Abstract
MicroRNAs have been proposed to be a class of biomarkers of disease as expression levels are significantly altered in various tissues and body fluids when compared to healthy controls. As such, the detection and quantification of microRNAs is imperative. While many methods have been established for quantification of microRNAs, they typically rely on time consuming handling such as RNA extraction, purification, or ligation. Here we describe a novel method for quantification of microRNAs using direct amplification in body fluids without upstream sample preparation. Tested with a point-of-care device (termed Gene-Z), the presence of microRNA promotes base-stacking hybridization, and subsequent amplification between two universal strands. The base-stacking approach, which was achieved in <60 min, provided a sensitivity of 1.4 fmol per reaction. Tested in various percentages of whole blood, plasma, and faeces, precision (coefficient of variation = 2.6%) was maintained and comparable to amplification in pristine samples. Overall, the developed method represents a significant step towards rapid, one-step detection of microRNAs.
Keywords: MicroRNA quantification, Isothermal amplification, Point-of-care diagnostics, Direct amplification
1 Introduction
Point-of-care (POC) nucleic acids-based methods and technologies have the potential to offer minimally invasive alternatives to biopsies and routine examinations allowing early detection and therapeutics. MicroRNAs (short, non-coding RNA molecules) are one such potential marker of disease and cancer with varied expression levels in tissues (Esquela-Kerscher and Slack 2006; Gaur et al. 2007), faeces (Link et al. 2012; Link et al. 2010), saliva (Michael et al. 2010; Shao et al. 2012), and blood (Schwarzenbach et al. 2014) between patient samples and healthy controls; depending on the disease (Ruepp et al. 2010). For example, increased expression of miR-30b, miR-29b, miR-142-2p, miR-144, miR-203, and miR-223 (> eight fold in some instances) has been observed in oral squamous samples collected from cell carcinoma patients compared to healthy controls (Manikandan et al. 2016). In serum, miR-141 expression levels up to 46 fold between patients with prostate cancer and healthy controls (Mitchell et al. 2008). Thousands of microRNAs have been identified in humans (Griffiths-Jones et al. 2008; Griffiths-Jones et al. 2006; Griffiths-Jones 2004) and efforts to associate microRNAs to various types of cancer and disease is extensive and ongoing (Ruepp et al. 2010).
Methods to quantify microRNA require superior limit of detection, large dynamic range, and precision (Tricoli and Jacobson 2007). Existing amplification-based methods for measurement of microRNAs include stem-loop reverse transcription polymerase chain reaction (RT-PCR) (Chen et al. 2005) and reverse transcription-free PCR (Lu et al. 2011). Isothermal approaches such as rolling circle amplification (Harcourt and Kool 2012; Liu et al. 2013; Zhou et al. 2010), loop-mediated isothermal amplification (LAMP) (Li et al. 2011), exponential amplification reaction (EXPAR) (Wang et al. 2014; Zhang and Zhang 2012), and others have also been described for microRNA (Table 1). Isothermal approaches have the advantage of simplicity in terms of constant temperature and high amplicon yields (Mori et al. 2001), which allow for quantification with relatively simpler devices (e.g. turbidity meters, Illumigene, NucleSENSE easyQ, Gene-Z). However, upstream sample preparation for microRNA is challenging. This is because amplification-based techniques typically require RNA isolation by skilled personnel in a centralized laboratories. However, isothermal polymerases (e.g. Bst) are more robust and less impacted by inhibitory substrates compared to PCR polymerases (Kostic et al. 2015; Stedtfeld et al. 2014). Thus, an isothermal direct amplification approach has the potential to reduce analysis time and costs, without isolation and purification, and is therefore well suited for use outside of laboratories with specialized infrastructures (Njiru 2012).
Table 1.
Summary of selected isothermal amplification approaches for measurement of microRNAs
| Validated microRNA(s) |
Sample processing required |
Reaction name; polymerase; detection |
Detection limit | Reaction volume | Dynamic range | Turn-around- Time (TAT) |
Incubation temperature(s) |
Reference |
|---|---|---|---|---|---|---|---|---|
| Isothermal approaches requiring RNA extraction prior to amplification of microRNAs | ||||||||
| miR-319a | RNA extraction, biotin-labeling | Poly (U) polymerase- mediated isothermal signal amplification; Poly(U); Electrochemical | 8.5 × 10−6 fmol (8.5 zmol) | 5 µl | 10–1000 fM | 4 h | 37 °C | (Zhou et al. 2016) |
| Random sequence | Probes spiked into biological samples | Enzyme- assisted target amplification; Klenow fragment; Colorimetric | 50 × 10−6 fmol (50 zmol) | 100 µl | 0.5 fM–1 nM | 2 h | 90 °C, 37 °C | (Yan et al. 2013) |
| miR-l miR-122, miR-150, miR-143, and let-7a | RNA Extraction; reverse transcription, phosphorylation, ligation | Isothermal ramification amplification (RAM); Bst; SYBR Green | 25 × 10−6 fmol (25 zmol) | 25 µl | 1 fM-10 nM | 8.5 h | 37 °C, 41–37 °C, 55 °C, 65 °C | (Yao et al. 2009) |
| let-7a | RNA extraction after ligation | Rolling circle amplification (RCA); Φ-29; probes | 10 fmol | 50 µl | 200 pM-10 nM | 30 h | 30 °C | (Harcourt and Kool 2012) |
| miR-21, let-7d | RNA extraction, ligation | RCA; Φ-29; SYBR Green | 20 × 10−6 fmol (20 zmol) | 20 µl | 1 fM-100 nM | 8 h | 37 °C, 30 °C | (Zhou et al. 2010) |
| N/A | RNA extraction | Branched RCA; Φ-29; bioluminescence | 0.1 fmol | 10 µl | 10 pM – 7.5 pM | 2.5 h | 37 °C | (Mashimo et al. 2011) |
| miR-1 | RNA extraction | Cascade RCA-NESA-DNAzyme amplification; Φ-29; Color change |
0.2 × 10−6 zmol (0.2 zmol) | 100 µl | 10 aM - 10 µM | 6 h | 37 °C | (Wen et al. 2012) |
| miR-16 | RNA extraction, ligation | RCA; Φ-29; Northern blot | 0.5 × 10−3 fmol (0.5 amol) | N/A | N/A | 10 h | 37 °C, 30 °C | (Jonstrup et al. 2006) |
| let-7a | RNA extraction | Loop-mediated isothermal amplification (LAMP); Bst; SYBR Green | 1 × 10−3 (1 amol) | 10 µl | 0.1 pM-0.1 µM | ~2 h | 55 °C | (Li et al. 2011) |
| let-7a | RNA extraction | Strand displacement amplification (SDA); Klenow fragment; SYBR Green | 16 × 10−6 fmol (16 zmol) | 10 µl | 16 fM-0.1 µM | 90 min | 37 °C | (Shi et al. 2014) |
| Isothermal approaches employing direct amplification of microRNAs | ||||||||
| miR-486-5p | Demonstrated directly and after RNA extraction | Hairpin probe RCA; Φ-29; SYBR Green 11 | 0.5 × 10−6 (0.5 amol) | 50 µl | 0.2 fM-1 nM | 4 h | 35 °C | (Li et al. 2013) |
| let-7a | Demonstrated directly and after RNA extraction | Cascade signal amplification; Klenow fragment; Molecular beacon | 1 fmol direct; 0.1 × 10−6 fmol after RNA extraction | 10 µl | 10 aM- 5 nM | 80 min | 37 °C | (Ma et al. 2014) |
| miR-141 | Demonstrated directly and after RNA extraction | LAMP; Bst; SYT082 | 1.4 fmol | 10 µl | 14 aM – 14 fM | 60 min | 65 °C | This study |
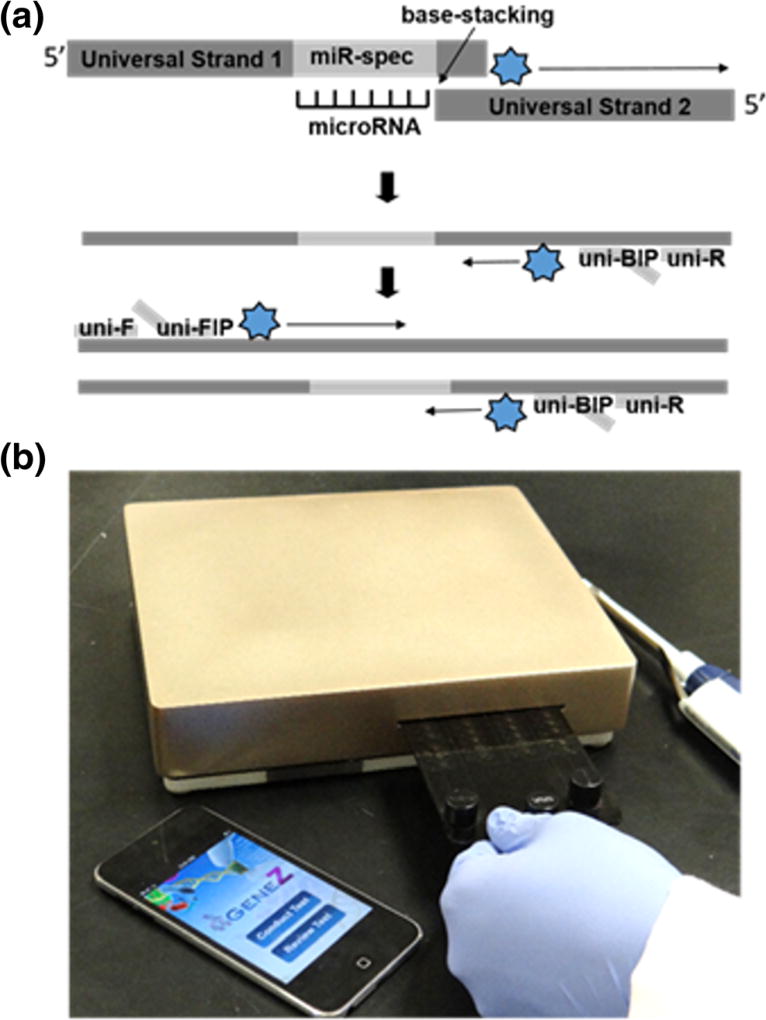
The developed isothermal technique for direct microRNA detection is a variant of a qPCR-based approach reported by Lu and coauthors (Lu et al. 2011) and enhanced by Yu and coauthors (Yu et al. 2013). In detail, the presence of microRNA promotes a base-stacking, hybridization of two universal strands and amplification under isothermal conditions (Fig. 1a). The developed approach for microRNAs was demonstrated in a conventional thermal cycler and a compact, low-cost real time isothermal amplification device (termed Gene-Z) that uses disposable microfluidic cards (Fig. 1b; Stedtfeld et al. 2012). The direct amplification method was also tested in mouse body fluids (whole blood, plasma, and faeces) using real time fluorescence and confirmed via gel electrophoresis.
Fig. 1.
a Schematic description of base-stacking microRNAs isothermal amplification. b Real-time, Gene-Z and microfluidic chip for POC microRNA quantification
2 Methods and materials
Single-stranded universal template sequences were obtained from Integrated DNA Technologies (Coralville, IA) as HPLC-purified 4 nmole ultramers and rehydrated in nuclease-free water. Loop-mediated isothermal amplification primers specific to the universal strands, were designed using Primer Explorer Software V4 and obtained from Integrated DNA Technologies (Table 2). A 10X primer mix was created with 16 µM forward inner primer (FIP) and backward inner primer (BIP) and 2 µM forward (F) and reverse (R) primers. The final 2X reaction included 1X primer mix, 2X Isothermal Buffer (New England Biolabs; Ipswich, MA), 0.28 mM dNTPs (Invitrogen; Carlsbad, CA), 1.6 mM Betaine solution (Sigma-Aldrich; St. Louis, MO), 12 mM MgSO4 (New England Biolabs; Ipswich, MA) and sterile water (Thermo Fisher Scientific; Waltham, MA). A final 10 µl isothermal amplification reaction contained 1X reaction mix, 16 units Bst 2.0 Polymerase (New England Biolabs, Ipswich, MA), 20 µM SYTO82 orange fluorescent nucleic acid stain (Invitrogen), 3.75% formamide (Sigma-Aldrich; St. Louis, MO), 4 µg bovine serum albumin (BSA; New England Biolabs; Ipswich, MA), 0.4% Pluronic F-68 (Life Technologies; Carlsbad, CA), 1 µl clinical sample or water, 0.25 µM universal strands, and 1 µl microRNA.
Table 2.
Isothermal microRNA assay sequences, including universal strands, primers and oligos used in validation experiments
| Name | Sequence 5′-3′ |
|---|---|
| miR-141 Specific Strand 1* | TGCTTAATGCTTTGATCGGCCTTGAGCACCATAAGGCAACCACCACAGAAGTATTTAAATGG GATGGGGAAAAAAGGCTATTCCCAG CCATCTTTACCAGACAGTGTTA GGTCG |
| miR-92 Specific Strand 1* | TGCTTAATGCTTTGATCGGCCTTGAGCACCATAAGGCAACCACCACAGAAGTATTTAAATGG GATGGGGAAAAAAGGCTATTCCCAG ACAGGCCGGGACAAGTGCAATA GGTCG |
| miR-141 Specific Strand 1* (Lower GC Content Overhang) | TGCTTAATGCTTTGATCGGCCTTGAGCACCATAAGGCAACCACCACAGAAGTATTTAAATGG GATGGGGAAAAAAGGCTATTCCCAG CCATCTTTACCAGACAGTGTTA TGTCG |
| miR-141 Specific Strand 1* (7 bp Overhang) | TGCTTAATGCTTTGATCGGCCTTGAGCACCATAAGGCAACCACCACAGAAGTATTTAAATGG GATGGGGAAAAAAGGCTATTCCCAG CCATCTTTACCAGACAGTGTTA GGTCGCA |
| miR-141 Specific Strand 1* (4 bp Overhang) | TGCTTAATGCTTTGATCGGCCTTGAGCACCATAAGGCAACCACCACAGAAGTATTTAAATGG GATGGGGAAAAAAGGCTATTCCCAG CCATCTTTACCAGACAGTGTTA GGTC |
| Universal Strand 2 | CACTTCCTTAGACATGAGCTATACGACGAGCTAAATCTTGATCGCCTAGGGTCATGTTCTTCGACC |
| Universal Strand 2 (Lower GC Content Overhang) | CACTTCCTTAGACATGAGCTATACGACGAGCTAAATCTTGATCGCCTAGGGTCATGTTCTT CGACA |
| Universal Primer F3 | TGCTTAATGCTTTGATCGG |
| Universal Primer B3 | CACTTCCTTAGACATGAGCT |
| Universal Primer FIP | CTGGGAATAGCCTTTTTTCCCCACTTGAGCACCATAAGGCAA |
| Universal Primer BIP | AAGAACATGACCCTAGGCGAATACGACGAGCTAAATCTTGA |
| Has-miR-141-3p | UAACACUGUCUGGUAAAGAUGG |
| Has-miR-29a-3p | UAGCACCAUCUGAAAUCGGUUA |
| Has-miR-92 miR-8 Family** | UAUUGCACUUGUCCCGGCCUGU |
| Has-miR-141-3p | UAACACUGUCUGGUAAAGAUGG |
| Has-miR-200a-3p | UAACACUGUCUGGUAACGAUGU |
| Has-miR-200b-3p | UAAUACUGCCUGGUAAUGAUGA |
| Has-miR-200c-3p | UAAUACUGCCGGGUAAUGAUGGA |
| Has-miR-429 | UAAUACUGUCUGGUAAAACCGU |
Underline represents microRNA-complementary regions. Italics represents overhangs
Underline represents nucleotides that differ from miR-141-3p.
Reactions in the commercial real-time PCR machine were conducted in 96-well plates. Cycling included 58 s incubation at 65 °C followed by a plate read, and repeated for 60 cycles. For reactions in the microfluidic chip, primers were dispensed into wells, dried at 70 °C for 5 min, and sealed with optical adhesive film (Stedtfeld et al. 2015) (Applied Biosystems; Foster City, CA). Pluronic and BSA were not used in experiments conducted in the microfluidic chip. Reaction mixture was injected into the chip and the chip was inserted into Gene-Z™. After reaching 65 °C, fluorescence was measured every 16 s for 60 min (Stedtfeld et al. 2012). The sample is then added to the reaction mixture, mixed and then injected into the chip using a pipette. Once completed, raw data is emailed from the iPod to a PC for data processing and analysis.
Gene-Z performance has been demonstrated for many applications including bacterial pathogens important to water safety, human health, and assessing bioremediation performance in contaminated acquifers (Kostic et al. 2015; Stedtfeld et al. 2014). The work described here is its first demonstration for microRNA detection. Features of this real time device include: i) simple microfluidic chips consisting of up to 64 reaction wells each with 1 to 20 µl (Stedtfeld et al. 2015) ii) real time monitoring of amplification in less than 1 h, iii) potential for wireless communication, automated data processing and reporting using a smartphone user interface, and iv) a hand-held format with internal rechargeable battery.
Experiments involving samples collected from mice were conducted in compliance with relevant laws and institutional guidelines, under Animal Use Form (AUF) Approval No. 02/14–030-00. Fecal pellets were collected, frozen, and stored at −80 °C. prior to direct amplification, fecal pellets were hydrated with nuclease free water, crudely lysed using a pestle, and vortexed for 1 min to homogenize. A portion of the whole blood was used for collection of plasma, via centrifugation at 2000 × g for 10 min.
To verify true positive amplification in blood samples spiked with microRNA, reactions were also analyzed using 1% agarose gel electrophoresis in 1 × TAE buffer at 110 Vat room temperature for 1 h. The gels were stained with SYBR safe, and images were captured using a smartphone.
All data was analyzed by calculating the signal to noise ratio (SNR). For the commercial real time PCR machine, the SNR was defined as a ratio between the differences in signal from the mean background to the difference in signal from the maximum signal. The amplification time was thus defined as the time the SNR crossed a threshold of 0.1. The amplification time (Tt) was defined as the time where the signal crossed a threshold of 5. The difference in amplification time between the target assay and the no template control (NTC) was defined as ΔTt.
3 Results
Base-stacking isothermal amplification method
The isothermal amplification method includes two universal strands, approximately 100 and 80 nucleotides in length, one that has a complementary region to the microRNA of interest (Fig. 1a). The other is a universal strand that can be used for detection of any microRNA. Directly adjacent to the microRNA-specific region at the 5′ end of the left strand is a five nucleotide long overhang sequence that is complementary to the first five nucleotides on the 3′ of the right strand. When the target microRNA is present in the reaction, it stabilizes the heterodimer (based on nearest neighbor base stacking interactions), thus allowing the binding of the overhangs to occur. When it is not present, annealing conditions are not favored – especially at the high reaction temperature (65 °C), thus resulting in a significant delay in amplification time. Primers for the isothermal amplification were designed according to requirements for a one h loop-mediated isothermal amplification (LAMP) (Mori et al. 2001; Notomi et al. 2000), which utilizes Bst polymerase, an enzyme minimally inhibited by complex sample components (Koloren et al. 2011; Stedtfeld et al. 2015; Stedtfeld et al. 2014). The forward primer (F) and forward-inner-primers (FIP) have the same sequences as universal strand 1, so are unable to bind and begin amplifying until the overhang connection has allowed the formation of its complementary strand. The same is true for the reverse primer (R) and the backward-inner primer (BIP) for universal strand 2 (Table 2).
Method optimization
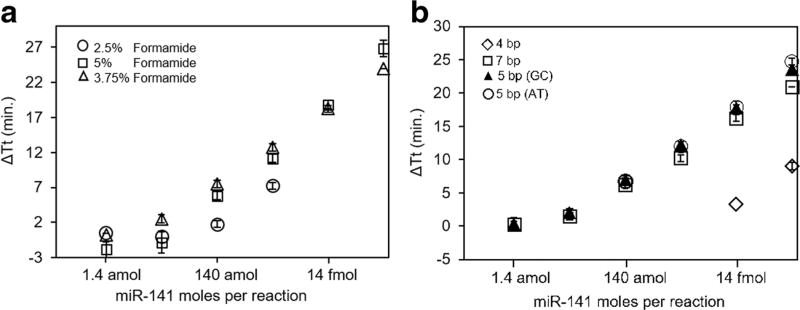
Two aspects of the isothermal amplification reaction chemistry were systematically tested including overhang length and reaction chemistry. An overhang length of five nucleotides was determined to the optimal length to increase ΔTt (Fig. 2a). This overhang length was similarly observed to be optimal for the reverse transcription-free qPCR method developed by Lu and coauthors (Lu et al. 2011). The addition of formamide in DNA amplification reactions lowers the melting temperature of DNA by ~2.4 °C/mol of formamide and is destabilizing (Blake and Delcourt 1996). In our system, it greatly increased the ΔTt at 3.75% (Fig. 2b) but increased the amplification time at concentrations higher than 5%. Formamide concentrations above 7% resulted in no amplification. In addition, loop primers, which are typically used to decrease the amplification time (Nagamine et al. 2002), were not used as they also decreased the ΔTt.
Fig. 2.
Optimization of the base-stacking microRNA isothermal amplification approach. a Standard curves with formamide concentrations of 2.5%, 3.75%, and 5%. b Standard curves obtained with various overhang lengths (four, five, and seven nucleotides) and five base overhang with AT content or GC content. No amplification was observed at 7% and above. Points represent average and error bars are standard deviation of three technical replicates
Sensitivity and specificity
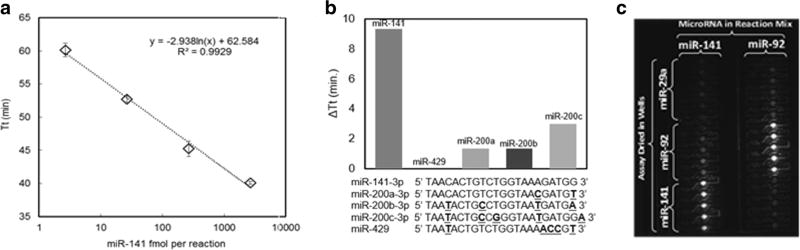
To validate the optimized method, a dilution series of miR-141 was prepared with four concentrations. Tested on a commercial real time cycler (used isothermally), the dynamic range was 1.4 fmol – 1.4 pmol per 10 µl reaction (Fig. 3a). No template control (NTC) amplified after 65 min but amplification time compared to 1.4 fmol was significantly higher (p value <0.05). Thus a detection limit of 1.4 fmol was observed using this technique with the miR-141 assay.
Fig. 3.
Sensitivity and specificity of base-stacking isothermal amplification method targeting miR-141. a Dynamic range of isothermal amplification assay with miR-141 from 1.4–1400 fmol/reaction. b Specificity of the miR-141 assay when tested with closely related members of the miR-8 family (14 fmol each). Points represent average and error bars are standard deviation of three technical replicates. c Specificity is shown via imaging the Gene-Z microfluidic chip with a CCD
Specificity is crucial to the development of microRNA assays. Some amplification-based strategies that employ intercalating dyes may be unable to correctly differentiate one nucleotide mismatches, particularly when located near the 5′ end (Shen et al. 2015). To assess specificity among closely related microRNAs, a universal strand specific to miR-141 was tested with members of the miR-8 family, which differ by 1–6 nucleotides, including miR-200a, miR-429, miR-200a, miR-200b, and miR-200c (Fig. 3b). The concentration of microRNAs used for the specificity experiments was 14 fmol. There was no difference between the no template control (NTC) and the assay targeting miR-429, while the ΔTt ranged from 1.3 to 3 min. A ΔTt of 10 min was observed for miR-141 compared to the NTC. Thus, the assay is indeed specific to miR-141.
Serial dilutions of microRNA were also tested on the Gene-Z device. The chip was divided into 8 groups (n = 4 wells each) and was preloaded with a ten-fold dilution series of the miR-141, and one group (n = 4 wells) was loaded without template to serve as the NTC. Results in Gene-Z were similar to the conventional thermal cycler with a significantly different ΔTt down to 1.4 fmol per reaction compared to the NTC. In a specificity experiment on the microfluidic chip used in Gene-Z, assays targeting miR-141 and miR-92 amplified in respective wells in which target microRNA was loaded. (Fig. 3c). Thus specificity and sensitivity were maintained for assays tested using the Gene-Z device.
Direct amplification of microRNA from body fluids
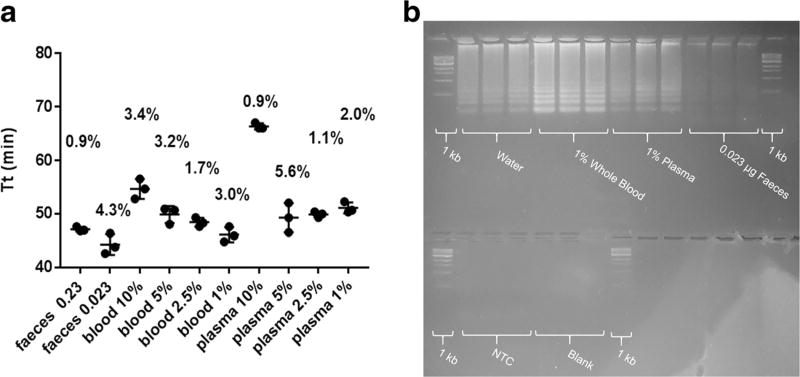
To test the suitability of the developed methodology for use with clinical samples, miR-141 was spiked into hydrated faeces, whole blood, and plasma. The amount of body sample used per reaction was also tested for each matrix. Complete inhibition was only observed at higher concentrations of faeces (23 and 2.3 µg/µl; Fig. 4a). Complete inhibition was not observed for any of the tested concentrations of whole blood and plasma though a slight delay in amplification time was observed in reactions with more blood and plasma. The NTC control was also delayed in higher concentrations of blood and plasma per reaction indicating that assay sensitivity was not influenced by direct amplification. For samples spiked with miR-141, gel electrophoresis of the reaction after 48 min of incubation further confirms correct amplification in body sample matrices (Fig. 4b). Though still positive amplification product, the signal intensity of the amplicons on the gel was lower for faeces compared to the signal intensities for amplicons for whole blood and plasma.
Fig. 4.
Precision and gel electrophoresis verifying utility of direct isothermal amplification in body samples spiked with miR-141. a Amplification time (Tt) results after spiking 140 fmol miR-141 into different body sample matrices. Final concentration indicates amount of body matrix added to the amplification reaction (feaces indicate µg per µl). Error bars indicate standard deviation of three techinal replicates and CV indicates coeffiecint of variation (%). b Gel electrophoresis 48 min of incubation of base-stacking isothermal amplification for body fluids spiked with or without miR-141
4 Discussion
The described method appears to be well-suited for detection of microRNA via direct amplification from unprocessed biological samples, as Bst polymerase is less influenced by inhibition compared to qPCR (Koloren et al. 2011; Stedtfeld et al. 2015; Stedtfeld et al. 2014). To our knowledge, this is the most rapid method (< 60 min) described for direct quantitative detection of microRNAs in body fluids (Table 1). The time to result was mainly achieved by eliminating sample processing and RNA extraction. A majority of previously described methods for microRNA require RNA extraction, ligation, and other steps that involve opening tubes and sample manipulation (Table 1). Two other studies described direct isothermal amplification; one of which required an incubation time of 4 h (Li et al. 2013). The other method appears to be comparable in terms of hands-on-time, and was tested with cell extracts from lung; however, both precision and detection limits were questionable in that replication was not described, nor was a subsequent proof of amplification other than real-time fluorescence curves (Li et al. 2013).
A sensitivity of 1.4 fmol per reaction was observed using our base-stacking isothermal amplification method, which is comparable to other direct isothermal amplification studies (Table 1). Although sensitivity is important, it is not the most vital factor determining the clinical utility of a microRNA assay. For example, a single cell of human tissue may contain anywhere from 100,000–500,000 copies (0.5–1 amol) of total microRNAs or 0–40,000 copies (0–0.1 amol) of a single microRNA (Liang et al. 2007). This suggests that only ~103 cells would be required to measure microRNAs in higher abundance. Using a larger sample volume is always a possibility with human samples. For example, to measure 2.38 × 106 copies (4 amol) miR-224 per µl urine required to distinguish Type 1 diabetes mellitus patients from healthy controls (Bacon et al. 2015), approximately 350 µl of urine will be required.
Precision is a more important factor as the fold difference between diseased and cohort samples may be smaller for some microRNA/disease associations. Targeting mR-141 with our method, the slope of the standard curve was 6.74 min per 10 fold difference in template concentration. This slope is 2 times higher compared to conventional methods of qPCR (which is typically 3.3 cycles per 10 fold change in template concentration). This allows greater distinction between levels of microRNA. The coefficient of variation among all tested samples was also low (mean CV = 2.6%), which is near or below the CV observed in other studies using isothermal amplification following DNA extraction (Bosward et al. 2016; Brotons et al. 2016) or directly from groundwater samples (Stedtfeld et al. 2016).
Implications of having a direct isothermal approach for measuring microRNAs are far reaching. For example, oral cancer is among the leading causes of death in India due to use of chewing tobacco and betel nuts (Sen et al. 2002; Sinha et al. 2016). Thus, a simple microRNA-based test for early detection of oral cancer may influence timely treatment. Although the benefit of prostate specific antigen (PSA) assay are being questioned routinely, more than 30 million PSA tests are still conducted in the United States alone (Andriole et al. 2009). MicroRNA markers such as miR-141 reported by Mitchell and coauthors appears to be an extremely viable marker for prostate cancer (Mitchell et al. 2008). Using a rapid and easy to use direct amplification method, marker validation studies could potentially be extended to larger populations at a significantly lower cost.
Based on the review of literature presented in Table 1, an important observation was related to the usefulness of a NTC. It was noted that nearly all isothermal microRNA amplification approaches result in NTC amplification over an extended incubation period (Li et al. 2011; Ma et al. 2014). Thus, to establish the clinical utility of isothermal methods at the limit of detection, inclusion of NTC along with the standard deviations of NTC as well as the lowest concentration standard is critical. This information was not always available in many reported studies and must be included as part of any isothermal amplification method development for microRNAs.
5 Conclusions and prospective
The <60 min time to results achieved with the base-stacking isothermal amplification method is among the most rapid methods currently known for quantification of microRNAs. Direct amplification of microRNAs from blood, plasma, sputum, or other body fluids could reduce the necessity for hands-on training as well as need for sophisticated instruments. Combined with comparable sensitivities (Ma et al. 2014) and an inexpensive and quantitative device (Gene-Z; Stedtfeld et al. 2012), this method highlights the potential for microRNAs- based diagnostics under limited resource settings. Further validation is warranted using clinical specimens to establish clinical sensitivity, specificity, performance, and ruggedness under field conditions.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences Superfund Research Program (NIEHS SRP P42ES004911) and the twenty-first Century Michigan Economic Development Corporation (GR-476 PO 085P3000517).
Footnotes
Compliance with ethical standards
Ethical approval All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
References
- Andriole G, Crawford D, Robert G, Buys SS, Chia D, Ph D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ, Weissfeld JL, Yokochi LA, Brien BO, Clapp JD, Rathmell JM, Riley TL, Hayes RB, Kramer BS, Izmirlian G, Miller AB, Pinsky PF, Prorok PC, Gohagan JK, Berg CD. N. Engl. J. Med. 2009;360:1310. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon S, Engelbrecht B, Schmid J, Pfeiffer S, Gallagher R, McCarthy A, Burke M, Concannon C, Prehn J, Byrne M. Genes (Basel) 2015;6:399. doi: 10.3390/genes6020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake RD, Delcourt SG. Nucleic Acids Res. 1996;24:2095. doi: 10.1093/nar/24.11.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosward KL, House JK, Deveridge A, Mathews K, Sheehy PA. J. Dairy Sci. 2016;99:2142. doi: 10.3168/jds.2015-10073. [DOI] [PubMed] [Google Scholar]
- Brotons P, de Paz HD, Esteva C, Latorre I, Munoz-Almagro C. Expert. Rev. Mol. Diagn. 2016;16:125. doi: 10.1586/14737159.2016.1112741. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Nat. Rev. Cancer. 2006;6:259. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Cancer Res. 2007;67:2456. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. Nucleic Acids Res. 2004;32:D109. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. Nucleic Acids Res. 2006;34:D140. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. Nucleic Acids Res. 2008;36:D154. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt EM, Kool ET. Nucleic Acids Res. 2012;40:1. doi: 10.1093/nar/gkr1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonstrup SP, Koch J, Kjems J. RNA. 2006;12:1747. doi: 10.1261/rna.110706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koloren Z, Sotiriadou I, Karanis P. Ann. Trop. Med. Parasitol. 2011;105:607. doi: 10.1179/2047773211Y.0000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic T, Ellis M, Williams MR, Stedtfeld TM, Kaneene JB, Stedtfeld RD, Hashsham SA. Appl. Microbiol. Biotechnol. 2015;99:7711. doi: 10.1007/s00253-015-6774-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li Z, Jia H, Yan J. Chem. Commun. 2011;47:2595. doi: 10.1039/c0cc03957h. [DOI] [PubMed] [Google Scholar]
- Li Y, Liang L, Zhang CY. Anal. Chem. 2013;85:11174. doi: 10.1021/ac403462f. [DOI] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A. Cancer Epidemiol. Biomark. Prev. 2010;19:1766. doi: 10.1158/1055-9965.EPI-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A, Becker V, Goel A, Wex T, Malfertheiner P. PLoS One. 2012;7:1. doi: 10.1371/journal.pone.0042933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li L, Duan L, Wang X, Xie Y, Tong L, Wang Q, Tang B. Anal. Chem. 2013;85:7941. doi: 10.1021/ac401715k. [DOI] [PubMed] [Google Scholar]
- Lu Z, Duan D, Cao R, Zhang L, Zheng K, Li J. Chem. Commun. (Camb.) 2011;47:7452. doi: 10.1039/c1cc10442j. [DOI] [PubMed] [Google Scholar]
- Ma C, Han D, Shi C. Chem. Commun. 2014;50:3799. doi: 10.1039/c3cc49841g. [DOI] [PubMed] [Google Scholar]
- Manikandan M, Deva Magendhra Rao AK, Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R, Munirajan AK. Mol. Cancer. 2016;15:28. doi: 10.1186/s12943-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo Y, Mie M, Suzuki S, Kobatake E. Anal. Bioanal. Chem. 2011;401:221. doi: 10.1007/s00216-011-5083-3. [DOI] [PubMed] [Google Scholar]
- Michael A, Bajracharya SD, Yuen PST, Zhou H, Star RA, Illei GG, Alevizos I. Oral Dis. 2010;16:34. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10513. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Nagamine K, Tomita N, Notomi T. Biochem. Biophys. Res. Commun. 2001;289:150. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Nagamine K, Hase T, Notomi T. Mol. Cell. Probes. 2002;16:223. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Njiru ZK. PLoS Negl. Trop. Dis. 2012;6:1. doi: 10.1371/journal.pntd.0001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Kowarsch A, Schmidl D, Buggenthin F, Brauner B, Dunger I, Fobo G, Frishman G, Montrone C, Theis FJ. Genome Biol. 2010;11:R6. doi: 10.1186/gb-2010-11-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Nishida N, Calin Ga, Pantel K. Nat. Rev. Clin. Oncol. 2014;11:145. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- Sen U, Sankaranarayanan R, Mandal S, Ramanakumar AV, Parkin DM, Siddiqi M. Int. J. Cancer. 2002;100:86. doi: 10.1002/ijc.10446. [DOI] [PubMed] [Google Scholar]
- Shao C, Yu Y, Yu L, Pei Y, Feng Q, Chu F, Fang Z, Zhou Y. Arch. Oral Biol. 2012;57:1012. doi: 10.1016/j.archoralbio.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Shen Y, Tian F, Chen Z, Li R, Ge Q, Lu Z. Biosens. Bioelectron. 2015;71:322. doi: 10.1016/j.bios.2015.04.057. [DOI] [PubMed] [Google Scholar]
- Shi C, Liu Q, Ma C, Zhong W. Anal. Chem. 2014;86:336. doi: 10.1021/ac4038043. [DOI] [PubMed] [Google Scholar]
- Sinha DN, Abdulkader RS, Gupta PC. Int. J. Cancer. 2016;138:1368. doi: 10.1002/ijc.29884. [DOI] [PubMed] [Google Scholar]
- Stedtfeld RD, Liu Y-C, Stedtfeld TM, Kostic T, Kronlein MR, Srivannavit O, Khalife WT, Tiedje JM, Gulari E, Hughes M, Etchebarne B, Hashsham SA. Biomed. Microdevices. 2015;17:89. doi: 10.1007/s10544-015-9994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld RD, Stedtfeld TM, Kronlein M, Seyrig G, Steffan RJ, Cupples AM, Hashsham AM. Environ. Sci. Technol. 2014;48:13855. doi: 10.1021/es503472h. [DOI] [PubMed] [Google Scholar]
- Stedtfeld RD, Stedtfeld TM, Samhan F, Kanitkar YH, Hatzinger PB, Cupples AM, Hashsham SA. J. Microbiol. Methods. 2016;131:61. doi: 10.1016/j.mimet.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld RD, Tourlousse DM, Seyrig G, Stedtfeld TM, Kronlein M, Price S, Ahmad F, Gulari E, Tiedje JM, Hashsham SA. Lab Chip. 2012;12:1454. doi: 10.1039/c2lc21226a. [DOI] [PubMed] [Google Scholar]
- Tricoli JV, Jacobson JW. Cancer Res. 2007;67:4553. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang K, Lv Z, Zhu X, Zhu L, Zhou F. Biosens. Bioelectron. 2014;57:91. doi: 10.1016/j.bios.2014.01.058. [DOI] [PubMed] [Google Scholar]
- Wen Y, Xu Y, Mao X, Wei Y, Song H, Chen N, Huang Q, Fan C, Li D. Anal. Chem. 2012;84:7664. doi: 10.1021/ac300616z. [DOI] [PubMed] [Google Scholar]
- Yan C, Jiang C, Jiang J, Yu R. Anal. Sci. 2013;29:605. doi: 10.2116/analsci.29.605. [DOI] [PubMed] [Google Scholar]
- Yao B, Li J, Huang H, Sun C, Wang Z, Fan Y, Chang Q, Li S, Xi J. RNA. 2009;15:1787. doi: 10.1261/rna.1555209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-Y, Yin B-C, Ye B-C. Chem. Commun. 2013;49:8247. doi: 10.1039/c3cc44125c. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang CY. Anal. Chem. 2012;84:224. doi: 10.1021/ac202405q. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Huang Q, Gao J, Lu J, Shen X, Fan C. Nucleic Acids Res. 2010;38:e156. doi: 10.1093/nar/gkq556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yin H, Li J, Li B, Li X, Ai S, Zhang X. Biosens. Bioelectron. 2016;79:79. doi: 10.1016/j.bios.2015.12.009. [DOI] [PubMed] [Google Scholar]