Understanding both genetic and environmental factors contributing to risk and resilience for psychiatric disorders is critical for developing better approaches to prevention and intervention. Recent research indicates many genetic and environmental factors contribute to risk and resilience across diagnostic boundaries (Belsky et al., 2007, Caspi et al., 2014, Caspi et al., 2003, Kim-Cohen and Gold, 2009). These findings shift the focus of research towards intermediate phenotypes associated with cross-disorder psychological functioning. One promising phenotype to consider in understanding psychopathology is emotion dysregulation (Bradley et al., 2011a, Cicchetti et al., 1995, Gross, 2002, Gross and Munoz, 1995, John and Gross, 2004).
Emotion dysregulation reflects deficits in the ability to regulate intense, negative, and shifting emotional states and is seen as a transdiagnostic process that is linked to increased risk for the development and maintenance of a range of psychopathology, including depression and posttraumatic stress disorder (PTSD; Aldao et al., 2010, Berenbaum et al., 2003, Bradley et al., 2011b, Kring, 2008). While it is strongly related to the presence of negative affect, it is in fact a distinct construct representative of problems with the regulation of those negative emotions (Bradley et al., 2011). Traumatic experiences, especially in early life, appear to put individuals at greater risk for the development of emotion regulation difficulties and subsequent psychopathology (Alink et al., 2009, Horwitz et al., 2001, Kim and Cicchetti, 2010, Southam-Gerow and Kendall, 2002). Despite growing evidence of the importance of emotion dysregulation across psychiatric conditions, there remains a great deal to understand about how this cross-disorder risk factor may contribute to psychiatric symptoms.
Only a limited number of studies have investigated genetic associations with emotion dysregulation and similar constructs. These have primarily focused on candidate genes already associated with a range of psychiatric and stress-related conditions, such as the oxytocin receptor gene (OXTR; Bradley et al., 2011b, Kim et al., 2011), the serotonin transporter gene (5-HTT; Canli and Lesch, 2007, Hariri and Holmes, 2006), catechol-o-methyltransferase (COMT; Drabant et al., 2006), and monoamine oxidase A (MAOA; Buckholtz and Meyer-Lindenberg, 2008, Williams et al., 2009). There are limitations to candidate gene studies, including the need for a priori hypotheses and subjective decisions in what genes to examine. Another method used to study genetic associations are genome wide association studies (GWAS), which use an unbiased approach to provide an evaluation of common genetic variation across the genome and can identify genetic risk loci. To date, there have been no GWAS with emotion dysregulation. However, a number of recent studies performed with major psychiatric disorders have shown evidence of cross-disorder risk genes (Green et al., 2010, Hodgkinson et al., 2004). As part of the Psychiatric Genomics Consortium (PGC), the largest genome-wide analysis of psychiatric illness thus far, investigators found initial evidence that specific SNPs are significantly associated with cross-disorder risk of both childhood-onset and adult-onset psychiatric conditions (Smoller et al., 2013). The investigators also examined shared genetic etiology and found evidence for shared genetic variation across schizophrenia, bipolar disorder, major depressive disorder, autism spectrum disorders, and attention-deficit/hyperactivity disorder (Kendler et al., 2013). In addition, a meta-analysis of GWAS studies for neuroticism, a personality trait characterized by strong negative emotions and associated with emotion dysregulation, found evidence for a genetic locus that has been associated with major depressive disorder, bipolar disorder, and schizophrenia (de Moor et al., 2015). Because emotion dysregulation has been implicated as a risk factor and component of a range of psychiatric conditions, gaining a better understanding of genetic risk loci associated with emotion dysregulation could help to inform cross-disorder risk more generally.
In this study, we present the results of an initial GWAS of emotion dysregulation, demonstrating sex-specific differences in the genetic architecture of this phenotype. Because of the data suggesting that emotion dysregulation is a phenotype that cuts across psychiatric disorders, we then conducted post-hoc analysis examining whether the SNPs identified in the GWAS were associated with current symptom levels of major depressive disorder and PTSD, as well as lifetime history of suicide attempt. Next, we performed additional genomic analyses demonstrating that these SNPs are likely functional in that they are associated with differential regulation of methylation (via meQTL analyses). Finally, using gene set enrichment analyses, we examined pathways related to the top genes within the entire GWAS of emotion dysregulation, to more broadly determine biological processes involved.
Methods and Materials
Participants
A total of 2600 African American adults (aged 18–65 years; mean age = 39, 70% female) were enrolled as part of a larger study investigating genetic risk for stress-related disorders. The sample was predominantly low income with approximately 70% of participants unemployed and 85% reporting a household monthly income less than $2000. Participants were recruited from the general medical clinics of a publicly funded hospital, as detailed previously (Gillespie et al., 2009). Individuals were deemed eligible for participation if they could give informed consent and understand English, as determined by a study researcher. Study procedures were approved by the institutional review board of Emory University School of Medicine. After signing the informed consent approved by the Emory Institutional Review Board, an initial interview was administered by trained research assistants.
Phenotype Measures
Emotion Dysregulation Scale (EDS)
The EDS is a 12-item self-report scale to measure the severity of current emotion dysregulation symptoms (Bradley et al., 2011a, Powers et al., 2015). Items are scored on a 7-point Likert scale ranging from 1 (“Not true”) to 7 (“Very true”). Items assess domains of emotional experiencing (e.g., “Emotions overwhelm me”), cognition (e.g., “When I’m upset, everything feels like a disaster or crisis”), and behavior (e.g., “When my emotions are strong, I often make bad decisions”). The internal consistency of the EDS was high (α=0.94). Average total score for the overall sample was 37.19 (SD=21.08, range=12-84) with similar scores across sex; for males, mean=35.04, SD=20.20; for females, mean=38.10, SD=21.38.
Modified PTSD Symptom Scale (mPSS)
The mPSS (Falsetti et al., 1993) is a psychometrically valid 17-item self-report measure assessing frequency of PTSD symptoms over the prior two weeks. It distinguishes among re-experiencing, avoidance, and hyperarousal symptom clusters of PTSD. Current PTSD diagnosis was determined based on DSM-IV-TR (APA, 2000) criteria. In the overall sample, 769 (30.7%) met for current PTSD (males only, n=220, 29%).
Beck Depression Inventory-II (BDI-II)
The BDI-II (Beck et al., 1996) is a widely used, 21-item self-report measurement of depressive symptoms. In the present study, current depression diagnosis was determined based on DSM-IV-TR (APA, 2000) criteria. In the overall sample, 753 (29.1%) met for current depression (males only, n=201, 26.1%). In addition to the BDI, participants were also asked to self-report any history of suicide attempts (overall sample, n=335, 13.1%; males only, n=66, 8.7%).
Traumatic Events Inventory (TEI)
The TEI is a lifetime assessment of different categories of traumas based on a yes/no answer for natural disasters, accidents, life-threatening illnesses, military combat, witnessing a murder or assault of a family member or close friend, sexual/physical assaults, and childhood abuse for a total of 21 items (Gillespie et al., 2009; Schwartz et al., 2005). Overall level of trauma exposure variable reflects the sum of the different types of traumatic events experienced or witnessed by the participant (overall sample: mean = 4.50, SD = 3.28, range=0-19; in males: mean=5.24, SD=3.29; in females: mean=4.19, SD=3.23). Within the overall sample, 40% of individuals reported exposure to child abuse (in males, n=250; in females, n=772).
Genotyping and Quality Control
Participants provided a saliva sample and/or blood sample. DNA was extracted from saliva in Oragene collection vials (DNA Genotek Inc, Ontario, Canada) using the DNAdvance kit (Beckman Coulter Genomics, Danvers, MA), while DNA from blood was extracted using either the E.Z.N.A. Mag-Bind Blood DNA Kit (Omega Bio-Tek, Inc., Norcross, GA) or ArchivePure DNA Blood Kit (5 Prime, Inc, Gaithersburg, MD). Genome-wide SNP genotyping was conducted on approximately 5000 subjects using Illumina’s Omni1-Quad BeadChip, which interrogates 1,011,219 individual SNPs. Genotypes were called using Illumina’s GenomeStudio software. We used PLINK (Purcell et al., 2007) to perform quality-control (QC) analyses on the genetic data. In brief, initial QC involved removing samples with very low call rates and those outside acceptable levels of heterozygosity (−0.25<Fhet>0.25); the remaining samples were recalled in GenomeStudio. We then removed SNPs with call rates less than or equal to 98%, and a frequency of less than 0.01, and individuals with greater than 2% missing data. We further identified and removed related individuals by using PLINK to estimate the proportion of identity by descent (IBD) for each pair of individuals. Among pairs of individuals with an IBD proportion > 0.12 (indicating cousins or a closer relation), we removed the individual in each pair with the higher rate of missing genotype data. Using data (autosomes only) pruned in PLINK, we performed principal-component analysis (PCA) to infer axes of ancestry and remove outlier subjects. Based on PCA, we retained those African-American individuals who fell within three standard deviations of the medians of the first and second principal components in our sample. After completion of QC and PCA, our sample consisted of 3814 African-American individuals genotyped for 883,511 SNPs. Of the individuals included in the genetic sample (N=3814), 68% had EDS data and were included in the GWAS (N=2600). When comparing the genotyped individuals to the non-genotyped individuals, no significant differences in demographic measures relevant to this study were found; furthermore, the full genetic sample and analytic samples were similar.
DNA methylation
Blood samples were collected from a subset of participants in EDTA vacuum tubes (Overall sample, N=2600; subsample for methylation in males, N=107). DNA was extracted using the Puregene Genomic DNA kit (Invitrogen). Samples were resolved on a 1% agarose gel to verify that the DNA was of high molecular weight (at least 2kb) and quantified using PicoGreen (Invitrogen). DNA methylation was interrogated for each sample using the HumanMethylation450 BeadChip (Illumina). Briefly, 1 μg of DNA was converted with sodium bisulfite, amplified, fragmented, and hybridized according to the manufacturer’s instructions. Beta values were generated with BeadStudio and were set to missing (no call) if detection p-values exceeded .001. All samples had probe detection call rates <95% and an average intensity value of either <50% of the experiment-wide sample mean or <2,000 arbitrary units (AU). CpG sites with missing data for >10% of samples were excluded from analysis. Normalization of probe distribution and background differences between Type I and type II probes was conducted using Beta Mixture Quantile Normalization (BMIQ; Teschendorff et al., 2013), and ComBat was used to account for sources of technical variations including batch and positional effects (Johnson, Li, & Rabinovic, 2007).
Statistical Approach
Using the statistical package PLINK (Purcell et al., 2007), we regressed continuous score on the EDS on allele count, assuming an additive model (0, 1, or 2 copies of risk allele) and the top ten principal components (Lin and Zhoa, 2009, Price et al., 2006) of genome-wide data as covariates separately in males (N=774) and females (N=1826). Previous research suggests that biological substrates of psychiatric disorders in men and women may be different (Harrison and Tunbridge, 2008, Williams and Gatt, 2009). Additionally, a number of recent GWAS, linkage, and methylation studies have demonstrated sex-specific genome-wide significance across a number of medical and psychiatric disorders (Chesi et al., 2015; Sharma et al., 2015; Werling et al., 2014; and Xu et al., 2014). We have also found sex-specific effects of genes on various psychiatric outcomes including PTSD (Ressler et al., 2011) and depression (Heim et al., 2009), and since emotion dysregulation is a transdiagnostic factor related to both of these psychiatric conditions, we ran separate GWAS for males and females.
Additional posthoc analyses were conducted using logistic regressions to examine the unique predictive value of the GWAS identified SNP associated with emotion dysregulation and psychiatric conditions of interest. The emotion dysregulation measure was positively skewed. However, the level of skewness and kurtosis (overall sample: skewness = 0.58, kurtosis = -0.87, males: skewness = 0.67, kurtosis = -0.73, females: skewness = 0.54, kurtosis = -0.92) fell within acceptable parameters for the sample size (Tabachnick and Fidell, 2001). Correlations between emotion dysregulation and the other phenotype variables were examined and all were significantly correlated at p < .001: For EDS and current depression (BDI-II), rpb = 0.54; for EDS and current PTSD (mPSS), r = 0.43, for EDS and lifetime suicide attempt, rpb = 0.25. Logistic regression models were built in two steps to identify the unique predictive value of emotion dysregulation above and beyond the effect of demographic variables and SNP. More specifically, Model 1 included demographic variables (i.e., age education level, and income level) and SNP in predicting psychiatric outcome variables. Then, Model 2 included demographic variables, SNP, and emotion dysregulation in predicting psychiatric outcome variables.
Next, in males only, we examined methylation quantitative trait loci (meQTL) in this cohort by applying the approach described previously (Smith et al., 2014) to the HumanMethylation450 data from the parent study; rs6602398 genotype was tested for association with the proportion of methylation for the 5 CpG sites in IL2RA. Linear regressions were used to model methylation of each CpG site as a linear function of genotype, controlling for the cellular proportions (CD4, CD8, CD14, CD56, and CD19) and the first 3 principal components from the GWAS. Multiple test correction was not used since the purpose of this analysis is to extend the genome-wide corrected SNP association analysis and methylation of adjacent CpG sites may be correlated and may not represent independent tests.
We also examined the association signal of GWAS markers in a set of genes categorized by biological pathways, assuming that emotion dysregulation, which may represent cross-disorder risk, may result from a number of genes which disrupt one or more pathways. To reduce bias, we applied a statistical method to identify overrepresented pathways in a single GWAS dataset. Analyses were conducted using WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/; Wang et al., 2013, Zhang et al., 2005), a web-based gene set analysis toolkit which uses genes as the input. It incorporates published information about biochemical pathways taken from the MolecularSignatures Database (MSigDB; http://software.broadinstitute.org/gsea/msigdb/index.jsp). We first used Gene Set Enrichment Analysis (GSEA) which was developed for microarray gene expression analysis (Subramanian et al., 2005) but has also been used for GWAS (Jia, Wang, Meltzer, & Zhao, 2010). Using the male sample GWAS SNPs, we included the genes for all SNPs with a cut-off p-value of 1x10−4. Even if more than one SNP was identified for a gene, the gene was only included in the analysis once. Then, we ran a disease enrichment analysis, which examined the list of genes identified through GSEA against a database of genes that past research has found to be associated with specific diseases and provides a list of disease pathways that are “over-represented” by the identified list of genes from MSigDB curated gene sets (http://software.broadinstitute.org/gsea/msigdb/index.jsp). It then compares the number of genes in the above list that are on a given disease to the total number of genes on the pathway overall. More specifically, GSEA takes an a prioi set of genes S, and determines whether the members of S are randomly distributed through L or primarily found at the top or bottom. An “enrichment score” (ES) is calculated to determine the degree to which a set S is overrepresented at the extremes (top or bottom) of the entire ranked list L; this is calculated with a running-sum statistic of “hits” (when S genes are present in L) and “misses” (genes encountered in L that are not in S): Phit – Pmiss. Statistical significance of the ES is based on a permutation approach, comparing ES with ESNULL based on randomly assigned phenotypes (see Subramanian et al., 2005 for a full description of this approach and all statistical methodology). We also ran a Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) pathway enrichment analysis. Multiple testing correction was performed using the Benjamini-Hochberg method (Benjamini and Hochberg, 1995). All significant results at adjusted p<.01 are presented. See Figure 1 for a full depiction of the analytic approach and relative sample sizes for each analysis run.
Figure 1.
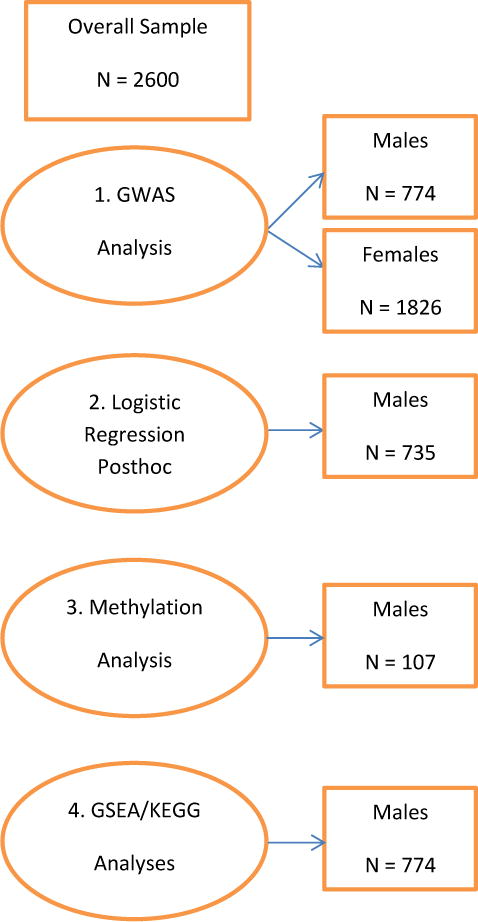
Representation of analytic approach and relative sample sizes for each analysis conducted. GSEA = Gene Set Enrichment Analysis; KEGG = Kyoto Encyclopedia of Genes and Genomes.
Results
GWAS with Emotion Dysregulation
We found no evidence to suggest inflation of the association test statistics in the sample (genomic inflation factor λ=1.00; Supplementary Figure 1). Using a cutoff level of 5x10−8, we did not find any SNPs significantly associated with emotion dysregulation in females. In contrast, in the male cohort, we found that rs6602398, a SNP that resides within IL2RA at chromosomal position 10p15.1, was significantly associated with EDS (Figure 2A, additive model: N=774, β (SE)=14.9 (2.6); p=1.1×10−8)1. Further, a peak in IL2RA shows several SNPs that were also associated with the outcome (Figure 2B). SNP rs6602398 had a minor allele frequency (MAF) of 0.04, and a HWE p-value of 1 in males.
Figure 2.
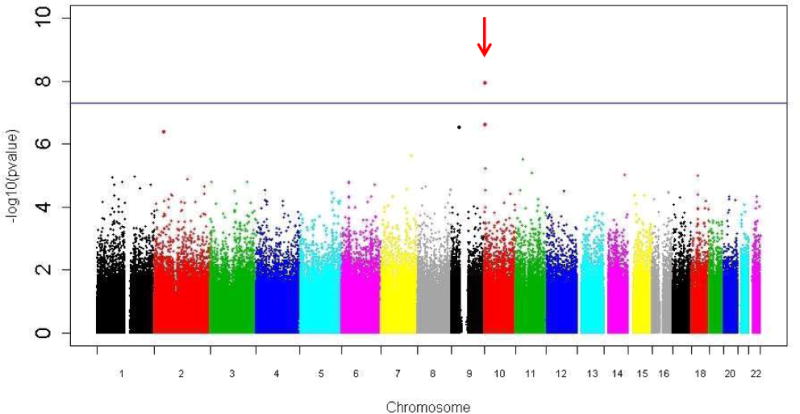
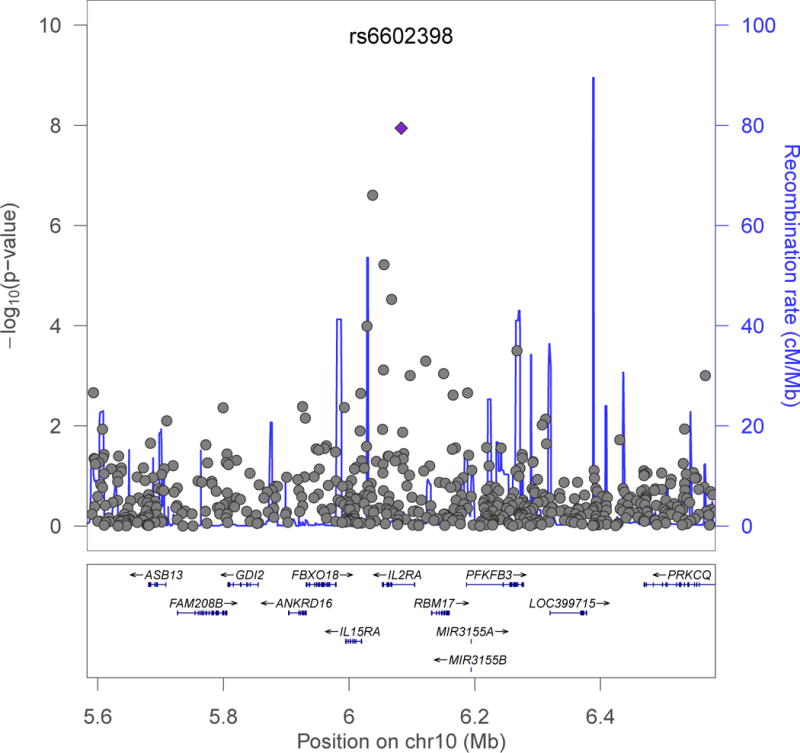
rs6602398, on chromosome 10, is associated with emotion dysregulation in males.
a) Manhattan plot for emotion dysregulation in the sample (African American males only, N=774) shows genome-wide associated SNP rs6602398 (β (SE)=14.9 (2.6); p=1.1×10−8).
b) Regional plot of chromosome 10 showing a peak of associated SNPs associated with emotion dysregulation in IL2RA (significant SNP rs6602398 is shown by a diamond) in males. Plot generated with LocusZoom (Prium et al., 2010).
Associations of Emotion Dysregulation and other Psychiatric Conditions with IL2RA SNPs
Logistic regression analyses were conducted in males to determine the association between EDS-associated SNP (rs6602398) and psychiatric symptoms (i.e., current depression, current PTSD, and lifetime history of suicide attempt). As shown in Table 1, rs6602398 was significantly associated with current depression and PTSD, but not lifetime suicide attempt. When emotion dysregulation symptoms were included in the model, the associations between rs6602398 and current depression and PTSD were no longer significant.
Table 1.
Associations among current PTSD, current depression, and lifetime history of suicide attempt with EDS-associated SNP in males using logistic regression models
| BasePair Position+ | Closest Gene | Model 1 | Model 2 | |||||
|---|---|---|---|---|---|---|---|---|
| B | Exp(B) | OR (95% CI) | B | Exp(B) | OR (95% CI) | |||
| Depression | ||||||||
| rs6602398 | 6082953 | IL2RA | 0.98*** | 2.67 | 1.55 – 4.59 | 0.22 | 1.25 | 0.66 – 2.35 |
| PTSD | ||||||||
| rs6602398 | 6082953 | IL2RA | 0.73** | 2.07 | 1.23 – 3.48 | 0.09 | 1.10 | 0.61 – 1.96 |
| Suicide Attempt History | ||||||||
| rs6602398 | 6082953 | IL2RA | 0.60 | 1.82 | 0.86 – 3.86 | -0.02 | 1.05 | 0.44 – 2.18 |
BasePair position is from Build GRCh37.1
N=735;
p < .001,
<.01
Model 1) Final step adjusting for age, education level, and income level, with rs6602398 SNP.
Model 2) Final step adjusting for age, education level, income level, and emotion dysregulation (EDS) with rs6602398 SNP.
Methylation Analyses with EDS-associated SNPs
One way in which a genetic variant can influence gene expression is through association with the nearby epigenetic structure, which may regulate gene expression. We examined the relationship between the EDS-associated SNP in IL2RA (rs6602398) and methylation of IL2RA CpG sites. In a subset of 107 GTP males, rs6602398 was associated with methylation of a CpG site (cg11733245) in IL2RA (p<.05). These results were not corrected for multiple testing.
Gene and Disease Enrichment Analyses
Using GSEA with the male sample GWAS SNPs, the results showed a list of 51 identified genes (see supplemental materials). Those 51 genes were then run in a disease enrichment analysis to determine potential disease pathways that are “over-represented” by the identified genes. Using this technique and setting a cutoff of p<.01 after correction for multiple tests, ten psychiatric disease pathways were identified (see Table 2). Similarly, using those 51 genes we also conducted a KEGG pathway enrichment analysis. We found one pathway with an adjusted probability of p<.01: the Calcium Signaling Pathway (number of reference genes in pathway = 174, number of study genes = 4, expected number of genes in pathway = 0.43, ratio of enrichment = 9.40, adjusted p-value = 0.008).
Table 2.
Disease Enrichment Analysis Results
| Disease Pathway | Number of Reference Genes in Disease Pathway | # of Genes in Disease Pathway (Expected Number of Genes) | Ratio of Enrichment | Adjusted p-value |
Associated Genes |
|---|---|---|---|---|---|
| Anxiety Disorder | 137 | 6 (.33) | 17.91 | 8.06 x 10−5 | DISC1, NLGN1, TACR1, NRG3, CACNA1C, GABRA5 |
| Unspecified Disturbance of Conduct | 95 | 5 (.23) | 21.53 | .0001 | NRG3, VWC2L, GABRA5, TTL, ARHGAP15 |
| Bipolar Disorder | 286 | 7 (.70) | 10.01 | .0002 | NRG2, DISC1, NLGN1, TACR1, NRG3, CACNA1C, GABRA5 |
| Autistic Disorder | 125 | 5 (.31) | 16.36 | .0003 | DISC1, NLGN1, STS, CACNA1C, GABRA5 |
| Autism Spectrum Disorder | 138 | 5 (.34) | 14.82 | .0004 | DISC1, NLGN1, STS, CACNA1C, GABRA5 |
| Eating Disorder | 161 | 5 (.39) | 12.70 | .0006 | DISC1, NLGN1, TACR1, NRG3, GABRA5 |
| Schizophrenia | 310 | 6 (.76) | 7.92 | .0012 | NRG2, DISC1, NRG3, CACNA1C, GABRA5, SYN3 |
| Mental Disorders | 503 | 7 (1.23) | 5.69 | .002 | DISC1, NLGN1, NRG3, CACNA1C, GABRA5, SYN3, VLDLR |
| Mood Disorders | 253 | 5 (.57) | 8.70 | .003 | DISC1, NLGN1, TACR1, CACNA1C, GABRA5 |
| Depression | 169 | 4 (.41) | 9.68 | .007 | DISC1, NLGN1, TACR1, CACNA1C |
Discussion
Through GWAS, we found that emotion dysregulation is associated at genome-wide level significance in a sex-specific manner, with a SNP in IL2RA in men. This is the first study to our knowledge that has found a relationship between emotion dysregulation and IL-2, a pro-inflammatory cytokine that is involved in the systemic inflammatory response. Previous GWAS have shown a relationship between IL2RA and several medical disorders including asthma, multiple sclerosis (MS), rheumatoid arthritis, and inflammatory bowel disease (Bouzid et al., 2013, Ferreira et al., 2014, Hinks et al., 2012, Metasanz et al., 2012, Purcell et al., 2007). There are also a number of studies showing overlap of risk for MS and risk for psychiatric disorders including MDD, bipolar disorder, and schizophrenia (Andreassen et al., 2014, Iacovides and Adreoulakis, 2011, Joffe et al., 1987, Patten et al., 2003, Siegert and Abernethy, 2005). Studies have also shown associations between IL-2 polymorphisms and schizophrenia (Paul-Samojedny et al., 2013; Schwarz et al., 2006; Watanabe et al., 2008). Growing evidence suggests that inflammation may be a common path that explains why autoimmune diseases and certain psychiatric disorders co-occur (Leonard and Maes, 2012, Miller et al., 2002).
There are a number of studies showing an association of cross diagnosis psychiatric illnesses and inflammatory markers. For example, a recent review found that that suicidal behavior, a behavior that is associated with many psychiatric conditions, was associated with elevations of IL-2, IL-6, IL-8 and TNF-α (Serafini et al., 2013). Another review of inflammatory profiles in depression, psychosis, and bipolar disorder found that there is overwhelming evidence to suggest that inflammatory abnormalities are a feature of many psychiatric conditions (Baumeister et al., 2014). Research also suggests that early life stress may increase risk of chronic inflammation, particularly among individuals with depression (Muller and Bechter, 2013).
Although interesting, it is unclear why the GWAS findings were found only in men. Findings are mixed, but there is some evidence of sex differences in inflammatory markers (Rohleder et al., 2001, Toker et al., 2005). One important factor in understanding these sex differences may be sex hormones. Women are more likely to develop certain autoimmune disorders that may be associated with inflammation, such as rheumatoid arthritis, but the severity of the diseases varies with sex hormone status (e.g., pregnancy, menopause; Da Silva and Hall, 1992, Lahita, 1996, Silva, 1999). The variability in age and sex hormone status of our female participants may have impacted the ability to find significant genetic risk results for females; more research is needed to determine whether our findings are in fact sex specific or may be influenced by hormones in a way that could not be measured with the present study. These data suggest that as we continue to examine mechanisms underlying emotion regulation, we need to be aware that such biological pathways may often be different across the sexes.
Our results also showed that there were significant associations between the identified EDS-related SNP and psychiatric conditions in this traumatized population. More specifically, rs6602398 was significantly associated with current depression and PTSD. In both logistic regression analyses, when emotion dysregulation was included in the model, the associations between rs6602398 and these outcome variables were no longer significant. It is possible that high levels of emotion dysregulation could serve as an intermediate phenotype between this risk gene and certain psychiatric conditions, although it is impossible to know causality based on the present findings using cross-sectional data. Mediational models should be examined in future longitudinal studies.
Our findings also suggest that this SNP may influence emotion dysregulation through a combination of genetic and epigenetic mechanisms that influence IL2RA expression. Methylation is influenced by SNPs across the genome, and rs6602398 genotype associated with methylation of a CpG site in IL2RA. However, it is important to note that there were a limited number of subjects with available methylation data, and multiple test corrections were not performed for this analysis. DNA methylation can alter transcriptional regulation, and histone modifications and DNA methylation are increasingly appreciated to be dynamically regulated with gene activation and repression.
Our gene enrichment analyses also suggest that the genes most associated with emotion dysregulation have previously been found to be associated with psychiatric disorders including anxiety disorders, mood disorders, schizophrenia, disturbance of conduct, autism, and eating disorders. This is consistent with our data showing that emotion dysregulation may help to explain the associations between SNPs of IL2RA and depressive symptoms, PTSD symptoms, and suicide attempt risk. Lastly, in our KEGG pathway enrichment analyses we found an overrepresentation of genes from the calcium signaling pathway, which has also been associated with risk for many psychiatric disorders including major depressive disorder, bipolar disorder and schizophrenia (Erk et al., 2010, Lancaster et al., 2014). Variants of the calcium signaling pathway have also been associated with a range of structural and functional brain circuitry, most notably emotion processing and executive functioning (Bigos et al., 2010).
Several study limitations are worth noting. Given the cross-sectional nature of this study and the use of retrospective reports, we cannot make assertions about causality or time of onset for PTSD symptoms, depressive symptoms, or emotion dysregulation. Therefore, prospective, longitudinal studies are required to examine the temporal onset of emotion dysregulation, PTSD and depressive symptomatology. We also focused on self-report measures of emotion dysregulation, current PTSD, and current depression. There are limitations to what an individual can correctly identify in themselves regarding psychiatric symptoms and it would be beneficial for future research to incorporate more thorough and objective measures of these constructs. Additionally, the sample size of our male group was small, and it is critical that these results be replicated in additional independent samples before any clear determinations about the implications of these findings can be made. Also, the methylation analyses used to supplement the genome-wide corrected SNP association analysis did not include multiple test correction. Finally, our sample was largely low income and African American. However, this weakness is balanced by the public health importance of studying these variables in an often under-researched and under-served population with such high rates of trauma exposure as well as mental and physical health problems. Individuals living in low income, inner-city environments are at particularly high risk for exposure to childhood abuse, multiple traumatic events, and trauma-related psychopathology (Gillespie et al., 2009, Schwartz et al., 2005). Therefore, this population may be a particularly interesting group with which to examine genetic associations with emotion dysregulation and how that might help to elucidate cross-disorder genetic vulnerability. Furthermore, there are very limited mental health resources available for individuals in this population, and it is even more critical that we continue to study factors that might inform evidence-based treatments for such groups.
Overall, the combination of our findings on emotion dysregulation, via GWAS identifying the IL2RA gene which is associated in prior research with a number of medical disorders that have high comorbidity with psychiatric disorders, the association between the EDS-related SNP in IL2RA and depressive and PTSD symptoms, and the gene enrichment findings suggesting that the SNPs closely associated with emotion dysregulation show overrepresentation with genes associated with risk for multiple psychiatric disorders and the calcium signaling pathway, together provide initial convergent evidence into genetic risk loci associated with emotion dysregulation. Clearly, emotion dysregulation is important in a wide range of psychiatric conditions, and this risk factor needs to be understood further at the genetic and epigenetic levels, as well as in understanding underlying mechanisms mediating sex-specific differences, as we continue to improve understanding of cross-disorder risk more generally.
Supplementary Material
Footnotes
After imputing IL2RA using IMPUTE2, SNP rs6602398 was still the most significant SNP in the gene.
References
- Aldao A, Nolen-Hoeksema S, Schweizer S. Emotion-regulation strategies across psychopathology: A meta-analytic review. Clinical Psychology Review. 2010;30:217–37. doi: 10.1016/j.cpr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Alink L, Cicchetti D, Kim J, Rogosh F. Mediating and moderating processes in the relation between maltreatment and psychopathology: Mother-child relationship quality and emotion regulation. Journal of Abnormal Child Psychology. 2009 doi: 10.1007/s10802-009-9314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen O, Harbo H, Wang Y, Thompson W, Schork A, Mattingsdal M, et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: Differential involvement of immune-related gene loci. Molecular Psychiatry. 2014 Jan 28; doi: 10.1038/mp.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychological Association. Diagnostic and statistical manual-text revision (DSM-IV-TRim, 2000) American Psychiatric Association; 2000. [Google Scholar]
- Baumeister D, Russel A, Pariante C, Modelli V. Inflammatory biomarker profiles of mental disorders and their relation to clinical, social, and lifestyle factors. Social Psychiatry and Psychiatric Epidemiology. 2014;49:841–9. doi: 10.1007/s00127-014-0887-z. [DOI] [PubMed] [Google Scholar]
- Beck A, Steer R, Brown G. Manual for the Beck Depression Inventory-II. San Antonio, TX: Psychological Corporation; 1996. [Google Scholar]
- Belsky J, Bakermans-Kranenburg MJ, Van IJzendoorn MH. For better and for worse differential susceptibility to environmental influences. Current Directions in Psychological Science. 2007;16:300–4. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- Berenbaum H, Raghaven C, Huynh-Nhu L, Vernon L, Gomez J. A taxonomy of emotional disturbances. Science and Practice. 2003;10:206–26. [Google Scholar]
- Bigos KL, Mattay VS, Callicott JH, Straub RE, Vakkalanka R, Kolachana B, et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Archives of general psychiatry. 2010;67:939–45. doi: 10.1001/archgenpsychiatry.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzid D, Amouri A, Fourati H, Marques I, Abida O, Tahri N, et al. Polymorphisms in the IL3RA and IL2RB genes in inflammatory bowel disease risk. Genet Test Mol Biomarkers. 2013;17:833–9. doi: 10.1089/gtmb.2013.0291. [DOI] [PubMed] [Google Scholar]
- Bradley B, DeFife J, Guarnaccia C, Phifer J, Fani N, Ressler K. Emotion dysregulation and negative affect: Association with psychiatric symptoms. Journal of Clinical Psychiatry. 2011a;72:685–91. doi: 10.4088/JCP.10m06409blu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B, Westen D, Mercer K, Binder E, Jovanovic T, Crain D, et al. Association between childhood maltreatment and adult emotional dysregulation in a low-income, urban, African American sample: Moderation by oxytocin receptor gene. Development and Psychopathology. 2011b;23:439–52. doi: 10.1017/S0954579411000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholtz J, Meyer-Lindenberg A. MAOA and the neurogenetic architecture of human aggression. Trends in Neuroscience. 2008;31:120–9. doi: 10.1016/j.tins.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Canli T, Lesch K. Long story short: The serotonin transporter in emotion regulation and social cognition. Nature Neuroscience. 2007;10:1103–9. doi: 10.1038/nn1964. [DOI] [PubMed] [Google Scholar]
- Caspi A, Houts RM, Belsky DW, Goldman-Mellor SJ, Harrington H, Israel S, et al. The p Factor One General Psychopathology Factor in the Structure of Psychiatric Disorders? Clinical Psychological Science. 2014;2:119–37. doi: 10.1177/2167702613497473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–9. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Chesi A, Mitchell JA, Kalkwarf HJ, Bradfield JP, Lappe JM, et al. A trans-ethnic genome-wide association study identifies gender specific loci influencing pediatric aBMD and BMC at the distal radius. Hum Mol Genet. 2015;24(17):5053–9. doi: 10.1093/hmg/ddv210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti D, Ackerman B, Izard C. Emotions and emotion regulation in developmental psychopathology. Development and Psychopathology. 1995;7:1–10. [Google Scholar]
- Da Silva J, Hall G. The effects of gender and sex hormones on outcome in rheumatoid arthritis. Baillière’s clinical rheumatology. 1992;6:193–219. [PubMed] [Google Scholar]
- De Moor MH, Van Den Berg SM, Verweij KJ, Krueger RF, Luciano M, et al. Meta-analysis of genome-wide association studies for neuroticism, and the polygenic association with major depressive disorder. JAMA psychiatry. 2015;72(7):642–650. doi: 10.1001/jamapsychiatry.2015.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabant E, Hariri A, Meyer-Lindenberg A, Munoz K, Mattay V, Kolachana B, et al. Catechol O-methyltransferase Val158Met genotype and neural mechanisms related to affective arousal and regulation. Archives of General Psychiatry. 2006;63:1396–406. doi: 10.1001/archpsyc.63.12.1396. [DOI] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Schnell K, von Boberfeld CO, Esslinger C, Kirsch P, et al. Brain function in carriers of a genome-wide supported bipolar disorder variant. Archives of general psychiatry. 2010;67:803–11. doi: 10.1001/archgenpsychiatry.2010.94. [DOI] [PubMed] [Google Scholar]
- Falsetti S, Resnick H, Resick P, Kilpatrick D. The modified PTSD symptom scale: A brief self-report measure of posttraumatic stress disorder. Behaviour Therapist. 1993;16:161. [Google Scholar]
- Ferreira M, Matheson M, Tang C, Granell R, Ang W, Hui J, et al. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J Allergy Clin Immunol. 2014;133:1564–71. doi: 10.1016/j.jaci.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie C, Bradley B, Mercer K, Smith A, Conneely K, Gapen M, et al. Trauma exposure and stress-related disorders in inner city primary care patients. General Hospital Psychiatry. 2009;31:505–14. doi: 10.1016/j.genhosppsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E, Grozeva D, Jones I, Kirov G, Caesar S, et al. The bipolar disorder risk aleele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Molecular Psychiatry. 2010;15:1016–22. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. Emotion regulation: Affective, cognitive, and social consequences. Psychophysiology. 2002;39:281–91. doi: 10.1017/s0048577201393198. [DOI] [PubMed] [Google Scholar]
- Gross J, Munoz R. Emotion regulation and mental health. Clin Psychol Sci Prac. 1995;2:151–64. [Google Scholar]
- Hariri A, Holmes A. Genetics of emotional regulation: The role of the serotonin transporter in neuroal function. TRENDS in Cognitive Sciences. 2006;10:182–91. doi: 10.1016/j.tics.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Harrison P, Tunbridge E. Catechol-O-Methyltransferase (COMT): A gene contributing to sex differences in brain function, and to sexual dimorphism in the predisposition to psychiatric disorders. Neuropsychopharmacology. 2008;33:3037–45. doi: 10.1038/sj.npp.1301543. [DOI] [PubMed] [Google Scholar]
- Heim C, Bradley B, Mletzko TC, Deveau TC, Musselman DL, et al. Effect of childhood trauma on adult depression and neuroendocrine function: sex-specific moderation by CRH receptor 1 gene. Frontiers in behavioral neuroscience. 2009;3 doi: 10.3389/neuro.08.041.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinks A, Cobb J, Sudman M, Eyre S, Martin P, Flynn E, et al. Investigation of rheumatoid arthritis susceptibility loci in juvenile idiopathis arthritis confirms high degress of overlap. Ann Rheum Dis. 2012;71:1117–21. doi: 10.1136/annrheumdis-2011-200814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson C, Goldman D, Jaeger J, Persaud S, Kane J, Lipsky R, et al. Disrupted in schizophrenia 1 (DISC1): Association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am J Hum Genet. 2004;75:862–72. doi: 10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz A, Widom C, McLaughlin J, White H. The impact of childhood abuse and neglect on adult mental health: A prospective study. Journal of Health and Social Behavior. 2001;42:184–202. [PubMed] [Google Scholar]
- Iacovides A, Adreoulakis E. Bipolar disorder and resembling special psychpathological manifestations in multiple sclerosis: A review. Current Opinion in Psychiatry. 2011;24:336–40. doi: 10.1097/YCO.0b013e328347341d. [DOI] [PubMed] [Google Scholar]
- Jia P, Wang L, Meltzer H, Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophrenia research. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffe R, Lippert G, Gray T, Sawa G, Horvath Z. Mood disorder and multiple sclerosis. Arch Neurol. 1987;44:376–8. doi: 10.1001/archneur.1987.00520160018007. [DOI] [PubMed] [Google Scholar]
- John O, Gross J. Healthy and unhealthy regulation: Personality processes, individual differences, and life span development. Journal of Personality. 2004:1301–34. doi: 10.1111/j.1467-6494.2004.00298.x. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kendler K, Wray N, et al. Genetic relationship between five psychiatric disorders estmiated from genome-wide SNPs. Nature Genetics. 2013;459 doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Cohen J, Gold AL. Measured gene–environment interactions and mechanisms promoting resilient development. Current Directions in Psychological Science. 2009;18:138–42. [Google Scholar]
- Kim H, Sherman D, Mojaverian T, Sasaki J, Park J, Suh E, et al. Gene-culture interaction: Oxytocin receptor polymorphism (OXTR) and emotion regulation. Social Psychological and Personality Science. 2011;2:665–72. [Google Scholar]
- Kim J, Cicchetti D. Longitudinal pathways linking child maltreatment, emotion regulation, peer relations, and psychopathology. Journal of Child Psychol Psychiatry. 2010;51:706–16. doi: 10.1111/j.1469-7610.2009.02202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kring A. Emotion disturbances as transdiagnostic processes in psychopathology. In: Lewis M, Hayiland-Jones J, Barrett L, editors. Handbook of emotion. New York: Guilford Press; 2008. pp. 691–705. [Google Scholar]
- Lahita RG. Predisposing factors to autoimmune disease. International journal of fertility and women’s medicine. 1996;42:115–9. [PubMed] [Google Scholar]
- Lancaster T, Heerey E, Mantripragada K, Linden D. CACNA1C risk variant affects reward responsiveness in healthy individuals. Translational psychiatry. 2014;4:e461. doi: 10.1038/tp.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard B, Maes M. Mechanistic explanations how cell-mediated immune activation, inflammation, and oxidative and nitrosative stress pathways and their sequels and concomitants play a role in the pathophysiology of unipolar depression. Neurscience and Biobehavioral Reviews. 2012;36:764–85. doi: 10.1016/j.neubiorev.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Lin S, Zhoa H. Handbook on analyzing human genetic data: Computational approaches and software. New York: Springer; 2009. [Google Scholar]
- Metasanz F, Gonzalez-Perez A, Lucas M, Sanna S, Gayan J, Urcelay E, et al. Genome-wide association study of multiple sclerosis confirms a novel locus at 5p13.1. PLoS One. 2012;7:e36140. doi: 10.1371/journal.pone.0036140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Stetler C, Carney R, Freedland K, Banks W. Clinical depression and inflammatory risk markers for coronary heart disease. Am J Cardiol. 2002;90:1279–83. doi: 10.1016/s0002-9149(02)02863-1. [DOI] [PubMed] [Google Scholar]
- Muller N, Bechter K. The mild encephalitis concept for psychiatric disorders revisited in light of current psychoneuroimmunological findings. Neurology, Psychiatry and Brain Research. 2013;19:87–101. [Google Scholar]
- Patten S, Beck C, Williams M, Barbui M, Metz M. Major depression in multiple sclerosis: A population-based perspective. Neurology. 2003;61:1524–7. doi: 10.1212/01.wnl.0000095964.34294.b4. [DOI] [PubMed] [Google Scholar]
- Paul-Samojedny M, Owczarek A, Kowalczyk M, Suchanek R, Palacz M, et al. Association of interleukin 2 (IL-2), interleukin 6 (IL-6), and TNF-alpha (TNFα) gene polymorphisms with paranoid schizophrenia in a Polish population. The Journal of neuropsychiatry and clinical neurosciences. 2013;25(1):72–82. doi: 10.1176/appi.neuropsych.12020021. [DOI] [PubMed] [Google Scholar]
- Powers A, Stevens J, Fani N, Bradley B. Construct validity of a short, self report instrument assessing emotional dysregulation. Psychiatry research. 2015;225:85–92. doi: 10.1016/j.psychres.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Prium R, Welch R, Sanna S, Teslovich T, Chines P, Glidt T, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Mercer K, Bradley B, Jovanovic T, Mahan A, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470(7335):492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohleder N, Schommer NC, Hellhammer DH, Engel R, Kirschbaum C. Sex differences in glucocorticoid sensitivity of proinflammatory cytokine production after psychosocial stress. Psychosomatic medicine. 2001;63:966–72. doi: 10.1097/00006842-200111000-00016. [DOI] [PubMed] [Google Scholar]
- Schwartz A, Bradley B, Sexton M, Sherry A, Ressler K. Posttraumatic stress disorder among African Americans in an innery city mental health clinic. Psychiatric Services. 2005;56:212–6. doi: 10.1176/appi.ps.56.2.212. [DOI] [PubMed] [Google Scholar]
- Schwarz MJ, Krönig H, Riedel M, Dehning S, Douhet A, et al. IL-2 and IL-4 polymorphisms as candidate genes in schizophrenia. European archives of psychiatry and clinical neuroscience. 2006;256(2):72–6. doi: 10.1007/s00406-005-0603-9. [DOI] [PubMed] [Google Scholar]
- Serafini G, Pompili M, Serretti E, Stefani H, Palermo M, Coryell W, et al. The role of inflammatory cytokines in suicidal behavior: A systematic review. European Neuropsychopharmacology. 2013;23:1672–86. doi: 10.1016/j.euroneuro.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Sharma S, Londono D, Eckalbar WL, Gao X, Zhang D, et al. A PAX1 enhancer locus is associated with susceptibility to idiopathic scoliosis in females. Nat Commun. 2015;6:6452. doi: 10.1038/ncomms7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R, Abernethy D. Depression in multiple sclerosis: A review. J Neurol Neurosurg Psychiatry. 2005;76:469–75. doi: 10.1136/jnnp.2004.054635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J. Sex hormones and glucocorticoids: interactions with the immune system. Annals of the New York Academy of Sciences. 1999;876:102–18. doi: 10.1111/j.1749-6632.1999.tb07628.x. [DOI] [PubMed] [Google Scholar]
- Smith AA, Huang Y-T, Eliot M, Houseman EA, Marsit CJ, Wiencke JK, et al. A novel approach to the discovery of survival biomarkers in glioma using a joint analysis of DNA methylation and gene expression. Epigenetics. 2014;9 doi: 10.4161/epi.28571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller J, Kendler K, Craddock N, et al. Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet. 2013;381:1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southam-Gerow M, Kendall P. Emotion regulation and understanding: Implications for child psychopathology and therapy. Clinical Psychology Review. 2002;22:189–222. doi: 10.1016/s0272-7358(01)00087-3. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, et al. Gene set enrichment analysis: A knowledge-based approach for interpresting genome-wide expression profiles. PNAS. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabachnick BG, Fidell LS. Using multivariate statistics. 2001 [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, et al. beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–96. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker S, Shirom A, Shapira I, Berliner S, Melamed S. The association between burnout, depression, anxiety, and inflammation biomarkers: C-reactive protein and fibrinogen in men and women. Journal of occupational health psychology. 2005;10:344. doi: 10.1037/1076-8998.10.4.344. [DOI] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT Analysis Toolkit (WebGestalt) Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Nunokawa A, Shibuya M, Kaneko N, Nawa H, et al. Association study of interleukin 2 (IL2) and IL4 with schizophrenia in a Japanese population. European archives of psychiatry and clinical neuroscience. 2008;258(7):422–7. doi: 10.1007/s00406-008-0813-z. [DOI] [PubMed] [Google Scholar]
- Werling DM, Lowe JK, Luo R, Cantor RM, Geschwind DH. Replication of linkage at chromosome 20p13 and identification of suggestive sex-differential risk loci for autism spectrum disorder. Mol Autism. 2014;5(1):13. doi: 10.1186/2040-2392-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Gatt J, Kuan S, Dobson-Stone C, Palmer D, Paul R, et al. A polymorphism of the MAOA Gene is associated with emotional brain markers and personality traits on an antisocial index. Neuropsychopharmacology. 2009;34:1797–809. doi: 10.1038/npp.2009.1. [DOI] [PubMed] [Google Scholar]
- Xu H, Wang F, Liu Y, Yu Y, Gelernter J, et al. Sex-biased methylome and transcriptome in human prefrontal cortex. Hum Mol Genet. 2014;23(5):1260–70. doi: 10.1093/hmg/ddt516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


