Abstract
A method has been developed for the direct determination of agmatine in bacterial culture supernatants using isotope dilution ultra performance liquid chromatography (UPLC)-tandem mass spectrometry (UPLC-MS/MS). Agmatine determination in bacterial supernatants is comprised of spiking culture or isolate supernatants with a fixed concentration of uniformly labeled 13C5,15N4-agmatine (synthesized by decarboxylation of uniformly labeled 13C6,15N4-arginine using arginine decarboxylase from Pseudomonas aeruginosa) as an internal standard, followed by derivatization with 4-fluoro-7-nitro-2,l,3-benzoxadiazole (NBDF) to improve the reversed-phase chromatographic retention characteristics of agmatine, as well as the selectivity and sensitivity of UPLC-MS/MS detection of this amine in complex biologically derived mixtures. Intrasample precisions for measurement of agmatine in culture supernatants average 4.1 % (relative standard deviation). Calibration curves are linear over the range 5 nM to 10 μM, and the detection limit is estimated at 1.5 nM. To demonstrate the utility of the method, agmatine levels in supernatants of overnight cultures of wild-type (UCBPP-PA14), as well as arginine decarboxylase and agmatine deiminase mutant strains of P. aeruginosa strain UCBPP-PA14 were measured. This method verified that the mutant strains are lacking the specific metabolic capabilities to produce and metabolize agmatine. In addition, measurement of agmatine in supernatants of a panel of clinical isolates from patients with cystic fibrosis revealed that three of the P. aeruginosa isolates hyper-secreted agmatine into the supernatant, hypothesized to be a result of a mutation in the aguA gene. Because agmatine has potential inflammatory activities in the lung, this phenotype may be a virulence factor for P. aeruginosa in the lung environment of cystic fibrosis patients.
Keywords: Metabolite, Metabolite profiling, Disease, Biomarker
Introduction
The polyamines putrescine, cadaverine, spermine, and spermidine are ubiquitous components of the cellular milieu in most forms of life and play vital roles in cellular division and DNA synthesis [1, 2]. Polyamine synthesis is initiated through conserved pathways including the ornithine decarboxylase pathway (ODC) and arginine decarboxylase pathway (ADC). The ADC pathway is more utilized in plants and bacteria than in mammals [3], but the discovery of agmatine (decarboxylated arginine) in humans in 1994 has generated much interest in this molecule [4], Agmatine has been shown to function as a neurotransmitter interacting with α2-adrenergic, nicotinic, serotonin, and imidazoline receptors in the central nervous system and may block N-methyl-D-aspartate receptor channels [5–7]. Agmatine has also been demonstrated to be an inhibitor of NOS-2 with far-reaching biological implications including cytoprotection during oxidative stress [8, 9]. Avast arena of potential therapeutic applications is being explored given these mechanisms of action [5].
Because agmatine is also a bacterial metabolite, our work-ing group has been focused on its role at the host-pathogen interface in chronic lung infections. In this complex environ-ment, agmatine’s biological roles may be under a competitive dynamic between the host immune system and pathogens. It is known that agmatine has both immunomodulatory properties and can induce a biofilm in the lung pathogen Pseudomonas aeruginosa [10]. Furthermore, agmatine has been measured in human sputum and its levels correlated with disease state [11]. The ability to accurately measure agmatine has been a critical component of this research.
Agmatine has been traditionally quantified employing liq-uid chromatography (LC) with fluorescence detection [12, 13], capillary electrophoresis [14], and immunoassays [15], all of which lack the sensitivity and selectivity of a tandem mass spectrometry-based approach. Additionally, the short-lived nature of agmatine in biologically relevant matrices re-quires rapid, high-throughput analysis with the use of isotopi-cally labeled internal standards to accurately quantify this compound. Non-mass spectrometry (MS) methods for amine quantification such as those listed above are time-consuming, labor-intensive, and/or rely on antibodies, which can suffer from lack of specificity for small molecule targets. Two pre-vious studies using LC/MS-based methods for measurement of agmatine in biologically derived samples have been report-ed [16, 17]. The first study does not employ an internal stan-dard for quantification to compensate for such detrimental effects as sample handling losses, analyte instability, ioniza-tion variability, source contamination, or matrix suppression [16]. The second study employs a time-consuming solid phase extraction and derivatization protocol prior to analysis, and employs non-agmatine internal standards [17]. Due to the lim-itations of previously reported methods, and in order to accu-rately measure ADC activity in P aeruginosa, wc have devcl-oped a rapid (10 min/analysis) isotope dilution ultra perfor-mancc liquid chromatography (UPLC)-tandcm mass spec-trometry (UPLC-MS/MS) method for the quantification of agmatine in bacterial cell extracts. This work details the UPLC-MS/MS method development and its application to the targeted analysis of agmatine in bacterial culture and clin-ical isolate supernatants.
Materials and methods
Reagents and sample materials
J.T. Baker Ultra LC/MS-grade acetonitrile (ACN) and water were purchased from VWR International (Radnor, PA). LC/MS Ultra-grade formic acid (Fluka) was purchased from Sigma-Aldrich (Saint Louis, MO, USA). Roswell Park.
Memorial Institute (RPMI) media (Gibco), pBAD Directional TOPO® Expression Kit, and ProBond™ Purification System were purchased from Invitrogcn (Carlsbad. CA, USA). DL- dithiothreitol (DTT), MgS04, and 13C6, 15N4-L-arginine hydrochloride were purchased from Sigma-Aldrich (St. Louis, MO, USA). LB media (MP Biomedicals), GC-Rich PCR system (Roche), 4-(2-hydroxyethyl)-l -pipcrazineethancsulfonic acid (HEPES) at pH 8.4, 2-propanol (Fisher Chemical), and borate buffer at pH 9.5 (R1CCA Chemical) were purchased from Fisher Scientific (Pittsburgh, PA, USA). The Quick Blunting kit and restriction enzymes (EcoRI, EcoRV, HindIII, SphI) were purchased from New England Biosciences (Ipswich, MA, USA). 4-Fluoro-7-nitro-2,l,3-bcnzoxadiazole (NBDF) was purchased from Dojindo Laboratories (Rockville, MD, USA). Amicon Ultra 3 kDa MW cutoff centrifugal filters and Ultrafree-MC GV centrifugal filters were purchased from Millipore (Milford, MA, USA). Polypropylene auto-sampler tubes with PTFE/silicone septa were purchased from Waters (Milford, MA, USA).
Acquisition and growth of P. aeruginosa isolates
The clinical microbiology lab at the University of Minnesota Medical Center hospital isolated and identified isolates of P aeruginosa from the sputum of patients with cystic fibrosis. These isolates were all de-identified and used under an exemption from the UMN institutional review board. P aeruginosa strain PA 14, its agmatine mutants, and the P aeruginosa clinical isolates were either grown in LB broth medium or RPMI as indicated in the text. All growth occured at 37°C with orbital shaking for liquid cultures at 225 rpm. RPMI was used for mass spectrometry analysis of spent media as it is a defined medium without added agmatine and did not cause the same degree of analyte signal suppression as did LB broth.
Creation of a speA deletion mutant of P. aeruginosa
The speA gene was amplified from P. aeruginosa PA 14 genomic DNA using forward primer 5′-TTGTTGACCTGGCC CGTCGA-3′ and reverse primer 5′-GGGAAGCGGAAATG AAGGGG-3′ and inserted into pEX18-Ap utilizing the native EcoRI and HindIII sites within the PCR fragment [18]. To create the speA knockout the plasmid was digested with SphI and EcoRV, followed by conversion to blunt ends with the NEB ‘Quick blunting’ system. This removed 744 bp near the center of speA leaving flanking regions of 1035 bp and 842 bp on either side. The final construct was transformed into the mating E. coli strain SM10 and subsequently mated with PA 14 with mutant colonies selected and treated as previously described [18]. The genomic DNA of the resulting mutants was screened via PCR using the primers for speA amplification. All PCR reactions were performed with the GC-Rich PCR system (Roche).
Expression of speA and synthesis of 13C5,15N4-agmatine
The speA gene was amplified from the chromosome of the P aeruginosa isolate PA 14 using forward primer 5′-CACC ATGGCCGCTCGACGGACT-3′ and reverse primer 5′- GGACAGGTACGCCGAGCGG-3′ and then cloned into the pBAD Directional TOPO vector as described by the manufacturer (Invitrogen). The speA gene product was expressed using the arabinose promoter intrinsic to the pBAD vector and purified using the imparted 6-His tag and a nickel embedded agarose slurry per manufacturer instructions (Invitrogen). The purified protein was reacted with a 10 mM concentration of 13C6,15N4-L-arginine hydrochloride (Sigma), 100 mM HEPES pH 8.4, 5 mM MgS04, 1 mM DTT, 0.04 mM pyridoxal phosphate at 37°C for 30 min [19]. The reaction was terminated by heat inactivation at 80°C for 10 minutes and the reaction was filtered through a 3 kDa MW cutoff filter from Millipore.
Sample preparation and derivatization for UPLC-MS/MS analysis
Samples for analysis by UPLC-MS/MS were spiked with a fixed volume of the isotopically labeled agmatine internal standard described above. One hundred microliters of this spiked sample was mixed with 200 μL of ice cold isopropanol and chilled to −20°C for 5–8 hours. The sample was then centrifuged at 21.000 × g and the supernatant was separated from the proteinacious pellet to a Amicon Ultra 3 kDa MW cutoff centrifugal filter (Millipore). This filter was centrifuged at 14,000 × g for 4–6 hours to recover at least 100 μL of filtrate. To 100 μL of this filtrate, 15 μL of borate buffer (pH 9.5) was added and followed by 15 μL of 10 mM NBDF in acetonitrile. The sample was mixed and immediately placed at 60°C for 10 min. After incubation the sample was placed on ice, then treated with 20 μL of 0.3 % formic acid within 2 min to stabilize the NBD-derivatized analytes. The sample was centrifuged for 5 min at 21,000 × g, and the supernatant was then centrifuged through an Ultrafrec-MC GV filter (Millipore) for final particulate removal before transfer to a polypropylene auto-sampler tube with PTFE/silicone septa (Waters).
Analysis of agmatine concentration in biological samples
A Waters Acquity UPLC coupled to a Waters triple quadrupole mass spectrometer (Acquity TQD) was used for separation and detection of agmatine. A Waters HSS T3 2.1 mm × 100 mm column (1.7 μm particles) at 35°C was used during the following 10 min gradient separation with A: water containing 0.1 % formic acid and B: ACN containing 0.1 % formic acid, at a flow rate of 0.4 mL/min: 3 % B, 0 to 2.0 min; 3 % B to 48 % B, 2.0 to 3.0 min; 48 % B. 3.0 to 5.0 min; 48 % B to 97 % B, 5.0 to 5.5 min; 97 % B, 5.5 to 7.5 min; 97 % B to 3 % B, 7.5 to 7.8 min; and 3 % B, 7.8 to 10.0 min. By directly infusing NBD-agmatinc (C11H15N7O3) and NBD-internal standard, cone voltages and collision energies for each selected reaction monitoring (SRM) transition were optimized. The transitions that produced the highest sensitivity for the determination of each analyte were selected for quantification: NBD-agmatine, 294.2 to 235.0; NBD-13C5, 15N4-agmatine, 303.2 to 240.1 (Fig. 1). Dwell time for each transition was 0.05 s. For electrospray ionization tandem mass spectrometry (ESI-MS/MS) in positive ionization mode, parameters were as follows: capillary, 3.5 kV; cone, 40 V; extractor, 3 V; rf lens, 0.3 V; source temperature, 100°C; dcsolvation temperature, 350°C; desolvation flow, 1000 L/h; cone gas flow, 20 L/h; low-mass resolution (Ql), 15 V; high-mass resolution (Ql), 15 V; ion energy (Ql), 0.3 V; entrance, −5 V; exit, 1 V; collision energy, 20 V; low-mass resolution (Q2), 15 V; high-mass resolution (Q2), 15 V; and ion energy (Q2), 3.5 V.
Fig. 1.
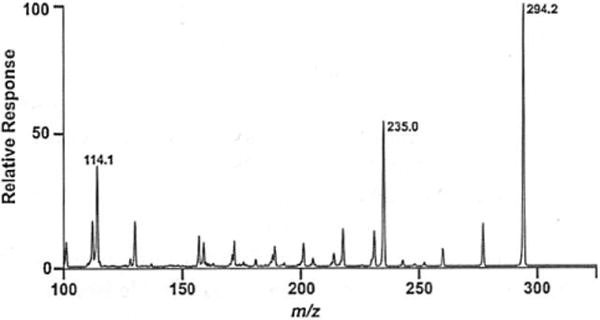
Product ion (MS/MS) scan of precursor mlz 294.2 (NBD-agmatine). Direct infusion of NBD-agmatine was employed to optimize cone voltages and collision energies for each selected reaction monitoring transition. The transition 294.2 to 235.0 was selected as the SRM transition that would produce the highest sensitivity for the detennination of agmatine in complex biologically derived mixtures. A similar infusion experiment was performed to determine the corresponding SRM transitions for 13C5.15N4-NBD-agmatine (not shown)
Method validation
Spike recoveries (accuracy) and associated analytical precisions were assessed by quantification of agmatine in samples prepared by the addition of 10 nM, 100 nM, 1 μM, and 10 μM agmatine to cell supernatants from cultures of P. aeruginosa that did not contain endogenous agmatine. For standardization, eight levels of calibration mixtures ranging from 0 to 10 μM were prepared for agmatine and 13C5,15N4-agmatine to achieve eight different response ratios for agmatine in the mixtures. These solutions were then analyzed by UPLC-MS/MS, and the data were subjected to a linear least squares analysis with the Waters Targetlynx™ software program. The peak area ratios of analyte/intemal standard were then used in conjunction with the calibration curves to determine the concentration of agmatine in the samples. Limits of detection (LOD) and quantitation (LOQ) were calculated by determining the signal-to-noise values for samples spiked with 50 nM agmatine and extrapolating to the concentration at which the signal-to-noise value was 3 for LOD or 10 for LOQ.
Analysis of agmatine secrctor strains
Clinical isolates PA002. PA004, PA005, PA006, and PA016, along with PA 14 wild-type (WT) and the PA 14 aguA∷Gm, Δagu2ABCA’ mutant [10] that cannot break down agmatine, were tested for their ability to grow on minimal medium with either agmatine or glutamate/ammonium sulfate as the sole carbon and nitrogen source. The strains were grown for 2 weeks on 0.8 % agarose minimal medium: 60 mM K2HPO4, 33 mM KH2PO4, and 195 μM MgSO4 supplemented with either 10 mM agmatine or 10 mM glutamate with 3.75 mM ammonium sulfate. The agarose liquid base was autoclaved then kept at 60°C, while the other ingredients were dissolved in water and filter sterilized using a 0.2-μm pore size filter, brought to 60°C, then added to the autoclaved agarose. Isolates PA002 and PA016 were chosen as positive controls for agmatine metabolism among the clinical isolates. They were also used as comparison for growth rate on a plate since isolate PA002 grows slowly similar to isolates PA004-006, while PA016 grows rapidly and swarms, comparable with PA014 and the aguA∷Gm, Δagu2ABCA’ mutant.
Results
Synthesis of the stable isotopically labeled 13C5,15N4-aginatine internal standard
Previous methods to measure agmatine have used various internal standards, including our own methods which included the use of homoagmatine [10]. Homoagmatine, however, can be difficult to synthesize and lacks stability for repeated use. The use of a stable isotopically labeled analog of the target molecule as an internal standard for quantification has long held a number of analytical advantages including but not limited to, compensation for sample handling losses, analyte instability, ionization variability, source contamination, and matrix suppression. Isotopically labeled agmatine is not commercially available, though a variety of isotopically labeled forms of arginine arc. Therefore, to synthesize a stable isotopically labeled form of agmatine for use as an internal standard in agmatine measurements, a method was developed that utilized the bacterial arginine decarboxylase (ADC) pathway of P. aeruginosa to decarboxylate uniformly labeled 13C6,15N4- arginine to produce 13C5,15N4-agmatine. The product of the speA gene in P. aeruginosa produces the enzyme ADC which has been purified and biochemically characterized previously [19]. However, this work was completed before modem molecular biology tools were available, and thus our purification technique was substantially different than previously described. As described in the “Materials and methods,” arginine decarboxylase from R aeruginosa strain PA 14 was cloned, overexpressed, and purified as shown in Fig. 2. The expected size of the speA gene product from P aeruginosa strain PA 14 is 70.6 kDa. The cloning strategy imparts a 6-His C-tcrminal tag (3 kDa) which aids in purification. The purified speA enzyme was then reacted with an excess of 13C6,15N4-L-arginine hydrochloride to produce stable isotopically labeled 13C5,15N4-agmatine. Successful production of 13C5,15N4-agmatine was verified by UPLC-MS/MS analysis (Fig. 3).
Fig. 2.
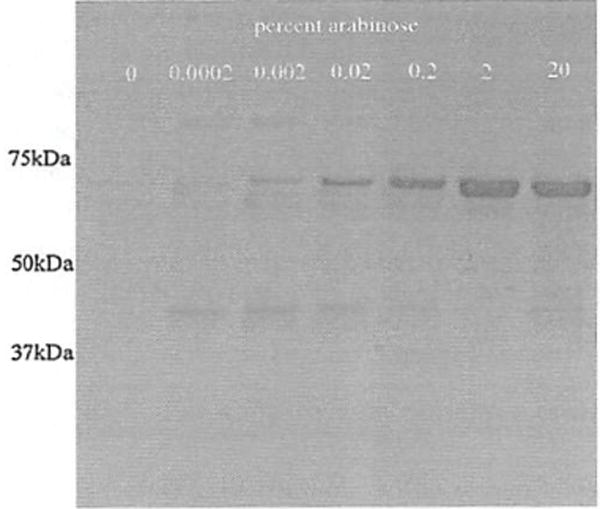
The expression and purification of arginine decarboxylase from P. aeruginosa. E. coli strain TOP 10 harboring the speA gene within the pBAD cloning vector was induced with varying amounts of arabinose and the cell lysates were purified using the imparted 6-His tag and a nickel agarose column described in the methods. The eluted samples were analyzed by SDS-PAGE 10% acrylamide gels and stained with Coomassie dye. The eluate from the 2% arabinose condition was used for synthesis of isotopically labeled agmatine
Fig. 3.
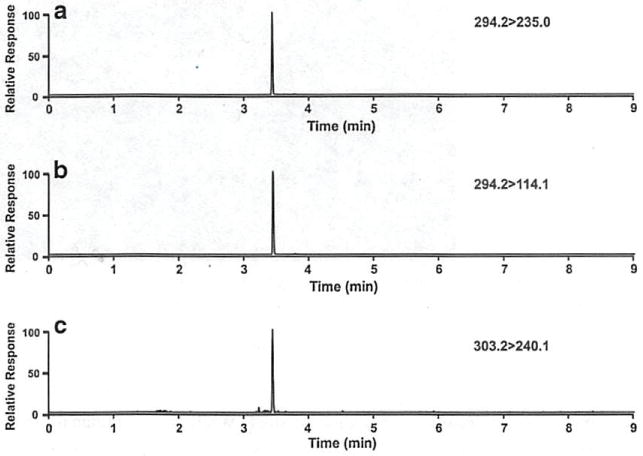
UPLC-MS/MS measurement of agmatine in bacterial culture supernatants. SRM transitions of NBD-agmatine employed for (a) quantification of agmatine. (b) identity verification of agmatine, (c) detection of the uniformly labeled 13C5,15N4-NBD-agmatine internal standard for isotope dilution quantification of agmatine
Analysis of agmatine by UPLC-MS/MS
In this study, a method was developed to rapidly quantify NBDF-derivatized agmatine in cell extracts using UPLC- MS/MS. A variety of gradient chromatography parameters, including column selection, flow rate, and mobile phase composition, were optimized to select the chromatographic separation that enabled a rapid 10 min separation and detection of agmatine.
Waters Acquity HSS-T3 C18 2.1 × 100 mm, BEH Shield RB18 C18 2.1 × 100 mm, and BEH C18 2.1 × 100 mm columns were tested. The best separation was achieved using a Waters Acquity HSS T3 C18 2.1 × 100 mm column. Flow rates of 0.4 to 0.8 mL/min were tested, and 0.4 mL/min was found to be the optimal flow rate from the standpoint of chromatographic resolution combined with minimal analysis time. Representative SRM chromatograms for NBD-agmatine and NBD-13C5,15N4-agmatine are shown in Fig. 3a–c. All calibration curves were linear over the range of 5 nM- 10 μM. Correlation coefficients ranged from 0.984 to 0.997 and averaged 0.990. To evaluate the accuracy of the method, agmatine was spiked into blank cell extracts at concentrations of 10 nM, 100 nM, 1 μM, and 10 μM prior to sample processing. UPLC-MS/MS was performed, and concentrations of analytes were calculated from calibration curves for agmatine. Precisions of the spike recovery sample set (see Electronic supplementary material (ESM) Table S1) ranged from 1.2 % relative standard deviation (RSD) to 10.0 % RSD and averaged 5.5% RSD. Recoveries (accuracies) of agmatine ranged from 94 to 100% and averaged 96.5%.
Validation of P aeruginosa mutants of arginine decarboxlyase pathway
To describe the functional outcomes of agmatine metabolism in P. aeruginosa, all the genes responsible for agmatine synthesis and metabolism were mutated, and every combination of these mutations was subjected to agmatine analysis by UPLC-MS/MS. As shown in Fig. 4, these mutants were grown in agmatine-free RPMI or RPMI supplemented with 10 μM agmatine overnight, and the remaining agmatine in the supernatants was measured. Endogenous ADC function is most clearly confirmed in the agmatine deiminase (AgDI) mutants that synthesize agmatine but do not metabolize it, resulting in agmatine secretion into the growth medium.
Fig. 4.
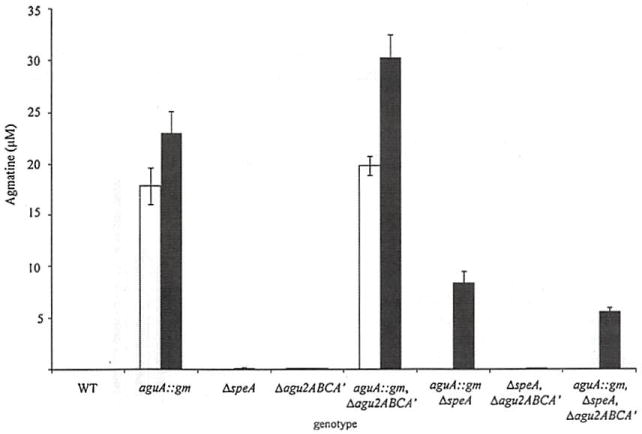
The agmatine metabolic phenotype of agmatine pathway mutants. The agmatine pathway mutants were grown overnight in RPMI (white bars) or RPMI supplemented with 10 μM agmatine (black bars). Error bars represent standard deviation of three measurements
This phenotype is negated by mutation of speA. While the genotypes of the AguA∷gm and Δagu2ABCA’ were previously described [10], confirmation of the further mutation of speA is shown in Fig. 5. As previously described, the AgDI enzymes of the agu2ABCA’ operon (Agu2A and Agu2A’) do little to impact agmatine values as they are not as metabolically active as aguA during planktonic growth [10]. Combined, these data validate the functions of the gene products involved in agmatine synthesis and metabolism.
Fig. 5.
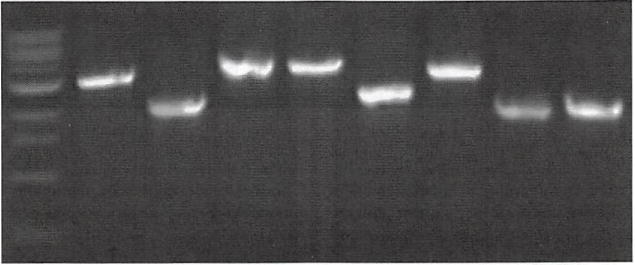
Continuation of speA internal deletion by PCR. PCR products from the speA F and speA R primers are shown for the selected mutants used in this work. The mutagenesis strategy removes 744 bp from the middle of the gene to render its protein product nonfunctional. Lanes 1, 1 kB ladder; 2, PA14 (WT); 3. ΔspeA: 4. aguA:gm\ 5. Δagu2ABCA’; 6. aguA:gm, ΔaspeA; 7, aguA:gm. Δagu2ABCA’: 8. Δagu2ABCA’ ΔspeA: 9. aguA:gm. Δagu2ABCA’, ΔspeA
Discovery of clinical isolates harboring aguA mutations
To validate the utility of agmatine measurement in bacterial supernatants, 31 clinical isolates of P. aeruginosa obtained from the sputum of patients with cystic fibrosis were analyzed. Results are illustrated in Fig. 6. Isolates PA004, PA005, and PA006 are all from different patients and visually distinct, but each demonstrates both a secretion of agmatine and an inability to metabolize agmatine, suggesting these are mutants of the aguBA operon, which is genetically intact in all isolates we have tested and likely ubiquitous in all P. aeruginosa [10]. The inability to metabolize agmatine by these clinical isolates was further validated by a complete lack of growth on agmatine minimal media which only contains agmatine as the carbon and nitrogen source. We have used this approach to describe lab created agmatine mutants in our prior work [10]. One explanation for the agmatine secretion phenotype of isolates PA004, PA005, and PA006 would be a loss of function of the aguA gene. Investigation of this hypothesis is the subject of a separate study beyond the scope of the current report.
Fig. 6.
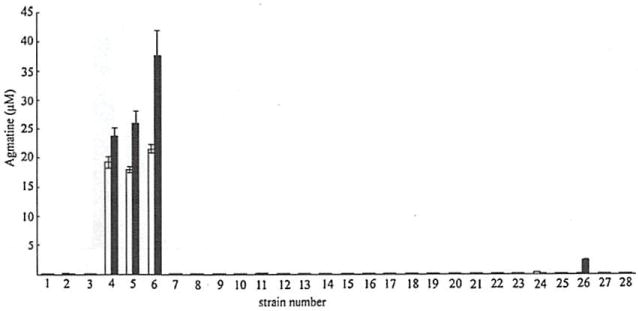
Agmatine production and consumption by clinical strains. Twenty-eight clinical isolates of P. aeruginosa were grown overnight with or without agmatine and 100 μL of the resultant supernatants were analyzed for agmatine. White bars represent cultures grown without agmatine; black bars represent cultures grown with 10 μM agmatine
Discussion
Agmatine is not a widely recognized molecule; however, it is critical to cellular processes in widely diverse species [4, 20–23]. It has a clearly established role in prokaryotes as an intermediary in the synthesis of polyamines, but numerous higher-order purposes are being proposed in eukaryotes including its role as a neurotransmitter, vasoactive mediator, immunomodulatory agent, and inhibitor of nitric oxide synthase [5]. While techniques to measure agmatine have been developed before, most lack the sensitivity and selectivity of LC/MS-bascd approaches and require time consuming, labor-intensive sample preparation protocols, while others, including reported LC/MS-based methods employ no or poorly chosen internal standards. There are numerous advantages to using isotope dilution LC/MS/MS analysis of agmatine, including equality in degradation, matrix behavior, labeling behavior, and solubility during extraction. In an effort to create the most effective internal standard for use in agmatine quantification, the arginine decarboxylase pathways of P aeruginosa were explored in this work, and a cloned and overexpressed ADC was employed to synthesize stable isotopically labeled agmatine (13C5,15N4-agmatine). With the resulting isotope dilution UPLC-MS/MS method developed here, we analyzed the outcomes of various mutations in the synthesis and metabolism of agmatine in P. aeruginosa. This technique validated many presumed functions of the genes in this system as well as their interactions with each other. These findings include the synthesis of agmatine by speA and its removal by aguA, and that naturally occurring or laboratory-induced mutations of aguA result in a robust secretion of agmatine into the extracellular matrix. The purpose of agmatine secretion by these mutants will be the focus of future work; as it appears, this mutation is being retained in the P aeruginosa populations of patients with cystic fibrosis. Given agmatine’s higher-order functions, it is possible bacterially secreted agmatine alters the local inflammatory milieu to afford some benefit to the pathogen. Mutations of the AgDI pathways in the laboratory pathogen PA 14 show altered virulence in animal models of pneumonia [11]. As cystic fibrosis patients typically maintain the same P aeruginosa isolates for decades, tracking patients with these infections may offer insight into further biological roles of agmatine in disease. The outcome of these infections with agmatine hyper-secreting isolates could be monitored by measuring agmatine in their lung secretions with the UPLC-MS/MS method reported here, and subsequently correlated with other markers of inflammation and infection.
Acknowledgments
This research was supported by NIH NHLBI P30 HL101311-01 and NIH K08 PA-10-059 to BJW.
Footnotes
Conflicts of interest The authors report no conflicts of interest.
References
- 1.Wortham BW, Patel CN, Oliveira MA. Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv Exp Med Biol. 2007;603:106–115. doi: 10.1007/978-0-387-72124-8_9. [DOI] [PubMed] [Google Scholar]
- 2.Gugliucci A. Polyamines as clinical laboratory tools. Clin Chim Acta. 2004;344:23–35. doi: 10.1016/j.cccn.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 3.Janowitz T, Kneifel H, Piotrowski M. Identification and characterization of plant agmatine iminohydrolase, the last missing link in polyamine biosynthesis of plants. FEBS Lett. 2003;544:258–61. doi: 10.1016/s0014-5793(03)00515-5. [DOI] [PubMed] [Google Scholar]
- 4.Li G, Regunathan S, Barrow CJ, Eshraghi J, Cooper R, Reis DJ. Agmatine: an endogenous clonidine-displacing substance in the brain. Science. 1994;263:966–969. doi: 10.1126/science.7906055. [DOI] [PubMed] [Google Scholar]
- 5.Piletz JE, Aricioglu F, Cheng JT, Fairbanks CA, Gilad VH, Haenisch B, Halaris A, Hong S, Lee JE, Li J, Liu P, Molderings GJ, Rodrigues AL, Satriano J, Seong GJ, Wilcox G, Wu N, Gilad GM. Agmatine: clinical applications after 100 years in translation. Drug Discov Today. 2013;18:880–893. doi: 10.1016/j.drudis.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 6.Wade CL, Eskridge LL, Nguyen HO, Kitto KF, Stone LS, Wilcox G, Fairbanks CA. Immunoneutralization of agmatine sensitizes mice to micro-opioid receptor tolerance. J Pharmacol Exp Ther. 2009;331:539–46. doi: 10.1124/jpet.109.155424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reis DJ, Regunathan S. Agmatine: an endogenous ligand at imidazoline receptors is a novel neurotransmitter. Ann NY Acad Sci. 1999;881:65–80. doi: 10.1111/j.1749-6632.1999.tb09343.x. [DOI] [PubMed] [Google Scholar]
- 8.Regunathan S, Piletz JE. Regulation of inducible nitric oxide synthase and agmatine synthesis in macrophages and astrocytes. Ann NY Acad Sci. 2003;1009:20–29. doi: 10.1196/annals.1304.002. [DOI] [PubMed] [Google Scholar]
- 9.Mun CH, Lee WT, Park KA, Lee JE. Regulation of endothelial nitric oxide synthase by agmatine after transient global cerebral ischemia in rat brain. Anat Cell Biol. 2010;43:230–40. doi: 10.5115/acb.2010.43.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams BJ, Du RH, Calcutt MW, Abdolrasulnia R, Christman BW, Blackwell TS. Discovery of an operon that participates in agmatine metabolism and regulates biofilm formation in Pseudomonas aeruginosa. Mol Microbiol. 2010;76:104–119. doi: 10.1111/j.1365-2958.2010.07083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulson NB, Gilbertsen AJ, Dalluge JJ, Welchlin CW, Hughes J, Han W, Blackwell TS, Laguna TA, Williams BJ. The arginine decarboxylase pathways of host and pathogen interact to impact inflammatory pathways in the lung. PLoS One. 2014 doi: 10.1371/joumal.pone.0111441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y, Halaris AE, Piletz JE. Determination of agmatine in brain and plasma using high-performance liquid chromatography with fluorescence detection. J Chromatogr B Biomed Sci Appl. 1997;691:277–286. doi: 10.1016/s0378-4347(96)00458-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhao SL, Feng YZ, LeBlanc MIL, Piletz JE, Liu YM. Quantitation of agmatine by liquid chromatography with laser-induced fluorescence detection. Anal Chim Acta. 2002;470:155–161. [Google Scholar]
- 14.Zhao S, Wang B, Yuan H, Xiao D. Determination of agmatine in biological samples by capillary electrophoresis with optical fiber light-emitting-diodc-induced fluorescence detection. J Chromatogr A. 2006;1123:138–141. doi: 10.1016/j.chroma.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 15.Huisman H, Wynveen P, Nichkova M, Kellerman G. Novel ELISAs for screening of the biogenic amines GABA, glycine, beta-phenylethylamine, agmatine, and taurine using one derivatization procedure of whole urine samples. Anal Client. 2010;82:6526–6533. doi: 10.1021/ac100858u. [DOI] [PubMed] [Google Scholar]
- 16.Song Y, Quan Z, Evans JL, Byrd EA, Liu YM. Enhancing capillary liquid chromatography/tandem mass spectrometry of biogenic amines by pre-column derivatization with 7-fluoro-4-nitrobenzoxadiazole. Rapid Commun Mass Spectrom. 2004;18:989–994. doi: 10.1002/rcm.1437. [DOI] [PubMed] [Google Scholar]
- 17.Ubhi BK, Davenport PW, Welch M, Riley J, Griffin JL, Connor SC. Analysis of chloroformate-derivatised amino acids, dipeptides and polyamines by LC-MS/MS. J Chromatogr B. 2013;934:79–88. doi: 10.1016/j.jchromb.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld HJ, Roberts J. Arginine decarboxylase from a Pseudomonas species. J Bacteriol. 1976;125:601–607. doi: 10.1128/jb.125.2.601-607.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder S, Wirth JJ, Bitonti AJ, McCann PP, Kierszenbaum F. Biochemical evidence for the presence of arginine decarboxylase activity in Trypanosoma cruzi. J Parasitol. 1992;78:371–374. [PubMed] [Google Scholar]
- 21.Minie Z, Herve G. Arginine metabolism in the deep sea tube worm Riftia pachyptila and its bacterial endosymbiont. J Biol Chem. 2003;278:40527–40533. doi: 10.1074/jbc.M307835200. [DOI] [PubMed] [Google Scholar]
- 22.Satriano J. Agmatine: at the crossroads of the arginine pathways. Ann NY Acad Sci. 2003;1009:34–43. doi: 10.1196/annals.1304.004. [DOI] [PubMed] [Google Scholar]
- 23.Yanagisawa H. Agmatine deiminase from maize shoots: purification and properties. Phytochemistry. 2001;56:643–647. doi: 10.1016/s0031-9422(00)00491-x. [DOI] [PubMed] [Google Scholar]


