Abstract
CONTEXT:
ICU shows increasing incidence of infection associated with the use of invasive procedures for the diagnostic purpose as well as the indiscriminate use of antibiotics. Pseudomonas aeruginosa and Acinetobacter species are “very successful” pathogen and the emergence of the Metallo-β-Lactamases (MBL) is becoming a therapeutic challenge.
AIMS:
To isolate the Nonfermenting Gram negative bacilli from the ICU samples. To identify the metallo betalactamase producers and to detect the bla gene presence among the Pseudomonas aeruginosa and Acinetobacter baumannii.
SETTINGS AND DESIGN:
The Nonfermenting Gram negative bacilli isolates from the ICU samples were taken over for 5 years (2009-2014) in a tertiary care hospital.
METHODS AND MATERIALS:
The isolates of Pseudomonas species and Acinetobacter species were confirmed by API analyser and processed according to standard procedures. Detection of the MBL producers were done by E strip method and subjected for bla gene detection by PCR method.
RESULTS:
In our study a total of 195 isolates of NFGNB were obtained from various ICU. Of these MBL producers, 26 % were Pseudomonas aeruginosa and 25 % were Acinetobacter baumannii. The subtypes of blaVIM MBL producing P.aeruginosa were 26%. The predominant gene coding for MBL activity in A.baumannii were found to be blaOXA gene 11.9%. The gene accession numbers were KF975367, KF975372.
CONCLUSIONS:
We have to control the development and dissemination of these superbugs among the ICU's.
Keywords: Acinetobacter baumanni, bla genes, ICU, Metallo-β-lactamases, Pseudomonas aeruginosa
Introduction
Isolation of nonfermenters from the clinical specimens obtained from Intensive Care Unit (ICU) shows that increasing incidence of infection associated with the use of invasive procedures, indiscriminate use of antibiotics, inadequate sterilization, and immune compromised condition due to lifestyle disease have also contributed.[1]
Among the nonfermenters, Pseudomonas aeruginosa is inherently resistant and Acinetobacter species capable of surviving in various environmental conditions are adapted at acquiring resistance.[2,3] The digestive tracts of patients within ICUs often serve as reservoirs for multidrug-resistant (MDR) isolates.[4,5]
P. aeruginosa resistance is a global disease burden[6] and it is a therapeutic challenge.[7] The Acinetobacter baumanii complex is emerging multidrug resistant nosocomial and community acquired pathogen. The incidence of infection by these species among the patients receiving the mechanical ventilation are quite increasing.[8] These organisms are “very successful” pathogen which possesses both acquired and intrinsic mechanisms of resistance to various classes of antibiotics.[9,10,11]
Infections in the ICU patients were commonly associated with ventilator-associated pneumonia, urinary tract infection, and bacteremia caused by MDR organism Gram-negative bacilli with increasing morbidity and mortality.[12,13] The emergence of the metallo-β-lactamases (MBL) is becoming a therapeutic challenge.[12] Antimicrobial resistance pattern has emerged as an important determinant of the outcome for patients in the ICUs.[14]
In our study, drug-resistant isolates in the ICUs were detected and the gene encoding carbapenem resistance in P. aeruginosa and Acinetobacter baumannii was identified. The resulting sequences were compared with those available in GenBank.
Methods
All the suspected colonies of the NFGNB were identified by Gram staining, colony characteristics, oxidase test, motility, and standard biochemical reactions, and further confirmation of the species was carried out by API analyzer. The study was carried out in a tertiary care hospital (2009–2014). All the organisms identified were tested for the susceptibility according to the standard Clinical and Laboratory Standards Institute guidelines.[15] The sensitivity pattern of first- and second-line drugs was tested.
For Pseudomonas species, the following 15 drugs were used: amikacin (Ak-30 μg), aztreonam (Az-30 μg), colistin (Cl-10 μg), ciprofloxacin (Cip-5 μg), ceftazidime (Caz-5 μg), cefepime (Cpm-5 μg), carbenicillin (Cb-100 μg), gentamicin (G-10 μg), imipenem (Imp-10 μg), meropenem (Mr-10 μg), netilmicin (Net-30 μg), ofloxacin (Of-5 μg), piperacillin-tazobactam (Pit-100 μg/10 μg), polymyxin B (Pb-300 units), tobramycin (Tb-10 μg).
For Acinetobacter species, amikacin (Ak-30 μg), cefepime (Cpm-5 μg), ceftazidime (Caz-5 μg), ciprofloxacin (Cf-5 μg), cefotaxime (Ce-5 μg), colistin (E strip), cotrimoxazole (Cot-5 μg), gentamicin (G-10 μg), imipenem (IMP-10 μg), meropenem (Mr-10 μg), piperacillin-tazobactam (Pt-100 μg/10 μg), and polymyxin B (E strip) were used, and in case of urine samples, nitrofurantoin (Nit-300 μg) disks were used.
The study was confined to the MBL-producing P. aeruginosa and A. baumannii species. The antibiotic discs used in our study were purchased from HiMedia. The E strip was purchased from HiMedia, Biomerieux, and Radianz biotechnologies. Screening for MBL production was done in imipenem-resistant isolates by the E strip method using the ceftazidime and ceftazidime + ethylenediaminetetraacetic acid.[16]
The MBL-producing resistant strains of P. aeruginosa were screened for the blagenes – blaVIM,KPC,NDM,IMP[1,17,18,19,20,21] [Table 1]. For A. baumannii, blaVIM,IMP,OXA,NDM genes [Table 2] were carried out.[1,18,21,22,23]
Table 1.
The primers used for bla gene detection in Pseudomonas aeruginosa
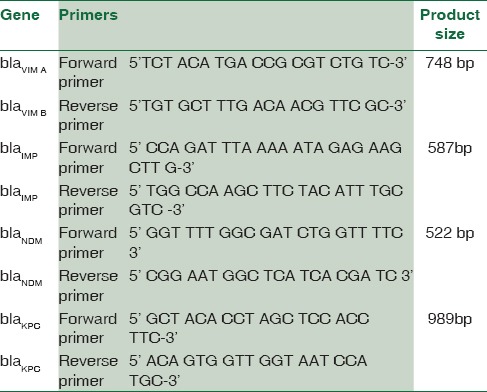
Table 2.
The primers used for bla gene detection in Acinetobacter baumannii

Polymerase chain reaction amplification
The reaction conditions were as follows: predenaturation at 94°C for 2 min, followed by 30 amplifications cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min 30 sec, with final extension step of 72°C for 5 min. The cycling parameters for the blaIMP,VIM,NDM,OXA genes were as follows: initial denaturation: 94°C for 3 min, denaturation: 94°C for 1 min, annealing: 58°C for 1 min 35 cycles, extension: 72°C for 1 min, final extension: 72°C for 5 min. After screening for the MBL, the positive PCR products were sequenced. Sequencing the amplified products, the BLAST results were analyzed.
Results
The nonfermenters isolated from ICU were found to be notorious as there were possibilities of drug-resistant strains being horizontally spread among the patients. In our study, a total of 195 isolates of NFGNB were obtained from various ICUs. Among them, 61 (31.2%) were Pseudomonas spp and 134 (68.8) were Acinetobacter spp. Among the 84 isolates of NFGNB, 32 (38%) were P. aeruginosa and 61.9% A. baumannii were isolated from surgical ICU. Distribution of NFGNB – P. aeruginosa and A. baumannii in different ICUs is shown in [Table 3]. Among 61 P. aeruginosa from ICU patients, 19 (31.1%) were from males and 42 (68.8%) were from females. Distribution of P. aeuginosa from ICU among different sexes is shown in Chart 1. Among 134 A. baumannii from ICU patients, 89 (66.4%) were from males and 45 (33.5%) were from females [Chart 2].
Table 3.
Distribution of P.aeruginosa and A.baumannii in different ICUs
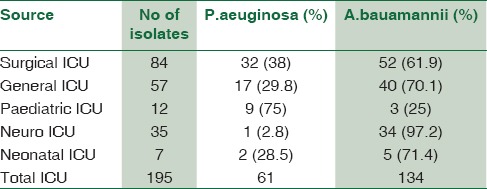
Chart 1.
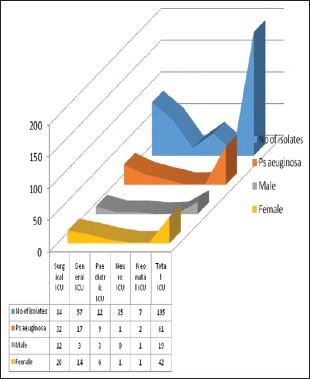
Distribution of Pseudomonas aeruginosa from Intensive Care Unit among different sexes
Chart 2.
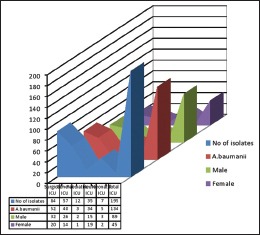
Distribution of Acinetobacter baumannii from Intensive Care Unit among different sexes
Among 195 NFGNB isolates from ICU, 89 (45.6%) were drug resistant. Out of these, 26 (13.33%) were P. aeruginosa and 63 (32.3%) were A. baumannii. Overall, the MDR isolates from ICU were 33.33%. The MBL producers from ICU were 49 (25.12%) [Table 4]. Of these MBL producers, 16 (26.22%) were P. aeruginosa and 33 (24.62%) were A. baumannii [Table 5 and Chart 3].
Table 4.
Drug resistance in ICU isolates
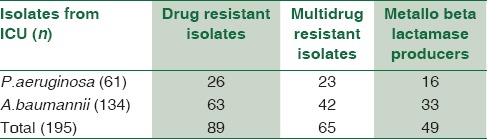
Table 5.
Showing MBL positive by Estrip method

Chart 3.
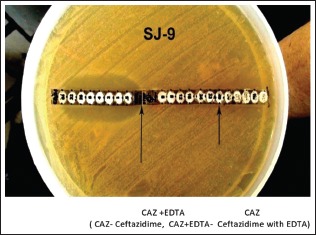
MIC E strip
The maximum numbers of MBL producers were in surgical ICU followed by general ICU. Among ICUs, 6.1% of isolates were from pediatric ICU and one isolate of P. aeruginosa was MBL producer [Table 6]. Among the MBL producers in ICU, P. aeruginosa was obtained from 11 males and 5 females and A. baumannii was isolated from 13 males and 20 females.
Table 6.
Distribution in Paediatric ICU
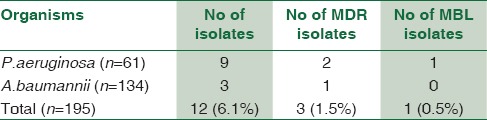
MDR and MBL producers were more from general ICU and surgical ICU. Among the 63 drug-resistant A. baumannii, 42 (66.6%) were multidrug resistant and 33 (52.3%) were MBL producer.
Clinical sources of the MBL-producing P. aeruginosa are shown in Table 7. The subtypes of blaVIM MBL-producing P. aeruginosa were 26% and strains of P. aeruginosa from ICU were negative for other blaKPC,NDM,IMP genes. Distribution of all three subtypes of MBL-producing P. aeruginosa was as follows: 13.1% blaVIM-4, 9.8% blaVIM–5, and 3.2% blaVIM–38 strains [Chart 4].
Table 7.
Clinical source of P.aeruginosa with blaVIM gene subtypes
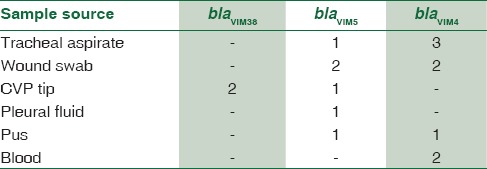
Chart 4.
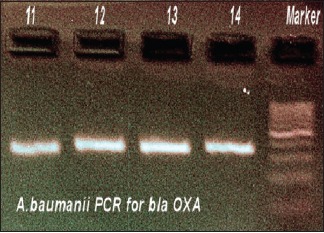
Polymerase chain reaction for Acinetobacter baumannii blaOXA
In MBL-positive A. baumannii, blaVIM gene was demonstrated in 4.4% strains, blaOXA gene was seen in 11.9%, and blaIMP gene was seen in 5.2%. Thus, the predominant gene coding for MBL activity was found to be OXA. Distribution of genes responsible for MBL activity in A. baumannii and its clinical source is shown in Table 8. The resulting sequences were compared with those available in GenBank (www.ncbi.nih.gov/BLAST) and the gene accession numbers were KF975367, KF975372.
Table 8.
Clinical source of A.baumannii with bla gene subtypes

Discussion
There is an increase in infection caused by the MBL-producing NFGNB in the ICUs, along with the significant morbidity and mortality. The incidence of infection in ICUs, especially the nosocomial infections, is a rising trend with a spectrum of clinical conditions. They may be in the range from impaired immunity, lapse in the sterilization, use of various invasive devices, and procedure to indiscriminate use of antibiotics.
A study by Aliskan et al.[24] showed that there was a decrease in susceptibility pattern of A. baumannii and P. aeruginosa isolates from the ICU samples. In our study, maximum P. aeruginosa and A. baumannii were from tracheal aspirates, followed by wound swab which was in concordance with study of Jaggi et al.[25]
In a study by Orrett, 17.3% of P. aeruginosa were from ICU.[26] The prevalence of Acinetobacter species from various parts of our country was 3%,[27] 4.5%,[28] 9.6% in West Bengal.[29] In our study, the prevalence of P. aeruginosa (31.2%) and A. baumannii (68.8%) in ICU was higher when compared with above study. Among the A. baumannii strains isolated from ICU, 65%–70% were resistant and they were not in concordance with our study[3,30,31] which is higher when compared to our study.
The percentage of MDR A. baumannii isolates increased from 4% to 55% and 2%–8% in P. aeruginosa isolates. According to Yan et al., 56.7% and 58.3% of P. aeruginosa were found to be imipenem resistant.[3,32]
Many studies have reported <50% of resistance to imipenem and meropenem in P. aeruginosa. Imipenem resistance according to Livermore[33] was 77.5% and Lone et al.[34] was 25.6%. Tan[35] reported that 9.6% carbapenem-resistant P. aerugionsa and 27.2% carbapenem-resistant P. aerugionsa were from ICU reported by Hsu et al.[36] In our study, carbapenem-resistant Acinetobacter spp. isolated from ICU were 25% and lesser than the resistance pattern (69%) reported by Tan.[35]
A study by Hsu et al.[36] showed that carbapenem resistance of Acinetobacter was 49.6%. Lagatolla et al.[37] showed that 70% of carbapenem-resistant P. aeruginosa were MBL producers. A study by Kabbaj et al.[38] showed that, among 57.4% imipenem-resistant isolates of Acinetobacter bauamnii, 74% were MBL producers and in concordance with our study. An Indian study stated that MBL producers among the A. baumannii were 70.9%,[39] and another study reported that 21%[40] of A. baumannii were MBL producers.
Tanzinah Nasrin showed the high level of MBL producers isolated from ICU unlike our study. Studies from the Indian subcontinent have shown the blaIMP1 gene carried by meropenem-resistant isolates.[41] Our study confirmed the presence of bla gene (blaVIM26% and blaOXA12%) among the isolates of P. aeruginosa and A. baumannii from the ICU samples and comparable with the study of Gautam et al.,[30] 25% prevalence of NDM-1 A. baumannii in ICU isolates.
Conclusion
We have to control the development and dissemination of these superbugs among the ICUs. Insight into the incidence of these superbugs alarms the need of every institution to have the interventional strategies to prevent these infections. The prevalence in ICU emphasizes the need for early detection of beta-lactamases-producing organisms.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgement
My sincere gratitude goes to Dr. Sundararaj.T, (Ret) Prof and Head of Dept of Microbiology, Dr. A.L. Mudaliar Post- Graduate Institute of Basic Medical Sciences. Tharamani, Chennai & Director of JASMN Education and Foundation, JASMN Laboratory, who has guided and spent valuable time during the course of our work.
References
- 1.Murray PR, Rosenthal KS, Pfaller MA, editors. Text Book of Medical Microbiology-Pseudomonas and Related Organisms. 5th ed. Vol. 34. Elsevier; 2005. pp. 357–65. [Google Scholar]
- 2.Towner KJ. Plasmid and transposon behaviour in Acinetobacter. In: Towner KJ, Bergogne-Berezin E, Fewson CA, editors. The Biology, Physiology, Industrial Relevance. New York: Plenum Press; 1991. pp. 149–67. [Google Scholar]
- 3.Cisneros JM, Rodríguez-Baño J, Fernández-Cuenca F, Ribera A, Vila J, Pascual A, et al. Risk-factors for the acquisition of imipenem -Resistant Acinetobacter baumannii in Spain: A nationwide study. Clin Microbiol Infect. 2005;11:874–9. doi: 10.1111/j.1469-0691.2005.01256.x. [DOI] [PubMed] [Google Scholar]
- 4.Corbella X, Pujol M, Ayats J, Sendra M, Ardanuy C, Domínguez MA, et al. Relevance of digestive tract colonization in the epidemiology of nosocomial infections due to multiresistant Acinetobacter baumannii. Clin Infect Dis. 1996;23:329–34. doi: 10.1093/clinids/23.2.329. [DOI] [PubMed] [Google Scholar]
- 5.Afzal-Shah M, Livermore DM. Worldwide emergence of carbapenem-resistant Acinetobacter spp. J Antimicrob Chemother. 1998;41:576–7. doi: 10.1093/jac/41.5.576. [DOI] [PubMed] [Google Scholar]
- 6.Paramythiotou E, Lucet JC, Timsit JF. Acquistion of multidrug resistant Pseudomonas aeruginosa in patients in Intensive Care Units: Role of antibiotics with antipseudomonal activity. Clin Infect Dis. 2003;38:676–7. doi: 10.1086/381550. [DOI] [PubMed] [Google Scholar]
- 7.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talbot GH, Bradley J, Edwards JE, Jr, Gilbert D, Scheld M, Bartlett JG, et al. Bad Bugs need drugs: An update on the development pipeline from the antimicrobial availability task force of the infectious diseases society of America. Clin Infect Dis. 2006;42:657–68. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 9.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA, et al. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–84. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice LB. Challenges in identifying new antimicrobial agents effective for treating infections with Acinetobacter baumannii and Pseudomonas aeruginosa. Clin Infect Dis. 2006;43(Suppl 2):S100–5. doi: 10.1086/504487. [DOI] [PubMed] [Google Scholar]
- 11.Garnacho-Montero J, Amaya-Villar R. Multi resistant Acinetobacter baumannii infections: Epidemiology and management. Curr Opin Infect Dis. 2010;23:332–9. doi: 10.1097/QCO.0b013e32833ae38b. [DOI] [PubMed] [Google Scholar]
- 12.Irfan S, Zafar A, Guhar D, Ahsan T, Hasan R. Metallo-beta-lactamase-producing clinical isolates of Acinetobacter species and Pseudomonas aeruginosa from intensive care unit patients of a tertiary care hospital. Indian J Med Microbiol. 2008;26:243–5. doi: 10.4103/0255-0857.42035. [DOI] [PubMed] [Google Scholar]
- 13.Oberoi L, Singh N, Sharma P, Aggarwal A. ESBL, MBL and ampc β lactamases producing superbugs - Havoc in the intensive care units of Punjab India. J Clin Diagn Res. 2013;7:70–3. doi: 10.7860/JCDR/2012/5016.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohanasundaram KM. Retrospective analysis of the incidence of nosocomial infection in the ICU-associated risk factors and microbiological profile. J Clin Diagn Res. 2010;4:33789–2. [Google Scholar]
- 15.Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2010. Clinical and Laboratory Standards Institute. Performance Standards for the Antimicrobial Susceptibility Testing. CLSI Document M100.S20. [Google Scholar]
- 16.Segal H, Elisha BG. Use of E test MBL strips for the detection of carbapenemases in Acinetobacter baumannii. J Antimicrob Chemother. 2005;56:598. doi: 10.1093/jac/dki265. [DOI] [PubMed] [Google Scholar]
- 17.Giakkoupi P, Xanthaki A, Kanelopoulou M, Vlahaki A, Miriagou V, Kontou S, et al. VIM-1 metallo-beta-lactamase-producing Klebsiella pneumoniae strains in Greek hospitals. J Clin Microbiol. 2003;41:3893–6. doi: 10.1128/JCM.41.8.3893-3896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong F, Xu XW, Song WQ, Lü P, Yu SJ, Yang YH, et al. Characterization of multidrug-resistant and metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates from a paediatric clinic in China. Chin Med J (Engl) 2008;121:1611–6. [PubMed] [Google Scholar]
- 19.Gutiérrez O, Juan C, Cercenado E, Navarro F, Bouza E, Coll P, et al. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob Agents Chemother. 2007;51:4329–35. doi: 10.1128/AAC.00810-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juan C, Beceiro A, Gutiérrez O, Albertí S, Garau M, Pérez JL, et al. Characterization of the new metallo-beta-lactamase VIM-13 and its integron-borne gene from a Pseudomonas aeruginosa clinical isolate in Spain. Antimicrob Agents Chemother. 2008;52:3589–96. doi: 10.1128/AAC.00465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, et al. Cloning and characterization of bla VIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–90. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yum JH, Yi K, Lee H, Yong D, Lee K, Kim JM, et al. Molecular characterization of metallo-beta-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: Identification of two new integrons carrying the bla (VIM-2) gene cassettes. J Antimicrob Chemother. 2002;49:837–40. doi: 10.1093/jac/dkf043. [DOI] [PubMed] [Google Scholar]
- 23.Sundararaj T, Sundararaj A. Microbiology Laboratory Manual. 4th ed. Vol. 1. A. Sundararaj; 2005. pp. 39–40. [Google Scholar]
- 24.Alişkan H, Colakoǧlu S, Turunç T, Demiroǧlu YZ, Erdoǧan F, Akin S, et al. Four years of monitoring of antibiotic sensitivity rates of Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from patients in Intensive Care Unit and inpatient clinics. Mikrobiyol Bul. 2008;42:321–9. [PubMed] [Google Scholar]
- 25.Jaggi N, Sissodia P, Sharma L. Acinetobacter baumannii isolates: The epidemiology, antibiogram and the nosocomial status which were studied over a 25 months period in a tertiary care hospital in India. BMC Proc. 2011;5:291. [Google Scholar]
- 26.Orrett FA. Antimicrobial susceptibility survey of Pseudomonas aeruginosa strains isolated from clinical sources. J Natl Med Assoc. 2004;96:1065–9. [PMC free article] [PubMed] [Google Scholar]
- 27.Dash M, Padhi S, Pattnaik S, Mohanty I, Misra P. Frequency, risk factors, and antibiogram of Acinetobacter species isolated from various clinical samples in a tertiary care hospital in Odisha, India. Avicenna J Med. 2013;3:97–102. doi: 10.4103/2231-0770.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rit K, Saha R. Multidrug-resistant Acinetobacter infection and their susceptibility patterns in a tertiary care hospital. Niger Med J. 2012;53:126–8. doi: 10.4103/0300-1652.104379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joshi SG, Litake GM, Satpute MG, Telang NV, Ghole VS, Niphadkar KB, et al. Clinical and demographic features of infection caused by Acinetobacter species. Indian J Med Sci. 2006;60:351–60. [PubMed] [Google Scholar]
- 30.Gautam V, Mewara A, Raj A, Gupta V, Singla N, Ray P, et al. High prevalence of New Delhi metallo-β-lactamase in Acinetobacter calcoaceticus-A. Baumannii complex at two tertiary care centres in North India. Indian J Med Microbiol. 2014;32:455–6. doi: 10.4103/0255-0857.142231. [DOI] [PubMed] [Google Scholar]
- 31.Abbo A, Navon-Venezia S, Hammer-Muntz O, Krichali T, Siegman-Igra Y, Carmeli Y, et al. Multidrug-resistant Acinetobacter baumannii. Emerg Infect Dis. 2005;11:22–9. doi: 10.3201/eid1101.040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan JJ, Ko WC, Tsai SH, Wu HM, Wu JJ. Outbreak of infection with multidrug resistant Klebsiella pneumonia carrying bla IMP -8 in a university medical centre in Taiwan. J Clin Microbiol. 2001;39:4433–9. doi: 10.1128/JCM.39.12.4433-4439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: Our worst nightmare? Clin Infect Dis. 2002;34:634–40. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 34.Lone R, Shah A, Kadri SM, Lone S, Shah F. Nosocomial multidrug resistant Acinetobacter infections-clinical findings, risk factors and demographic characteristics. Bangladesh J Med Microbiol. 2009;3:34–8. [Google Scholar]
- 35.Tan TT. “Future” threat of Gram negative resistance in Singapore. Ann Acad Med Singapore. 2008;37:884–90. [PubMed] [Google Scholar]
- 36.Hsu LY, Tan TY, Jureen R, Koh TH, Krishnan P, Tzer-Pin Lin R, et al. Antimicrobial drug resistance in Singapore hospitals. Emerg Infect Dis. 2007;13:1944–7. doi: 10.3201/eid1312.070299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagatolla C, Tonin EA, Monti-Bragadin C, Dolzani L, Gombac F, Bearzi C, et al. Endemic carbapenem-resistant Pseudomonas aeruginosa with acquired metallo-beta-lactamase determinants in European hospital. Emerg Infect Dis. 2004;10:535–8. doi: 10.3201/eid1003.020799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabbaj H, Seffar M, Belefquih B, Akka D, Handor N, Amor M, et al. Prevalence of Metallo betalactamases producing Acinetobacter baumannii in a Moroccan hospital. ISRN Infect Dis. 2013;2013:154921. [Google Scholar]
- 39.Uma Karthika R, Srinivasa Rao R, Sahoo S, Shashikala P, Kanungo R, Jayachandran S, et al. Phenotypic and genotypic assays for detecting the prevalence of metallo-beta-lactamases in clinical isolates of Acinetobacter baumannii from a South Indian tertiary care hospital. J Med Microbiol. 2009;58:430–5. doi: 10.1099/jmm.0.002105-0. [DOI] [PubMed] [Google Scholar]
- 40.Anil VK, Vishnu SP, Kavitha R, Shamusul D. The phenotypic detection of carbapenemase in the meropenem resistant Acinetobacter calcoaceticus-baumannii complex in a tertiary care hospital in S.I. J Clin Diagn Res. 2011;5:223–6. [Google Scholar]
- 41.Azim A, Dwivedi M, Rao PB, Baronia AK, Singh RK, Prasad KN, et al. Epidemiology of bacterial colonization at Intensive Care Unit admission with emphasis on extended-spectrum beta-lactamase-and metallo-beta-lactamase-producing Gram-negative bacteria – An Indian experience. J Med Microbiol. 2010;59:955–60. doi: 10.1099/jmm.0.018085-0. [DOI] [PubMed] [Google Scholar]


