Summary
CRISPR-Cas systems adapt their immunological memory against their invaders by integrating short DNA fragments into clustered regularly interspaced short palindromic repeat (CRISPR) loci. While Cas1 and Cas2 make up the core machinery of the CRISPR integration process, various class I and II CRISPR-Cas systems encode Cas4 proteins for which the role is unknown. Here, we introduced the CRISPR adaptation genes cas1, cas2, and cas4 from the type I-D CRISPR-Cas system of Synechocystis sp. 6803 into Escherichia coli and observed that cas4 is strictly required for the selection of targets with protospacer adjacent motifs (PAMs) conferring I-D CRISPR interference in the native host Synechocystis. We propose a model in which Cas4 assists the CRISPR adaptation complex Cas1-2 by providing DNA substrates tailored for the correct PAM. Introducing functional spacers that target DNA sequences with the correct PAM is key to successful CRISPR interference, providing a better chance of surviving infection by mobile genetic elements.
Keywords: CRISPR adaptation, Cas4, spacer acquisition, type I-D CRISPR-Cas system
Graphical Abstract
Highlights
-
•
Cas4 facilitates the integration of PAM-compatible spacers
-
•
Spacer length variation is dictated by Cas1-2
-
•
Cas4 shortens spacer length
-
•
Cas4-selected PAMs license type I-D CRISPR interference
Kieper et al. demonstrate that the ubiquitous protein Cas4 assists Cas1 and Cas2 in the selection of new CRISPR spacers with a PAM licensing efficient CRISPR interference.
Introduction
Microbes require updating their adaptive immune repertoire to keep up with ever-changing mobile genetic elements (MGEs) such as bacteriophages and conjugative plasmids. The prokaryotic CRISPR-Cas system is an adaptive immune system that uses clustered regularly interspaced short palindromic repeats (CRISPRs) and their associated proteins (Cas) (Jansen et al., 2002, Mojica et al., 2005, Barrangou et al., 2007). In a process termed CRISPR adaptation, microbes integrate short sequences from MGEs into their CRISPR array (Amitai and Sorek, 2016, Sternberg et al., 2016, Jackson et al., 2017). This array then becomes a source for small RNAs (i.e., CRISPR RNA [crRNA]) that guide Cas nuclease complexes to their target (van der Oost et al., 2014, Marraffini, 2015, Mohanraju et al., 2016).
CRISPR-Cas systems are grouped into two major classes that each hold several types and multiple subtypes, and remarkably, all contain cas1 and cas2 genes (Koonin et al., 2017). In the type I-E system of Escherichia coli, cas1 and cas2 are necessary and sufficient to mediate expansion of the CRISPR array (Yosef et al., 2012). However, next to cas1 and cas2, a number of other cas genes in class I and II systems have been directly linked to the spacer integration process, suggesting that CRISPR adaptation across different systems has different requirements (Koonin et al., 2017). Type II-B (Cas9), type V (Cas12a), and most type I (I-A, I-B, I-C, I-D, and I-U) CRISPR-Cas systems contain cas4 genes in conserved gene clusters with cas1 and cas2 genes, while in some systems, cas4 is fused with cas1 (I-B, I-U, and V-B) (Hudaiberdiev et al., 2017). Deletion of cas4 from the I-A type abrogated CRISPR adaptation in a Sulfolobus islandicus strain overexpressing csa3, a regulator of cas gene expression (Liu et al., 2017), while deletion of cas4 in type I-B revealed that cas4 is essential for CRISPR adaptation against HHPV-2 in Haloarcula hispanica (Li et al., 2014). Additionally, interaction between the Cas1/2 fusion protein, Csa1, and Cas4 of the archaeal type I-A system was found in vitro (Plagens et al., 2012). These findings suggest a strong functional association of Cas4 and the Cas1 and Cas2 adaptation proteins. Despite the conservation of the cas4 gene among these highly diverse CRISPR-Cas systems, a functional role for Cas4 has not been shown in vivo. Early biochemical studies have found different Cas4 proteins as monomers, dimers, and decamers and containing either [2Fe-2S] or [4Fe-4S] iron-sulfur clusters (Zhang et al., 2012, Lemak et al., 2013, Lemak et al., 2014). Furthermore, Cas4 proteins were shown to be active nucleases with catalytic domains belonging to the PD-DEXK phosphodiesterase superfamily (Hudaiberdiev et al., 2017). It was suggested that the observed catalytic activities play a role in either the generation or the processing of spacer precursors; i.e., DNA substrates that are used by Cas1 and Cas2 to form spacers (Zhang et al., 2012, Lemak et al., 2013, Lemak et al., 2014). Recently, Rollie et al. showed in vitro that Cas4 cleaves 3′ overhangs of prespacer substrates containing protospacer adjacent motifs (PAMs) (Rollie et al., 2018).
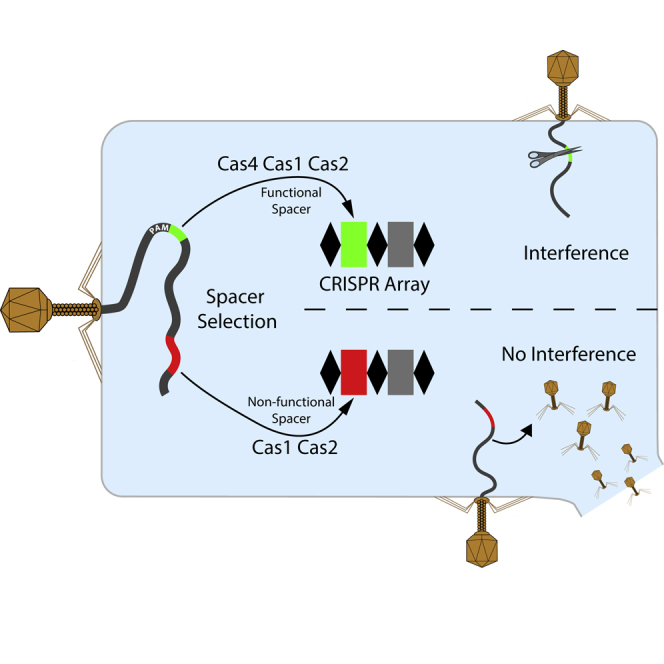
Obtaining new spacers that target an invading DNA sequence with a correct PAM is central to the success of CRISPR adaptation. The PAM is a short sequence motif (Deveau et al., 2008, Mojica et al., 2009, Shah et al., 2013) that is required for crRNA-effector complexes such as Cascade, Cas9, and Cas12a to find their target DNA and avoid targeting host CRISPR arrays (Mohanraju et al., 2016). Only when a new spacer has been selected from a target adjacent to a PAM can CRISPR interference efficiently take place. In type II systems, Cas9 assists Cas1-2 to select PAM-compliant spacers (Heler et al., 2015), but it remains unknown what other factors also contribute to PAM selection.
Here, we have determined the biological role of Cas4 by employing in vivo spacer acquisition assays in a heterologous E. coli host. We show that the type I-D adaptation proteins Cas1 and Cas2 from the cyanobacterium Synechocystis sp. 6803 are necessary and sufficient to integrate spacers into the CRISPR array. However, providing cas4 results in a significant enrichment of new spacers with PAM motifs that support CRISPR interference in the type I-D CRISPR-Cas system of the native host Synechocystis. Altogether, our results demonstrate that Cas4 enhances functional memory formation, which increases the chance of surviving infections by MGEs.
Results
The Cas1-Cas2 Complex Integrates Spacers Independently of Cas4
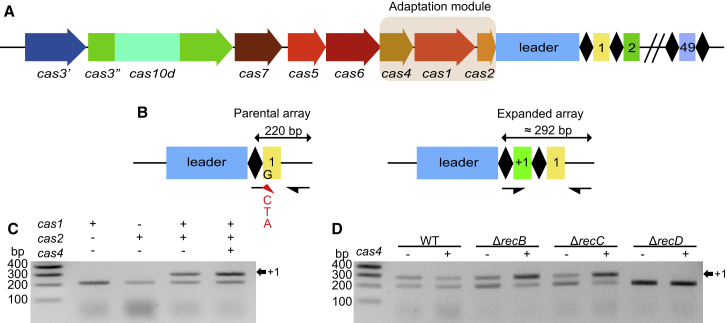
To determine the minimal requirements for spacer acquisition in the type I-D system, we cloned cas4, cas1, and cas2 genes from Synechocystis (Figure 1A) into T7-based expression vectors. A minimal CRISPR array with one repeat was obtained by cloning the full-leader sequence followed by the first repeat and the leader proximal spacer (Sp1) of the type I-D system into the pACYC-Duet1 vector system. The resulting plasmids were co-transformed into a wild-type (WT) E. coli K12 strain (BW25113) devoid of T7 RNA polymerase. This setup ensured constitutive and low expression levels of the adaptation genes from Synechocystis. We first tested the ability of the cells to integrate new spacers into the minimalized CRISPR array either in the presence or absence of the cas4-1-2 genes. With a sensitive PCR approach (Figure 1B), spacer acquisition was readily detectable in the presence of cas1-2 regardless of the presence of the cas4 gene (Figure 1C). Further, deletion of either cas1 or cas2 abolished spacer integration, indicating that the combination of cas1 and cas2 is necessary and sufficient to mediate the integration of new spacers (Figure 1C). The detection of expanded CRISPR arrays in E. coli K12 demonstrates that spacers were acquired even though E. coli is not the natural host of the type I-D system. Consequently, the type I-D adaptation module does not rely on any specific host factors only present in Synechocystis.
Figure 1.
Genetic Requirements of Type I-D CRISPR Adaptation
(A) Overview of the type I-D CRISPR-Cas locus as found on the Synechocystis 6803 pSYSA megaplasmid. The putative adaptation module consisting of cas4-1-2 is highlighted in light brown. Downstream of cas2 is the leader sequence in blue, followed by the I-D array consisting of 37-bp repeats interspaced by 49 spacers with a statistical mode length of 35 bp (Figure S2C).
(B) Degenerate primer PCR used for the detection of spacer acquisition (Heler et al., 2015). The 3′ end of the forward primer mix mismatches the first 5′ nucleotide of spacer 1 (indicated in red). Spacer integration restores complementarity allowing for efficient amplification. For more sensitive detection, the amplicon of expanded arrays was extracted and subjected to a second round of PCR (see Figure S1A).
(C) Co-expression of cas1 and cas2 is necessary and sufficient for the integration of new spacers.
(D) Assessing spacer integration in WT E. coli K12 and different recBCD mutant backgrounds in the presence or absence of cas4. The presence of cas4 enhances spacer integration in the ΔrecB and ΔrecC genotypes, while spacer integration is below the detection limit of this PCR (described in B) in the ΔrecD mutant regardless of the presence of cas4.
Cas4 Enhances Spacer Acquisition in the Absence of the RecBCD Complex
Next, we were interested in knowing whether the observed integration was dependent on the presence of host factors in our heterologous expression system. Since the E. coli strain used is not the natural host of the type I-D system, CRISPR adaptation by I-D Cas1-2 does not rely on cyanobacterial factors that are only present in Synechocystis. For the type I-E system of E. coli, a role for the RecBCD complex has been proposed in generating spacer precursors during double-stranded DNA break repair at stalled replication forks (Ivančić-Baće et al., 2015, Levy et al., 2015). The Synechocystis genome contains cyanobacterial orthologs of E. coli RecB and RecD, but RecC appears to be absent (Cassier-Chauvat et al., 2016). Hence, we sought to assess spacer integration in E. coli ΔrecB, ΔrecC, and ΔrecD mutant backgrounds from the KEIO collection (Baba et al., 2006). While we observed no difference in spacer acquisition frequencies for pCas1-2 in recB and recC deletion mutants, integration of spacers in the recD mutant was greatly reduced (Figure 1D) but could still be detected with the sensitive spacer detection approach (Figure S1B). Interestingly, when we supplied cas4 in the recB and recC deletion backgrounds, we observed a relative increase of array expansion (Figure 1D). The results demonstrate that Cas1-2 is the core requirement for type I-D adaptation as has been found for type I-E (Nuñez et al., 2014, Yosef et al., 2012). The presence of Cas4 seems to facilitate uptake of spacers in the absence of RecB or RecC, which is consistent with competing pathways for the generation of spacer precursors.
Cas4 Influences Spacer Length
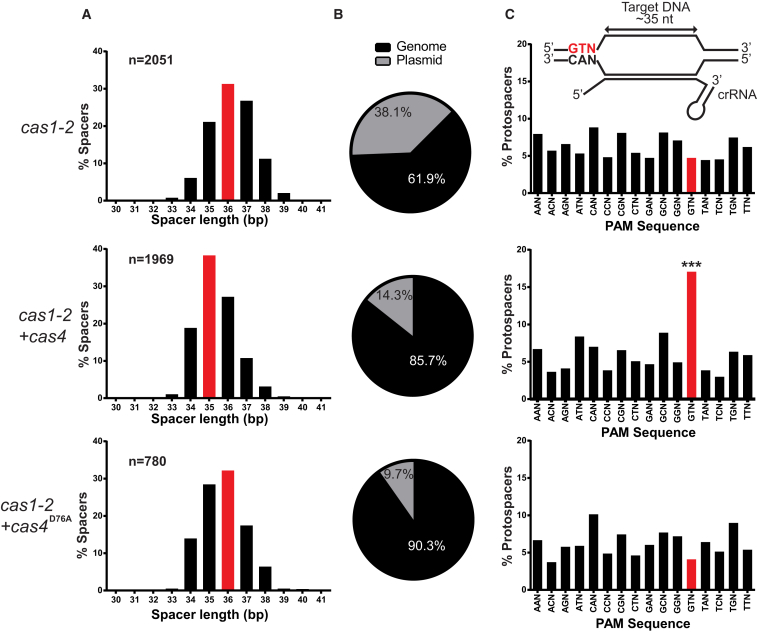
To understand the nature and origin of newly acquired spacers in the presence or absence of cas4, we subjected amplicons of expanded I-D arrays to next-generation sequencing. Analysis of novel spacers in the absence of cas4 revealed that spacers of 36 bp length were incorporated most frequently (Figure 2A). This length deviates by one nucleotide from the spacer length found in the native CRISPR array of Synechocystis in which the statistical mode of spacer length is 35 bp (Figure S2C). Interestingly, when cas4 was supplied to the system, the mode of spacer length was restored to 35 bp. To assess if Cas4 activity was responsible for the change in spacer length, we created an active site mutant in the RecB-domain by substituting a divalent metal-ion binding aspartic acid for alanine (i.e., D76 corresponding to D99 in Sso0001) (Zhang et al., 2012). When this mutant was introduced in strains containing pCas1-2, the same spacer length mode was observed as when cas4 was absent, showing that the catalytic activity of Cas4 influences spacer length. Furthermore, it suggests that Cas4 is involved in processing spacer precursors (i.e., prespacers) before they are integrated into the CRISPR array.
Figure 2.
Analysis of Spacers Acquired by Type I-D CRISPR Adaptation
(A) Spacer length distribution in cells harboring different combinations of cas genes. The variation in spacer size does not depend on the presence of cas4 but is solely dependent on the Cas1-2 adaptation complex. The presence of cas4 restores the statistical mode of spacer length as found in the native I-D array (35 bp; Figure S2C). Statistical mode of spacer length is indicated with red bars.
(B) Origin of newly acquired spacers. The type I-D adaptation proteins acquire mostly from genomic DNA.
(C) Percentage of protospacers with different PAMs. The presence of cas4 significantly increases the incorporation of spacers that match protospacers with the consensus GTN PAM, while no significant enrichment of PAMs is observed for spacers acquired by Cas1-2 alone or in conjunction with the Cas4D76A mutant. n, number of analyzed spacer sequences; significance level α = 0.001.
New Spacers Are Mostly Genome Derived
Next, we mapped the unique spacer sequences to the E. coli BW25113 genome as well as to the plasmids harbored by the cells. Approximately 60% of the spacers that were acquired in the absence of the cas4 mapped to the genome (Figure 2B). We observed increased numbers of spacers targeting the lacI gene, which is present both on the plasmid and the genome. Spacers were also preferentially acquired from the chromosomal replication terminus terC (Figure S3). The enrichment of spacers at the replication terminus is similar to what has been observed previously for type I-E (Levy et al., 2015) and suggests that the I-D Cas1-2 adaptation complex can use DNA degradation products from RecBCD as substrates for new spacers. When we supplied WT or mutant cas4, spacer acquisition from the genome further increased to 85% and 90%, respectively. However, the preferential uptake of spacers from terC was lost (Figure S3). We observed no orientation bias of the newly integrated spacers for either strand of the genome (Table S3). Although E. coli is not the native host of the I-D CRISPR system, the results are consistent with the notion that the adaptation proteins of I-D use prespacer substrates from abundant DNA sources in the cell (in this case, the genome).
Cas4 Facilitates Selection of Spacers with a Specific PAM
In order to determine which PAMs had been selected during spacer acquisition, we mapped the unique spacers to their targets and retrieved their flanking sequences. This revealed that in the absence of cas4 no particular sequence motifs were enriched in the flanking regions of the target. Interestingly, when we analyzed upstream flanking sequences of targets from spacers acquired in the presence of cas4, we observed that spacers with GTN PAMs were significantly enriched (Figure 2C). This GTN PAM matched the previously predicted PAM for I-D systems (Shah et al., 2013). When we introduced cas4D76A, the enrichment of GTN PAMs was no longer observed, indicating that the metal-ion coordinating residue, which is likely important for catalytic activity of Cas4, is also essential for PAM selection. Cas1-2 alone displays no inherent PAM selection preference. In order to assess whether inactivation of the recB gene would reduce background levels of spacers derived from RecBCD products, we subjected the expanded arrays from the recB mutant to high-throughput sequencing. Although this genetic background did not abolish background spacer integration, the presence of cas4 further increased GTN-PAM-compliant spacers (Figures S2A and S2B; Table S3).
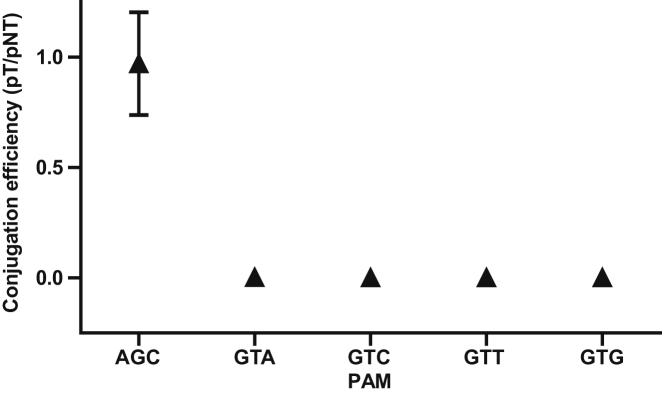
GTN Is a Functional PAM in the Native Type I-D Host Synechocystis
To test whether the GTN PAM enriched in the presence of cas4 licenses CRISPR interference in Synechocystis, we performed interference assays using a conjugative plasmid containing a protospacer matching spacer 1 of the type I-D array. The protospacer was flanked by one of the four GTN PAMs (GTA, GTC, GTG, or GTT) and carried gentamicin resistance for selection. Compared to a non-target control plasmid, we observed a dramatic reduction in the numbers of transconjugants with each of the four possible GTN PAMs (i.e., no transconjugants for GTC, GTG, and GTT and 1 for the GTA PAM) (Figure 3). In contrast, plasmids containing protospacers flanked by AGC PAMs resulted in the same conjugation efficiency as found for the non-target control. We conclude that the type I-D system in Synechocystis is active and provides efficient CRISPR interference with GTN PAMs.
Figure 3.
Conjugation Efficiency of Target Plasmids Carrying either AGC-Protospacer1 or GTN-Protospacer1 in Synechocystis 6803
The AGC PAM does not license interference, while plasmids containing the observed GTN PAM are efficiently cleared. Data points represent mean ± SE (n = 3).
Discussion
Microbes face a number of challenges when they update their CRISPR memory. First, how can they select new spacers from invading elements while preventing sampling from their own genome? Second, how can they maintain a balance between spacer uptake and turnover? Third, how can they select spacers that give functional CRISPR interference? Here, we addressed the last question and show that the highly ubiquitous Cas4 protein present in type I, II, and V systems helps to integrate spacers targeting DNA sequences with PAMs that support CRISPR interference. Because crRNA-effector complexes such as Cascade, Cas9, and Cas12a are critically reliant on PAMs to find their target DNA and to avoid host CRISPR arrays, the selection of PAM-compliant spacers enhances the success rate of CRISPR interference and promotes clearance of invader DNA from cells (Mohanraju et al., 2016). Apart from influencing the PAM, we found that the statistical mode of spacer length was shifted by 1 nt to shorter spacers, suggesting a role for Cas4 in processing spacer substrates before or during integration. The variable spacer size itself is dictated only by Cas1-2 from type I-D. While structural constraints of the Cas1-2 complex from type I-E, and presumably also of the Cas1-2/3 complex from type I-F, act like a molecular ruler that predetermines a fixed spacer length of predominantly 32 nt (Nuñez et al., 2015, Wang et al., 2015, Fagerlund et al., 2017, Rollins et al., 2017), the integration complex of the type I-D system likely displays plasticity and enables incorporation of spacers that vary in size by 5 or 6 nucleotides. This spacer size variation is observed not only in type I-D but also in type I-B (Li et al., 2017) and many CRISPR systems containing cas4 genes. Our data are consistent with a model in which the nuclease activities of Cas4 tailor prespacer substrates for the Cas1-2 adaptation machinery during the integration of new spacers.
The cas4 gene has long been implicated in CRISPR adaptation. Many cas4 genes have been found adjacent to cas1 and cas2, and in some cases, fusions between cas4 and cas1 have been observed (Hudaiberdiev et al., 2017, Koonin et al., 2017). The cas4 gene was shown to be essential for CRISPR adaptation in the type I-B system of Haloarcula (Li et al., 2014). Interestingly, a Campylobacter bacteriophage containing a cas4 gene was reported to promote acquisition of self-targeting spacers in the Campylobacter type II-C system (Hooton and Connerton, 2015), which is in line with our finding that Cas4 promotes the integration of spacer from abundant DNA populations in the cell. The Cas4 protein has been observed in a complex with a Cas1-Cas2 fusion protein and Csa1 in the Sulfolobus type I-A system and this complex was coined Cascis after CRISPR-associated complex for integration of spacers (Plagens et al., 2012). The Cas4 protein of the Bacillus halodurans type I-C system forms a tight heterohexameric complex with Cas1 consisting of two Cas1 dimers and two Cas4 subunits (Lee et al., 2018). Although different catalytic activities have been assigned (Zhang et al., 2012, Lemak et al., 2013, Lemak et al., 2014, Hudaiberdiev et al., 2017), the biological role of Cas4 has remained elusive. Only recently it was shown that Cas4 nuclease activity participates in PAM-dependent cleavage of 3′ overhangs of prespacers (Rollie et al., 2018). Furthermore, Lee et al. demonstrate that this PAM-dependent cleavage of prespacers ensures that only functional spacers are integrated into the CRISPR array (Lee et al., 2018). This sequence specific cleavage is in line with the findings presented in this study in which Cas4-derived spacers are shorter and enriched in functional GTN PAMs.
While Cas4 may aid the generation of spacers with the correct PAM in a number of CRISPR-Cas systems, some other Cas proteins have been found to influence PAM selection as well. The crRNA-guided effector complex Cas9 present in type II-A systems is required for spacer acquisition (Wei et al., 2015) and helps to select new spacers with a correct PAM (Heler et al., 2015). Next to Cas1-2, the integration of new spacers in type II-A requires the toroidal DNA binding protein Csn2, a protein known to interact with Cas1 (Heler et al., 2015, Ka et al., 2016).
Other ways to improve taking up spacers with the correct PAM include primed CRISPR adaptation (Datsenko et al., 2012, Swarts et al., 2012), which appears to be a general feature of type I systems only (Li et al., 2014, Rao et al., 2016, Staals et al., 2016, van Houte et al., 2016). In contrast to naive spacer acquisition, primed CRISPR adaptation uses preexisting spacer matches to trigger updates of the CRISPR memory against that target. Apart from Cas1-2, the priming process requires the presence of a crRNA-effector complex (e.g., Cascade) and the DNA nuclease Cas3. It seems that the frequency of acquiring functional spacers is much higher during priming than during naive spacer acquisition (Jackson et al., 2017). This can be partly explained by considering that functional spacers confer a selective advantage to the host when the interference machinery is present. On the molecular level, the increased frequency of functional spacers during priming may be explained by the observation that the Cas3 nuclease cleaves target DNA in a PAM-compatible manner to fuel the Cas1-2 adaptation machinery with suitable DNA substrates for integration (Künne et al., 2016).
Taken together, a picture has emerged that it is important for microbes to acquire functional instead of randomly selected spacers in their CRISPR arrays and that there is a variety of ways in which CRISPR systems can accomplish this. The conserved component Cas4, which is present in about half of all CRISPR subtypes, appears to be a Cas protein dedicated to the task of facilitating the integration of functional spacers during CRISPR adaptation.
Experimental Procedures
Bacterial Strains and Growth Conditions
E. coli strains DH5α, BW25113 (WT), JW2788 (BW25113 ΔrecB), JW2790 (BW25113 ΔrecC), and JW2787 (BW25113 ΔrecD) were grown in lysogeny broth (LB) at 37°C and continuous shaking at 180 rpm or grown on LB agar plates (LBA) containing 1.5% (wt/vol) agar. Synechocystis 6803 was cultivated as described previously (Scholz et al., 2013). When required, the media were supplemented with 100 μg/mL ampicillin, 50 μg/mL spectinomycin, 25 μg/mL chloramphenicol, and 7.5 μg/mL gentamicin (see Table S1 for plasmids and corresponding selection markers).
Plasmid Construction and Transformation
Plasmids used in this study are listed in Table S1. All cloning steps were performed in E. coli DH5α. Primers described in Table S2 were used for PCR amplification of the type I-D CRISPR-Cas locus (cas4, cas1, cas2, and leader-repeat-spacer1) from Synechocystis cell material using the Q5 high-fidelity Polymerase (New England Biolabs). PCR amplicons were subsequently cloned into Berkeley MacroLab ligation-independent cloning (LIC) vectors (http://qb3.berkeley.edu/macrolab/addgene-plasmids/) using either LIC or into the pACYCDuet-1 vector system (Novagen, EMD Millipore) using conventional restriction-ligation cloning. The cas4D76A mutant (Zhang et al., 2012) was obtained using a PCR-based mutagenesis using primers listed in Table S2. The conjugative plasmid pVZ322 used in the interference study was obtained by fusing the 5′ PAM (GTA, GTT, GTG, GTC, and AGC)-protospacer1 3′ sequence in-frame with a gentamicin resistance cassette upstream of its stop codon using inverse PCR using primers listed in Table S2. The gentamicin resistance cassette with and without the PAM-protospacer sequence (pT and pNT, respectively) was then assembled with the linearized pVZ322 backbone. All plasmids were verified by Sanger sequencing (Macrogen Europe, Amsterdam, the Netherlands; GATC Biotech, Konstanz, Germany). Bacterial transformations were either carried out by electroporation (2.5 kV, 25 mF, 200 V) using a ECM 630 electroporator (BTX Harvard Apparatus) or using chemically competent cells prepared according to manufacturer’s manual (Mix&Go, Zymo research). Electrocompetent cells were prepared following a protocol adapted from Gonzales et al. (2013). Transformants were selected on LBA supplemented with appropriate antibiotics.
In Vivo Spacer Acquisition Assay
E. coli BW25113 and E. coli mutant strains JW2788 (BW25113 ΔrecB), JW2790 (BW25113 ΔrecC), and JW2787 (BW25113 ΔrecD) were transformed with pCas1-2, pCRISPR, and pCas4, pCas4D76A, or the pEmp control plasmid (Table S1). Cultures were inoculated from single colonies and passaged once after 24 hr of growth at 37°C and continuous shaking at 180 rpm. 200 μL of cells was harvested by centrifugation and resuspended in 50 μL MilliQ water. Subsequently, 2 μL cell suspension was subjected to spacer detection PCR using a forward primer annealing in the 3′ end of the CRISPR repeat of pCRISPR but mismatching the first nucleotide of spacer 1 (degenerated primer mix; Heler et al., 2015) and a reverse primer annealing in the vector backbone (Table S2). When higher sensitivity was required, amplicons of expanded pCRISPR arrays were separated from parental pCRISPR array amplicons using the BluePippin automated agarose-electrophoresis system (3% agarose gel cassette, SageScience). The extracted expanded CRISPR array amplicons were then subjected to an additional PCR reaction using the same degenerated primer mix but a different reverse primer matching spacer 1.
Next-Generation Sequencing and Statistical Analysis
After validation of PCR amplicons by gel electrophoresis and clean up with the GeneJET PCR Purification kit (Thermo Fisher Scientific), the samples were analyzed using Invitrogen Qubit fluorometric quantification. Samples were prepared for sequencing with the Nextera XT DNA Library Preparation Kit (Illumina) and each library individually barcoded with the Nextera XT Index Kit v2 SetA (Illumina). Libraries were pooled equally and spiked with ∼5% of the PhiX control library (Illumina) to artificially increase the genetic diversity before sequencing on a Nano flowcell (2 × 250 base paired-end) with an Illumina MiSeq. Image analysis, base calling, de-multiplexing, and data quality assessments were performed on the MiSeq instrument. FASTAQ files generated by the MiSeq were analyzed by pairing and merging the reads using Geneious 9.0.5 and subsequently extracting newly acquired spacers by identifying the 3′ end of the degenerate primer and the 5′ end of the single repeat present in the parental pCRISPR. Unique spacer sequences were mapped to the chromosome and the replicons carried by the corresponding strains with the BLAST-function of Geneious 9.0.5.
Statistical Tests
To infer the likelihood of finding a certain distribution of PAMs we used a binomial test, where we estimated the likelihood of the observed frequency of each PAM in the case of a randomly distributed PAM-pool (likelihood per PAM: 1/16). As we performed the test multiple times on the same dataset, we used a Bonferroni correction to decrease the probability of a type I error. Spacer size preference was tested by using a bootstrapping resampling method with replacement. 10,000 bootstrap resamples were generated from each observed dataset (each of similar size to the observed dataset). The statistical mode of a certain spacer size within these resamples represented the likelihood of observing this mode in the observed dataset.
Synechocystis Interference Assay
Synechocystis 6803 contains on its megaplasmid pSYSA a type I-D and two type III CRISPR-Cas systems (III-D and III-B) (Scholz et al., 2013). Synechocystis I-D interference assays were performed as described previously (Behler et al., 2018) with a Synechocystis 6803 derivative strain with 16 instead of 49 spacers (spacers 1–14 and 48–49 retained) in its I-D CRISPR array (Trautmann et al., 2012). Conjugation assays were performed using the self-replicating conjugative vector pVZ322 and the gentamicin resistance cassette for selection. Target plasmids with a number of different PAMs were constructed containing the target of spacer 1 of the I-D CRISPR array. Plasmids were conjugated into Synechocystis by triparental mating as described previously (Scholz et al., 2013). Briefly, overnight cultures of the helper strain E. coli J53/RP4 and the donor strain E. coli DH5α with the plasmid of interest were diluted and incubated for 2.5 h at 37°C with shaking at 180 rpm. For each conjugation, an optical density 600 (OD600) of 7.0 of the plasmid-bearing and helper cultures were harvested, resuspended in LB and combined. The mixed culture was incubated for 1 hr at 30°C without shaking. In parallel, a Synechocystis culture with an OD750 of 1.0 was harvested and combined with the mixed culture of the plasmid-bearing and helper culture. The pellet was resuspended and placed on a sterile filter. After overnight incubation at 30°C, the filter was rinsed and 30 μL of the resulting cell suspension was plated on BG11 agar plates containing 7.5 μg/mL gentamicin. Transconjugants were counted after further incubation at 30°C for 2 weeks. Mean values of conjugation efficiency and corresponding standard errors were calculated by dividing the number of transconjugants obtained with the target plasmids (pT) by the number of transconjugants obtained with the non-target control plasmid (pNT). Experiments were performed in biological triplicates and in parallel with the control plasmid.
Acknowledgments
The authors thank members of the Brouns and Hess lab for helpful discussions and feedback on the manuscript and M. Niessen for helpful initial contributions. LIC cloning vectors were a kind gift from Scott Gradia (UC Berkeley, CA, USA). S.J.J.B. thanks the funding sources FOM (Projectruimte 15PR3188-2), the European Research Council (starting grant [Stg] 638707), the Netherlands Organization for Scientific Research (VIDI Grant 864.11.005), and a startup grant from Delft University of Technology. F.L.N. is supported by the Netherlands Organization for Scientific Research (NWO) Veni grant 016.Veni.181.092. W.R.H. acknowledges financial support from the German Research Foundation (DFG) program FOR1680 “Unravelling the Prokaryotic Immune System” (grant HE 2544/8-2).
Author Contributions
S.N.K., C.A., and S.J.J.B. designed research. S.N.K., C.A., J.B., R.E.M., F.L.N., A.C.H., and J.N.A.V. performed research. S.N.K., C.A., J.B., R.E.M., F.L.N., J.N.A.V., W.R.H., and S.J.J.B. analyzed data. S.N.K., C.A., and S.J.J.B. wrote the paper with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: March 27, 2018
Footnotes
Supplemental Information includes three figures and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.02.103.
Data and Software Availability
The accession number for the sequencing data reported in this paper is European Nucleotide Archive: PRJEB25213.
Supplemental Information
References
- Amitai G., Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Behler J., Sharma K., Reimann V., Wilde A., Urlaub H., Hess W.R. The host-encoded RNase E endonuclease as the crRNA maturation enzyme in a CRISPR-Cas subtype III-Bv system. Nat. Microbiol. 2018;3:367–377. doi: 10.1038/s41564-017-0103-5. [DOI] [PubMed] [Google Scholar]
- Cassier-Chauvat C., Veaudor T., Chauvat F. Comparative Genomics of DNA Recombination and Repair in Cyanobacteria: Biotechnological Implications. Front. Microbiol. 2016;7:1809. doi: 10.3389/fmicb.2016.01809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Deveau H., Barrangou R., Garneau J.E., Labonté J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund R.D., Wilkinson M.E., Klykov O., Barendregt A., Pearce F.G., Kieper S.N., Maxwell H.W.R., Capolupo A., Heck A.J.R., Krause K.L. Spacer capture and integration by a type I-F Cas1-Cas2-3 CRISPR adaptation complex. Proc. Natl. Acad. Sci. USA. 2017;114:E5122–E5128. doi: 10.1073/pnas.1618421114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales M.F., Brooks T., Pukatzki S.U., Provenzano D. Rapid protocol for preparation of electrocompetent Escherichia coli and Vibrio cholerae. J. Vis. Exp. 2013;80:50684. doi: 10.3791/50684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R., Samai P., Modell J.W., Weiner C., Goldberg G.W., Bikard D., Marraffini L.A. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton S.P.T., Connerton I.F. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front. Microbiol. 2015;5:744. doi: 10.3389/fmicb.2014.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudaiberdiev S., Shmakov S., Wolf Y.I., Terns M.P., Makarova K.S., Koonin E.V. Phylogenomics of Cas4 family nucleases. BMC Evol. Biol. 2017;17:232. doi: 10.1186/s12862-017-1081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivančić-Baće I., Cass S.D., Wearne S.J., Bolt E.L. Different genome stability proteins underpin primed and naïve adaptation in E. coli CRISPR-Cas immunity. Nucleic Acids Res. 2015;43:10821–10830. doi: 10.1093/nar/gkv1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J. CRISPR-Cas: Adapting to change. Science. 2017;356:356. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- Jansen R., Embden J.D.A.V., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Ka D., Lee H., Jung Y.-D., Kim K., Seok C., Suh N., Bae E. Crystal structure of Streptococcus pyogenes Cas1 and its interaction with Csn2 in the type II CRISPR-Cas system. Structure. 2016;24:70–79. doi: 10.1016/j.str.2015.10.019. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künne T., Kieper S.N., Bannenberg J.W., Vogel A.I.M., Miellet W.R., Klein M., Depken M., Suarez-Diez M., Brouns S.J.J. Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol. Cell. 2016;63:852–864. doi: 10.1016/j.molcel.2016.07.011. [DOI] [PubMed] [Google Scholar]
- Lee H., Zhou Y., Taylor D.W., Sashital D.G. Cas4-dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol. Cell. 2018;70 doi: 10.1016/j.molcel.2018.03.003. Published online March 27, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak S., Beloglazova N., Nocek B., Skarina T., Flick R., Brown G., Popovic A., Joachimiak A., Savchenko A., Yakunin A.F. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J. Am. Chem. Soc. 2013;135:17476–17487. doi: 10.1021/ja408729b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak S., Nocek B., Beloglazova N., Skarina T., Flick R., Brown G., Joachimiak A., Savchenko A., Yakunin A.F. The CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis contains a [2Fe-2S] cluster: crystal structure and nuclease activity. Nucleic Acids Res. 2014;42:11144–11155. doi: 10.1093/nar/gku797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wang R., Zhao D., Xiang H. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Gong L., Zhao D., Zhou J., Xiang H. The spacer size of I-B CRISPR is modulated by the terminal sequence of the protospacer. Nucleic Acids Res. 2017;45:4642–4654. doi: 10.1093/nar/gkx229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Liu Z., Ye Q., Pan S., Wang X., Li Y., Peng W., Liang Y., She Q., Peng N. Coupling transcriptional activation of CRISPR-Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res. 2017;45:8978–8992. doi: 10.1093/nar/gkx612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V., van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- Mojica F.J., Díez-Villaseñor C., García-Martínez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Nuñez J.K., Kranzusch P.J., Noeske J., Wright A.V., Davies C.W., Doudna J.A. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez J.K., Harrington L.B., Kranzusch P.J., Engelman A.N., Doudna J.A. Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature. 2015;527:535–538. doi: 10.1038/nature15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A., Tjaden B., Hagemann A., Randau L., Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C., Guyard C., Pelaz C., Wasserscheid J., Bondy-Denomy J., Dewar K., Ensminger A.W. Active and adaptive Legionella CRISPR-Cas reveals a recurrent challenge to the pathogen. Cell. Microbiol. 2016;18:1319–1338. doi: 10.1111/cmi.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollie C., Graham S., Rouillon C., White M.F. Prespacer processing and specific integration in a Type I-A CRISPR system. Nucleic Acids Res. 2018;46:1007–1020. doi: 10.1093/nar/gkx1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins M.F., Chowdhury S., Carter J., Golden S.M., Wilkinson R.A., Bondy-Denomy J., Lander G.C., Wiedenheft B. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc. Natl. Acad. Sci. USA. 2017;114:E5113–E5121. doi: 10.1073/pnas.1616395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz I., Lange S.J., Hein S., Hess W.R., Backofen R. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS ONE. 2013;8:e56470. doi: 10.1371/journal.pone.0056470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S.A., Erdmann S., Mojica F.J., Garrett R.A. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals R.H.J., Jackson S.A., Biswas A., Brouns S.J.J., Brown C.M., Fineran P.C. Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system. Nat. Commun. 2016;7:12853. doi: 10.1038/ncomms12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S.H., Richter H., Charpentier E., Qimron U. Adaptation in CRISPR-Cas Systems. Mol. Cell. 2016;61:797–808. doi: 10.1016/j.molcel.2016.01.030. [DOI] [PubMed] [Google Scholar]
- Swarts D.C., Mosterd C., van Passel M.W.J., Brouns S.J.J. CRISPR interference directs strand specific spacer acquisition. PLoS ONE. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann D., Voss B., Wilde A., Al-Babili S., Hess W.R. Microevolution in cyanobacteria: re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 2012;19:435–448. doi: 10.1093/dnares/dss024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houte S., Ekroth A.K., Broniewski J.M., Chabas H., Ashby B., Bondy-Denomy J., Gandon S., Boots M., Paterson S., Buckling A., Westra E.R. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature. 2016;532:385–388. doi: 10.1038/nature17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li J., Zhao H., Sheng G., Wang M., Yin M., Wang Y. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell. 2015;163:840–853. doi: 10.1016/j.cell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Wei Y., Terns R.M., Terns M.P. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I., Goren M.G., Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kasciukovic T., White M.F. The CRISPR associated protein Cas4 Is a 5′ to 3′ DNA exonuclease with an iron-sulfur cluster. PLoS ONE. 2012;7:e47232. doi: 10.1371/journal.pone.0047232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.