregulated with age similar to its response to MS in young animals (Fig. 5e,f).
Disease aspects and biofunctions associated with mHTT suggest distinct responsiveness to enriched environment and maternal separation
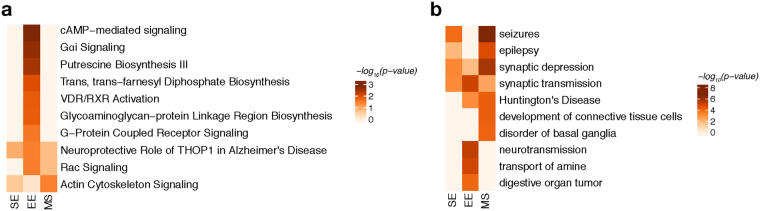
For several genes, EE as well as MS modulated the mHTT-induced gene expression disturbances (Figs 4 and 5), while other genes remained altered irrespective of the environment. Using pathway analysis tools, we investigated this environmental modulation further. Few pathways were detected to varying significance across all environmental conditions such as actin cytoskeleton signalling and Rac signalling (Fig. 6a). Additional pathways originated mostly from the comparison in EE including cAMP signalling, VDR/RXR activation, and G-protein coupled receptor signalling (Fig. 6a).
Figure 6.
Shared and distinct characteristics in pathways and disease aspects for BACHD rats after environmental enrichment and maternal separation. (a) Canonical pathway analysis identified significantly enriched cellular pathways among DEGs derived in the tested environmental conditions for BACHD rats. Significance values colour-coded and hierarchically clustered representing likeliness of pathway participation. (b) Disease aspects and biological functions predicted to be affected based on DEGs derived in the tested environmental conditions. Significance values colour-coded and hierarchically clustered representing likeliness of downstream effects on disease aspects.
Exploring these pathways with respect to disease aspects in TG animals, we found seizures, epilepsy, and synaptic depression to be more significant for MS DEGs compared to the SE or EE groups (Fig. 6b). In contrast, synaptic transmission and neurotransmission were more significant in EE, while they seemed less involved in SE or MS (Fig. 6b), agreeing with previously described protective effects of environmental enrichment20.
Validation of specific candidate genes on protein level
To quantify protein levels of NR1D1, mGluR4 (GRM4), and SCN4B which showed environment-dependent gene expression changes Western blot assays were performed (Supplementary Fig. S6). Additionally, protein levels of mHTT were quantified even though mHTT gene expression was not significantly affected by any environmental condition (Supplementary Fig. S6).
Although not significant, NR1D1 protein levels showed similar tendencies to those observed for the respective mRNA with increased expression in TGEE compared to TGSE and WTEE (Supplementary Fig. S6a). Opposite to the gene expression results, protein levels of mGluR4 (GRM4) were significantly lower in TG than in WT rats irrespective of the environmental condition (Supplementary Fig. S6a). Statistical analyses showed a genotype but no environment effect for mGluR4 protein levels. Moreover, no significant environment or genotype effects were detected for SCN4B (Supplementary Fig. S6a).
Intriguingly, protein levels of mHTT were reduced in TGEE relative to TGSE (Supplementary Fig. S6a) while they were comparable between TGMS and TGSE (Supplementary Fig. S6b) which suggests a specific, potentially protective, effect of environmental enrichment on mHTT protein levels.
Discussion
Taken together, the overall aim of this study was to assess environmental effects on striatal gene expression in young BACHD rats. Our results show that mHTT-induced gene expression changes (i) correlated between younger and older animals, (ii) were dependent on the environment, and (iii) were partially modulated against and with their trend of dysregulation at later ages through environmental enrichment and maternal separation, respectively.
Specifically, we observed a dominant effect of mHTT on striatal gene expression changes across the tested environmental conditions. Combining differentially expressed genes from these comparisons were enriched for Gene Ontology terms that agree with previously reported disturbances of mHTT on cellular vesicle transport and secretion as well as synaptic transmission and vesicle exocytosis40–42.
Further, we found changes in gene expression induced by mHTT at young age to agree with disturbances observed in later stages of the disease in this model. Differentially expressed genes found in young and older BACHD rats suggest that these genes may serve as characteristic markers of disease unfolding. Candidates in this overlap such as Syt10, a vesicular Ca2+-sensor with neuroprotective potential43, and Nts a modulator of dopaminergic transmission known to be dysregulated in HD37, were downregulated and point at aspects of the disease that are disturbed early on.
Support for the idea that disease aspects are already detectable at this young age can be derived from the significant overlap of DEGs with some of the most informative HD consensus modules from co-expression meta-analyses11. Furthermore, these disturbances seem characteristic for the disease as consensus module data from HD mouse models largely agree with gene expression changes in BACHD rats.
Also on cell type level, our findings support the critical role of D2 neurons for the developing pathology in this disease model, agreeing with this cell type’s high vulnerability in HD44, and strengthen the emerging concept that also glial-cell populations are affected. Glia impairments have been described in R6/2 mice45 and overexpression of mHTT in glia cells has been shown to worsen the HD phenotype in N171–82Q mice46, suggesting a role of glia cells in HD pathology to be further followed up functionally in BACHD rats.
Both tested environmental conditions partially modulated the mHTT-induced disturbances. Environmental enrichment affected gene expression changes against the consensus module trend suggesting a preventive effect in disease progression. Intriguingly, this prevention exhibited not only an ameliorating effect on some disturbed genes, but also led to stronger expression changes for other genes. Despite being counter-intuitive, the latter modulation type seems to represent a response of the system to the EE in order to prevent disturbances appearing later in time. Among the genes perceiving an amelioration of their disturbed expression was Scn4b, encoding the 4β subunit of a voltage dependent sodium channel implicated in neurite growth47. Its downregulation in young and old BACHD rats agrees with previous reports on impaired Snc4b expression in R6/2 mice and HD patients already at a pre-symptomatic stage47 suggesting a critical role of this gene in the degenerative processes in HD. Grm4, on the other hand, was among the genes responding to the EE by stronger upregulation. It encodes for the glutamate metabotropic receptor 4, and is a hub gene in consensus module M25, whose genes are downregulated and overrepresented for glutamate receptor signalling11. In addition, Grm5, encoding a receptor of the same protein family, has been linked to HD-like impairments in Hdh(Q111/Q111) mice48 and implicated in hippocampal effects of environmental enrichment49. The observed upregulation of Grm4 in TGEE animals might, thus, be part of known protective responses of EE23, and are in line with previously reported findings of metabotropic receptors in neuroprotection50.
In contrast to the EE, maternal separation aggravated disturbed gene expression of some genes towards their later stage trend of dysregulation in young BACHD rats. Plk2, for example, was among the genes that were further downregulated by MS in the context of mHTT. Activation of Plk2 has been shown in response to early insults such as prenatal stress and neonatal seizures suggesting a role in hippocampal synaptic downscaling and supporting the idea that Plk2 might be protective51. In keeping, Plk2 was increased in WT animals subjected to maternal separation. BACHD rats, in contrast, showed decreased expression that was further decreased after MS, in line with a general aggravation of HD markers upon stress experiences27,28,52. This aggravating effect further suggests greater sensitivity of BACHD rats to stressful experiences agreeing with hyperactivity of the hypothalamic-pituitary-adrenal axis in HD patients53.
Phenotypically, we observed lower body weights for rats in the MS group as it has been previously reported as a consequence of maternal separation54 as well as in response to stressors in adult rats55,56 suggesting body weight loss to reflect stress here, too. We did not assess direct physiological stress responses (e.g. body temperature or stress hormones) to limit manipulations of the animals. Measuring stress hormone levels when rats were sacrificed would have potentially allowed assessing basal stress activity57,58. Yet, they have also been found comparable to control levels58–61.
Behavioural tests may also allow assessing MS effects and are essential to link gene expression changes to phenotypic consequences in the context of environmental factors. Here, we however sought to minimize disturbances of the animals through behavioural testing and handling as they may confound gene expression changes62. Moreover, only part of the behavioural phenotypes described for BACHD rats have been examined at such early age29–32,63,64. Follow-up studies that consider later time points will enable investigations whether behavioural and physiological characteristics in the BACHD rat model depend on environmental factors. In order to determine preventive and aggravating aspects of environmental factors in the unfolding of the disease, rats would have to be kept in standardized environments for longer periods. Having data from such later time points will then allow resolving disease progression and environmental impact in greater detail and facilitate analyses of their interplay with respect to temporal delays or accelerations on HD pathology. To additionally investigate whether there are any EE-driven rehabilitative effects or reversals of dysfunctions animals could first be housed in SE or MS until developing initial signs of pathology and then migrating them to the EE.
On a molecular level, typical markers such Bdnf and Il1b showed very low expression in striatal tissue and did not allow deriving statistically significant results. Other genes including Oprk1, Nrxn1, and Nfia38,39 that are known to be responsive to stress showed trends of changed expression in WT rats after maternal separation. However, no significantly expressed genes were identified comparing TGMS/TGSE or WTMS/WTSE, possibly related to general timings in the developmental gene regulatory program as many essential genes for striatal function and relevant in the context of HD may not yet be fully expressed during the first two postnatal weeks65. Additionally, housing animals in groups of four under standard conditions after the stress paradigm may have masked early stress effects as enriched conditions can even reverse them36.
Agreeing with previous reports on few differentially expressed genes in the striatum upon environmental enrichments in the context of mHTT9, we identified seven distinctively activated genes specifically in BACHD rats. Among them was Nr1d1 encoding a transcription factor of the nuclear receptor subfamily 1 (Rev-erbα) that is involved in regulating circadian rhythm and metabolic processes66. Given the metabolic deficits of young BACHD rats including obesity and altered levels of energy metabolism factor64, upregulation of Nr1d1 in BACHD rats upon EE may hint at relevant processes as to how environmental enrichment affects metabolic phenotypes. Collecting metabolic parameters known to be altered in BACHD rats29,64 will help to follow up this gene-environment interaction.
To reveal whether gene expression changes were present at protein level too, we measured protein levels of NR1D1, mGluR4 (GRM4), and SCN4B for which environment-dependent gene expression changes were shown and partially confirmed mRNA results on protein level.
Such discrepancies between mRNA and protein levels have been observed for a variety of proteins and may originate from both biological as well as methodological aspects67. A poor correlation between mRNA and protein levels in our samples may be explained by post-translational mechanisms regulating protein turnover and abundance67. These mechanisms may explain the opposite pattern of changes for Grm4 mRNA and its protein. Although not in line with mRNA changes, this effect is interesting given the neuroprotective function of mGluR4 and other group III metabotropic glutamate receptors68,69. Therefore, mGlur4 downregulation may represent a potential mechanism mediating neuropathological alterations in BACHD rats.
Alternative reasons for the weak correlation between mRNA and protein changes could result from technical limitations. The lower sensitivity of Western blot analysis relative to RNA-seq and qPCR, and the high variability among Western blot replicates may have limited the detection of small changes. This could in part explain the lack of significant EE effects on NR1D1 and SCN4B protein levels, despite the similar pattern of mRNA and protein levels.
By affecting post-translational events and other regulatory mechanisms, the environment might also affect protein levels without altering mRNA expression. Accordingly, EE downregulated mHTT protein without altering its mRNA levels. The underlying mechanisms of EE on mHTT in BACHD rats will be interesting to follow up considering the central role of mHTT.
In summary, results of this study show disturbances on striatal gene expression by mHTT in BACHD rats that are partially dependent on environmental condition early in life. Bivalent modulations of disturbed genes in response to environmental enrichment or maternal separation may, thus, contribute to explain their opposing phenotypic effects in HD and related neurodegenerative diseases. In order to better connect these disease aspects with disturbances on gene expression level, it will be critical to also interrogate protein abundancies, physiological parameters, and behavioural phenotypes.
Materials and Methods
Animals, study design and environmental treatments
Experiments were performed in BACHD rats and WT littermates. The BACHD rat model was generated using a BAC containing the whole human HTT locus with 20 kb upstream and 50 kb downstream flanking genomic sequences including regulatory elements29. WT HTT exon 1 was replaced by mHTT exon 1 carrying 97 mixed CAA/CAG repeats flanked by two LoxP sites29.
All animals were bred on a Sprague Dawley background by pairing heterozygous BACHD males with wildtype females (Charles River, Germany). At the day of birth, indicated as postnatal day (PD) 0, all litters were culled to eight pups ensuring equal numbers of females and males similar to previous maternal separation experiments58,59,70.
In order to avoid separating littermate pups during maternal separation and to keep comparable conditions for all experimental groups, a litter, including both male and female pups, was randomly allocated to one of three environmental paradigms: standard environment (SE), maternal separation (MS), and enriched environment (EE) (Supplementary Fig. S1). Each environmental paradigm (n = 32) included four groups of WT males, WT females, TG males, TG females, (n = 8 per group), totalling to 96 animals. To avoid variability of the data based on hormonal influences in cycling female rats only male rats were used for RNA-seq which was performed on tissue from six rats that were randomly chosen from each of the six environmental groups (n = 36).
The maternal separation protocol was adapted from previous studies58,61,71. In brief, from PD 1 to 14, pups were separated from their mother for 4 h daily starting at the beginning of the light phase. Separated pups were placed in glass containers leaned with paper and nesting material and transported to a different room to prevent communication between pups and dams. In order to maintain comparable temperatures (32–33 °C) between pup environment in the home cage and the nest during separation, pups were kept in containers placed in a warm water bath.
At PD 16, rats were genotyped following established protocols29. At PD 21, rats were weaned. Rats of the EE group were housed in groups of eight animals in larger cages (1222 × 580 × 443 mm) enriched with bedding, nesting material, toys, and objects of different material, colour, and shape for the entire duration of the experiment. Novel objects were introduced every 3–4 days. Rats in other experimental groups were housed in groups of four in standard type IV cages (598 × 380 × 200 mm) equipped with bedding and nesting material. Control animals were left undisturbed for the entire experiment except for cage cleaning and body weight measurements that were collected weekly from weaning throughout the experiment in all experimental groups. All rats were housed with same-gender littermates of mixed genotype and maintained in a room with constant temperature (22 ± 1 °C), humidity (55 ± 10%), and a regular 12 h light/dark cycle (lights on/off at 7 AM/7 PM). Food and water were provided ad libitum.
At two months of age (PD 59–60), animals were sacrificed by live decapitation before extracting their brains for dissection. Brain samples were frozen in liquid nitrogen and stored at –80 °C.
RNA from eight additional male rats of the same line (4 WT and 4 BACHD) was used to measure gene expression of selected candidate genes at 12 months of age. These rats were bred in a separate cohort and left undisturbed except for cage cleaning until being sacrificed. Their housing and room conditions were the same as for SE groups except for additional tissue paper in the cage. Protocols applied for live decapitation as well as brain collection, dissection, and storage were as described above. This extra cohort of animals included only WT and TG rats housed in SE. This assay does not represent a second time point in the experiment and was solely used to compare changes at young and later age.
All procedures strictly adhered international standards for the care and use of laboratory animals and were approved by the local Animal Welfare and Ethics committee of the Country Commission Tübingen, Germany (TVA HG 2/15 and TVA HG 6/13).
Experimental procedures
RNA isolation
Total RNA and DNA were simultaneously extracted from striatal samples of 2-month-old rats using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) following the manufacturer’s protocol. Total RNA was extracted from striatal samples of 12-month-old rats with RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol.
RNA sequencing
The polyadenylated fraction of RNA isolated from striatal tissue of 2-month-old rats (n = 6 animals in each of the 6 experimental groups) was used for RNA-seq. Quality was assessed with an Agilent 2100 Bioanalyzer. Samples with high RNA integrity number (RIN >8) were selected for library construction. Using the TruSeq Stranded RNA Sample Prep Kit (Illumina) and 100 ng of total RNA for each sequencing library, poly(A) selected paired-end sequencing libraries (136 bp read length) were generated according to the manufacturer’s instructions. All libraries were sequenced on an Illumina HiSeq 2500 platform at a depth of ~15–25 million reads each. Library preparation and sequencing procedures were performed by the same individual, and a design aimed to minimize technical batch effects was chosen. Fastq files and raw counts are available through GEO (GSE107259).
Reverse transcription-quantitative PCR (RT-qPCR)
RNA-seq results were validated using RT-qPCR with primers specific for Nr1d1, Snc4b, Grm4, Col11a1, and Plk2 genes (see Supplementary Table S1 for primers sequences). 100 ng of total RNA was used for the reverse transcription reaction (QuantiTect Reverse Transcription kit, Qiagen) following the manufacturer’s instructions. The resulting cDNA was diluted (1:20) and 2 µl were used for qPCR assays, mixed with primers (0.25 µM) and SYBR green master mix (Qiagen). Expression was calculated relative to the mean of the WTSE group according to the Pfaffl model72 after normalization to the geometric mean relative expression of three reference genes (Pgk1, Gapdh, and Eif4a2 for 2-month-old rats. Pgk1, Gapdh, and Ywhaz for 12-month-old rats). Expression stabilities of the reference genes were assessed using Normfinder73 and Genorm74.
Protein extraction and Western blotting
Homogenates of rat striata were obtained by homogenising brain hemispheres in RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate and 1% Triton X-100, containing cOmplete® protease inhibitor cocktail and PhosSTOP™ phosphatase inhibitors (both Roche)) using an ULTRA-TURRAX® disperser (VWR). Samples were ultrasonicated for 3 s using a SONOPULS ultrasonic homogenizer (Bandelin) at 10% power. Homogenates were incubated for 30 min on ice, mixed with glycerol to a final concentration of 10% and stored at −80 °C for further analysis. Protein concentrations of RIPA homogenates were measured spectrophotometrically using Bradford reagent (Bio-Rad Laboratories).
Protein blotting was performed according to standard procedures. Briefly, 25 µg of total protein were mixed with 4 × LDS sample buffer (1 M Tris Base pH 8.5, 2 mM EDTA, 8% LDS, 40% glycerol, and 0.025% phenol red) in a 3:1 ratio and supplemented with 100 mM dithiothreitol. After heat-denaturing for 10 min at 70 °C, protein samples were electrophoretically separated using 4–12% Bolt® Bis-Tris gradient gels, 10% Bis-Tris gels, and 7% NuPAGE® Tris-Acetate gels (both Thermo Fisher Scientific) with respective electrophoresis buffers, MES buffer (50 mM MES, 50 mM Tris Base pH 7.3, 0.1% SDS, 1 mM EDTA), MOPS buffer (50 mM MOPS, 50 mM Tris Base pH 7.7, 0.1% SDS, 1 mM EDTA) or Tris-Acetate running buffer (50 mM Tricine, 50 mM Tris Base pH 8.25, 0.1% SDS). Proteins were transferred on Amersham™ Protran™ Premium 0.2 µm nitrocellulose or Amersham™ Hybond™ P 0.2 polyvinyliden fluoride membranes (both GE Healthcare) using Bicine/Bis-Tris transfer buffer (25 mM Bicine, 25 mM Bis-Tris pH 7.2, 1 mM EDTA, 15% methanol) and a TE22 Transfer Tank (Hoefer) at 250 mA for 1.5–2 h.
After transfer, membranes were blocked for 1 h with 5% skim milk powder (Merck) in TBS (Tris-buffered saline) at room temperature, and probed overnight at 4 °C with the following primary antibodies diluted in TBS-T (TBS with 0.1% Tween 20): rabbit anti-huntingtin (1:1000; clone D7F7, #5656, Cell Signaling), rabbit anti-metabotropic glutamate receptor 4 (1:250; ab53088, Abcam), rabbit anti-NR1D1 (1:500; clone EPR10376, ab174309, Abcam), rabbit anti-SCN4B (1:1000; ab80539, Abcam), mouse anti-polyglutamine expansion diseases marker (1:2000; clone 5TF1–1C2, MAB1574, EMD Millipore), and rabbit anti-vinculin (1:1000; clone E1E9V, #13901, Cell Signaling). Afterwards, blots were washed with TBS-T and incubated at room temperature for 1 h with the HRP-conjugated secondary antibody goat anti-rabbit (1:10,000; ab97051, Abcam) or with the respective secondary IRDye antibodies goat anti-mouse 680LT, goat anti-mouse 800CW, and goat anti-rabbit 800CW (all 1:10,000; LI-COR Biosciences). After final washing with TBS-T, chemiluminescence and fluorescence signals were detected using the LI-COR ODYSSEY® FC and quantified with Image Studio 4.0 software (both LI-COR Biosciences).
Bioinformatics
Quality control, alignment, and expression analysis
Read quality of RNA-seq data in fastq files was assessed using FastQC (v0.11.4)75 to identify sequencing cycles with low average quality, adaptor contamination, or repetitive sequences from PCR amplification. Reads were aligned using STAR (v2.5.2b)76 allowing gapped alignments to account for splicing against a custom-built genome composed of the Ensembl Rattus norvegicus genome v87 and the human HTT transgene. Alignment quality was analyzed using samtools (v1.1)77 and visually inspected in the Integrative Genome Viewer (v2.3.67)78. Msantd1 was manually excluded from further downstream analyses as its differential expression likely results from its genomic sequence being present in the 50 kb 3′ flanking region of the transgenic HTT BAC construct. Normalized read counts for all genes were obtained using DESeq2 (v1.16.1)79. Transcripts covered with less than 50 reads were excluded from the analysis leaving 12,639 genes for determining differential expression in each of the pair-wise comparisons between experimental groups.
The 2 × 3 factorial design of the experiment was captured in a generalized linear model describing expression (t) as a function of genotype (g), the environment (e), and their interaction (g × e). Surrogate variable analysis (sva, v3.24.4) was applied to minimize unwanted variation between samples80. Given that differences in transcript abundances in brain tissue are often small in magnitude and in vivo RNA-seq data are deemed to be more variable81, we set |log2 fold-change |≥0.3 and adjusted p-value ≤ 0.1 to determine differential expression, as computationally predicted candidates down to the lower end of these thresholds could be confirmed in qPCR assays.
Gene-level abundances were derived from DESeq2 as normalized read counts and used for calculating the log2-transformed expression changes underlying the k-means clustering with ratios computed relative to the mean expression in WTSE. Raw counts provided by DESeq2 also went into calculating nRPKMs (normalized Reads Per Kilobase per Million total reads) as a measure of relative gene expression as motivated before82. The DESeq2 sizeFactors served in scaling estimated abundances derived from Salmon (v0.7.2)83 when determining transcript-level compositions of individual genes. Transcript-level differential expression was further explored and verified with kallisto (v0.43.0)84.
Gene annotation, cell type enrichments, and pathway analyses
ClusterProfiler (v3.4.4) was employed to identify overrepresented Gene Ontology terms and associated cellular functions in sets of differentially expressed genes (DEGs)85. Terms with at least three annotated genes were queried and reported with their adjusted p-value ≤ 0.1. All Gene ID conversions were done using biomaRt Bioconductor package (v2.32.1) querying v87 of the Ensembl database.
Canonical pathways between sets of DEGs as well as predicted relationships between DEGs and disease aspects/biofunctions were derived from Ingenuity Pathway Analysis (IPA, v01–12, Qiagen). For the latter analysis module, only functions with at least three associated genes and at least one activating/inhibiting relationship are reported.
Expression data for 12-month-old animals are based on data files provided with Yu-Taeger et al.12. Cell type-specific attributions are based on RNA-seq data derived with Laser Capture Microdissection from main cellular populations of the striatum isolated from transgenic mice expressing eGFP in astrocytes (Glt1-eGFP86), microglia (Cx3cr1-eGFP87), Drd2 medium spiny neurons (Drd2-eGFP88), and Tomato fluorescent protein in Drd1 medium spiny neurons (Drd1-Tomato89) (unpublished data, Déglon lab). Enrichments of DEGs were calculated against 13 striatal consensus modules defined in Langfelder et al.11, of which 5 modules contain down- and 8 modules upregulated genes.
Statistical analysis
Body weight data were analyzed in a three-way ANOVA with Graph Pad Prism (v7) using genotype as within-subject and age as well as environmental treatment as between-subject factors. Alpha was set at 0.05. ANOVA analyses were performed separately for the EE and MS because of different time spans of the treatments. SE groups served as control for both treatment conditions. Body weight results are shown in Supplementary Fig. S2.
For RT-qPCR data, two-way ANOVA was applied to test for genotype, environment, and their interaction followed by Tukey’s correction for multiple comparisons. Expression differences in the 12-month-old cohorts were tested with unpaired two-tailed t-test using significance thresholds of p < 0.05.
For Western blot data for NR1D1, mGluR4, and SCN4B, two-way ANOVA was applied to test for genotype, environment, and their interaction followed by Tukey’s correction for multiple comparisons. Gene expression differences in RT-qPCR data in the 12-month-old cohorts as well as EE and MS effects on mHTT protein expression in Western blot data were tested with unpaired two-tailed t-test using significance thresholds of p < 0.05.
Electronic supplementary material
Acknowledgements
We thank Patrycja Bambynek-Dziuk and Celina Tomczak for their help with setting up the breeding and performing the genotyping. Furthermore, we thank the core facility c.ATG for preparing the libraries and sequencing the samples. We acknowledge the support through the Deutsche Forschungsgemeinschaft and the Open Access Publishing Fund of the University of Tübingen. This Project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 643417 (ModelPolyQ) to HPN as well as funding from the Interdisciplinary Centre for Clinical Research (IZKF) Tübingen to JMSH. JMSH was funded by a Margarete-von-Wrangell fellowship supported by the Ministry of Science, Research and the Arts Baden-Württemberg, and ZW by a scholarship from the German Academic Exchange Service (DAAD).
Author Contributions
A.N., J.M.S.H., and H.P.N. conceived and designed the research. A.N., Z.W., and L.Y.T. performed the experiments and analysed the qPCR as well as body weight data. J.J.W. and Z.W. ran and analysed the Western blots. TH and JMSH analysed the RNA-seq data. N.D. contributed cell type-specific data. A.N., T.H., H.P.N., and J.M.S.H. wrote the paper. All authors read and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Arianna Novati, Thomas Hentrich, Huu Phuc Nguyen and Julia M. Schulze-Hentrich contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24243-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huu Phuc Nguyen, Email: hoa.nguyen@med.uni-tuebingen.de.
Julia M. Schulze-Hentrich, Email: julia.schulze-hentrich@uni-tuebingen.de
References
- 1.Bates GP. History of genetic disease: the molecular genetics of Huntington disease - a history. Nat Rev Genet. 2005;6:766–773. doi: 10.1038/nrg1686. [DOI] [PubMed] [Google Scholar]
- 2.Waldvogel HJ, Kim EH, Tippett LJ, Vonsattel JP, Faull RL. The Neuropathology of Huntington’s Disease. Curr Top Behav Neurosci. 2015;22:33–80. doi: 10.1007/7854_2014_354. [DOI] [PubMed] [Google Scholar]
- 3.Vonsattel JP, et al. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Kumar A, Vaish M, Ratan RR. Transcriptional dysregulation in Huntington’s disease: a failure of adaptive transcriptional homeostasis. Drug Discov Today. 2014;19:956–962. doi: 10.1016/j.drudis.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha JH. Transcriptional signatures in Huntington’s disease. Prog Neurobiol. 2007;83:228–248. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas EA, et al. In vivo cell-autonomous transcriptional abnormalities revealed in mice expressing mutant huntingtin in striatal but not cortical neurons. Hum Mol Genet. 2011;20:1049–1060. doi: 10.1093/hmg/ddq548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn A, et al. Mutant huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington’s disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 9.Benn CL, et al. Environmental enrichment reduces neuronal intranuclear inclusion load but has no effect on messenger RNA expression in a mouse model of Huntington disease. J Neuropathol Exp Neurol. 2010;69:817–827. doi: 10.1097/NEN.0b013e3181ea167f. [DOI] [PubMed] [Google Scholar]
- 10.Becanovic K, et al. Transcriptional changes in Huntington disease identified using genome-wide expression profiling and cross-platform analysis. Hum Mol Genet. 2010;19:1438–1452. doi: 10.1093/hmg/ddq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langfelder P, et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat Neurosci. 2016;19:623–633. doi: 10.1038/nn.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu-Taeger L, Bonin M, Stricker-Shaver J, Riess O, Nguyen HH. Dysregulation of gene expression in the striatum of BACHD rats expressing full-length mutant huntingtin and associated abnormalities on molecular and protein levels. Neuropharmacology. 2017;117:260–272. doi: 10.1016/j.neuropharm.2017.01.029. [DOI] [PubMed] [Google Scholar]
- 13.Reitz C, Mayeux R. Alzheimer disease: epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem Pharmacol. 2014;88:640–651. doi: 10.1016/j.bcp.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingre C, Roos PM, Piehl F, Kamel F, Fang F. Risk factors for amyotrophic lateral sclerosis. Clin Epidemiol. 2015;7:181–193. doi: 10.2147/CLEP.S37505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ascherio A, Schwarzschild MA. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol. 2016;15:1257–1272. doi: 10.1016/S1474-4422(16)30230-7. [DOI] [PubMed] [Google Scholar]
- 16.Mo C, Hannan AJ, Renoir T. Environmental factors as modulators of neurodegeneration: insights from gene-environment interactions in Huntington’s disease. Neurosci Biobehav Rev. 2015;52:178–192. doi: 10.1016/j.neubiorev.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Zajac MS, et al. Wheel running and environmental enrichment differentially modify exon-specific BDNF expression in the hippocampus of wild-type and pre-motor symptomatic male and female Huntington’s disease mice. Hippocampus. 2010;20:621–636. doi: 10.1002/hipo.20658. [DOI] [PubMed] [Google Scholar]
- 18.Spires TL, et al. Environmental enrichment rescues protein deficits in a mouse model of Huntington’s disease, indicating a possible disease mechanism. J Neurosci. 2004;24:2270–2276. doi: 10.1523/JNEUROSCI.1658-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pang TY, Du X, Zajac MS, Howard ML, Hannan AJ. Altered serotonin receptor expression is associated with depression-related behavior in the R6/1 transgenic mouse model of Huntington’s disease. Hum Mol Genet. 2009;18:753–766. doi: 10.1093/hmg/ddn385. [DOI] [PubMed] [Google Scholar]
- 20.Nithianantharajah J, Barkus C, Murphy M, Hannan AJ. Gene-environment interactions modulating cognitive function and molecular correlates of synaptic plasticity in Huntington’s disease transgenic mice. Neurobiol Dis. 2008;29:490–504. doi: 10.1016/j.nbd.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Lazic SE, et al. Neurogenesis in the R6/1 transgenic mouse model of Huntington’s disease: effects of environmental enrichment. Eur J Neurosci. 2006;23:1829–1838. doi: 10.1111/j.1460-9568.2006.04715.x. [DOI] [PubMed] [Google Scholar]
- 22.Wood NI, et al. Responses to environmental enrichment differ with sex and genotype in a transgenic mouse model of Huntington’s disease. PLoS One. 2010;5:e9077. doi: 10.1371/journal.pone.0009077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Dellen A, Blakemore C, Deacon R, York D, Hannan AJ. Delaying the onset of Huntington’s in mice. Nature. 2000;404:721–722. doi: 10.1038/35008142. [DOI] [PubMed] [Google Scholar]
- 24.Renoir T, et al. Differential effects of early environmental enrichment on emotionality related behaviours in Huntington’s disease transgenic mice. J Physiol. 2013;591:41–55. doi: 10.1113/jphysiol.2012.239798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo C, Renoir T, Pang TY, Hannan AJ. Short-term memory acquisition in female Huntington’s disease mice is vulnerable to acute stress. Behav Brain Res. 2013;253:318–322. doi: 10.1016/j.bbr.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 26.Wood NI, Glynn D, Morton AJ. “Brain training” improves cognitive performance and survival in a transgenic mouse model of Huntington’s disease. Neurobiol Dis. 2011;42:427–437. doi: 10.1016/j.nbd.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Mo C, Renoir T, Hannan AJ. Effects of chronic stress on the onset and progression of Huntington’s disease in transgenic mice. Neurobiol Dis. 2014;71:81–94. doi: 10.1016/j.nbd.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Mo C, et al. High stress hormone levels accelerate the onset of memory deficits in male Huntington’s disease mice. Neurobiol Dis. 2014;69:248–262. doi: 10.1016/j.nbd.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Yu-Taeger L, et al. A novel BACHD transgenic rat exhibits characteristic neuropathological features of Huntington disease. J Neurosci. 2012;32:15426–15438. doi: 10.1523/JNEUROSCI.1148-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adjeroud N, et al. Reduced impact of emotion on choice behavior in presymptomatic BACHD rats, a transgenic rodent model for Huntington Disease. Neurobiol Learn Mem. 2015;125:249–257. doi: 10.1016/j.nlm.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Abada YS, Nguyen HP, Schreiber R, Ellenbroek B. Assessment of motor function, sensory motor gating and recognition memory in a novel BACHD transgenic rat model for huntington disease. PLoS One. 2013;8:e68584. doi: 10.1371/journal.pone.0068584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abada YS, Nguyen HP, Ellenbroek B, Schreiber R. Reversal learning and associative memory impairments in a BACHD rat model for Huntington disease. PLoS One. 2013;8:e71633. doi: 10.1371/journal.pone.0071633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardon MC, Rattray I. What do we know about the long-term consequences of stress on ageing and the progression of age-related neurodegenerative disorders? Neurosci Biobehav Rev. 2008;32:1103–1120. doi: 10.1016/j.neubiorev.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 34.de Kloet ER, Sibug RM, Helmerhorst FM, Schmidt MV. Stress, genes and the mechanism of programming the brain for later life. Neurosci Biobehav Rev. 2005;29:271–281. doi: 10.1016/j.neubiorev.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Sale A, Berardi N, Maffei L. Environment and brain plasticity: towards an endogenous pharmacotherapy. Physiol Rev. 2014;94:189–234. doi: 10.1152/physrev.00036.2012. [DOI] [PubMed] [Google Scholar]
- 36.van Praag H, Kempermann G, Gage FH. Neural consequences of environmental enrichment. Nat Rev Neurosci. 2000;1:191–198. doi: 10.1038/35044558. [DOI] [PubMed] [Google Scholar]
- 37.Chen JY, Wang EA, Cepeda C, Levine MS. Dopamine imbalance in Huntington’s disease: a mechanism for the lack of behavioral flexibility. Front Neurosci. 2013;7:114. doi: 10.3389/fnins.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papale LA, Madrid A, Li S, Alisch RS. Early-life stress links 5-hydroxymethylcytosine to anxiety-related behaviors. Epigenetics. 2017;12:264–276. doi: 10.1080/15592294.2017.1285986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Granholm L, et al. The expression of opioid genes in non-classical reward areas depends on early life conditions and ethanol intake. Brain Res. 2017;1668:36–45. doi: 10.1016/j.brainres.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Smith R, Brundin P, Li JY. Synaptic dysfunction in Huntington’s disease: a new perspective. Cell Mol Life Sci. 2005;62:1901–1912. doi: 10.1007/s00018-005-5084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozas JL, Gomez-Sanchez L, Tomas-Zapico C, Lucas JJ, Fernandez-Chacon R. Presynaptic dysfunction in Huntington’s disease. Biochem Soc Trans. 2010;38:488–492. doi: 10.1042/BST0380488. [DOI] [PubMed] [Google Scholar]
- 42.Brandstaetter H, Kruppa AJ, Buss F. Huntingtin is required for ER-to-Golgi transport and for secretory vesicle fusion at the plasma membrane. Dis Model Mech. 2014;7:1335–1340. doi: 10.1242/dmm.017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woitecki AM, et al. Identification of Synaptotagmin 10 as Effector of NPAS4-Mediated Protection from Excitotoxic Neurodegeneration. J Neurosci. 2016;36:2561–2570. doi: 10.1523/JNEUROSCI.2027-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehrlich ME. Huntington’s disease and the striatal medium spiny neuron: cell-autonomous and non-cell-autonomous mechanisms of disease. Neurotherapeutics. 2012;9:270–284. doi: 10.1007/s13311-012-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang R, Diaz-Castro B, Looger LL, Khakh BS. Dysfunctional Calcium and Glutamate Signaling in Striatal Astrocytes from Huntington’s Disease Model Mice. J Neurosci. 2016;36:3453–3470. doi: 10.1523/JNEUROSCI.3693-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradford J, et al. Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. J Biol Chem. 2010;285:10653–10661. doi: 10.1074/jbc.M109.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oyama F, et al. Sodium channel beta4 subunit: down-regulation and possible involvement in neuritic degeneration in Huntington’s disease transgenic mice. J Neurochem. 2006;98:518–529. doi: 10.1111/j.1471-4159.2006.03893.x. [DOI] [PubMed] [Google Scholar]
- 48.Ribeiro FM, et al. Metabotropic glutamate receptor 5 knockout promotes motor and biochemical alterations in a mouse model of Huntington’s disease. Hum Mol Genet. 2014;23:2030–2042. doi: 10.1093/hmg/ddt598. [DOI] [PubMed] [Google Scholar]
- 49.Buschler A, Manahan-Vaughan D. Metabotropic glutamate receptor, mGlu5, mediates enhancements of hippocampal long-term potentiation after environmental enrichment in young and old mice. Neuropharmacology. 2017;115:42–50. doi: 10.1016/j.neuropharm.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 50.Flor PJ, Battaglia G, Nicoletti F, Gasparini F, Bruno V. Neuroprotective activity of metabotropic glutamate receptor ligands. Adv Exp Med Biol. 2002;513:197–223. doi: 10.1007/978-1-4615-0123-7_7. [DOI] [PubMed] [Google Scholar]
- 51.Sun H, Kosaras B, Klein PM, Jensen FE. Mammalian target of rapamycin complex 1 activation negatively regulates Polo-like kinase 2-mediated homeostatic compensation following neonatal seizures. Proc Natl Acad Sci USA. 2013;110:5199–5204. doi: 10.1073/pnas.1208010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du X, et al. The influence of the HPG axis on stress response and depressive-like behaviour in a transgenic mouse model of Huntington’s disease. Exp Neurol. 2015;263:63–71. doi: 10.1016/j.expneurol.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Aziz NA, et al. Increased hypothalamic-pituitary-adrenal axis activity in Huntington’s disease. J Clin Endocrinol Metab. 2009;94:1223–1228. doi: 10.1210/jc.2008-2543. [DOI] [PubMed] [Google Scholar]
- 54.Iwasaki S, Inoue K, Kiriike N, Hikiji K. Effect of maternal separation on feeding behavior of rats in later life. Physiol Behav. 2000;70:551–556. doi: 10.1016/S0031-9384(00)00305-X. [DOI] [PubMed] [Google Scholar]
- 55.Lenglos C, Mitra A, Guevremont G, Timofeeva E. Sex differences in the effects of chronic stress and food restriction on body weight gain and brain expression of CRF and relaxin-3 in rats. Genes Brain Behav. 2013;12:370–387. doi: 10.1111/gbb.12028. [DOI] [PubMed] [Google Scholar]
- 56.Harris RB, et al. Weight loss in rats exposed to repeated acute restraint stress is independent of energy or leptin status. Am J Physiol Regul Integr Comp Physiol. 2002;282:R77–88. doi: 10.1152/ajpregu.2002.282.1.R77. [DOI] [PubMed] [Google Scholar]
- 57.Marais L, van Rensburg SJ, van Zyl JM, Stein DJ, Daniels WM. Maternal separation of rat pups increases the risk of developing depressive-like behavior after subsequent chronic stress by altering corticosterone and neurotrophin levels in the hippocampus. Neurosci Res. 2008;61:106–112. doi: 10.1016/j.neures.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 58.Hulshof HJ, et al. Maternal separation decreases adult hippocampal cell proliferation and impairs cognitive performance but has little effect on stress sensitivity and anxiety in adult Wistar rats. Behav Brain Res. 2011;216:552–560. doi: 10.1016/j.bbr.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 59.Wigger A, Neumann ID. Periodic maternal deprivation induces gender-dependent alterations in behavioral and neuroendocrine responses to emotional stress in adult rats. Physiol Behav. 1999;66:293–302. doi: 10.1016/S0031-9384(98)00300-X. [DOI] [PubMed] [Google Scholar]
- 60.Plotsky PM, et al. Long-term consequences of neonatal rearing on central corticotropin-releasing factor systems in adult male rat offspring. Neuropsychopharmacology. 2005;30:2192–2204. doi: 10.1038/sj.npp.1300769. [DOI] [PubMed] [Google Scholar]
- 61.Plotsky PM, Meaney MJ. Early, postnatal experience alters hypothalamic corticotropin-releasing factor (CRF) mRNA, median eminence CRF content and stress-induced release in adult rats. Brain Res Mol Brain Res. 1993;18:195–200. doi: 10.1016/0169-328X(93)90189-V. [DOI] [PubMed] [Google Scholar]
- 62.Rubin TG, Gray JD, McEwen BS. Experience and the ever-changing brain: what the transcriptome can reveal. Bioessays. 2014;36:1072–1081. doi: 10.1002/bies.201400095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manfre G, et al. The BACHD Rat Model of Huntington Disease Shows Specific Deficits in a Test Battery of Motor Function. Front Behav Neurosci. 2017;11:218. doi: 10.3389/fnbeh.2017.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clemensson EK, Clemensson LE, Fabry B, Riess O, Nguyen HP. Further investigation of phenotypes and confounding factors of progressive ratio performance and feeding behavior in the BACHD rat model of Huntington disease. PLoS One. 2017;12:e0173232. doi: 10.1371/journal.pone.0173232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novak G, Fan T, O’Dowd BF, George SR. Striatal development involves a switch in gene expression networks, followed by a myelination event: implications for neuropsychiatric disease. Synapse. 2013;67:179–188. doi: 10.1002/syn.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Solt LA, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 68.Williams CJ, Dexter DT. Neuroprotective and symptomatic effects of targeting group III mGlu receptors in neurodegenerative disease. J Neurochem. 2014;129:4–20. doi: 10.1111/jnc.12608. [DOI] [PubMed] [Google Scholar]
- 69.Betts MJ, O’Neill MJ, Duty S. Allosteric modulation of the group III mGlu4 receptor provides functional neuroprotection in the 6-hydroxydopamine rat model of Parkinson’s disease. Br J Pharmacol. 2012;166:2317–2330. doi: 10.1111/j.1476-5381.2012.01943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slotten HA, Kalinichev M, Hagan JJ, Marsden CA, Fone KC. Long-lasting changes in behavioural and neuroendocrine indices in the rat following neonatal maternal separation: gender-dependent effects. Brain Res. 2006;1097:123–132. doi: 10.1016/j.brainres.2006.04.066. [DOI] [PubMed] [Google Scholar]
- 71.Wang Q, Shao F, Wang W. Maternal separation produces alterations of forebrain brain-derived neurotrophic factor expression in differently aged rats. Front Mol Neurosci. 2015;8:49. doi: 10.3389/fnmol.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 74.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andrews, S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
- 76.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maze I, et al. Analytical tools and current challenges in the modern era of neuroepigenomics. Nat Neurosci. 2014;17:1476–1490. doi: 10.1038/nn.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srinivasan K, et al. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun. 2016;7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 85.Yu G, Wang LG, Han Y, He Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Regan MR, et al. Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci. 2007;27:6607–6619. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/MCB.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 89.Ade KK, Wan Y, Chen M, Gloss B, Calakos N. An Improved BAC Transgenic Fluorescent Reporter Line for Sensitive and Specific Identification of Striatonigral Medium Spiny Neurons. Front Syst Neurosci. 2011;5:32. doi: 10.3389/fnsys.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.